Abstract
Background.
Human platelet antigens (HPA) are determinant in several platelet-specific alloimmune disorders, such as neonatal alloimmune thrombocytopenia, post-transfusion purpura and platelet transfusion refractoriness. The distribution of HPA systems in the Malaysian population is not known. Defining the patterns of HPA systems provides a basis for risk assessment and management of the above complications.
Materials and methods.
The aim of this study was to investigate the distribution of HPA -1 to -6 and -15 in the three major ethnic groups (Malay, Chinese and Indian) in the Malaysian population. A total of 600 random donor samples, 200 from each of the three ethnic groups, were genotyped by means of real time polymerase chain reaction (PCR) with hydrolysis probes and PCR-restriction fragment length polymorphism (PCR-RFLP).
Results.
The most common genotype observed in this study was HPA-1a/1a-2a/2a-3a/3b-4a/4a-5a/5a-6a/6a-15a/15b (17%) followed by HPA-1a/1a-2a/2a-3a/3a-4a/4a-5a/5a-6a/6a-15a/15b (14.33%). The allele frequencies of HPA in Malays and Chinese were found to be similar those of other East and South-East Asian populations, while those of Indians were comparable to the frequencies found in Europeans.
Conclusion.
The results of this study have been useful for determining the distribution of HPA polymorphisms in this region and for potential clinical implications.
Keywords: HPA, genotyping, Malay, Southeast Asia
Introduction
Human platelet antigens (HPA) are immunogenic polymorphic forms of platelet membrane glycoproteins. The nomenclature of HPA currently recognises 21 HPA biallelic systems, all of which except one are defined by a single amino acid substitution, generally caused by a single nucleotide polymorphism (SNP) in the gene encoding the relevant membrane glycoprotein (http://www.ebi.ac.uk/ipd/hpa/). Alloimmune responses to HPA-1 to HPA-6 and HPA-15 have been reported to contribute to clinical complications of varying severity among different populations1,2. Alloimmunisation may occur through exposure to non-self HPA, commonly during pregnancy, in which case it can cause neonatal alloimmune thrombocytopenia, or via blood transfusion, when it leads to the risk of post-transfusion purpura and platelet transfusion refractoriness.
With the characterisation of the molecular basis of HPA polymorphisms and rapid developments in sensitive polymerase chain reaction (PCR) techniques, HPA molecular typing has virtually replaced serological typing3. High-throughput PCR techniques such as allele-specific PCR, melting curve analysis, and 5’-nuclease assays have also enabled widespread population typing and comprehensive HPA allele frequencies have been reported in diverse populations using these techniques4. Among Asian populations, considerable prevalence data are available for Chinese people and, to a lesser extent, Indian populations5–8. Allele frequency data among Malays are, however, lacking. Malaysia is a multiethnic country with a population size of about 28.3 million. The majority of the inhabitants are Malays (54.4%), Chinese (25.0%) and Indians (7.5%) (http://www.statistics.gov.my). The three major ethnic groups each originate from different parts of Asia: the Indo-Malay Archipelago in South-East Asia, China in East Asia, and India in South Asia respectively. The mixture of the predominant races within South and East Asia provided us a unique opportunity to study HPA allele frequencies in Asian populations simultaneously. We aimed to characterise HPA allele frequencies for seven of the clinically relevant HPA systems among our population with the hope of providing an informative background of HPA polymorphisms in Asia and consequently enabling risk assessment in multi-ethnic environments.
Materials and methods
Study population
Blood samples were collected into Vacutainer® (Becton-Dickinson, Franklin Lakes, NJ, USA) tubes containing ethylenediaminetetraacetic acid (EDTA). Samples were collected from 600 blood donors, equally divided between the three major ethnic groups (Malay, Chinese, and Indian). All participants in this study were unrelated individuals of Malaysian nationality and were grouped according to their self-reported ethnicities. Informed consent was obtained from all subjects and the study was approved by the institution’s review board.
DNA extraction
Genomic DNA was isolated from the EDTA-anticoagulated whole blood samples using either phenol-chloroform extraction or by spin-column utilising the QIAamp DNA blood mini kit (Qiagen, Hilden, Germany). Procedures were performed according to the manufacturer’s instructions. The concentration of each DNA sample was standardised through serial dilution so that each working volume (2.25 μL) contained 10 ng of DNA.
Genotyping of HPA 1–5, 15 by single nucleotide polymorphism genotyping assay
Genotyping for HPA-1 -5 and -15 was performed on a Roche LightCycler 480 II real-time PCR system (Roche, Mannheim, Germany) using TaqMan® SNP genotyping assays (Applied Biosystems, Foster City, CA, USA). Primers and probes (Table I) were either obtained as pre-designed SNP genotyping assays or were custom designed using the Custom TaqMan Assay Design Tool (https://www.appliedbiosystems.com/tools/cadt/)9. Tests were performed as recommended and under the thermal conditions stated in the protocol provided by the manufacturer: a temperature hold at 95 °C for 10 minutes, followed by a two-step cycle consisting of a 15-second denaturation step at 92 °C and a 1 minute annealing/extension step at 60 °C, repeated for 40 cycles. Results were analysed with LightCycler 480® Software Version 1.5 (Roche). All genotyping assays were designed to target SNP as identified in the Immuno Polymorphism Database (http://www.ebi.ac.uk/ipd/hpa/) and according to their reference in the dbSNP database.
Table I.
Assay ID and corresponding TaqMan® SNP Genotyping Assay probe sequences used for HPA-1 to -5 and -15 detection.
| Assay ID | dbSNP No. | Probe sequence [VIC/FAM] | Specificity |
|---|---|---|---|
| 818008_30 | rs5918 | GCTCCTGTCTTACAGGCCCTGCCTC[C/T]GGGCTCACCTCGCTGTGACCTGAAG | HPA-1 |
| 11442703_10 | rs6065 | AAGACCCTGCCCCCAGGGCTCCTGA[C/T]GCCCACACCCAAGCTGGAGAAGCTC | HPA-2 |
| 3017440_10 | rs5911 | CGGGTGAATGGGGGAGGGGCTGGGG[C/A]TGGGCAGCCCCCAGTCCACCTGGGG | HPA-3 |
| 11416452_20 | rs5917 | GGTACCAAGCTGGCCACCCAGATGC[A/G]AAAGCTCACCAGTAACCTGCGGATT | HPA-4 |
| 29770100_20 | rs10471371 | GTTTATTCTCAACATGGGAGTCAGG[A/G]TGATCTTTTGACAAATTAAATGGAA | HPA-5 |
| 3226894_10 | rs10455097 | TATATTTATTATCTTGACTTCAGTT[A/C]CAGGATTTACCAAGAATTTGAAGTA | HPA-15 |
HPA-6 genotyping by polymerase chain reaction restriction fragment length polymorphism
HPA-6 wa s genotyped by an alternative method to TaqMan® SNP genotyping using PCR-RFLP with the MvaI restriction enzyme, as part of an earlier study. The SNP-containing region was amplified using TopTaq PCR Master Mix (Qiagen) with the following primer sequences; forward 5′-CTGGCTGGCTGGGATCCCAGTG-3′ and reverse 5′-CCCTGCAGTTCTCCTCACCTGAG-3′10. The thermal cycling conditions involved an initial denaturation step for 3 minutes at 94 °C followed by a three-step cycle consisting of denaturation at 94 °C for 30 seconds, annealing at 60 °C for 30 seconds, and extension at 72 °C for 1 minute, repeated for 30 cycles. The amplified product with a size of 240 bp was treated with FastDigest MvaI restriction enzyme (Fermentas, Hanover, MD, USA) at 37 °C for 5 minutes. The final RFLP products were resolved in 2% agarose gel stained with ethidium bromide, and visualised under ultraviolet light.
Statistical analysis
Genotype and allele frequencies were determined through direct counting. The validity of the Hardy-Weinberg equilibrium for each of the HPA system was tested based on heterozygosity using the population genetics software Arlequin11. A chi square (χ2) test or Fisher’s exact test was performed to compare the genotype values and allele frequencies between populations. We used an alpha level of 0.05 for all statistical tests. Bonferroni’s adjusted P-values were applied when multiple hypothesis testing was performed, for example for analysis between different population groups. Hierarchical clustering was performed using the seven HPA systems tested and their corresponding frequencies for the “a” alleles in the three Malaysian groups and 26 chosen worldwide populations as shown in Table II. Euclidean distance was used as the similarity measure between alleles and populations. The distance tree was constructed using clustering with the unweighted pair-group method with an arithmetic mean. Cluster 3.0 software and Java TreeView (Eisen Lab, University of California, Berkeley) were used for the analysis and visualisation of the results, respectively. Principal component analysis was also performed to summarise the distribution of populations according to their continents of origin based on the HPA allele frequencies using PAST statistical software12.
Table II.
Frequency distribution of HPA alleles in various populations worldwide.
| Population | n |
HPA frequency distribution
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 2a | 2b | 3a | 3b | 4a | 4b | 5a | 5b | 6a | 6b | 15a | 15b | ||
| Malay (This study) | 200 | 0.9750 | 0.0250 | 0.9625 | 0.0375 | 0.5025 | 0.4975 | 0.9950 | 0.0050 | 0.9500 | 0.0500 | 0.9925 | 0.0075 | 0.5150 | 0.4850 |
| Malaysian Chinese (This Study) | 200 | 1.0000 | 0.0000 | 0.9675 | 0.0325 | 0.5725 | 0.4275 | 0.9975 | 0.0025 | 0.9825 | 0.0175 | 0.9825 | 0.0175 | 0.4975 | 0.5025 |
| Malaysian Indian (This Study) | 200 | 0.8850 | 0.1150 | 0.9600 | 0.0400 | 0.6200 | 0.3800 | 0.9975 | 0.0025 | 0.9400 | 0.0600 | 0.9950 | 0.0050 | 0.4075 | 0.5925 |
| America | |||||||||||||||
| Argentinean23 | 192 | 0.8780 | 0.1220 | 0.8750 | 0.1250 | 0.6120 | 0.3880 | 1.0000 | 0.0000 | 0.9270 | 0.0730 | 1.0000 | 0.0000 | 0.5110 | 0.4890 |
| Amerindian Toba23 | 27 | 1.0000 | 0.0000 | 0.9440 | 0.0560 | 0.3890 | 0.6110 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 0.6850 | 0.3150 |
| Brazilian (White)24 | 400 | 0.7800 | 0.2200 | 0.9000 | 0.1000 | 0.6300 | 0.3700 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | - | - | - | - |
| Brazilian (Black)24 | 150 | 0.9030 | 0.0970 | 0.8100 | 0.1900 | 0.6660 | 0.3340 | 1.0000 | 0.0000 | 0.8760 | 0.1240 | - | - | - | - |
| Brazilian (Amerindian)24 | 70 | 1.0000 | 0.0000 | 0.8210 | 0.1790 | 0.7570 | 0.2430 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | - | - | - | - |
| Africa | |||||||||||||||
| Algerian25 | 485 | 0.8347 | 0.1653 | 0.8347 | 0.1653 | 0.6296 | 0.3704 | 1.0000 | 0.0000 | 0.8431 | 0.1569 | - | - | 0.5300 | 0.4700 |
| Beninese22 | 154 | 0.8960 | 0.1040 | 0.7080 | 0.2920 | 0.6790 | 0.3210 | 1.0000 | 0.0000 | 0.8180 | 0.1820 | 1.0000 | 0.0000 | 0.6460 | 0.3540 |
| Cameroonian22 | 118 | 0.9070 | 0.0930 | 0.7630 | 0.2370 | 0.6140 | 0.3860 | 1.0000 | 0.0000 | 0.7460 | 0.2540 | 1.0000 | 0.0000 | 0.6910 | 0.3090 |
| Congolese22 | 125 | 0.9040 | 0.0960 | 0.7760 | 0.2240 | 0.5960 | 0.4040 | 1.0000 | 0.0000 | 0.7320 | 0.2680 | 1.0000 | 0.0000 | 0.7010 | 0.2990 |
| Central African22 | 110 | 1.0000 | 0.0000 | 0.6070 | 0.3930 | 0.5000 | 0.5000 | 1.0000 | 0.0000 | 0.5950 | 0.4050 | 1.0000 | 0.0000 | 0.6980 | 0.3020 |
| Moroccan26 | 107 | 0.7480 | 0.2520 | 0.8180 | 0.1820 | - | - | - | - | - | - | - | - | - | - |
| 104 | - | - | - | - | 0.6820 | 0.3180 | - | - | - | - | - | - | - | - | |
| 108 | - | - | - | - | - | - | 1.0000 | 0.0000 | - | - | 1.0000 | 0.0000 | - | - | |
| 112 | - | - | - | - | - | - | - | - | 0.8610 | 0.1390 | - | - | - | - | |
| South African (Bantu Speaker)27 | 150 | 0.8905 | 0.1095 | 0.8135 | 0.1865 | 0.6995 | 0.3005 | 1.0000 | 0.0000 | 0.6835 | 0.3165 | - | - | 0.8075 | 0.1925 |
| South African (Indian)27 | 150 | 0.8930 | 0.1070 | 0.9305 | 0.0695 | 0.6695 | 0.3305 | 1.0000 | 0.0000 | 0.9370 | 0.0630 | - | - | 0.4395 | 0.5605 |
| South African (White)27 | 150 | 0.8205 | 0.1795 | 0.9005 | 0.0995 | 0.5735 | 0.4265 | 1.0000 | 0.0000 | 0.8870 | 0.1130 | - | - | 0.5405 | 0.4595 |
| Tunisian28 | 90 | 0.7500 | 0.2500 | - | - | 0.6940 | 0.3060 | - | - | 0.7790 | 0.2210 | - | - | - | - |
| Europe | |||||||||||||||
| Austrian29 | 911 | 0.8520 | 0.1480 | - | - | - | - | - | - | - | - | - | - | - | - |
| 907 | - | - | 0.9180 | 0.0820 | - | - | - | - | - | - | - | - | - | - | |
| 906 | - | - | - | - | 0.6120 | 0.3880 | - | - | - | - | - | - | - | - | |
| 100 | - | - | - | - | - | - | 1.0000 | 0.0000 | - | - | 1.0000 | 0.0000 | - | - | |
| 931 | - | - | - | - | - | - | - | - | 0.8920 | 0.1080 | - | - | - | - | |
| 254 | - | - | - | - | - | - | - | - | - | - | - | - | 0.5 | 0.5 | |
| British30 | 134 | 0.8400 | 0.1600 | 0.9250 | 0.0750 | 0.6270 | 0.3730 | 1.0000 | 0.0000 | 0.9140 | 0.0860 | 1.0000 | 0.0000 | 0.5240 | 0.4760 |
| Croatian31,32 | 219 | 0.8540 | 0.1460 | 0.8900 | 0.1100 | 0.5750 | 0.4250 | - | - | 0.8950 | 0.1050 | - | - | - | - |
| 558 | - | - | - | - | - | - | - | - | - | - | - | - | 0.5300 | 0.4700 | |
| Czech33 | 235 | 0.8300 | 0.1700 | 0.9000 | 0.1000 | 0.5900 | 0.4100 | - | - | 0.9300 | 0.0700 | - | - | - | - |
| Danish34 | 557 | 0.8300 | 0.1700 | - | - | - | - | - | - | - | - | - | - | - | - |
| 163 | - | - | 0.9200 | 0.0800 | 0.6300 | 0.3700 | - | - | - | - | - | - | - | - | |
| 131 | - | - | - | - | - | - | 1.0000 | 0.0000 | - | - | - | - | - | - | |
| 427 | - | - | - | - | - | - | - | - | 0.9200 | 0.0800 | - | - | - | - | |
| 100 | - | - | - | - | - | - | - | - | - | - | 1.0000 | 0.0000 | - | - | |
| 100 | - | - | - | - | - | - | - | - | - | - | - | - | 0.5000 | 0.5000 | |
| French35 | 6192 | 0.8480 | 0.1520 | - | - | 0.6200 | 0.3800 | - | - | 0.8740 | 0.1260 | - | - | - | - |
| 525 | - | - | 0.9200 | 0.0800 | - | - | - | - | - | - | - | - | - | - | |
| 1242 | - | - | - | - | - | - | - | - | - | - | - | - | 0.4550 | 0.5450 | |
| German36 | 1583 | 0.8400 | 0.1600 | - | - | - | - | - | - | - | - | - | - | - | - |
| 1576 | - | - | 0.9300 | 0.0700 | - | - | - | - | - | - | - | - | - | - | |
| 1562 | - | - | - | - | 0.6000 | 0.4000 | - | - | - | - | - | - | - | - | |
| 1643 | - | - | - | - | - | - | - | - | 0.9200 | 0.0800 | - | - | - | - | |
| Italian37 | 144 | 0.8500 | 0.1500 | 0.8900 | 0.1100 | 0.6100 | 0.3900 | 1.0000 | 0.0000 | 0.9000 | 0.1000 | 1.0000 | 0.0000 | - | - |
| Lebanese38 | 205 | 0.8100 | 0.1900 | - | - | - | - | - | - | - | - | - | - | - | - |
| Macedonian39 | 122 | 0.8650 | 0.1350 | - | - | - | - | - | - | - | - | - | - | - | - |
| 132 | - | - | 0.8520 | 0.1480 | - | - | - | - | - | - | - | - | - | - | |
| 115 | - | - | - | - | 0.5780 | 0.4220 | - | - | - | - | - | - | - | - | |
| 126 | - | - | - | - | - | - | - | - | 0.9090 | 0.0910 | - | - | - | - | |
| Norwegian40 | 105 | 0.8670 | 0.1330 | 0.9430 | 0.0570 | 0.4710 | 0.5290 | 1.0000 | 0.0000 | 0.9290 | 0.0710 | - | - | 0.4950 | 0.5050 |
| Poland41 | 135 | 0.8240 | 0.1760 | - | - | - | - | - | - | - | - | - | - | - | - |
| 211 | - | - | 0.9010 | 0.0990 | - | - | - | - | - | - | - | - | - | - | |
| 130 | - | - | - | - | 0.5820 | 0.4180 | - | - | - | - | - | - | - | - | |
| 103 | - | - | - | - | - | - | 1.0000 | 0.0000 | - | - | - | - | - | - | |
| 166 | - | - | - | - | - | - | - | - | 0.9440 | 0.0560 | - | - | - | - | |
| 300 | - | - | - | - | - | - | - | - | - | - | - | - | 0.4850 | 0.5150 | |
| Slovenian42 | 152 | 0.8320 | 0.1680 | 0.8990 | 0.1010 | 0.6670 | 0.3330 | - | - | 0.8920 | 0.1080 | - | - | - | - |
| 90 | - | - | - | - | - | - | - | - | - | - | - | - | 0.5270 | 0.4630 | |
| Spanish43 | 727 | 0.8100 | 0.1900 | - | - | - | - | - | - | - | - | - | - | - | - |
| 462 | - | - | 0.9000 | 0.1000 | - | - | - | - | - | - | - | - | - | - | |
| 662 | - | - | - | - | 0.6500 | 0.3500 | - | - | - | - | - | - | - | - | |
| 264 | - | - | - | - | - | - | 1.0000 | 0.0000 | - | - | - | - | - | - | |
| 454 | - | - | - | - | - | - | - | - | 0.8800 | 0.1080 | - | - | - | - | |
| 254 | - | - | - | - | - | - | - | - | - | - | 1.0000 | 0.0000 | - | - | |
| 385 | - | - | - | - | - | - | - | - | - | - | - | - | 0.4740 | 0.5200 | |
| Swiss44 | 500 | 0.8090 | 0.1910 | 0.9180 | 0.1090 | 0.5910 | 0.4070 | 0.9970 | 0.0030 | 0.9340 | 0.0660 | - | - | - | - |
| Welsh45 | 392 | 0.8253 | 0.1747 | 0.9018 | 0.0982 | 0.6071 | 0.3929 | 1.0000 | 0.0000 | - | - | - | - | - | - |
| 339 | - | - | - | - | - | - | - | - | 0.9027 | 0.0973 | - | - | - | - | |
| 166 | - | - | - | - | - | - | - | - | - | - | 1.0000 | 0.0000 | 0.5241 | 0.4759 | |
| Asia | |||||||||||||||
| Indian5 | 1164 | 0.9244 | 0.0756 | 0.9979 | 0.0021 | 0.0100 | 0.9900 | 0.9953 | 0.0047 | 0.9562 | 0.0438 | 0.9923 | 0.0077 | - | - |
| Pakistani46 | 593 | 0.8850 | 0.1150 | 0.9200 | 0.0800 | 0.6900 | 0.3100 | 1.0000 | 0.0000 | 0.9000 | 0.1000 | - | - | 0.5900 | 0.4100 |
| Northeastern Thai15 | 300 | 0.9720 | 0.0280 | 0.9380 | 0.0620 | 0.5330 | 0.4670 | 1.0000 | 0.0000 | 0.9630 | 0.0370 | 0.9850 | 0.0150 | 0.4950 | 0.5050 |
| Thai14 | 500 | 0.9850 | 0.0150 | 0.9520 | 0.0480 | 0.5600 | 0.4400 | 1.0000 | 0.0000 | 0.9680 | 0.0320 | 0.9860 | 0.0140 | 0.4910 | 0.5090 |
| Indonesian7 | 107 | 0.9910 | 0.0090 | 0.9390 | 0.0610 | 0.5050 | 0.4950 | 1.0000 | 0.0000 | 0.9950 | 0.0050 | 0.9670 | 0.0330 | - | - |
| Vietnamese13 | 120 | 0.9860 | 0.0140 | 0.9530 | 0.0470 | 0.4860 | 0.5140 | 1.0000 | 0.0000 | 0.9720 | 0.0280 | 0.9860 | 0.0140 | 0.4770 | 0.5230 |
| Han Chinese6 | 1,000 | 0.9940 | 0.0060 | 0.9515 | 0.0485 | 0.5945 | 0.4055 | 0.9955 | 0.0045 | 0.9860 | 0.0140 | 0.9865 | 0.0135 | 0.5320 | 0.4680 |
| Taiwanese8 | 300 | 0.9967 | 0.0033 | 0.9600 | 0.0400 | 0.5750 | 0.4250 | 0.9983 | 0.0017 | 0.9850 | 0.0150 | 0.9633 | 0.0367 | 0.5380 | 0.4620 |
| Japanese20 | 73 | 0.9980 | 0.0020 | 0.9000 | 0.1000 | 0.7180 | 0.2820 | 0.9890 | 0.0110 | 0.9730 | 0.0270 | 0.9730 | 0.0270 | - | - |
| Korean19 | 200 | 0.9880 | 0.0120 | 0.9230 | 0.0770 | 0.5550 | 0.4450 | 0.9900 | 0.0100 | 0.9780 | 0.0220 | 0.9800 | 0.0200 | - | - |
| Oceania | |||||||||||||||
| French Polynesian13 | 81 | 0.9750 | 0.0250 | 0.9130 | 0.0870 | 0.5990 | 0.4010 | 1.0000 | 0.0000 | 0.9750 | 0.0250 | 0.9320 | 0.0680 | 0.4630 | 0.5370 |
| Australian47 | 185 | 0.8580 | 0.1420 | 0.9270 | 0.0730 | 0.6190 | 0.3810 | 1.0000 | 0.0000 | 0.9050 | 0.0950 | - | - | - | - |
Results
Results of genotyping for HPA-1 to -6 and -15, of the 600 samples are shown according to racial groups in Table III. All results were consistent with Hardy-Weinberg equilibrium except HPA-4 in Malays (P=0.003) among whom no heterozygous HPA-4a/4b individuals were observed but a single HPA-4b homozygous individual was identified. The most common genotype found in the subjects of this study was HPA-1a/1a-2a/2a-3a/3b-4a/4a-5a/5a-6a/6a-15a/15b (17%), followed by HPA-1a/1a-2a/2a-3a/3a-4a/4a-5a/5a-6a/6a-15a/15b (14.33%). The former has the highest prevalence among Malays (20%) and Chinese (23%), while the latter is most common among Indians (16%). Chinese subjects were also found to have a lower number of haplotype combinations (n=15) as compared to Malays and Indians (n=22 each) implying that the Malays and Indians in Malaysia may have more diverse origins as compared to the Chinese in the country.
Table III.
Gene frequencies of HPA-1 to -6 and -15 in the Malaysian population.
| System | Genotype | Malay | Chinese | Indian | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| No. | % | *p-value | Allele frequency | No. | % | *p-value | Allele frequency | No. | % | *p-value | Allele frequency | |||||
|
|
|
|
||||||||||||||
| a | b | a | b | a | b | |||||||||||
| HPA-1 | aa | 190 | 95.0 | 200 | 100.0 | 155 | 77.5 | |||||||||
| ab | 10 | 5.0 | 0 | 0.0 | 44 | 22.0 | ||||||||||
| bb | 0 | 0.0 | 1.000 | 0.9750 | 0.0250 | 0 | 0.0 | 1.0000 | 0.0000 | 1 | 0.5 | 0.482 | 0.8850 | 0.1150 | ||
| HPA-2 | aa | 185 | 92.5 | 188 | 94.0 | 185 | 92.5 | |||||||||
| ab | 15 | 7.5 | 11 | 5.5 | 14 | 7.0 | ||||||||||
| bb | 0 | 0.0 | 1.000 | 0.9625 | 0.0375 | 1 | 0.5 | 0.182 | 0.9675 | 0.0325 | 1 | 0.5 | 0.268 | 0.9600 | 0.0400 | |
| HPA-3 | aa | 49 | 24.5 | 66 | 33.0 | 78 | 39.0 | |||||||||
| ab | 103 | 51.5 | 97 | 48.5 | 92 | 46.0 | ||||||||||
| bb | 48 | 24.0 | 1.000 | 0.5025 | 0.4975 | 37 | 18.5 | 0.665 | 0.5725 | 0.4275 | 30 | 15.0 | 0.221 | 0.6200 | 0.3800 | |
| HPA-4 | aa | 199 | 99.5 | 199 | 99.5 | 199 | 99.5 | |||||||||
| ab | 0 | 0.0 | 1 | 0.5 | 1 | 0.5 | ||||||||||
| bb | 1 | 0.5 | 0.003 | 0.9950 | 0.0050 | 0 | 0.0 | 1.000 | 0.9975 | 0.0025 | 0 | 0.0 | 1.000 | 0.9975 | 0.0025 | |
| HPA-5 | aa | 180 | 90.0 | 193 | 96.5 | 177 | 88.5 | |||||||||
| ab | 20 | 10.0 | 7 | 3.5 | 22 | 11.0 | ||||||||||
| bb | 0 | 0.0 | 1.000 | 0.9500 | 0.0500 | 0 | 0.0 | 1.000 | 0.9825 | 0.0175 | 1 | 0.5 | 0.520 | 0.9400 | 0.0600 | |
| HPA-6 | aa | 197 | 98.5 | 193 | 96.5 | 198 | 99.0 | |||||||||
| ab | 3 | 1.5 | 7 | 3.5 | 2 | 1.0 | ||||||||||
| bb | 0 | 0.0 | 1.000 | 0.9925 | 0.0075 | 0 | 0.0 | 1.000 | 0.9825 | 0.0175 | 0 | 0.0 | 1.000 | 0.9950 | 0.0050 | |
| HPA-15 | aa | 50 | 25.0 | 46 | 23.0 | 36 | 18.0 | |||||||||
| ab | 106 | 53.0 | 107 | 53.5 | 91 | 45.5 | ||||||||||
| bb | 44 | 22.0 | 0.479 | 0.5150 | 0.4850 | 47 | 23.5 | 0.395 | 0.4975 | 0.5025 | 73 | 36.5 | 0.462 | 0.4075 | 0.5925 | |
p-value of Hardy-Weinberg test
Table II shows the distribution of HPA alleles found in our study together with the distributions reported for other populations worldwide. Results of hierarchical clustering and principal component analysis performed on selected populations are graphically represented in Figures 1 and 2, respectively.
Figure 1.
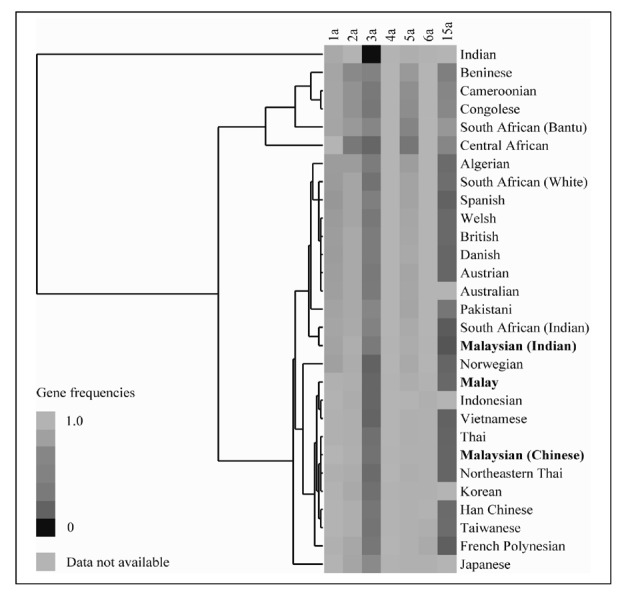
Heat-map showing gene frequencies for the “a” allele of HPA-1 to -6 and -15, together with a cluster dendrogram showing the relationships of various racial groups based on the frequency distribution.
Figure 2.
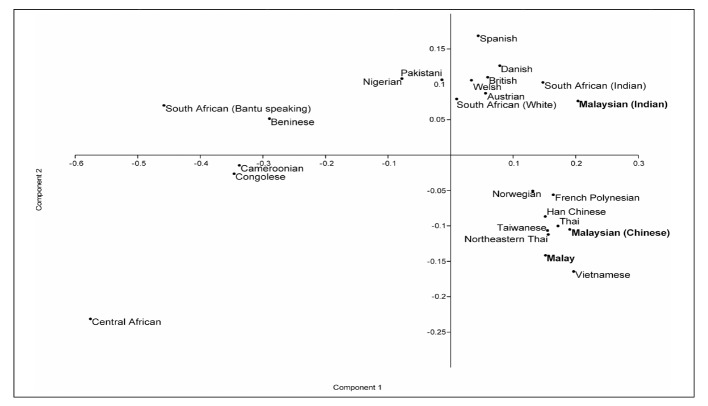
Principal component analysis based on the gene frequencies of HPA-1 to -5 and -15 showing the relationships among various racial groups worldwide. Contributions of the first and second components were 69.76% and 16.95%, respectively.
Discussion
HPA genotyping of donor blood and establishment of a donor registry for HPA is useful to manage platelet-specific alloimmunisation and facilitate the selection of matching HPA types for patients who possess rare alleles. Among all the HPA alleles tested, only HPA-4 showed significant disequilibrium, which was limited to Malays. Low heterozygosity usually implies a certain degree of inbreeding in a population. However, such a conclusion should not be drawn in this case because the HPA-4b allele is present at an extremely low frequency. It is interesting that the HPA-4b allele, commonly associated with Japanese and Koreans, has been found in mainland China and India but has not been observed in other South-East Asian populations such as the Indonesian7, Vietnamese13, and Thai14,15. It is possible that the HPA-4b allele has only recently been introduced to the Malay gene pool through admixture with other ethnic groups and it is not widely spread as yet in the gene pool. Most of the cases of platelet immunisation against HPA-4 have been reported in Japan with only two cases of neonatal alloimmune thrombocytopenia due to anti-HPA -4b reported having been reported in Caucasians16,17. Apart from HPA-4b, HPA-6b is also nearly exclusively found within the Asian population18.
The “b” allele generally occurred at low frequencies (<0.1) in all HPA systems tested except for HPA-1, 3 and -15. The HPA-1b allele frequency among Indians was 0.115, which was significantly higher than the frequency of 0.025 and 0 recorded among Malays and Chinese, respectively. The absence of the HPA-1b allele among Chinese subjects in our population reflects its extremely rare occurrence among East Asian populations, similar to that reported in many other studies6,8,19,20. When comparing HPA-1 genotypes between the three racial groups, all pairs were significantly different (Malay and Chinese, P=0.03; Malay and Indian, P<0.001; Chinese and Indian, P<0.001) indicating that each group has different risks of alloimmunisation caused by the HPA-1 system. It should, however, be noted that the risk of neonatal alloimmune thrombocytopenia is modulated not only by anti-HPA antibodies, but also by factors such as HLA type21.
Comparisons of each of the three Malaysian racial groups to each of the other populations using Fisher’s exact test as shown in Table II, revealed significant differences (P<0.001) for the HPA-2 and -5 allele frequencies to those in populations of African origin. The “b” alleles of these two systems were observed to be generally higher in Africa, and show a gradual decrease toward the east22. Lower frequencies of these alleles have been reported in Europeans and Asians.
The cluster dendr ogram illustrated in Figure 1 shows that Malays are clustered near to Indonesians and Vietnamese which are then joined by Thais and the other East Asian populations, including Chinese of Han and Malaysian origin, Koreans and Taiwanese. This would be consistent with the hypothesis of a central migratory origin of South-East Asian populations, possibly together with Polynesians who cluster closely together with the above groups. Malaysians of Indian ethnicity cluster together with the Indians of South Africa, consistent with a common point of emigration. Principal component analysis showed similar results although we limited the analysis to HPA-1 to -5 and -15 due to the non-availability of HPA-6 frequency data for many of the populations studied. As reported by Kulkarni et al.5, the allele frequencies of Indians, composed mainly of Maharashtrians, Parsis and various other native groups in northern India, is substantially different from those of all other populations due to the unusually high predominance of the HPA-3b allele in this group. This aberrancy remains unexplained. The clustering of related racial groups using a limited set of six to seven HPA markers reveals the utility of HPA polymorphisms in charting population movements.
In conclusion, this study provides comprehensive information on HPA allele distribution among the major ethnic groups in Malaysia. We show that the HPA allelic profile of Malays is closely linked to that of other ethnic groups within the South-East Asian region. This information can serve as an outline for future clinical research associated with platelet disorders and also form the basis of a databank on platelet antigen polymorphisms.
Acknowledgments
This work was supported in part by a Postgraduate Research Fund (N. PS245-2010B) from the University of Malaya.
Footnotes
The Authors declare no conflicts of interest.
References
- 1.Rozman P. Platelet antigens. The role of human platelet alloantigens (HPA) in blood transfusion and transplantation. Transpl Immunol. 2002;10:165–81. doi: 10.1016/s0966-3274(02)00063-1. [DOI] [PubMed] [Google Scholar]
- 2.Ghevaert C, Rankin A, Huiskes E, et al. Alloantibodies against low-frequency human platelet antigens do not account for a significant proportion of cases of fetomaternal alloimmune thrombocytopenia: evidence from 1054 cases. Transfusion. 2009;49:2084–9. doi: 10.1111/j.1537-2995.2009.02246.x. [DOI] [PubMed] [Google Scholar]
- 3.McBride SE. Real-time PCR assays for high-throughput human platelet antigen typing. In: Bugert P, editor. DNA and RNA Profiling in Human Blood Methods in Molecular Biology. New York: Humana Press; 2009. pp. 39–49. [DOI] [PubMed] [Google Scholar]
- 4.Curtis BR. Genotyping for human platelet alloantigen polymorphisms: applications in the diagnosis of alloimmune platelet disorders. Semin Thromb Hemost. 2008;34:539–48. doi: 10.1055/s-0028-1103365. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni B, Mohanty D, Ghosh K. Frequency distribution of human platelet antigens in the Indian population. Transfus Med. 2005;15:119–24. doi: 10.1111/j.0958-7578.2005.00561.x. [DOI] [PubMed] [Google Scholar]
- 6.Feng ML, Liu DZ, Shen W, et al. Establishment of an HPA-1- to -16-typed platelet donor registry in China. Transfus Med. 2006;16:369–74. doi: 10.1111/j.1365-3148.2006.00687.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu TC, Shih MC, Lin CL, et al. Gene frequencies of the HPA-1 to HPA-8w platelet antigen alleles in Taiwanese, Indonesian, and Thai. Ann Hematol. 2002;81:244–8. doi: 10.1007/s00277-002-0451-x. [DOI] [PubMed] [Google Scholar]
- 8.Shih MC, Liu TC, Lin IL, et al. Gene frequencies of the HPA-1 to HPA-13, Oe and Gov platelet antigen alleles in Taiwanese, Indonesian, Filipino and Thai populations. Int J Mol Med. 2003;12:609–14. doi: 10.3892/ijmm.12.4.609. [DOI] [PubMed] [Google Scholar]
- 9.Vega FMDL, Lazaruk KD, Rhodes MD, Wenz MH. Assessment of two flexible and compatible SNP genotyping platforms: TaqMan® SNP Genotyping Assays and the SNPlex™ Genotyping System. Mutation Res. 2005;573:111–35. doi: 10.1016/j.mrfmmm.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Amengual O, Atsumi T, Komano Y, et al. A polymorphism in human platelet antigen 6b and risk of thrombocytopenia in patients with systemic lupus erythematosus. Arthritis Rheum. 2007;56:2803–5. doi: 10.1002/art.22799. [DOI] [PubMed] [Google Scholar]
- 11.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–7. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 12.Hammer Ø, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4 [Google Scholar]
- 13.Halle L, Bach KH, Martageix C, et al. Eleven human platelet systems studied in the Vietnamese and Ma’ohis Polynesian populations. Tissue Antigens. 2004;63:34–40. doi: 10.1111/j.1399-0039.2004.00149.x. [DOI] [PubMed] [Google Scholar]
- 14.Kupatawintu P, Nathalang O, O-Chareon R, Patmasiriwat P. Gene frequencies of the HPA-1 to 6 and Gov human platelet antigens in Thai blood donors. Immunohematology. 2005;21:5–9. [PubMed] [Google Scholar]
- 15.Romphruk AV, Akahat J, Srivanichrak P, et al. Genotyping of human platelet antigens in ethnic Northeastern Thais by the polymerase chain reaction-sequence specific primer technique. J Med Assoc Thai. 2000;83:1333–9. [PubMed] [Google Scholar]
- 16.Ohto H, Miura S, Ariga H, et al. The natural history of maternal immunization against foetal platelet alloantigens. Transfus Med. 2004;14:399–408. doi: 10.1111/j.1365-3148.2004.00535.x. [DOI] [PubMed] [Google Scholar]
- 17.Puig N, Muñiz-Diaz E, Monteagudo E, et al. A second case of neonatal alloimmune thrombocytopenia by anti-HPA-4b (anti-Yuka) in a Caucasian family. Transfus Med. 1993;3:164–5. doi: 10.1111/j.1365-3148.1993.tb00058.x. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka S, Ohnoki S, Shibata H, et al. Gene frequencies of human platelet antigens on glycoprotein IIIa in Japanese. Transfusion. 1996;36:813–7. doi: 10.1046/j.1537-2995.1996.36996420760.x. [DOI] [PubMed] [Google Scholar]
- 19.Seo DH, Park SS, Kim DW, et al. Gene frequencies of eight human platelet-specific antigens in Koreans. Transfus Med. 1998;8:129–32. doi: 10.1046/j.1365-3148.1998.00138.x. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka S, Taniue A, Nagao N, et al. Simultaneous DNA typing of human platelet antigens 2, 3 and 4 by an allele-specific PCR method. Vox Sanguinis. 1995;68:225–30. doi: 10.1111/j.1423-0410.1995.tb02577.x. [DOI] [PubMed] [Google Scholar]
- 21.Stuge TB, Skogen B, Ahlen MT, et al. The cellular immunobiology associated with fetal and neonatal alloimmune thrombocytopenia. Transfus Apher Sci. 2011;41:53–9. doi: 10.1016/j.transci.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Halle L, Bigot A, Mulen-Imandy G, et al. HPA polymorphism in sub-Saharan African populations: Beninese, Cameroonians, Congolese, and Pygmies. Tissue Antigens. 2005;65:295–8. doi: 10.1111/j.1399-0039.2005.00360.x. [DOI] [PubMed] [Google Scholar]
- 23.De La Vega Elena CD, Nogues N, Fernandez Montoya A, et al. Human platelet-specific antigens frequencies in the Argentinean population. Transfus Med. 2008;18:83–90. doi: 10.1111/j.1365-3148.2007.00819.x. [DOI] [PubMed] [Google Scholar]
- 24.Castro V, Origa AF, Annichino-Bizzacchi JM, et al. Frequencies of platelet-specific alloantigen systems 1–5 in three distinct ethnic groups in Brazil. Eur J Immunogenet. 1999;26:355–60. doi: 10.1046/j.1365-2370.1999.00174.x. [DOI] [PubMed] [Google Scholar]
- 25.Brouk H, Halle L, Bertrand G, et al. Human platelet antigen allele frequencies in different Algerian populations. Tissue Antigens. 2010;75:673–8. doi: 10.1111/j.1399-0039.2009.01429.x. [DOI] [PubMed] [Google Scholar]
- 26.Ferrer G, Muniz-Diaz E, Aluja MP, et al. Analysis of human platelet antigen systems in a Moroccan Berber population. Transfus Med. 2002;12:49–54. doi: 10.1046/j.1365-3148.2002.00349.x. [DOI] [PubMed] [Google Scholar]
- 27.Foxcroft Z. Human platelet antigen (HPA) 1–5 and -15 frequencies in South African blood donors. Africa Sanguine. 2008;11:15. [Google Scholar]
- 28.Mojaat N, Halle L, Proulle V, et al. Gene frequencies of human platelet antigens in the Tunisian population. Tissue Antigens. 1999;54:201–4. doi: 10.1034/j.1399-0039.1999.540214.x. [DOI] [PubMed] [Google Scholar]
- 29.Holensteiner A, Walchshofer S, Adler A, et al. Human platelet antigen gene frequencies in the Austrian population. Haemostasis. 1995;25:133–6. doi: 10.1159/000217152. [DOI] [PubMed] [Google Scholar]
- 30.Jones DC, Bunce M, Fuggle SV, et al. Human platelet alloantigens (HPAs): PCR-SSP genotyping of a UK population for 15 HPA alleles. Eur J Immunogen. 2003;30:415–9. doi: 10.1111/j.1365-2370.2003.00426.x. [DOI] [PubMed] [Google Scholar]
- 31.Tomicic M, Bingulac-Popovic J, Drazic V, Hundric-Haspl Z. Frequency of HPA-15a and HPA-15b (Gov a/b) human platelet alloantigens in the Croatian population. Arch Med Res. 2006;37:172–4. doi: 10.1016/j.arcmed.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Pavic M, Zadro R, Coen Herak D, et al. Gene frequencies of platelet-specific antigens in Croatian population. Transfusion Medicine. 2010;20:73–7. doi: 10.1111/j.1365-3148.2009.00971.x. [DOI] [PubMed] [Google Scholar]
- 33.Korinkova P, Suttnar J, Vytiskova J, Stehlikova M. [Fetomaternal alloimmune thrombocytopenia--possibilities of diagnosis and exchange transfusion] Ceska Gynekol. 1999;64:28–31. [PubMed] [Google Scholar]
- 34.Steffensen R, Kaczan E, Varming K, Jersild C. Frequency of platelet-specific alloantigens in a Danish population. Tissue Antigens. 1996;48:93–6. doi: 10.1111/j.1399-0039.1996.tb02613.x. [DOI] [PubMed] [Google Scholar]
- 35.Merieux Y, Debost M, Bernaud J, et al. Human platelet antigen frequencies of platelet donors in the French population determined by polymerase chain reaction with sequence-specific primers. Pathol Biol. 1997;45:697–700. [PubMed] [Google Scholar]
- 36.Carlsson LE, Greinacher A, Spitzer C, et al. Polymorphisms of the human platelet antigens HPA-1, HPA-2, HPA-3, and HPA-5 on the platelet receptors for fibrinogen (GPIIb/IIIa), von Willebrand factor (GPIb/IX), and collagen (GPIa/IIa) are not correlated with an increased risk for stroke. Stroke. 1997;28:1392–5. doi: 10.1161/01.str.28.7.1392. [DOI] [PubMed] [Google Scholar]
- 37.Mazzucco L, Santi R, Contino L. HPA-1/6 allelomorphism study in juvenile stroke: the possible role of HPA-2b and HPA-5b. 6th European Symposium on Platelet, Granulocyte and Red Cell Immunobiology (Abstract book); Amsterdam. 2000. [Google Scholar]
- 38.Sabbagh AS, Taher AT, Zaatari GS, Mahfouz RAR. Gene frequencies of the HPA-1 platelet antigen alleles in the Lebanese population. Transfusion Medicine. 2007;17:473–8. doi: 10.1111/j.1365-3148.2007.00792.x. [DOI] [PubMed] [Google Scholar]
- 39.Pavkovic M, Petlichkovski A, Strezova A, et al. Gene frequencies of human platelet antigens in the Macedonian population. Tissue Antigens. 2006;67:241–6. doi: 10.1111/j.1399-0039.2006.00551.x. [DOI] [PubMed] [Google Scholar]
- 40.Randen I, Sorensen K, Killie MK, Kjeldsen-Kragh J. Rapid and reliable genotyping of human platelet antigen (HPA)-1, -2, -3, -4, and -5 a/b and Gov a/b by melting curve analysis. Transfusion. 2003;43:445–50. doi: 10.1046/j.1537-2995.2003.00354.x. [DOI] [PubMed] [Google Scholar]
- 41.Drzewek K, Brojer E, Zupanska B. The frequency of human platelet antigen (HPA) genotypes in the Polish population. Transfus Med. 1998;8:339–42. doi: 10.1046/j.1365-3148.1998.00164.x. [DOI] [PubMed] [Google Scholar]
- 42.Rozman P, Drabbels J, Schipper RF, et al. Genotyping for human platelet-specific antigens HPA-1, -2, -3, -4 and -5 in the Slovenian population reveals a slightly increased frequency of HPA-1b and HPA-2b as compared to other European populations. Eur J Immunogenet. 1999;26:265–9. doi: 10.1046/j.1365-2370.1999.00142.x. [DOI] [PubMed] [Google Scholar]
- 43.Muniz-Diaz E, Arilla M, Ibanez M, et al. Frequency of platelet alloantigens in the Spanish population. Sangre (Barc) 1993;38:289–93. [PubMed] [Google Scholar]
- 44.Boehlen F, Bulla O, Michel M, et al. HPA-genotyping and antiplatelet antibodies in female blood donors. Hematol J. 2003;4:441–4. doi: 10.1038/sj.thj.6200338. [DOI] [PubMed] [Google Scholar]
- 45.Sellers J, Thompson J, Guttridge MG, Darke C. Human platelet antigens: typing by PCR using sequence-specific primers and their distribution in blood donors resident in Wales. Eur J Immunogen. 1999;26:393–7. doi: 10.1046/j.1365-2370.1999.00176.x. [DOI] [PubMed] [Google Scholar]
- 46.Bhatti FA, Uddin M, Ahmed A, Bugert P. Human platelet antigen polymorphisms (HPA-1, -2, -3, -4, -5 and -15) in major ethnic groups of Pakistan. Transfus Med. 2010;20:78–87. doi: 10.1111/j.1365-3148.2009.00982.x. [DOI] [PubMed] [Google Scholar]
- 47.Bennett JA, Palmer LJ, Musk AW, Erber WN. Gene frequencies of human platelet antigens 1–5 in indigenous Australians in Western Australia. Transfus Med. 2002;12:199–203. doi: 10.1046/j.1365-3148.2002.00371.x. [DOI] [PubMed] [Google Scholar]