Abstract
Frequently, it is important to ascertain whether a molecule that is involved in one model of pain is also involved in other models of pain. Similarly, it may be important to determine whether a molecule involved in nociception in one tissue is also expressed in other tissues and to ascertain the degree of enrichment. Additionally, before initiating a complex set of experiments or purchasing an expensive immunoassay kit, it may be useful to obtain initial supporting evidence to justify the time and money. Is the transcript for the target receptor, protein, or peptide precursor present in, for example, the dorsal root ganglion? And, if present, how abundant is it? Here is where the power of PCR can be applied to obtain a quick but informative answer. In this chapter, we mainly detail the use of gel-based RT-PCR and also provide suggestions on tissue dissection and interpretation of results. The use of gel-based RT-PCR can address many of the questions of abundance or tissue specificity with a minimum of expense and time.
Keywords: RT-PCR, Nociception, mRNA, Gene regulation, Pain, Pirt, Dorsal root ganglion
1. Introduction
Although essential for host survival, pain can become unbearable especially when chronic. Accordingly, a great deal of money is expended for current pain relief medications and treatments, as well as invested to find newer, better treatments. A key to discovering better pain therapeutics inevitably involves understanding the molecules that constitute the pain circuits, starting from damaged peripheral tissue and the peripheral nervous system (1) and ending in cerebral cortex (2). The identification of TRPV1, a channel involved in transduction of nociceptive stimuli in peripheral nerve terminals (3), the expression of which is highly enriched in DRG, serves as an example of how new molecular insights can translate into new therapeutic approaches. For example, the intrathecal administration of the ultrapotent TRPV1 agonist, resiniferatoxin, selectively depletes TRPV1-expressing ganglionic neurons (4) and results in a dramatic reduction of pain from inflammation, cancer and arthritis in animals (5, 6). TRPV1 antagonists are also capable of controlling pain in experimental models although human clinical trials have encountered problems with drug-induced hyperthermia (7).
There are many known and likely many as yet unrecognized molecules that are involved in nociceptive transmission and the neural processing of pain in general. Depending on the cause of pain (e.g. inflammation or nerve injury), a different cascade of pain-inducing molecules appears to be activated, although there is considerable overlap. These molecular changes can occur in circulating and infiltrating leukocytes, resident cells of the affected peripheral tissues, in nerve endings and axons of dorsal root ganglia neurons, in various layers of the spinal cord and a wide variety of brain regions. Molecular alterations due to peripheral inflammation also occur in the choroid plexus, which makes the cerebrospinal fluid, and changes in secreted factors may influence brain function in a very broad fashion (8). The array and diversity of possible changes, therefore, underscore the range of questions that can be asked in attempting to unravel the roles of these molecules and neural circuits in various painful conditions.
For some of these questions, RT-PCR has proven to be a valuable tool in assessing the involvement of a particular gene in a persistent pain state. The focus of this chapter is to describe the advantages and comparative ease of using gel-based RT-PCR to establish the relative expression level of a particular molecule in a target tissue or its regulation during nociception.
2. Materials
2.1. RNA Isolation
Trizol (Invitrogen, Carlsbad, CA).
RNeasy Mini Kit (Qiagen, Valencia, CA).
ZR-Whole Blood Total RNA Kit™ (Zymo Research, Orange, CA).
Sonicator (Sonics and Materials, Inc., Newtown, CT) (see Note 1).
RNase-Free DNase Set (Qiagen).
2.2. RNA Quantitation
Quant-iT™ Ribogreen RNA assay kit (Invitrogen).
SpectraMax Gemini XS Fluorescent plate reader.
96-well plate.
2.3. RT-PCR
Access RT-PCR System (Promega, Madison, WI).
PCR Strip Tubes (Axygen, Union City, CA).
Robocycler (Strategene, La Jolla, CA, the thermal cycler we use in our lab).
Tomy Capsulefuge Model PMC-860 (Research Products International Corp., Mt. Prospect, IL).
2.4. Making Gels
2.5. Running Gel
60 well HLA Plate (Nunc, Thermo Fisher Scientific, Rochester, NY).
1 kb DNA Ladder Mix (Crystalgen, Plainview, NY) (see Note 6).
50× Tris–Acetate–EDTA Buffer (TAE).
Power Supply Model 250.
Casting Tray.
Ethidium Bromide 10 mg/ml (Invitrogen).
Blue/Orange 6× Loading Dye (Promega) (see Note 7).
Aerosol tips (see Note 8)
2.6. Obtaining and Quantitating Gel Image
FluorChem 8900 System (Alpha Innotech Corp, San Leandro, CA).
A connection to a PC.
ImageQuant 5.2 (Molecular Dynamics, Piscataway, NJ).
3.Methods
When deciding to use gel-based RT-PCR (also called traditional RT-PCR) it is important to understand the advantages and disadvantages of the technique, which in part can be explained by the mechanism of PCR. PCR amplifications can be broken down into three phases. The exponential phase occurs when the reaction components are in excess thereby theoretically allowing for a doubling of the PCR product per cycle. In the next phase, the amplification of the PCR product occurs linearly rather exponentially as the reagents are partially depleted. Plateau phase represents the point in which the amplifications have further slowed or have completely stopped.
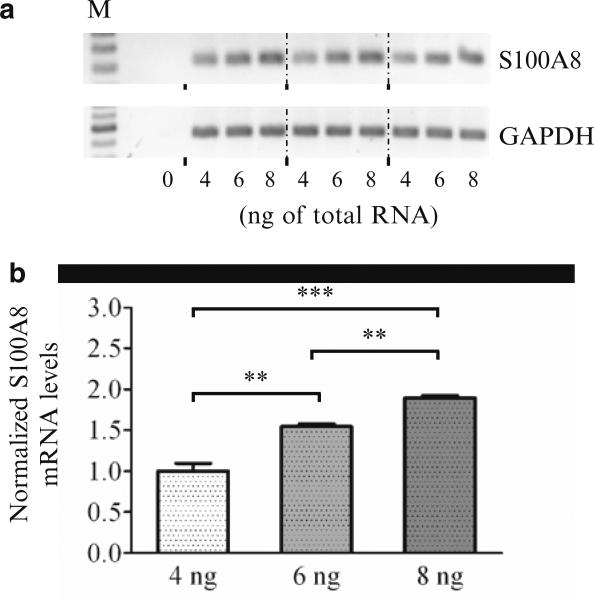
A minor disadvantage of gel-based RT-PCR is that the reaction generally must proceed into the linear phase before the product can be visualized. Thus, when comparing transcript expression in multiple samples, it is likely that samples that contain higher levels of transcript will enter the linear phase well before the other samples, which are still accumulating product exponentially. In this situation, when too many cycles are used, the gel-based approach can result in a reduced difference in expression between samples (i.e., by the time the lower-expressing samples are visualized the higher-expressing sample may reach a plateau). Conversely, an artificially magnified difference may be inferred if too few cycles are used. Moreover, in cases where the actual change in expression is extremely small, it may be possible that this technique may fail to detect those differences (see Fig. 1). Usually, after examination of the expression level in the first RT-PCR amplification by gel electrophoresis, a second amplification starting with a new aliquot of RNA, where the number of cycles is appropriately adjusted up or down, can place the results into the linear range for most studies.
Fig. 1.
Ability of gel-based RT-PCR to detect small differences in expression. (a) Total RNA from spinal cord was used to amplify S100A8 expression. In triplicate, either 4, 6 or 8 ng of total RNA from one sample was used per RT-PCR reaction. For normalization, GAPDH expression was separately determined by adding 8 ng of the same RNA for all nine reactions. The RT-PCR products for S100A8 and GAPDH were run in three blocks (4, 6, 8 ngfirst set, 4, 6, 8 ngsecond set, 4, 6, 8 ngthird set) on the gel rather than loading them in a 444, 666 and 888 pattern. The former arrangement is useful in case one side of the gel yields weaker signals than the other side due to non-uniformity of UV transillumination. (b) The RT-PCR data in (a) were quantified and the fold change is shown. Analysis demonstrated that 6 ng of input RNA resulted in a signal that was 1.54 times higher than that obtained when 4 ng of RNA was used (1.50 is the expected value) and 1.23 times lower when 8 ng was used (1.33 is expected). Additionally, the 8 ng samples resulted in a signal that was 1.89 times higher than that obtained with 4 ng (2.00 is expected). Thus, these data demonstrate that analysis by gel-based RT-PCR can be used to detect small changes in gene expression
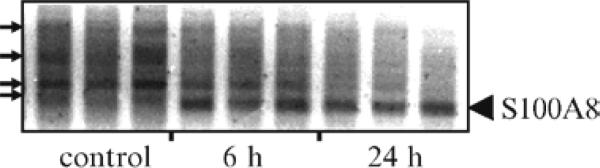
In contrast to gel-based RT-PCR, real time quantitative RT-PCR (qRT-PCR) allows for comparison of samples while each are in the exponential phase, thus, in theory, there is no biasing against samples that start with different expression levels. Despite the advantages of qRT-PCR, it is possible, however, to obtain different quantitative measurements even if performed in the same lab (9). One factor which can contribute to such variation is the failure to appreciate nonspecific amplicons. This is more likely to be seen in the simpler, less expensive SYBR green qRT-PCR assays as compared to Taqman or other qRT-PCR approaches. SYBR green is an intercalating dye and will stain any double-stranded DNA in the RT-PCR tube, thus both specific and nonspecific amplification products will provide signal to the fluorometric detector. Nonspecific amplification, which can vary from sample to sample, also can result in depletion of reaction components and a skewing of data. Although this implies that one should use primers that yield one amplicon (and no primer-dimers), this can sometimes be complicated. When the transcript of interest is expressed at sufficient levels in a sample, it is usually easy to obtain one dominant amplicon. However, when the transcript is expressed at low levels in the samples, it is possible that the primers will latch on to other transcripts giving one or more nonspecific products (see Fig. 2). Visualization of the reaction products on gels can aid in understanding the extent of this artifact. This is very important information to obtain since it can guide choices of qRT-PCR methodology and also be a determinant of one's commitment to the target protein itself: if the gene is expressed at very low levels, analysis of the transcript or protein will present challenges when using other methods such as in situ hybridization, western blot, or immunocytochemistry. When little or no prior data are available on the expression level of a particular gene in a specific tissue, a quick evaluation by RT-PCR, as outlined here, can be most informative (see Fig. 3).
Fig. 2.
Potential for amplification of nonspecific PCR-products when target gene is minimally or not expressed in samples. Gel shows PCR products after amplification of S100A8 in spinal cord samples taken from control animals or animals 6 or 24 h after hind paw inflammation. Generally, S100A8 is undetectable in spinal cord samples. However, due to an influx of neutrophils, which highly express S100A8, into the CNS vasculature, the spinal cord samples contain high levels of S100A8 after peripheral inflammation (13). Thus in the 6 and 24 h samples, high levels of S100A8 are detected on the gel. In the control samples, we failed, as expected, to detect an amplicon corresponding to S100A8. Instead, we detected several nonspecific amplicons that were not (or minimally) expressed in the 6 and 24 h samples. Failure to identify these nonspecific amplicons, in for example, the SYBR green qRT-PCR assays can in some cases lead to a misquantification of gene expression changes. Schemes to reduce nonspecific amplicons are described in Notes and Trouble Shooting sections. **P < 0.01 and **P < 0.001, as determined by Student's t-test
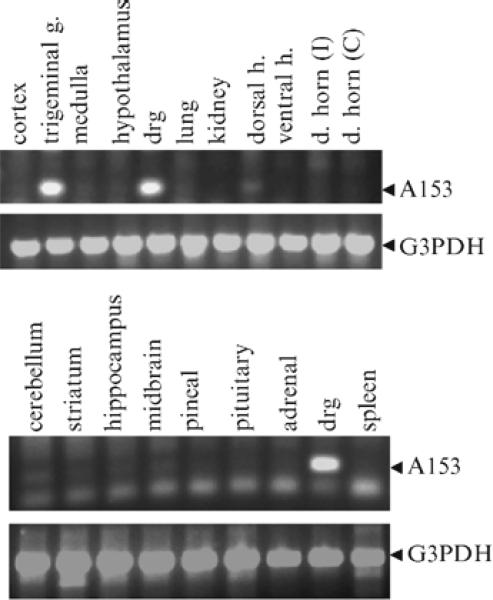
Fig. 3.
Gel-based RT-PCR analysis to determine expression profile of a gene in different tissues. After subtraction hybridization analysis (DRG-sciatic nerve genes), we identified numerous clones that were enriched in the DRG as compared to sciatic nerve. Clone A153 (recently identified as the phosphoinositide-binding protein, Pirt, (14) was tested for expression in numerous tissues. Pirt is highly expressed in DRG and trigeminal ganglia. In contrast, its expression is dramatically weaker (or undetectable) in CNS and was not detected at all in nonperipheral nervous tissues. Thirty-three cycles were used to amplify Pirt. GAPDH expression was used to show that the RNA extracted from each tissue sample was intact
Generally, the technique presented provides relative levels of expression. One approach for getting more quantitative data with gel-based RT-PCR is to generate an operationally-defined standard curve. In this case, different amounts (ng) of RNA from a given sample are amplified and run on a gel. The densitometric values are converted back to the amount of sample used. This curve can be used to examine issues of linearity with the amount of input RNA and the number of cycles of amplification and to compare transcript expression in test samples. It is possible to generate single stranded RNA as a template for making a real standard curve, but this will not be covered in this chapter.
The use of gel-based RT-PCR has several advantages. Assuming some of the equipment is already available, this technique is fairly inexpensive and does not require an expensive real-time PCR device to monitor each cycle (as compared to qRT-PCR). It is simple to perform and can be applied without the use of radioactive materials (as compared to Northern Blots). Below, we describe how to perform gel-based RT-PCR with emphasis on using this tool for semi-quantitative purposes.
3.1. Preparation of Samples
Dissection of tissue and tissue storage: Dissected tissues are frozen immediately on the bottom of a pre-labeled Eppendorf tube, stored at –80°C and care is taken to prevent thawing (see Note 9).
Before sonicating (or homogenizing) samples, push eppendorf tubes containing the entire set of samples deep into dry ice to prevent thawing. Immediately sonicate after the addition of homogenization buffer (done one sample at a time to prevent thawing of subsequent samples).
3.2. Isolation of RNA
Isolation of RNA can be achieved in numerous ways. Below are three common methods to extract RNA from tissue. Note that these are procedures used in our lab, and each kit provides instructions and troubleshooting tips for isolating RNA from tissues and cell cultures.
3.2.1. TRIzol Reagent
Sonicate every 50–100 mg of tissue sample in 1 ml of TRIzol Reagent.
Incubate at RT for 5 min.
Centrifuge at 12,000×g for 10 min at 2–8°C.
Transfer clear supernatant into a new tube.
Add 0.2 ml of chloroform per every 1 ml of TRIzol Reagent.
Shake vigorously by hand for 15 s and incubate at RT for 2–3 min.
Centrifuge at 12,000×g for 15 min at 2–8°C.
Gently collect the aqueous phase without disturbing the interphase into a new tube and add 0.5 ml of isopropyl alcohol per 1 ml of TRIzol Reagent.
Incubate at RT for 10 min.
Centrifuge at 12,000×g for 10 min at 2–8°C.
Remove supernatant and wash pellet with 1 ml of 75% ethanol per ml of TRIzol Reagent.
Vortex and centrifuge at 7,500×g for 5 min at 2–8°C.
Remove ethanol and let pellet air dry.
Add at least 40 μl of RNase-free water and mix thoroughly by pipetting.
For RT-PCR, it is especially important that the samples are not contaminated with DNA. Therefore, a DNase digestion step is highly recommended. Our DNA clean up protocol generally uses the RNeasy Mini Kit (see below), where the initial steps are the addition of 300 μl of Buffer RLT containing 1% β-mercapethanol to a tube containing an equal volume of the RNA sample and ethanol (whose concentration in this tube should be 70%). For example, 210 μl of 100% EtOH and 50 μl of RNase/DNase-free water are added to 40 μl of RNA sample followed by the addition of RLT. The rest of the steps are described in Subheading 3.2.2 (see *).
Store at –80°C. We generally freeze samples once before quantifying RNA.
3.2.2. RNeasy Mini Kit
Sonicate 20–30 mg of tissue in 600 ml of Buffer RLT containing 1% β-mercapethanol.
Centrifuge lysate at maximum speed with table-top centrifuge for 3 min.
Transfer supernatant into a new tube.
Add 1 volume of 70% ethanol and mix immediately by pipetting.
*Apply 700 μl of sample to a pre-labeled RNeasy mini column placed in a 2 ml collection tube.
Centrifuge for 15 s at 8,000×g at RT.
Discard flow-through.
Repeat steps 5–7 if the initial volume of sample was greater than 700 μl.
Add 350 μl of Buffer RW1.
Centrifuge for 15 s at 8,000×g at RT.
Add 80 μl of freshly prepared DNase I solution (prepared in buffer RDD).
Incubate at RT for 15 min.
Add 350 μl of Buffer RW1.
Centrifuge for 15 s at 8,000×g.
Discard flow-through.
Transfer column into new 2 ml collection tube.
Add 500 μl of RPE.
Centrifuge for 15 s at 8,000×g.
Discard flow-through. Add 500 μl of RPE.
Centrifuge for 2 min at 8,000×g.
Place column into new 2 ml collection tube.
Centrifuge at maximum speed for 1 min.
Transfer column into a pre-labeled RNase/DNase-free 1.5 ml tube (provided).
Add 40 μl of RNase/DNase-free water.
Centrifuge for 1 min at 8,000×g.
Store at –80°C.
3.2.3. ZR Whole-Blood Total RNA Kit™
Add 700 μl of ZR buffer to 100 μl of blood.
Mix and transfer to Zymo-spin IIIC™ column in a collection tube.
Centrifuge for 60 s at 12,000×g at RT.
Discard collection tube containing flow-through.
Add 400 μl of RNA prewash buffer to the column in a new collection tube.
Centrifuge for 60 s at 12,000×g.
Discard flow-through.
Add 400 μl of RNA wash buffer to column in a new collection tube.
Centrifuge for 60 s at 12,000×g.
Discard flow-through.
Transfer column into a 1.5 ml tube.
Add 50 μl of RNase/DNase-free water.
Centrifuge for 1 min at 8,000×g.
As describe above, it may be important to perform a DNase digestion step.
Store samples at –80°C.
3.3. Quantification of RNA by Fluorescence
A standard curve is generated with known concentrations of RNA (generally ranging from 0.0 to 1.0 ng/μl) totaling a volume of 250 μl in RNase free TE buffer (pH 7.4). After mixing, duplicates of each concentration are added to a standard 96 well plate at a volume of 100 μl.
RNA samples are diluted (generally 1:100 in RNase-free TE buffer). Per sample, 5 μl of diluted RNA is added to 95 μl of RNase-free TE buffer in the 96 well-plate. Again, this should be done in duplicate.
100 μl of diluted RiboGreen reagent is carefully, but quickly, added to the known standards and to the RNA samples.
The 96 well-plate is gently mixed, stored in dark, and read between 5 and 30 min with a fluorometic plate reader. It takes some time for the dye to intercalate thoroughly into the RNA strands.
3.4. Setup of RT-PCR Assay
If using the Access RT-PCR System, the reaction is set up as following in an RNase-free environment (see Note 10).
Gently mix, quick spin and pipette 21.0 μl into PCR tubes (see Note 11).
Add 8 ng of RNA (2 ng/μl) to each tube while on ice (see Note 12).
It should also be mentioned that the concentration of reagents (e.g. magnesium or primer concentrations) may need to be adjusted from that recommended in Table 1 (see Notes 13 and 14).
Also, it may be good to run a positive control (see Note 15) as well as a negative control (see Note 16) when analyzing the expression of a gene from RNA obtained from newly tested tissues.
Table1.
RT-PCR Reaction Mixture
| Reagents | Per reaction | Master mix 10 |
|---|---|---|
| DNAse/RNAse Free H2O | 8.5 μl | 85.0 μl |
| AMV/Tfl 5× buffer | 5.0 μl | 50.0 μl |
| dNTP (10 μM) | 0.5 μl | 5.0 μl |
| MgSO4 (25 mM) | 1.0 μl | 10.0 μl |
| AMV RT (5 U/μl) | 0.5 μl | 5.0 μl |
| Tfl DNA polymerase (5 U/μl) | 0.5 μl | 5.0 μl |
| Forward primer (10 μM) | 2.5 μl | 25.0 μl |
| Reverse primer (10 μM) | 2.5 μl | 25.0 μl |
| Total volume | 21.0 μl | 210.0 μl |
| 21.0 μl/tube |
3.5. RT-PCR Conditions
The RT-PCR reaction is carried out using a Robocycler thermal cycler according to the manufacturer's instructions.
One cycle (45 min at 45°C) is used for reverse transcription. This is followed by one cycle (2 min at 94°C) of transcriptase inactivation and 26–32 cycles of denaturation, annealing and extension (94°C for 30 s, 55°C for 1 min, and 68°C for 2 min; respectively). A final extension cycle is done at 68°C for 7 min.
Generally, housekeeping genes such as β-actin and GAPDH only require 21–23 cycles, whereas other genes may require 26–32 cycles.
3.6. Loading and Running Gels
Using a HLA tray, 5 μl of RT-PCR product is added to 1 μl of fresh 6× loading dye (see Fig. 4).
50× TAE buffer is diluted to 1× using double distilled H2O. This buffer is used for electrophoresis and to make 2% agarose gels (2 g of agarose/100 ml of buffer). For other options, see also (10).
Place gel in electrophoresis chamber and add TAE buffer to cover gel. Load sample to gel and apply 5 V/cm of gel. When the leading dye approaches the end of the gel, discontinue the current.
Soak the gel for about 5 min in TAE buffer containing 5 μg/ml ethidium bromide. For simplicity, ethidium bromide can also be added to fresh TAE buffer at a concentration of 0.5 μg/ml to make and run gels without the need of post-incubation. Gloves must be worn when using ethidium bromide as it is a mutagen. If the gel is soaked in ethidium bromide solution, the gel must be soaked in tap water for at least 5 min to remove excess ethidium bromide. The tap water must be discarded in proper chemical waste container, whereas the ethidium bromide solution can be reused as long as it is sealed and stored in the dark.
Fig. 4.
Mixing RT-PCR products and loading buffer and dye in HLA tray. We have found the HLA tray to be a useful, convenient and inexpensive piece of labware for the preparation of small-volume samples prior to loading on the gel
3.7. Inspecting the Gel and Acquiring Image
Ideally, the gel should have one major band at the correct MW. The appearance of primer-dimers is possible. It is acceptable to have some nonspecific bands as long as they are not close in MW to the target band, which could prevent quantification.
AlphaEase(FC) software (included with FluorChem 8900) allows for easy capture of a transilluminated gel image (see Note 17). Turn camera and UV transilluminator on and open software. After setting exposure time (auto-exposure or manual exposure), the image can be previewed and acquired. There are many devices of this type for digital capture of the gel image.
3.8. Quantification of the Image
The gel image is opened with ImageQuant 5.2 software.
A rectangle is drawn just large enough to encompass the largest band (or region of interest – ROI). The ROI is then copied and placed over the corresponding band in other lanes.
Select the auto volume report function under the analysis tool to obtain the volume (sum of the individual pixel intensities over the entire ROI) and the background volume.
The volume corresponding to the gene of interest is divided by the volume corresponding to the housekeeping gene to obtain a ratio of target gene to housekeeping gene.
Trouble-shooting (see Note 18).
Failure to obtain a product
Check mRNA integrity and RT-PCR procedure by amplifying another gene, e.g. GAPDH, β-actin or another commonly expressed gene
Check primer integrity by using mRNA from samples known to express the gene of interest
If mRNA and primers are good, a very low copy number may be in the test sample
Increase number of cycles for amplification.
Use nested PCR (i.e. amplify with one primer pair, take product and amplify again with a primer pair that is within the sequence of the first set of primers).
If mRNA is good and primers do not work
Increase MgSO4 concentration in RT-PCR reaction (e.g. to 1.5 mM final concentration)
Lower the annealing temperature
Pick primers from different region of mRNA
If unable to amplify any genes with test mRNA
Re-quantitate diluted and stock RNA concentrations
Ensure working concentration of 2 ng/μl
Ensure RT-PCR reagents are not contaminated by RNase
If unable to measure RNA, then sample may be degraded or contaminated:
Check how you dissected the tissue. Did it take too long? Was the dissected piece place in contact with condensation? Was there an unintentional freeze thaw?
Ensure water used for diluting RNA is RNase/DNase free
Ensure proper storage of RNA (–80°C)
Ensure working in RNase free environment, do not touch the tubes with your bare hands, the skin has enough RNase on it to degrade your sample.
Background problems on gel
Too many bands
Lower MgSO4 concentration Raise the annealing temperature
Pick primers from different region of mRNA
Background too high on gel
Soak in tap water
Change exposure time
If needed for quantification
Use lower cycle numbers to prevent saturation
Run standard curve
Acknowledgments
This research was supported by the Intramural Research Program, NIDCR, NIH, DHHS. We thank Dr. H.-Y. T. Yang for helpful comments.
Footnotes
Sonicating times and settings have to be determined for each tissue type. It is important to consider that incomplete or insufficient sonication can result in a low RNA yield.
For designing primers, we typically input mRNA sequences into primer design programs such as the program found on the following website: http://frodo.wi.mit.edu/. We choose 18–20 mers for primer size, optimal Tm of 60°C, and a GC concentration between 55 and 60%. A more detailed background into the design of primers can be found at http://www.premierbiosoft.com/tech_notes/PCR_Primer_Design.html.
We order primers from Operon, which sends back lyophilized primers along with a specification sheet indicating the nano-moles of oligonucleotide, Tm, molecular weight, and OD. This information is also labeled on the tubes. RNase/DNase free water is used to bring the primers to a concentration of 100 μM (thus for every 100 nmole, 1,000 μl of H2O is used). In a separate, autoclaved Eppendorf tube, an aliquot of the stock is diluted to a working concentration of 10 μM. Primers are stored at –20°C.
As described above, the RT-PCR amplicons are run on a 2.0% agarose gel and the resulting images can be used to quantitate the expression of genes. It is therefore important to obtain the highest signal-to-noise ratio as possible. This includes using primer pairs that result in one amplicon or in which the amplicon of interest is not in close proximity to nonspecific amplicons. It is also important to make sure the gel is void of impurities, such as from dust particulates or air bubbles, which can result in significant quantifiable signals. To reduce impurities, make sure that the agarose is sufficiently boiled till the point of no bubbles and to use a microwaveable, clean plastic lid to prevent unwanted particulates from falling into the agarose (which could occur if paper towels are used as a sealant). It is also important to pour the gel before bubbles reappear but not too hot as to cause warping of the casting tray. Also note that the lid should loosely cover the Erlenmeyer flask to prevent a buildup of pressure and spilling of agarose during microwaving. It may also be useful to microwave in intervals, with stirring in between, since a long continuous microwaving may also cause spilling. We use a 12 × 14 cm gel tray with slots for two sets of combs. 100 ml is poured for each run.
When using gels for quantification, the amount of space between samples is an important practical consideration. Adequate spacing between the teeth is needed for drawing rectangles around amplicons thus making it easier to perform the quantification. Also the shape of the tooth is a factor; if it is too square the product looks like a circle rather than a rectangle. Rectangles are also easier to line up with the molecular weight markers. We prefer using combs whose teeth are distanced at least 1.5 mm from each other. In addition, it may be good to obtain combs that are at least 1.5 mm thick and nearly 4 mm wide which allows for easy pipetting of sample in to wells (5 μl of RT-PCR product plus 1 μl of loading buffer).
Although DNA ladders are a valuable guide for product size, sometimes it may be unclear whether the amplicon is running in the correct position. Here, a positive control, or sequencing the product, may be needed for confirmation. Either of these additional verification tests can be invaluable if there is a spurious band being amplified that runs near the molecular weight of the expected band.
When loading, mix the samples and loading dye thoroughly with the pipettor. This ensures that the samples are as dense as possible. Also make sure that air is not trapped in the pipette tip which could result in air bubbles and an uneven amount of sample loaded which will affect between-lane comparisons. For sample loading, place the tip just inside the well, slowly expel, and then slowly raise the pipette tip. If one needs to step away while loading samples, it is imperative to tightly cover the HLA tray to prevent evaporation of the PCR product. Similarly, if one needs to rerun the PCR product, then store the PCR product at –20°C in tightly capped PCR tubes.
Aerosol tips are highly recommended during many of the steps listed in the Methods. These tips trap liquid and aerosol and thus reduce cross-contamination.
Freeze thaws will lyse the cell and organelles, liberating RNase which will then degrade the RNA. Thus, freeze thaws must be avoided. If sub-dissecting the brain or spinal cord (11), one might be tempted, after removal of the tissue, to perform the subdissection on a chilled surface such as a glass Petri dish on ice. However, condensation can be a problem. If the tissue piece is small and condensation occurs on the plate and comes into contact with the tissue sample, this can lyse the cells and the RNA within will rapidly degrade. At present, we perform dissections at room temperature. If we need to collect a large number of brain areas or perform a complicated dissection (i.e. separate removal of lamina I–II, dorsal columns, lamina X and lamina VII–IX from a trans-verse slice of spinal cord) keeping the tissue chilled is very helpful. Using chilled saline to lubricate the plate surface can help in these circumstances, but each piece of tissue should be frozen quickly. Each small piece of tissue is placed directly into a pre-labeled tube that is on dry ice. Ideally, 1.5 or 2 ml eppendorf tubes should be used due to the volume of reagents needed in subsequent steps for RNA isolation.
Although our lab has never encountered RNase contamination, we still consider this a threat. As a result, we periodically clean the work area and pipettors with RNaseZap or RNase Away. Also, to reduce chances of contamination from DNA sources, it may be helpful to use different pipettors for preparing RT-PCR and applying PCR products to gels.
If pipetting master mix with a single pipette tip, do not push the plunger of the pipettor beyond the first resistance point. Doing so may result in unequal volumes of master mix being loaded. Only push the plunger beyond the first resistance point while attempting to pipette excess solution back into the Master Mix tube.
When setting up RT-PCR reaction, add reagents and RNA to PCR tubes that are placed on ice in order to prevent reverse transcriptase activity.
Optimal MgSO4 concentration can increase the efficiency of reverse transcription and amplification. We generally use a final concentration of 1.0 mM MgSO4 in our initial assays. However, a failure to observe the correct amplicon or the identification of too many products with a set of primers (and template) may suggest the need to vary MgSO4 concentrations until the optimal MgSO4 concentration is attained. If no PCR product is observed, then MgSO4 should be increased. If the gel is full of unwanted bands, then MgSO4 concentration should be decreased, for example, to a final concentration of 0.5 mM. See also Trouble shooting tips below.
We generally use a final concentration of 1 μM for each primer. As with MgSO4 concentration, the concentration of primers can be optimized. However, our preference would be to design primers from other regions of the mRNA rather than change primer concentrations.
It is often important to have a positive control to ensure that the primers amplify the correct product. We always try to do this if possible. For example, if one designs primers for TRPV1 in order to determine whether the encoding gene is expressed in spinal cord, it would be useful for comparative purposes to use RNA extracted from trigeminal or dorsal root ganglia as a control because of the high abundance of this transcript in ganglia.
It is also advisable to run a reaction without RNA to ensure that there is no contamination, which could occur if the working area has been exposed to a plasmid containing the gene of interest or if the amplicon has contaminated the work area and/or pipettors.
It is important to obtain equivalent signals when running identical samples in different wells of a gel. Given, however, the nonuniformity of many UV transilluminators (12), this may be a challenge, especially if the repeated samples are loaded on opposite ends of the gel. Thus, before using gel-based RT-PCR for semiquantitative purposes, one should run identical samples across the entire gel to determine whether the signals are the same. If nonuniformity exists, then running the PCR product in blocks may be helpful (see Fig. 1). For example, with four different samples, it may be better to run them in an ABCD, ABCD, ABCD pattern instead of an AAA, BBB, CCC, DDD pattern. The former pattern would prevent samples AAA from being artifactually different from DDD if, for example, the left half of the gel yields weaker signals than the right half of the gel (due to nonuniformity of the transillumination).
Trouble shooting tips
References
- 1.Yang HY, Mitchell K, Keller JM, Iadarola MJ. Peripheral inflammation increases Scya2 expression in sensory ganglia and cytokine and endothelial related gene expression in inflamed tissue. J Neurochem. 2007;103:1628–1643. doi: 10.1111/j.1471-4159.2007.04874.x. [DOI] [PubMed] [Google Scholar]
- 2.Coghill RC, McHaffie JG, Yen YF. Neural correlates of interindividual differences in the subjective experience of pain. Proc Natl Acad Sci U S A. 2003;100:8538–8542. doi: 10.1073/pnas.1430684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 4.Olah Z, Szabo T, Karai L, Hough C, Fields RD, Caudle RM, Blumberg PM, Iadarola MJ. Ligand-induced dynamic membrane changes and cell deletion conferred by vanilloid receptor 1. J Biol Chem. 2001;276:11021–11030. doi: 10.1074/jbc.M008392200. [DOI] [PubMed] [Google Scholar]
- 5.Karai L, Brown DC, Mannes AJ, Connelly ST, Brown J, Gandal M, Wellisch OM, Neubert JK, Olah Z, Iadarola MJ. Deletion of vanilloid receptor 1-expressing primary afferent neurons for pain control. J Clin Invest. 2004;113:1344–1352. doi: 10.1172/JCI20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown DC, Iadarola MJ, Perkowski SZ, Erin H, Shofer F, Laszlo KJ, Olah Z, Mannes AJ. Physiologic and antinociceptive effects of intrathecal resiniferatoxin in a canine bone cancer model. Anesthesiology. 2005;103:1052–1059. doi: 10.1097/00000542-200511000-00020. [DOI] [PubMed] [Google Scholar]
- 7.Gavva NR. Body-temperature maintenance as the predominant function of the vanilloid receptor TRPV1. Trends Pharmacol Sci. 2008;29:550–557. doi: 10.1016/j.tips.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell K, Yang HY, Berk JD, Tran JH, Iadarola MJ. Monocyte chemoattractant protein-1 in the choroid plexus: a potential link between vascular pro-inflammatory mediators and the CNS during peripheral tissue inflammation. Neuroscience. 2009;158:885–895. doi: 10.1016/j.neuroscience.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali-Seyed M, Laycock N, Karanam S, Xiao W, Blair ET, Moreno CS. Cross-platform expression profiling demonstrates that SV40 small tumor antigen activates Notch, Hedgehog, and Wnt signaling in human cells. BMC Cancer. 2006;6:54. doi: 10.1186/1471-2407-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody JR, Calhoun ES, Gallmeier E, Creavalle TD, Kern SE. Ultra-fast high-resolution agarose electrophoresis of DNA and RNA using low-molarity conductive media. Biotechniques. 2004;37:598, 600, 602. doi: 10.2144/04374ST04. [DOI] [PubMed] [Google Scholar]
- 11.Yang HY, Wilkening S, Iadarola MJ. Spinal cord genes enriched in rat dorsal horn and induced by noxious stimulation identified by subtraction cloning and differential hybridization. Neuroscience. 2001;103:493–502. doi: 10.1016/s0306-4522(00)00573-x. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarti B, Louie M, Ratanaprayul W, Raval A, Gallagher S, Chakravarti DN. A highly uniform UV transillumination imaging system for quantitative analysis of nucleic acids and proteins. Proteomics. 2008;8:1789–1797. doi: 10.1002/pmic.200700891. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell K, Yang HY, Tessier PA, Muhly WT, Swaim WD, Szalayova I, Keller JM, Mezey E, Iadarola MJ. Localization of S100A8 and S100A9 expressing neutrophils to spinal cord during peripheral tissue inflammation. Pain. 2008;134:216–231. doi: 10.1016/j.pain.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 14.Kim AY, Tang Z, Liu Q, Patel KN, Maag D, Geng Y, Dong X. Pirt, a phosphoinositide-binding protein, functions as a regulatory subunit of TRPV1. Cell. 2008;133:475–485. doi: 10.1016/j.cell.2008.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]