Abstract
Here we describe the synthesis of new 7-substituted 8-aza-7-deazaadenosine ribonucleoside phosphoramidites and their use in generating major groove-modified duplex RNAs. A 7-ethynyl analog leads to further structural diversification of the RNA via post-automated RNA synthesis azide/alkyne cycloaddition reactions. In addition, we report preliminary studies on the effects of eight different purine 7-position modifications on RNA duplex stability and pairing specificity. Finally, the effect on RNAi activty of this type of modification at eight different positions in an siRNA guide strand has been explored. Analogs were identified with large 7-position substituents that maintain adenosine pairing specificity and are well-tolerated at specific positions in an siRNA guide strand.
Introduction
Nucleoside analogs incorporated into RNA have a variety of uses including probing RNA structure and function,1,2 exploring interactions between RNAs and proteins3 and imparting favorable properties on small interfering RNAs (siRNAs).4,5 Modifications to the natural structure of siRNAs are known to improve nuclease resistance,6–8 increase potency9–11 and reduce off-target effects12 including immune stimulation.13,14 In our previous studies, we showed that by changing the shape of RNA nucleobases while maintaining Watson-Crick base pairing one can generate fully active siRNAs with reduced undesirable protein binding and reduced immune stimulation.14,15 These earlier studies focused on modifications to purines that project substituents into the minor groove of the siRNA duplex.14,15 In this report, we explore an alternative purine modification strategy where the major groove edge (i.e. the Hoogsteen face) is modified. Inspection of the published crystal structures of Ago-RNA/DNA complexes suggest that the major-groove is largely free of contact with the nuclease of the RNA interference (RNAi) pathway.16–20 Therefore, the major groove is a logical location to introduce groups to modulate guide strand affinity and/or specificity for target RNA without interfering with Ago binding. This is particularly important in the search for guide strand modifications that reduce “miRNA-like” off-target effects that come about from binding to imperfectly matched off-target mRNAs.21,22 A previous report described the effects of pyrimidine C5-methyl and C5-propynyl modifications at multiple positions in the siRNA guide strand.4 In addition, we reported the effect of N2-alkylated 8-oxo-7,8-dihydro-2′-deoxyguanosine analogs at specific positions in the guide strand opposite A in the target, a pairing believed to place the N2 group in the major groove.23
However, a systematic study focusing on the effect of solitary major groove modifications at different guide strand positions has not been reported. Furthermore, for this study we chose to modify the purine 7-position because, like the C5 of pyrimidines, this site is located in the major groove of duplex structures and not involved in Watson-Crick base pairing.25 While 7-substituted 7-deazapurine 2′-deoxyribonucleosides have been used to modify major groove sites in duplex DNA,26 there are few examples of effective strategies to introduce these modifications into duplex RNA nor are there any analyses of their effects on RNA duplex stability, base pairing specificity or RNAi activity.27
Here we describe the synthesis of two new phosphoramidites useful for the modification of the duplex RNA major groove at adenosines and post-automated synthesis diversification via azide/alkyne cycloaddition reactions. We also report both the effects of eight structurally diverse purine 7-position modifications on duplex RNA stability and pairing specificity as well as RNAi activty of this type of modification at eight different positions in an siRNA guide strand.
Results and Discussion
Synthesis of RNAs containing 7-substituted 8-aza-7-deazaadenosine analogs
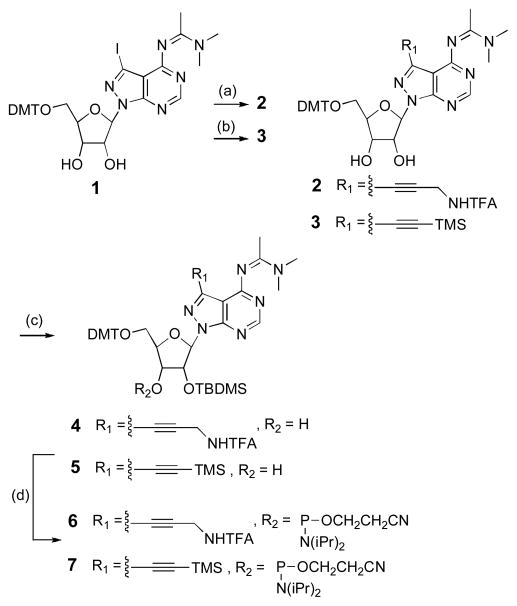
We previously reported the synthesis of 7-iodo-8-aza-7-deazaadenosine derivative 1 (Scheme 1).28 From this compound, derivatives 7-propargylamine 2 and 7-ethynyl 3 were obtained in good yields via Sonogashira couplings29,30 with the requisite protected alkynes. tert-Butyldimethylsilyl protection at the 2′-hydroxyls gave 4 and 5, which following phosphitylation at the 3′ positions yielded N,N-diisopropylamino β-cyanoethyl phosphoramidites 6 and 7. Both of these phosphoramidites coupled efficiently during standard automated RNA synthesis yielding oligoribonucleotides bearing either 7-ethynyl or 7-propargylamine substituted 8-aza-7-deazaadenosine (Figure 1). The identities of the resulting RNAs were confirmed by matrix assisted laser desorption/ionization mass spectrometry (MALDI-MS) analysis, which verified the removal of protecting groups from the ethynyl- and propargylamine-modified bases.
Scheme 1.
Synthesis of 7-propargylamine- (6) and 7-ethynyl- (7) phosphoramidites: (a) N′-(2-propynyl)-2″’,2‴,2″-trifluoroacetamide,24 Pd(PPh3)4, CuI, Et3N, DMF, rt, 91%. (b) ethynyltrimethylsilane, Pd(PPh3)4, CuI, Et3N, DMF, rt, 83%. (c) TBDMSCl, AgNO3, base, THF, rt, 36% (4), 40% (5). (d) 2-cyanoethyl-(N,N-diisopropylamino)chlorophosphite, DIPEA, THF, rt, 70% (6), 84% (7).
Figure 1.
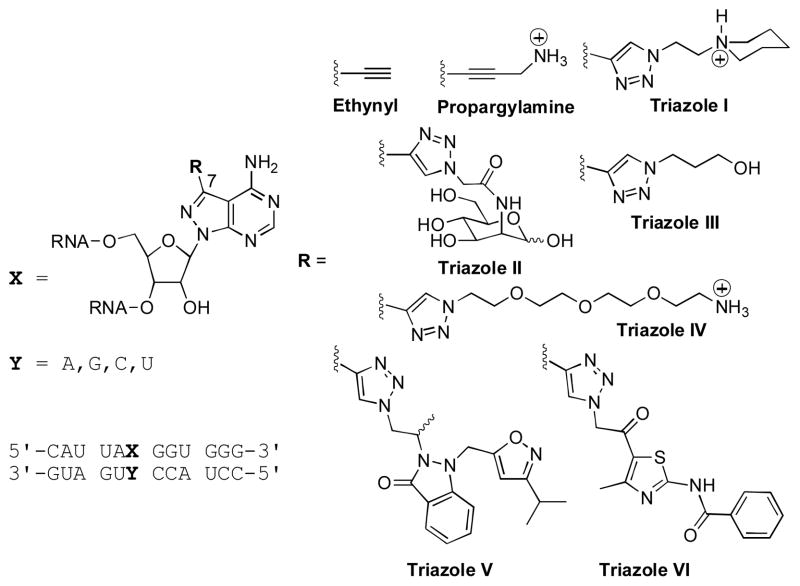
Eight purine 7-position modifications in duplex RNA.
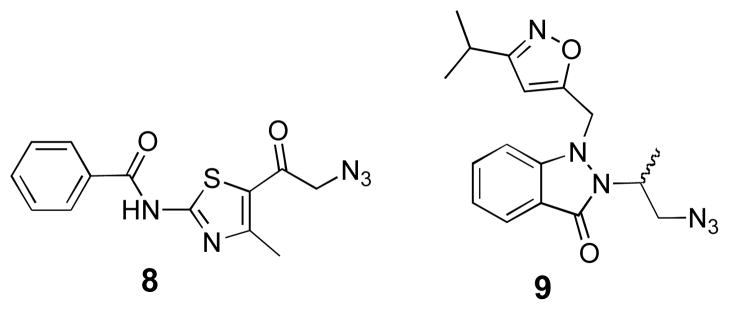
For further structural diversification at the 7-position, we explored the use of copper-catalyzed azide/alkyne cycloaddition (click) reactions31–34 with the oligonucleotide bearing 7-ethynyl-8-aza-7-deazaadenosine. Click products were generated by incubating an aqueous solution of the single-stranded RNA with tris-[1-(3-hydroxypropyl)-1H-[1,2,3]triazol-4-yl)methyl]amine (THPTA) ligand,35 CuSO4, sodium ascorbate and azide for 6.5 h at ambient temperature,36 with the exception of azide 8 which required heating to increase solubility. Azides were chosen that would generate triazoles with a variety of structural features (e.g. large, small, hydrophilic, charged, neutral and π-stacking). In particular, triazoles V and VI were chosen for their potential to simultaneously engage in π-stacking, hydrogen bonding, and hydrophobic contacts (Figure 1). Azides 8 and 9 were prepared from their corresponding bromoketone and bromoalkane, respectively (Figure 2).37,38
Figure 2.
Azides synthesized for their use in the copper-catalyzed azide/alkyne cycloaddition with oligonucleotides containing 7-ethynyl-8-aza-7-deazaadenosine.
Each click product was purified from the reaction mixtures by denaturing polyacrylamide gel electrophoresis (PAGE) and confirmed by MALDI-MS. Structures for the 8-aza-7-deazaadenosine analogs prepared by these approaches, including the 7-ethynyl, 7-propargylamine and the six new triazoles (I–VI), are shown in Figure 1.
Effect on RNA duplex stability and base pairing specificity
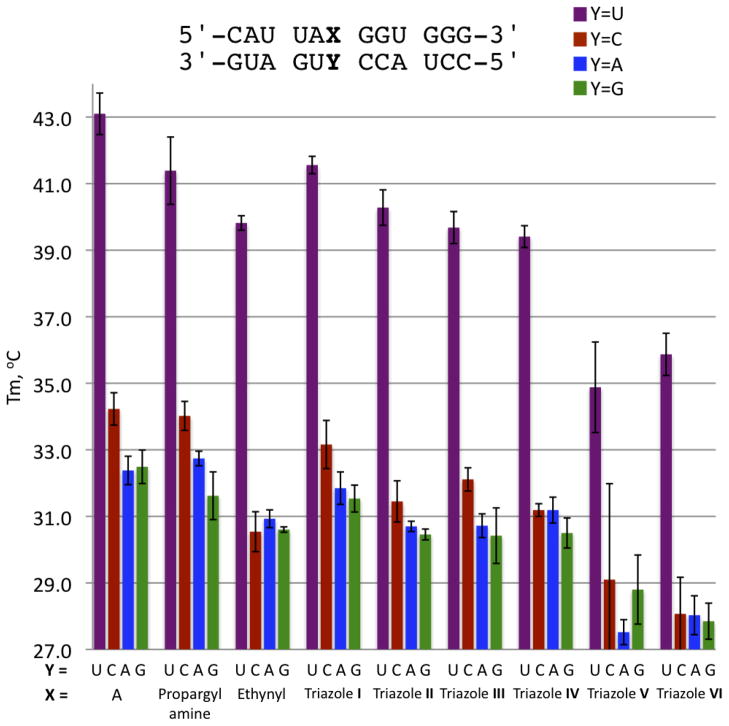
The effect of the new modifications on duplex stability was investigated via thermal denaturation (Tm) studies of a 12 base-pair (bp) RNA duplex (Figure 1, Figure 3). The nucleoside analogs were incorporated into the duplex opposite each of the four common bases (adenine (A), guanine (G), cytosine (C), and uracil (U)). Adenine was also incorporated in the same manner for comparison. Firstly, the Tm of each of the modifications opposite U was examined to determine the ability of the modifications to replace A in an A•U pair. The 12 bp duplex containing an A•U base pair had a measured Tm of 43.1 ± 0.6 °C under these conditions. Replacement of A with either the 7-propargylamine analog (Tm of 41.4 ± 1.0 °C) or the piperidine-containing triazole I (Tm of 41.6 ± 0.3 °C) lead to minimal destabilization of the 12 bp duplex (ΔTm < −2 °C). However, the more sterically demanding triazoles V and VI were substantially more destabilizing (ΔTm > −7 °C for a single modification in this 12 bp duplex). The 7-ethynyl derivative along with triazoles II–IV, were all slightly destabilizing relative to adenosine (ΔTm ~ −3 °C) (Figure 3, Supplementary Information Table 1).
Figure 3.
Thermal denaturation (Tm) of the 12-mer duplexes containing adenosine and the 7-substituted 8-aza-7-deazaadenosine analogues; propargylamine, ethynyl, and triazoles I–VI opposite the four natural bases.
As expected for modification of the purine Hoogsteen face, these alterations had little effect on the base pairing specificity. Indeed, even the highly destabilizing triazole VI modification showed selectivity for uracil over the other three nucleotides (ΔTm ~ −8 °C comparing match vs mismatches) that was similar to that observed for adenine (ΔTm ~ −10 °C match vs mismatches) (Figure 3, Supplementary Information Table 1).
Effects on RNA interference
To assess the effect of the C7-modifications on siRNA performance, we chose for analysis a sequence from the literature (PIK3CB) with a guide strand rich in adenosines such that several different positions could be tested with the analogs in hand (Figure 4A).39 The target sequence for the PIK3CB siRNA was inserted into the 3′-UTR of the Renilla luciferase sequence on the psiCHECK-2 vector as previously described.15 RNAi activity in HeLa cells was then measured at different siRNA concentrations as the ratio Renilla luciferase activity to control firefly luciferase encoded on the same plasmid. The siRNA guide strand was modified with either the 7-ethynyl analog or triazole I at positions 1, 3, 6, 10, 12, 15, 18 or 20. Triazole I was chosen for this initial study because of its large size and its minimal effect on duplex stability compared to adenosine (Figures 1 and 3). For each modified siRNA, we carried out a five-point concentration profile (0.01, 0.03, 0.1, 1, 10 nM) in the RNAi assay (Supplementary Figure 1). From these titrations, the concentration of 0.03 nM was chosen for comparison of knockdown activity for the different modified siRNAs (Figure 4B). We found that RNAi activity is altered by these structural changes in a position-dependent and, at least at one postion, a modification-dependent manner (Figure 4B). For instance, we found that both modfications tested were well-tolerated at positions 12 and 20 of the guide strand. On the other hand, a substantial decrease of knockdown potency is observed with either modification at positions 1, 3 or 10. These modifications at positions 6 or 18 moderately diminished potency. Interestingly, at position 15, triazole I is well tolerated with knockdown indistinguishable from the unmodified siRNA whereas 7-ethynyl at this location reduces potency. Thus, triazole I, bearing the N-ethylpiperidine, enhances potency over the ethynyl precursor, indicating that siRNA potency is sensitive to the structure of the major groove modification at guide position 15. Others have shown that minor groove modifications at guide position 15 can enhance RNAi activity.40 Together these results point to the important role of nucleotide structure at this position and suggest additional modifications here may further enhance activity. Also, since base pairing to nucleotides 13–16 of the guide has been shown to be a key factor in determining miRNA-like off-target effects, modulating the interaction between guide and target in this region is likely to control these effects.41
Figure 4.
(A) Sequence of siRNA used in this study showing sites of modification of the guide strand and the structures of the modifications tested. All siRNAs were prepared with a 5′-phosphorylated guide strand (p). (B) Knockdown activty of modified siRNAs. Activity is reported as the ratio of Renilla/firefly luciferase signal at 0.03 nM transfected siRNA in HeLa cells. buffer = no added siRNA, unmodified = siRNA with no modifications.
Our observation of decreased potency from either analog at position 1 is most likely due to binding changes with the Ago2 MID domain. The first nucleotide of the bound guide strand does not make contact with the target strand.16,17 Other types of modifications at guide position 1 are also known to decrease RNAi potency.42–44 The source of decreased potency from modification of positions 3, 6, 18 and 10 are unknown at this time, but the latter may be a result of modification of the guide/target duplex major groove directly across from the cleavage site of Ago2.17 In addition, the inhibitory effects seen from modification of guide position 3 or position 6 mirror Terrazas and Kool’s observation that C5-propynyl uridine modification at multiple positions in the 5′ half of the guide strand was detrimental to potency.4
Conclusions
We have synthesized and characterized RNA duplexes modified with different 7-substituted 8-aza-7-deazaadenosines, including the six triazoles formed via click reactions from the 7-ethynyl analog. This type of modification directs substitutents into the duplex RNA major groove. Analogs were identified bearing large major groove substituents that maintain duplex RNA stability and adenosine pairing specificity and are well-tolerated at specific positions in an siRNA guide strand. In particular, full RNA interference activity was maintained with two different modifications at positions 12 and 20 and with triazole I at position 15. Since major groove modifications are known to affect specificity of duplex formation,45 these analogs are interesting candidates for controlling miRNA-like off-target effects and the results of such studies will be reported in due course.
Experimental
General Synthetic Procedures
Glassware for all reactions were oven dried at 175 °C overnight and cooled in a desiccator prior to use. Reactions were carried out under an atmosphere of dry argon when anhydrous conditions were necessary. All reagents were purchased from commercial sources (Sigma/Aldrich or Fischer Scientific) and were used without further purification unless noted otherwise. Liquid reagents are introduced by oven-dried microsyringes. Tetrahydrofuran was dried in a solvent purification system that passes solvents through two columns of dry neutral alumina. Thin layer chromatography (TLC) was performed with Merck silica gel 60 F254 precoated TLC plates. Short and long wave visualization was performed with a Mineralight multiband ultraviolet lamp at 254 and 365 nm, respectively. Flash column chromatography was performed with Merck silica gel (Sorbent technologies, 60–200 mesh). Radial chromatography was preformed with Merck silica gel 60 PF 254 containing CaSO4. 1H, 13C, and 31P Nuclear Magnetic Resonance spectra of pure compounds were acquired with Varian VNMRs 600 and Mercury 300 spectrometers. Chemical shifts reported in parts per million (ppm) in the reference to a solvent peak. The abbreviations such as s, t, m, bs, dd, d stand for singlet, triplet, multiplet, broad singlet, doublet of doublets and doublet. High-resolution mass spectra were obtained at University of California, Davis Mass spectrometry facility.
4-{[(Dimethylamino)ethylidene]amino}-3-[N′-(2-propynyl)-2″,2″,2″-trifluoroacetamido]-1-(β-D-ribofuranosyl)-5′-O-(4,4′-dimethoxytriphenylmethyl)-1H-pyrazolo[3,4-d]pyrimidine (2)
A suspension of 128 (235 mg 0.307 mmol), Pd(PPh3)4 (36 mg, 0.03 mmol), and CuI (7.6 mg, 0.04 mmol) in anhydrous DMF (3 mL) was treated with N′-(2-propynyl)-2″,2″,2″-trifluoroacetamide24 (227 μL, 1.5 mmol) followed by anhydrous triethylamine (165 μL, 1.5 mmol). The mixture was stirred under Ar at room temperature. After the reaction was completed (TLC), the solvent was removed in vacuo, and the mixture was partioned between hexane:EtOAc (3:7, 100 mL) and water (100 mL). The organic layer was dried (Na2SO4), evaporated, and chromatographed on a flash silica gel column, eluting with MeOH:DCM (5:95) to give 2 (221mg, 91%) as a white foam. 1H NMR (CDCl3, 600 MHz): δ (ppm) 8.41 (s, 1H), 7.59 (bs, 1H), 7.39-7.15 (m, 9H), 6.80-6.77 (m, 4H), 6.37 (d, J = 6.0, Hz, 1H), 5.32 (t, J = 3.0, Hz, 1H), 4.92 (t, J = 6.0, Hz, 1H), 4.54 (t, J = 6.0, Hz, 1H), 4.43 (dd, J = 6.0, 18.0 Hz, 1H), 4.37 (dd, J = 6.0, 18.0 Hz, 1H), 4.2 (dd, J = 4.8, 9.6, Hz, 1H), 3.75 (s, 3H), 3.74 (s, 3H), 3.29 (dd, J = 4.8, 12.0 Hz, 1H), 3.22 (dd, J = 4.8, 12.0 Hz, 1H), 3.18 (s, 3H), 3.13 (s, 3H), 2.22 (s, 3H). 13C NMR (CDCl3, 150 MHz): δ (ppm) 164.4, 160.5, 160.4, 158.6, 157.0, 147.0, 138.4, 138.1, 132.3, 132.1, 130.6, 130.3, 129.8, 128.8, 115.2, 111.0, 90.8, 88.5, 88.1, 85.7, 75.9, 74.0, 66.0, 57.31, 57.30, 32.4, 19.1. ESIHRMS: calcd for C40H40F3N7O7 (M+H)+ 788.3020, obsd. 788.3018.
4-{[(Dimethylamino)ethylidene]amino}-3-[N′-(2-propynyl)-2″,2″,2″-trifluoroacetamido]-1-(β-D-ribofuranosyl)-5′-O-(4,4′-dimethoxytriphenylmethyl)-2′-O-(tert-butyldimethylsilyl)-1H-pyrazolo[3,4-d]pyrimidine (4)
To a stirred solution of 2 (217 mg, 0.275 mmol) and tert-butylchlorodimethylsilane (53.9 mg, 0.358 mmol) in freshly distilled THF (4 mL) was added AgNO3 (62.8 mg, 0.37 mmol) followed by DIEA (174 μL, 1 mmol) and continued stirring at room temperature for 12 h. The reaction was then diluted with EtOAc (25 mL), filtered, and washed with 5% aqueous NaHCO3 (1 × 30 mL). The organic portion was dried (Na2SO4), filtered, concentrated under reduced pressure. Purification by flash column chromatography on a silica gel EtOAc:hexane (30:70) as eluent to give 4 (87 mg, 36%). 1H NMR (CD2Cl2, 600 MHz): δ (ppm) 8.50 (s, 1H), 7.45-7.43 (m, 2H), 7.36-7.34 (m, 4H), 7.24- 7.21 (m, 2H), 7.18-7.15 (m, 1H), 7.07 (bs, 1H), 6.82-6.77 (m, 4H), 6.32 (d, J = 6.0, Hz, 1H), 5.30 (t, J = 3.0 Hz, 1H), 5.05 (t, J = 6.0 Hz, 1H), 4.41 (dd, J = 6.0, 18 Hz, 1H), 4.35 (dd, J = 6.0, 18 Hz, 1H), 4.28 (dd, J = 6.0, 12 Hz, 1H), 3.75 (s, 3H), 3.74 (s, 3H), 3.31 (dd, J = 6.0, 12 Hz, 1H), 3.20 (bs, 3H), 3.17 (dd, J = 6.0, 12 Hz, 1H), 3.14 (bs, 3H), 2.66 (d, J = 6.0 Hz, 1H), 2.22 (s, 3H), 0.81 (s, 9H), −0.01 (s, 3H), −0.16 (s, 3H). 13C NMR (150 MHz, CD2Cl2) δ 167.6, 163.8, 163.7, 162.2, 150.4, 141.8, 141.4, 135.7, 135.4, 133.8, 133.7, 133.1, 132.0, 118.5, 114.9, 93.8, 91.5, 91.4, 89.4, 80.3, 77.3, 69.4, 60.6, 59.2, 59.0, 58.8, 58.7, 58.5, 35.7, 30.8, 23.2, 22.3, 0.2, 0.0. The identity of 2′-O-TBDMS isomer (vs. 3′-O-TBDMS isomer) was confirmed by 2D-NMR (COSY). ESIHRMS: calcd for C46H54F3N7O7Si (M+H)+; 902.3884, obsd. 902.3862.
4-{[(Dimethylamino)ethylidene]amino}-3-[N′-(2-propynyl)-2″,2″,2″-trifluoroacetamido]-1-(β-D-ribofuranosyl)-5′-O-(4,4′-dimethoxytriphenylmethyl)-3′-O-[(2-cyanoethyl)(N,N-diisopropylamino) phosphino]-2′-O-(tert-butyldimethylsilyl)-1H-pyrazolo[3,4-d]pyrimidine (6)
N,N-Diisopropylethylamine (87 μL, 0.5 mmol), and 2-cyanoethyl-(N,N-diisopropylamino)chlorophosphite (103 μL, 0.435 mmol) were consequently added to a solution of 4 (157 mg, 0.174 mmol) in freshly distilled THF (1 mL). The resulting reaction mixture was stirred at room temperature for 2 h. It was then diluted with EtOAc (30 mL), filtered, and washed with 5% (w/v) aqueous NaHCO3 (2 × 15 mL). The organic portion was dried (Na2SO4), filtered, concentrated under reduced pressure. Purification was performed by radial chromatography (1 mm plate) on silica gel using EtOAc:hexane (40:60) as eluent to give 6 (134 mg, 70%). 31P NMR (CD2Cl2, 121 MHz): δ (ppm) 151.24, 150.78. ESIHRMS: calcd for C55H71N8O8PSi (M+H)+ 1102.4963, obsd. 1102.4956.
4-{[(Dimethylamino)ethylidene]amino}-3-[(trimethylsilyl)ethynyl]-1-(β-D-ribofuranosyl)-5′-O-(4,4′-dimethoxytriphenylmethyl)-1H-pyrazolo[3,4-d]pyrimidine (3)
A suspension of 128 (1.26 g, 1.64 mmol), Pd(PPh3)4 (0.48 g, 0.41 mmol), and CuI (0.08 g, 0.43 mmol) in anhydrous DMF (16 mL) was treated with ethynyltrimethylsilane (1.14 mL, 8.20 mmol) followed by anhydrous triethylamine (1.14 mL, 8.20 mmol). The mixture was stirred under Ar at room temperature. After the reaction was completed, the solvent was removed in vacuo, and the residue was chromatographed on a flash silica column, eluting with MeOH:DCM (0:100 → 3:97 → 5:95) to give 3 (1.01 g, 83%) as a foam. The progress of the reaction was monitored by following the disappearance of 1 with the use of a Q-Trap ESI-Mass spectrometer. 1H NMR (CD2Cl2, 300 MHz): δ (ppm) 8.37 (s, 1H), 7.44-7.41 (m, 2H), 7.31-7.13 (m, 7H), 6.78 (d, J = 2.4 Hz, 2H), 6.75 (d, J = 2.4 Hz, 2H) 6.38 (d, J = 3.9 Hz, 1H), 4.93 (t, J = 4.5 Hz, 1H), 4.51 (t, J = 5.1 Hz, 1H), 4.21 (dd, J = 9.1, 5.0 Hz, 1H), 3.75 (s, 3H), 3.74 (s, 3H), 3.35-3.20 (m, 5H), 3.12 (s, 3H), 2.12 (s, 3H), 0.26 (s, 9H). 13C NMR (CD2Cl2, 75 MHz): δ (ppm) 163.2, 162.0, 159.02, 158.98, 156.7, 155.2, 145.8, 136.48, 136.47, 130.64, 130.55, 129.9, 128.6, 128.3, 127.0, 113.5, 109.3, 99.2, 97.2, 89.5, 86.6, 84.1, 74.0, 72.2, 64.7, 55.7, 39.1, 38.9, 31.2, 17.6, 0.0. ESIHRMS: calcd for C40H47N6O6Si (M + H)+ 735.3326, obsd 735.3336.
4-{[(Dimethylamino)ethylidene]amino}-3-[(trimethylsilyl)ethynyl]-1-(β-D-ribofuranosyl)-5′-O-(4,4′-dimethoxytriphenylmethyl)-2′-O-(tert-butyldimethylsilyl)-1H-pyrazolo[ 3,4-d]pyrimidine (5)
Triethylamine (151 μL, 1.08 mmol) and tert-butylchlorodimethylsilane (0.10 g, 0.63 mmol) were consequently added to a solution of 3 (437 mg, 0.57 mmol) in anhydrous THF (10 mL). AgNO3 (0.11 g, 0.63 mmol) was added after stirring for 5 min. The resulting mixture was stirred under Ar at room temperature for 19 h. It was then diluted with EtOAc (25 mL), filtered, and washed with sat. NaHCO3 (1 × 30 mL). The organic portion was dried (Na2SO4), filtered, and concentrated under reduced pressure. Purification was carried out by flash column chromatography on silica gel, eluting with EtOAc:hexane (80:20) to give 5 (198 mg, 40%) as a yellow foam. 1H NMR (CD2Cl2, 300 MHz): δ (ppm) 8.52 (s, 1H), 7.50-7.47 (m, 2H), 7.38-7.31 (m, 4H), 7.29-7.23 (m, 2H), 7.20-7.14 (m, 1H), 6.82-6.76 (m, 4H), 6.32 (d, J = 4.7 Hz, 1H), 5.11 (t, J = 5.0 Hz, 1H), 4.28 (q, J = 4.7 Hz, 1H), 4.14 (dd, J = 8.2, 4.5 Hz, 1H), 3.76 (s, 3H), 3.75 (s, 3H), 3.39-3.16 (m, 8H), 2.67 (d, J = 5.1 Hz, 1H), 2.20 (s, 3H), 0.84 (s, 9H), 0.23 (s, 9H), 0.01 (s, 3H), −0.13 (s, 3H). 13C NMR (CD2Cl2, 75 MHz): δ (ppm) 163.4, 162.0, 159.1, 159.0, 157.2, 155.9, 145.8, 136.6, 136.5, 130.8, 130.7, 129.9, 128.7, 128.3, 127.0, 113.6, 109.6, 98.9, 97.3, 89.2, 86.7, 84.4, 75.2, 72.4, 64.6, 55.7, 39.0, 38.8, 26.0, 18.4, 17.5, 0.0, −4.6, −4.9. The identity of 2′-O-TBDMS isomer (vs. 3′-O-TBDMS isomer) was confirmed by 2D-NMR (COSY). ESIHRMS: calcd for C46H61N6O6Si2 (M + H)+ 849.4191, obsd 849.4201.
4-{[(Dimethylamino)ethylidene]amino}-3-[(trimethylsilyl)ethynyl]-1-(β-D-ribofuranosyl)-5′-O-(4,4′-dimethoxytriphenylmethyl)-3′-O-[(2-cyanoethyl)(N,N-diisopropylamino) phosphino]-2′-O-(tert-butyldimethylsilyl)- 1H-pyrazolo[3,4-d]pyrimidine (7)
N,N-Diisopropylethylamine (105 μL, 0.60 mmol) and 2-cyanoethyl-(N,N-diisopropylamino)chlorophosphite (45 μL, 0.20 mmol) were consequently added to a solution of 5 (80 mg, 0.10 mmol) in anhydrous DCM (1 mL). The resulting reaction mixture was stirred under Ar at room temperature for 10 h. It was then diluted with EtOAc (30 mL) and washed with sat. NaHCO3 (15 mL). The organic portion was dried with Na2SO4, filtered, and concentrated under reduced pressure. The residue was chromatographed on a flash silica column, eluting with EtOAc:hexane (80:20) to give 7 (88 mg, 84%) as a white foam. 31PNMR (CD2Cl2, 121 MHz): δ (ppm) 151.11, 149.67. ESIHRMS: calcd for C55H78N8O7PSi2 (M + H)+ 1049.5270, obsd 1049.5290.
N-(5-(2-azidoacetyl)-4-methylthiazol-2-yl)benzamide (8)
N-(5-(2-Bromoacetyl)-4-methylthiazol-2-yl)benzamide38 (50 mg, 0.148 mmol) was stirred with sodium azide (10 mg, 0.163 mmol) at room temperature in DMF (1 mL) for 3 hours. Then 10 mL of ice-cold water was added and the resulting precipitate was filtered. The off-white solid was purified by flash chromatography with EtOAc:hexane (1:1) yielding 8 as a white solid (37 mg, 85%). mp decomposition at 132 °C. IR (neat) nmax 3195, 2119, 1684, 1666, 1541, 1497, 1372, 1319, 1292, 1221, 910, 875 cm−1. 1H NMR (CDCl3, 600 MHz): δ (ppm) 7.92 (d, J = 7.8 Hz, 2H), 7.64 (t, J = 7.2 Hz, 1H), 7.51 (t, J = 7.2 Hz, 2H), 4.30 (s, 2H), 2.35 (s, 3H). 13C NMR (CDCl3, 150 MHz): δ (ppm) 186.7, 165.7, 160.9, 157.9, 133.8, 131.5, 129.4, 128.0, 121.5, 57.1, 18.3. ESIHRMS calcd for C13H11N5O2S (M + H)+ 301.0633, obsd 301.0713.
(±)-2-(1-Azidopropan-2-yl)-1-((3-isopropylisoxazol-5-yl)methyl)-1H-indazol-3(2H)-one (9)
(±)-2-(1-Bromopropan-2-yl)-1-((3-isopropylisoxazol-5-yl)methyl)-1H-indazol-3(2H)-one37 (149 mg, 394 mmol) was added to a 5–10 mL microwave vial and dissolved in DMF (2.0 mL). Sodium azide (30.7 mg, 473 mmol) was added and the vial was sealed and placed in an oil bath at 60 °C for 3 hours. The DMF was then removed under reduced pressure and the crude material was dissolved in EtOAc (30 mL). This solution was then washed with water (30 mL) and brine (30 mL), dried over sodium sulfate, and concentrated. This crude material was purified by flash chromatography with EtOAc:hexane (40:60) to afford 9 as a pale yellow solid (133 mg, 99%). mp 86–87 °C. IR (neat) nmax 2969, 2930, 2904, 2876, 2091, 1675, 1607, 1461, 1334, 1312, 1251, 1235 cm−1. 1H NMR (CDCl3, 600 MHz): δ (ppm) 7.73 (d, J = 7.8 Hz, 1H), 7.52 (ddd, J = 7.5, 7.5, 1.0 Hz, 1H), 7.22-7.14 (m, 2H), 5.57 (s, 1H), 4.88 (d, J = 16.9 Hz, 1H), 4.82 (d, J = 16.9 Hz, 1H), 4.21-4.14 (m, 1H), 4.03 (dd, J = 12.5, 8.7 Hz, 1H), 3.51 (dd, J = 12.5, 5.5 Hz, 1H), 2.82 (m, J = 6.9 Hz, 1H), 1.42 (d, J = 7.0 Hz, 3H), 1.08 (d, J = 6.9 Hz, 3H), 1.07 (d, J = 6.9 Hz, 3H). 13C NMR (CDCl3, 150 MHz): δ (ppm) 169.3, 166.4, 164.5, 150.9, 133.1, 124.2, 123.6, 120.2, 112.6, 101.4, 53.5, 53.4, 46.7, 26.5, 21.5, 21.5, 15.8. ESIHRMS calcd for C17H20N6O2 (M + H)+ 341.1648, obsd 341.1722.
Biochemical Procedures
Synthesis, purification and quantification of RNA 12-mer
RNA oligonucleotides were synthesized on a ABI 394 synthesizer (DNA/Peptide Core Facility, University of Utah, Salt Lake City) using 5′-DMTr protected β-cyanoethyl phosphoramidites on a 1.0 mmol scale with coupling times of 25 min for increased coupling efficiency of amidites 6 and 7. All RNAs were deprotected as previously described.46 The RNA oligonucleotides containing ethynyl and propargylamine modifications were gel-purified and quantified as previously described.14 The identity of the RNAs was confirmed by MALDI mass spectrometry.
List of mass values, [M+H]+, for the RNA containing propargylamine and ethynyl modifications: Propargylamine: Calc. 3929.5, Obs. 3929.5; Ethynyl: Calc. 3900.4, Obs. 3900.5.
Mass spectrometry analysis of RNA 12-mer
Mass spectra were obtained on an Applied Biosystems 4700 MALDI-TOF mass spectrometer operating in linear mode. Desalted samples (2 μL of 10 μM) were combined with an equal volume of matrix (either a 10:1 mixture of 50 mg/mL solution of 3-hydroxypicolinic acid in 1:1 acetonitrile: H2O and a 100 mg/mL solution of di-ammoniumhydrogen citrate in H2O or a saturated solution of 6-aza-2-thiothymine in 0.1 M aqueous dibasic ammonium citrate), and no more than 1 μL was applied to the target and air-dried. Mass spectra were recorded in the positive ionization mode and calibrated to an internal DNA standard of 4366.8 Daltons.
Click reactions on 12-mer RNAs
A dry pellet of 10–20 nmol pure (purified as described above) or 40–60 nmol crude ethynyl-containing RNA was dissolved in 1 μL of H2O, then treated sequentially with tris-[1-(3-hydroxypropyl)-1H-[1,2,3]triazol-4-yl)methyl]amine (THPTA) ligand35 (1 μL, 1 M in H2O), CuSO4 (1 μL, 100 mM in H2O), sodium ascorbate (1 μL, 1M in H2O) and 1 μL of the corresponding azide. Azides were prepared at the following concentrations: 1-(2-azidoethyl)- piperidine,47 50 mM in 0.5 M Tris-HCl, pH 8.0; N-azidoacetyl-D-mannosamine, 48 0.5 M in H2O; azides 8 and 9, 150 mM in DMSO; 3-azido-1-propanol and 11-azido-3,6,9-trioxaundecan-1-amine, 150 mM in H2O. The resulting reaction solutions were incubated at room temperature for 6.5 h except for the click reaction with azide 8, to which 1 μL more of DMSO was added and incubated at 35 °C to improve azide solubility. The reaction mixtures were diluted to 2x the original volume with PAGE loading buffer (80% formamide containing 10 mM EDTA). The 12-mer RNA was gel purified and quantified as above. Lyophilization gave white pellets, which were fully soluble in H2O. In the case of pure ethynyl-containing RNA, a single band migrated faster than the triazole-containing RNAs corresponded to the clicked product. The click reactions performed on the crude ethynyl-containing RNA exhibited comparable efficiency and gel-shift patterns to the pure reactions with ethynyl-containing RNA, though bands corresponding to by-products of incomplete coupling were also visible. MALDI-TOF analysis confirmed identity of the click products (as described above).
List of mass values, [M+H]+, for the triazole-containing RNAs: Triazole I: Calc. 4054.6, Obs. 4054.5; Traizole II: Calc. 4162.6, Obs. 4162.6; Triazole III: Calc. 4001.5, Obs. 4001.1; Triazole IV: Calc. 4118.7, Obs. 4118.5; Triazole V: Calc. 4240.8, Obs. 4042.8; Triazole VI: Calc. 4201.7, Obs. 4201.6.
Thermal melting (Tm) analysis
The thermal stability of the ethynyl, propargylamine and triazole-containing RNAs were analyzed in a 12 bp duplex of sequence derived from human glutamate receptor B subunit pre-mRNA using the following buffer conditions: 10 mM Tris-HCl, pH 7.8, 0.1 mM EDTA and 100 mM NaCl.36 The values reported in Supplementary Information Table 1 are an average of three denaturation experiments, with the experimental temperature range noted for each RNA duplex. The error bars in the graph (Figure 3) indicate ± standard deviation.
Synthesis, purification and quantification of 21-mer RNA
RNA oligonucleotides were synthesized as described above. 5′ Phosphorylation of siRNA guide strands was accomplished using the chemical phosphorylation reagent (Glenn Research Corporation). RNAs were deprotected, gel-purified and analyzed by MALDI-MS, as described above.
List of mass values, [M+H]+, for all ethynyl-modified siRNAs: (G = guide, indicates position of modification, calculated mass is the same for all siRNAs modified in this way) Calc. 6851.1, Observed: G1 = 6849.3; G3 = 6849.3; G6 = 6849.0; G10 = 6849.9; G12 = 6850.0; G15 = 6849.9; G18 = 6851.1; G20 = 6849.8.
Click reactions on siRNA guide strands
Reaction and purification of the ethynyl-modified siRNAs with Triazole I were executed is the same fashion as above with 15 nmol of crude RNA for each reaction. MALDI-MS analysis confirmed the identity of the click products.
List of mass values, [M+H]+, for all Triazole I-modified siRNAs: (G = guide, indicates position of modification, calculated mass is the same for all siRNAs modified in this way) Calc: 7005.3, Observed: G1 = 7007.4; G3 = 7005.9; G6 = 7004.2; G10 = 7002.5; G12 = 7006.4; G15 = 7006.4; G18 = 7003.6; G20 = 7003.6.
siRNA duplex formation
SiRNA duplex hybridization was accomplished by combining equal amounts of purified passenger and modified guide strands to a final concentration of 5 μM in 10 mM Tris-HCl, 50 mM KCl, pH 7.5. The samples were heated at 95 °C for 5 minutes followed by slow-cooling to room temperature over a period of approximately 2 h.
Cell culture
HeLa cells (ATCC) were grown at 37 °C in humidified 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% fetal bovine serum (FBS, GIBCO) and 100 Units mL−1 penicillin and 100 μg mL−1 streptomycin (GIBCO, 1x Pen Strep). The cells were maintained in exponential growth.
Transfection and RNAi activity assay
HeLa cells were reverse-transfected using siPORT NeoFX transfection reagent (Ambion) according to the manufacturer’s instructions. Cells grown in flasks to approximately 80–90% confluence then trypsinized (0.025% trypsin-EDTA, GIBCO) and diluted in fresh medium (DMEM, 10% FBS, 1x Pen Strep) to a concentration of 1 × 105 cells mL−1. The psiCHECK-2 plasmid (Promega), containing the reporter genes Renilla and firefly luciferase (hRluc and hluc+, respectively), was used as the vector. The PIK3CB siRNA target sequence was inserted in the 3′ UTR of the Renilla luciferase gene (psiCHECK-2-PIK3CB), between the NotI and XhoI restriction sites (see below), allowing this luciferase to be used as a reporter of siRNA potency, while the firefly luciferase was used as an internal control. Plasmid and siRNA cotransfections and 96 well plate assay were performed as previously described,14 with the exception that 20 ng per well of psiCHECK-2-PIK3CB was used for these assays. For the data in Figure 4, three separate assays were executed where all modifications were tested in the same 96 well plate, then averaged to give the values and standard deviations plotted. Sequence inserted between Xho1 and Not1 in psiCHECK-2-PIK3CB plasmid: 5′-GCACATCTCCTAAUATGAATCCTATCAGAA-3′
Supplementary Material
Acknowledgments
P.A.B. acknowledges the National Institutes of Health for financial support in the form of grant R01GM080784. The authors would like to acknowledge Erik Q. Fostvedt for helpful scientific discussions. J.M.I. thanks the Alfred P. Sloan Minority Ph.D. Program for fellowship support.
Footnotes
Electronic Supplementary Information available: 1H NMR, 13C NMR, and 31P NMR spectra of all synthesized compounds. See DOI: 10.1039/b000000x/
References
- 1.Hougland JL, Piccirilli JA, Daniel H. Methods in Enzymology. Academic Press; 2009. pp. 107–125. [DOI] [PubMed] [Google Scholar]
- 2.Rist MJ, Marino JP. Curr Org Chem. 2002;6:775. [Google Scholar]
- 3.Tor Y. Pure Appl Chem. 2009;81:263–272. [Google Scholar]
- 4.Terrazas M, Kool ET. Nucleic Acids Res. 2009;37:346–353. doi: 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watts JK, Deleavey GF, Damha MJ. Drug Discovery Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Layzer JM, McCaffrey AP, Tanner AK, Huang Z, Kay MA, Sullenger BA. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watts JK, Katolik A, Viladoms J, Damha MJ. Org Biomol Chem. 2009;7:1904–1910. doi: 10.1039/b900443b. [DOI] [PubMed] [Google Scholar]
- 8.Hernández AR, Peterson LW, Kool ET. ACS Chem Biol. 2012 doi: 10.1021/cb300174c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. J Med Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 10.Fisher M, Abramov M, Van Aerschot A, Rozenski J, Dixit V, Juliano RL, Herdewijn P. Eur J Pharmacol. 2009;606:38–44. doi: 10.1016/j.ejphar.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dande P, Prakash TP, Sioufi N, Gaus H, Jarres R, Berdeja A, Swayze EE, Griffey RH, Bhat B. J Med Chem. 2006;49:1624–1634. doi: 10.1021/jm050822c. [DOI] [PubMed] [Google Scholar]
- 12.Jackson AL, Linsley PS. Nat Rev Drug Discovery. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 13.Eberle F, Giessler K, Deck C, Heeg K, Peter M, Richert C, Dalpke AH. J Immunol. 2008;180:3229–3237. doi: 10.4049/jimmunol.180.5.3229. [DOI] [PubMed] [Google Scholar]
- 14.Peacock H, Fucini RV, Jayalath P, Ibarra-Soza JM, Haringsma HJ, Flanagan WM, Willingham A, Beal PA. J Am Chem Soc. 2011;133:9200–9203. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peacock H, Fostvedt E, Beal PA. ACS Chem Biol. 2010;5:1115–1124. doi: 10.1021/cb100245u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elkayam E, Kuhn C-D, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. Cell. 2012 doi: 10.1016/j.cell.2012.1005.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schirle NT, MacRae IJ. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall WS, Khvorova A. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 22.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kannan A, Fostvedt E, Beal PA, Burrows CJ. J Am Chem Soc. 2011;133:6343–6351. doi: 10.1021/ja2003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trybulski EJ, Zhang J, Kramss RH, Mangano RM. J Med Chem. 1993;36:3533–3541. doi: 10.1021/jm00075a007. [DOI] [PubMed] [Google Scholar]
- 25.Day RO, Seeman NC, Rosenberg JM, Rich A. Proc Natl Acad Sci U S A. 1973;70:849–853. doi: 10.1073/pnas.70.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding P, Wunnicke D, Steinhoff HJ, Seela F. Chem--Eur J. 2010;16:14385–14396. doi: 10.1002/chem.201001572. [DOI] [PubMed] [Google Scholar]
- 27.Piton N, Engels JW. Nucleosides Nucleotides Nucleic Acids. 2003;22:1661–1664. doi: 10.1081/NCN-120023107. [DOI] [PubMed] [Google Scholar]
- 28.Pokharel S, Jayalath P, Maydanovych O, Goodman RA, Wang SC, Tantillo DJ, Beal PA. J Am Chem Soc. 2009;131:11882–11891. doi: 10.1021/ja9034076. [DOI] [PubMed] [Google Scholar]
- 29.Chinchilla R, Najera C. Chem Soc Rev. 2011;40:5084–5121. doi: 10.1039/c1cs15071e. [DOI] [PubMed] [Google Scholar]
- 30.Sonogashira K, Tohda Y, Hagihara N. Tetrahedron Lett. 1975;50:4467–4470. [Google Scholar]
- 31.El-Sagheer AH, Brown T. Chem Soc Rev. 2010;39:1388–1405. doi: 10.1039/b901971p. [DOI] [PubMed] [Google Scholar]
- 32.Huisgen R, Guenter S, Leander M. Chem Ber. 1967:100. [Google Scholar]
- 33.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 34.Meldal M, Tornøe CW. Chem Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 35.Hong V, Presolski SI, Ma C, Finn MG. Angew Chem Int Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peacock H, Maydanovych O, Beal PA. Org Lett. 2010;12:1044–1047. doi: 10.1021/ol100019r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conrad WE, Fukazawa R, Haddadin MJ, Kurth MJ. Org Lett. 2011;13:3138–3141. doi: 10.1021/ol2010424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu GJ, Yoo CL, Yang B, Lodewyk MW, Meng L, El-Idreesy TT, Fettinger JC, Tantillo DJ, Verkman AS, Kurth MJ. J Med Chem. 2008;51:6044–6054. doi: 10.1021/jm800533c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butora G, Kenski DM, Cooper AJ, Fu W, Qi N, Li JJ, Flanagan WM, Davies IW. J Am Chem Soc. 2011;133:16766–16769. doi: 10.1021/ja2068774. [DOI] [PubMed] [Google Scholar]
- 41.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshikawa K, Ogata A, Matsuda C, Kohara M, Iba H, Kitade Y, Ueno Y. Bioconjug Chem. 2011;22:42–49. doi: 10.1021/bc100301w. [DOI] [PubMed] [Google Scholar]
- 43.Somoza A, Chelliserrykattil J, Kool ET. Angew Chem Int Ed Engl. 2006;45:4994–4997. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- 44.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetsky M, Rajeev KG, Egli M, Manoharan M. ACS Chem Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 45.Lin KY, Matteucci MD. J Am Chem Soc. 1998;120:8531–8532. [Google Scholar]
- 46.Maydanovych O, Easterwood LM, Cui T, Véliz EA, Pokharel S, Beal PA, Jonatha MG. Methods in Enzymology. Academic Press; 2007. pp. 369–386. [DOI] [PubMed] [Google Scholar]
- 47.Carboni B, Vaultier M, Carrié R. Tetrahedron Lett. 1988;29:1279–1282. [Google Scholar]
- 48.Muthana S, Yu H, Huang S, Chen X. J Am Chem Soc. 2007;129:11918–11919. doi: 10.1021/ja075736b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.