Abstract
Understanding how cells regulate microtubule nucleation during the cell cycle has been limited by the inability to directly observe nucleation from the centrosome. To view nucleation in living cells, we imaged GFP-tagged EB1, a microtubule tip-binding protein, and determined rates of nucleation by counting the number of EB1-GFP comets emerging from the centrosome over time. Nucleation rate increased 4-fold between G2 and prophase and continued to rise through anaphase and telophase, reaching a maximum of 7 times interphase rates. We tested several models for centrosome maturation, including γ-tubulin recruitment and increased centrosome size. The centrosomal concentration of γ-tubulin reached a maximum at metaphase, and centrosome size increased through anaphase, whereas nucleation remained high through telophase, implying the presence of additional regulatory processes. Injection of anti-γ-tubulin antibodies significantly blocked nucleation during metaphase but was less effective during anaphase, suggesting that a nucleation mechanism independent of γ-tubulin contributes to centrosome function after metaphase.
Centrosomes are the principal microtubule (MT) nucleating structures in animal cells, and their position dictates the orientation of MT arrays, contributing to interphase cell polarity and mitotic bipolar spindle assembly. The centrosome is composed of a pair of centrioles surrounded by pericentriolar material. The pericentriolar material is an organized fibrous lattice and is the site of MT nucleation (1–3).
Centrosomes undergo duplication and maturation as cells progress through the cell cycle (4). Electron microscopic analysis of the number of MTs assembled onto isolated centrosomes showed that the MT nucleation rate increased about 5-fold at mitosis (5–7). However, direct observation of MT nucleation in living cells has not been possible because the high density of MTs in the cell interior makes it difficult to image individual MTs as they emerge from the centrosome. Therefore, it is currently not known how the MT nucleation rate changes during mitotic progression in living cells.
In the experiments reported here, we used GFP-tagged EB1, a MT tip-binding protein, to visualize new MT plus ends as they emerge from the centrosome. Our data provide a quantitative measure of MT nucleation throughout the cell cycle in living cells.
Experimental Procedures
Cell Culture. A porcine kidney epithelial (LLCPK) stable cell line expressing EB1-GFP was grown as described (8). In this cell line, the expression level of EB1-GFP did not change MT dynamics or cell growth rate. Coverslips for live cell imaging were placed in Rose chambers (9) or grown directly on glass-bottom culture dishes (MatTek, Ashland, MA).
Confocal Microscopy. Live EB1-GFP-expressing LLCPK cells were imaged by using a ×63/1.4 numerical aperture (N.A.) plan apo objective on an inverted microscope (Zeiss Axiovert 200M) equipped with a Zeiss LSM510 META scan head as described (8). Live cell 12-bit image sequences (typically 30 images per cell) were acquired at 2-sec intervals with an average pixel exposure time of 1.3 μs.
Cells were also observed by using a Nikon Eclipse TE 300 microscope equipped with a ×100 phase, N.A. 1.4 objective lens, a spinning-disk confocal scan head (Perkin–Elmer), and a MicroMAX interline transfer cooled charge-coupled device camera (Roper Scientific, Trenton, NJ). All images (16-bit) were acquired by using a single-wavelength (488 nm) filter cube. Image acquisition was controlled by metamorph software (Universal Imaging, Media, PA). Time-lapse sequences were acquired at 2-sec intervals by using an exposure time of 0.3–0.7 sec. Image sequences captured with the laser scanning microscopy (LSM) or the spinning-disk confocal systems yielded nearly identical rates of MT nucleation.
Wide-Field Microscopy. Methanol-fixed cells (below) were examined by wide-field microscopy with a ×60/1.4 N.A. plan apo objective on an inverted microscope (Nikon TE300) (8). Twelvebit images were obtained by using metamorph and analyzed as described below.
Immunofluorescence. Cells were processed for immunofluorescence as described previously (8). Primary antibodies used were mouse anti-α-tubulin B512 (1:1000; Sigma-Aldrich), rabbit antidynein IC [1:250; gift of Kevin Vaughan (10), University of Notre Dame, Notre Dame, IN], rabbit anti-pericentrin (1:100, Babco, Richmond, VA), rabbit anti-γ-tubulin clone AK-15 (1:1,000, Sigma-Aldrich), and rabbit anti-TOGp (11). Secondary antibodies used were Cy5 and Alexa Fluor 568-conjugated goat anti-rabbit or anti-mouse (1:250; Cy5, Jackson ImmunoResearch; Alexa Fluor 568, Molecular Probes) and FITC-conjugated goat anti-mouse (1:100, Jackson ImmunoResearch).
Microinjection. EB1-GFP LLCPK cells were grown directly on glass-bottom culture dishes. Metaphase and anaphase cells were identified by phase-contrast microscopy and microinjected with 0.5 mg/ml (needle concentration) Cy3-conjugated rabbit anti-γ-tubulin antibody (Sigma-Aldrich) or Cy3-conjugated rabbit IgG (Jackson ImmunoResearch) in microinjection buffer. Additional metaphase cells were injected with 0.1 or 0.25 mg/ml anti-γ-tubulin antibody (data not shown). Immediately after injection, cells were imaged by LSM. Additional cells were injected with rhodamine tubulin and imaged as described (8).
Image Analysis. metamorph was used for image analysis. Time-lapse sequences were advanced frame by frame, and EB1-GFP comet emersion from the centrosome was counted. To determine the precision of the counting assay, the same sequence was five times. The repeated counts of the same sequence varied by 4%.
metamorph was used for fluorescence intensity measurements of γ-tubulin staining at the centrosome. A 1,280 × 1,024-pixel, 12-bit image was collected for each centrosome with an image exposure time of 100 ms and an 8× neutral density filter to attenuate illumination. Best focus was based on highest pixel intensity at the center of the centrosome. Imaging conditions were identical for all cell cycle stages, and no images were saturated as judged by the pixel intensity range (available 0–4,095, highest 2,966, lowest 360). The average pixel intensity reading within a 0.7-μm-diameter disk at the center of the centrosome was used, similar to methods used previously by Khodjakov and Rieder (12). In the same manner, we measured MT polymer density (α-tubulin immunofluorescent staining) by taking an average pixel intensity measurement within a 15 × 15-μm box at two different areas of the lamellar regions of G1 and G2 cells.
We also measured centrosome area (μm2). A region of interest (ROI) was drawn around the centrosome area (γ-tubulin immunofluorescent staining), and the area of the ROI was calculated by using metamorph.
Movies 1 and 2, which are published as supporting information on the PNAS web site, show EB1-GFP comets emerging from the centrosome and growing through the cytoplasm. G1/S (10 frames per sec) and metaphase (5 frames per sec) LLCPK cells are shown. Still frames corresponding to these movies are included in Fig. 2. Frames were collected at 2-sec intervals.
Fig. 2.
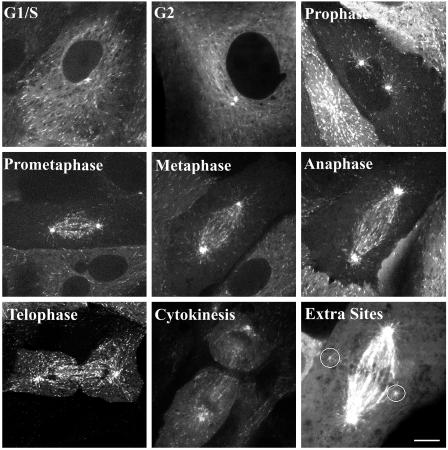
Representative images of EB1-GFP during different stages of the cell cycle. Digital image series from each cell cycle stage were used to count nucleation events per minute. For the telophase cell, only the left centrosome is in focus. In a subset of cells, extra nucleation sites were present (marked by circles). The image labeled Extra Sites was adjusted to show these dim extra nucleation sites. Digital movies of MT nucleation during G1/S and metaphase are available as Movies 1 and 2. (Scale bar, 10 μm.)
Results and Discussion
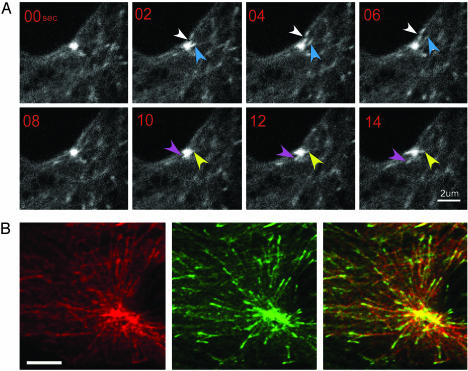
Using an LLCPK cell line stably expressing EB1-GFP (8), we found that EB1-GFP served as a marker for MT nucleation in living cells. EB1-GFP labeled the centrosome as well as MT tips (Fig. 1 A and B; refs. 13–16). In image sequences, EB1-GFP comets continually emerged from the centrosome. These EB1-GFP comets first appeared as a cloudy extension of the centrosome fluorescence, which condensed into a spot, and then moved away from the centrosome with the spot leading and a short comet-like tail trailing (Fig. 1A). To confirm that the EB1-GFP comets emerging from the centrosome were colocalized with newly polymerized MTs, we injected cells with rhodamine-tubulin, fixed them within 5 min, and examined the centrosome region by using confocal microscopy. Nearly all rhodamine-labeled MTs polymerized from the centrosome had a GFP-labeled tip (Fig. 1B), indicating that EB1-GFP emergence from the centrosome faithfully reflects an early step in MT growth that we will refer to as nucleation. To measure the rate of nucleation, we simply counted the number of EB1-GFP comets emerging from the centrosome over time. Our measurements are slight underestimates of the actual nucleation rate because EB1-GFP comets emerging directly above or below the bright centrosome were not detected.
Fig. 1.
EB1-GFP labels new MT ends as they emerge from the centrosome. (A) Time series of EB1-GFP comets emerging from a G1 centrosome. Each colored arrow follows a single EB1-GFP comet. Typically, each comet was visible for three to four frames. Images were collected by spinning disk confocal microscopy. (B) An interphase LLCPK cell stably expressing EB1-GFP was injected with rhodamine-tubulin and fixed <5 min later. Shown is a single LSM optical section. Both rhodamine tubulin and EB1-GFP brightly label each of the two closely spaced centrosomes in this cell. (Scale bar, 5 μm.)
To measure MT nucleation rate throughout the cell cycle, time-lapse image sequences of EB1-GFP-expressing cells were acquired by using either spinning-disk confocal microscopy or LSM. Fig. 2 shows representative images of cells at different stages of the cell cycle collected by spinning-disk confocal microscopy (see Movies 1 and 2). Mitotic stages were assigned based on phase contrast microscopy (data not shown). To differentiate between later stages of mitosis, we used the following criteria. Telophase was defined by the initiation of chromosome decondensation after chromosomes had reached the poles and the spindle had elongated. Telophase cells also had a prominent cleavage furrow (Fig. 2). Cytokinesis was defined by the nearly complete closure of the cleavage furrow (Fig. 2). At completion of cytokinesis, EB1-GFP-labeled MT tips no longer grew across the midbody region connecting dividing cells (data not shown).
Cells in G1 and early S phase contained a single centrosome that nucleated an average of 15 ± 2 MTs per min (Fig. 3). A slower nucleation rate (5.6 MTs per min) was measured in CHO cells by using GFP-CLIP-170 to mark MT ends (17). The rate of MT nucleation in interphase CHO cells transiently expressing GFP-EB1 was identical to that obtained in LLCPK cells (K. J. Salaycik and P.W., unpublished observations), indicating that the slower nucleation measured with CLIP-170 may represent a slower loading of CLIP-170 onto MT ends.
Fig. 3.
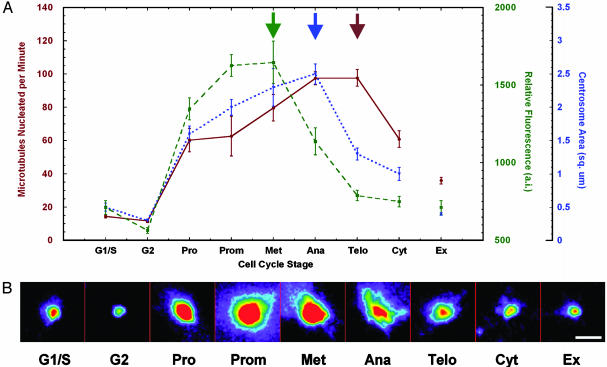
Rates of MT nucleation throughout the cell cycle do not correlate with changes in γ-tubulin recruitment to the centrosome or centrosome size. (A) Entry into mitosis corresponds with a 4-fold increase in nucleation rate. This rate continues to rise and reaches a maximum at anaphase and telophase. Rates were determined from image sequences acquired from 4–12 cells per cell cycle stage. γ-Tubulin staining intensity at the centrosome rose dramatically at prophase and peaked at metaphase. Centrosome area increased from prophase to anaphase. The area of the centrosome was determined by tracing the outline of γ-tubulin staining intensity. For both γ-tubulin staining intensity and centrosome area, 20–38 centrosomes were measured for each time point. Ex refers to extra nucleation sites present in a subset of cells. Data for extra sites were from metaphase cells. Data shown are mean ± SE. Arrows note maximum. (B) Representative images of γ-tubulin-stained centrosomes. Images were spectrally coded to represent staining intensity (red is the highest intensity). No saturated pixels were present in the original grayscale 12-bit images. (Scale bar, 3 μm.)
G2 cells contained two centrosomes, and each centrosome nucleated MTs at a rate similar to the G1 centrosome (12 ± 2 MTs per min); there was no significant difference in rates between the two G2 centrosomes (P < 0.05). All G2 cells were then fixed and stained for γ-tubulin. In all cases, γ-tubulin was present at each G2 centrosome (data not shown). These results indicate that the new daughter centrosome, replicated during S phase, is functionally mature by G2.
Given that each G2 centrosome nucleates MTs at the same rate, the combined nucleation rate of the two G2 centrosomes is approximately twice that in G1/S cells containing a single centrosome. Does the combined nucleation rate of the two G2 centrosomes result in an increase in MT polymer level? The density of MTs was not changed between G1/S and G2 [measured in anti-tubulin-stained cells (P < 0.05; data not shown)]. The percent of tubulin in polymer is also not changed between G1 and G2 in LLCPK cells (18). Because G2 cells are double the size of G1 cells, we suggest that the combined nucleation capacity of the two G2 centrosomes was necessary to maintain a constant MT density per unit area.
The MT nucleation rate showed a sharp rise when cells entered mitosis. In prophase cells, the nucleation rate at each centrosome was 4-fold higher than in G1/S or G2 centrosomes (Fig. 3). During metaphase, MT nucleation rate per centrosome was 5-fold higher than that in G1/S or G2, remarkably consistent with previous EM studies (5–7). Surprisingly, the nucleation rate did not peak at metaphase but remained high through anaphase and telophase (Fig. 3). During cytokinesis, the nucleation rate decreased significantly (P < 0.05) and fell to prophase levels (Fig. 3). We confirmed these observations by measuring the nucleation rate in CHO cells transiently expressing EB1-GFP. CHO cells had nucleation rates nearly identical to LLCPK cells at each cell cycle stage (data not shown).
The increased nucleation capacity of mitotic centrosomes may depend on recruitment of additional MT nucleating material from a cytoplasmic pool (1). It is generally thought that γ-tubulin, in a ring complex (γTuRC) with associated proteins, forms the nucleation sites on the pericentriolar matrix (19–21). Previous work has shown that γ-tubulin is recruited to the centrosome either abruptly at the beginning of mitosis (12) or gradually from G1 to metaphase (2).
To examine γ-tubulin recruitment to the centrosome in the EB1-GFP cell line, we stained fixed cells with an antibody to γ-tubulin and measured the fluorescent staining intensity at centrosomes throughout the cell cycle. The centrosome γ-tubulin staining intensity peaked at metaphase and then decreased significantly (P < 0.05) during anaphase and telophase (Fig. 3). Similar changes in γ-tubulin concentration at the centrosome were measured previously by using GFP-γ-tubulin (12). Comparison of the centrosomal γ-tubulin level and MT nucleation rates (Fig. 3) showed that the centrosome concentration of γ-tubulin dropped when nucleation remained high. Therefore, nucleation cannot be solely regulated by γ-tubulin recruitment to the centrosome.
Based on the shapes of centrosomes in sea urchin embryos, Mazia (22) proposed that folding and unfolding a lattice structure to hide or expose additional nucleation sites regulates the nucleating capacity of the centrosome. The best support for this model is the observation that the size of the centrosome increases at entry into mitosis (2, 12, 23). To examine whether changes in the size of the centrosome correlate with nucleation rate, we measured the area of the centrosome as indicated by γ-tubulin immunofluorescent staining (Fig. 3). Interphase centrosomes in G1/S had a mean area of 0.5 ± 0.3 μm2, and each G2 centrosome had an area of 0.3 ± 0.1 μm2. During prophase, the area of each centrosome increased to 5-fold the G2 area (1.6 ± 0.5 μm2). The area of each centrosome continued to increase, reaching a maximum area during metaphase and anaphase (8 times the G2 area). In telophase, centrosome area was significantly reduced (P < 0.05) to half the anaphase size, down to a 4-fold increase over G2 levels (1.3 ± 0.5 μm2). This decline in size continued during cytokinesis (1.0 ± 0.5 μm2). Thus, increases in centrosome area did not correlate with increases in nucleation rate throughout all stages of mitosis, suggesting that nucleation rate is not simply dependent on the size of the centrosome. It is important to note that we only measured the area of the centrosome and did not take into account changes in shape. As noted previously for sea urchin embryos (22), centrosomes appeared more oblong and less spherical during telophase (Fig. 3).
Additional evidence that the amount of γ-tubulin at the centrosome and the size of the nucleating structure do not correlate with nucleation rate came from observations of extra nucleation sites observed in about 25% of mitotic cells (Fig. 2, labeled EX). The extra nucleation sites were not observed in interphase cells but appeared as small particles located near the spindle in mitotic cells. We confined our observation of these sites to metaphase cells. Although we do not know whether these sites were true centrosomes or centrosomal satellites (24), the extra sites contained the centrosomal proteins γ-tubulin, pericentrin, TOGp, and dynein (data not shown). The rate of MT nucleation from the extra sites was approximately half that from the bona fide metaphase centrosomes and twice the rate of interphase centrosomes (Fig. 3). In contrast, the extra sites were similar to interphase centrosomes in γ-tubulin concentration and area (Fig. 3).
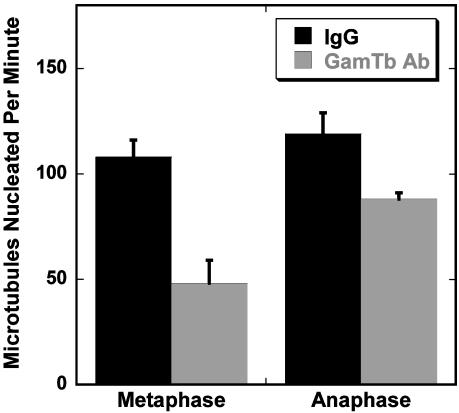
The above observations suggested that additional factors contribute to MT nucleation after anaphase onset. As a first step in characterizing possible differences in nucleation between metaphase and anaphase, we compared the ability of a function-blocking antibody to γ-tubulin (25) to inhibit nucleation from metaphase and anaphase centrosomes. Hannak et al. (26) recently described a γ-tubulin-independent nucleation pathway present during mitosis in Caenorhabditis elegans embryos. We reasoned that a γ-tubulin-independent pathway might be active during later stages of mitosis, allowing cells to maintain a high rate of nucleation when the γ-tubulin concentration at centrosomes declines. Injection of metaphase cells with Cy3-labeled anti-γ-tubulin caused a dose-dependent decrease in nucleation rate (data not shown). At the highest antibody concentration tested (0.5 mg/ml needle concentration), the nucleation rate was decreased to ≈50% of the rate in cells injected with a nonimmune IgG (Fig. 4). In 6 of 18 metaphase cells, antibody injection completely blocked nucleation. In contrast, anaphase cells injected with the same concentration of antibody showed a smaller (25%) reduction in nucleation rate (Fig. 4), and all injected cells (10/10) were able to nucleate MTs. These results suggest that cells in the late stages of mitosis have a γ-tubulin-independent pathway for MT nucleation.
Fig. 4.
Microinjection of an antibody to γ-tubulin blocks nucleation during metaphase but is less effective during anaphase. Cells were microinjected with a function-blocking γ-tubulin antibody coupled to Cy3 or with Cy3-rabbit IgG and then imaged by LSM. Injection of 0.5 mg/ml antibody (needle concentration) into metaphase cells resulted in a large decrease in nucleation rate. Anaphase cells injected with anti γ-tubulin showed a much smaller decrease in nucleation rate. Data shown are mean ± SE for 8–18 centrosomes for each condition. Injection alone did not change the nucleation rate. Mean nucleation rates for noninjected and control IgG-injected metaphase and anaphase cells were not significantly different (P < 0.05).
The mechanism responsible for centrosomal nucleation in the absence of γ-tubulin is not known but could be regulated by cell cycle-dependent changes in centrosomal proteins. For example, the rate of MT release from the centrosome is higher in mitotic cells (3, 27). Release may result from the activation of severing proteins (28), which could leave a MT stub attached to the nucleation site. These MT fragments could provide a new plus end for elongation. Centrosomal proteins are also highly phosphorylated at entry into mitosis (29), likely via CDK1 (30, 31), polo (32) and/or Randependent pathways (33). Although phosphorylation of most centrosomal proteins declines at anaphase onset (29), phosphorylation of a small subset of centrosomal proteins may be sufficient to maintain a high rate of nucleation after anaphase onset.
What might be the significance of maintaining a high rate of MT nucleation in anaphase and telophase? It is possible that high nucleation rates during anaphase and telophase are necessary to generate a large number of astral MTs that then form the new interphase MT arrays. In support of this possibility, the length and number of astral MTs has also been observed to increase during anaphase in mammalian cells (34). A high number of astral MTs also may be necessary to increase the probability that some MTs grow to sufficient length to make contact with sites at the cell cortex. Such interactions are thought necessary for spindle positioning and could contribute to cell cycle progression (35, 36).
Conclusions
Our direct observations of MT nucleation in living cells demonstrate that centrosomal nucleation of MTs increases dramatically at entry into mitosis and continues through telophase. In addition, we observed MT nucleation from noncentrosomal sites in many mitotic cells. The nature and significance of these sites is not known. Noncentrosomal nucleation has also been described from the iMTOCs of Schizosaccharomyces pombe (37). Our data show that nucleation rate is not correlated with changes in γ-tubulin concentration or centrosome size throughout all stages of mitosis, suggesting additional regulatory mechanisms. The assay we described here provides a mechanism to quantify MT nucleation in living cells and to decipher mechanisms controlling centrosome maturation.
Supplementary Material
Acknowledgments
We thank Frank Luca and Bob Skibbens for helpful discussions. This work was supported by National Institutes of Health Grants GM58025 (to L.C.) and GM59057 (to P.W.) and National Science Foundation Equipment Grant DBI-0200322 (to L.C.).
Abbreviations: LSM, laser scanning confocal microscopy; MT, microtubule.
References
- 1.Bornens, M. (2002) Curr. Opin. Cell Biol. 14, 25-34. [DOI] [PubMed] [Google Scholar]
- 2.Dictenberg, J. B., Zimmerman, W., Sparks, C. A., Young, A., Vidair, C., Zheng, Y., Carrington, W., Fay, F. S. & Doxsey, S. J. (1998) J. Cell Biol. 141, 163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, S. S. L. (1999) Int. Rev. Cytol. 187, 51-109. [DOI] [PubMed] [Google Scholar]
- 4.Palazzo, R. E., Vogel, J. M., Schnackenberg, B. J., Hull, D. R. & Wu, X. (2000) Curr. Top. Dev. Biol. 49, 449-470. [DOI] [PubMed] [Google Scholar]
- 5.Kuriyama, R. & Borisy, G. G. (1981) J. Cell Biol. 91, 822-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snyder, J. A. & McIntosh, J. R. (1975) J. Cell Biol. 67, 744-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telzer, B. R. & Rosenbaum, J. L. (1979) J. Cell Biol. 81, 484-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piehl, M. & Cassimeris, L. (2003) Mol. Biol. Cell 14, 916-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rusan, N. M., Tulu, U. S., Fagerstrom, C. & Wadsworth, P. (2002) J. Cell Biol. 158, 997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaughan, K. T. & Vallee, R. B. (1995) J. Cell Biol. 131, 1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassimeris, L., Gard, D., Tran, P. T. & Erickson, H. P. (2001) J. Cell Sci. 114, 3025-3033. [DOI] [PubMed] [Google Scholar]
- 12.Khodjakov, A. & Rieder, C. L. (1999) J. Cell Biol. 146, 585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berrueta, L., Kraeft, S. K., Tirnauer, J. S., Schuyler, S. C., Chen, L. B., Hill, D. E., Pellman, D. & Bierer, B. E. (1998) Proc. Natl. Acad. Sci. USA 95, 10596-10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison, E. E., Wardleworth, B. N., Askham, J. M., Markham, A. F. & Meredith, D. M. (1998) Oncogene 17, 3471-3477. [DOI] [PubMed] [Google Scholar]
- 15.Mimori-Kiyosue, Y., Shiina, N. & Tsukita, S. (2000) Curr. Biol. 10, 865-868. [DOI] [PubMed] [Google Scholar]
- 16.Tirnauer, J. S., Canman, J. C., Salman, E. D. & Mitchison, T. J. (2002) Mol. Biol. Cell 13, 4308-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Komarova, Y. A., Vorobjev, I. A. & Borisy, G. G. (2002) J. Cell Sci. 115, 3527-3539. [DOI] [PubMed] [Google Scholar]
- 18.Zhai, Y., Kronebusch, P. J., Simon, P. M. & Borisy, G. G. (1996) J. Cell Biol. 135, 201-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moritz, M., Zheng, Y., Alberts, B. M. & Oegema, K. (1998) J. Cell Biol. 142, 775-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman, W., Sparks, C. A. & Doxsey, S. J. (1999) Curr. Opin. Cell Biol. 11, 122-128. [DOI] [PubMed] [Google Scholar]
- 21.Job, D., Valiron, O. & Oakley, B. (2003) Curr. Opin. Cell Biol. 15, 111-117. [DOI] [PubMed] [Google Scholar]
- 22.Mazia, D. (1987) Int. Rev. Cytol. 100, 49-92. [DOI] [PubMed] [Google Scholar]
- 23.Rieder, C. L. (1990) Electron Microsc. Rev. 3, 269-300. [DOI] [PubMed] [Google Scholar]
- 24.Dammermann, A. & Merdes, A. (2002) J. Cell Biol. 159, 255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi, H. C., Palacios, M. J., McNamara, L. & Cleveland, D. W. (1992) Nature 356, 80-83. [DOI] [PubMed] [Google Scholar]
- 26.Hannak, E., Oegema, K., Kirkham, M., Gonczy, P., Habermann, B. & Hyman, A. A. (2002) J. Cell Biol. 157, 591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Belmont, L. D., Hyman, A. A., Sawin, K. E. & Mitchison, T. J. (1990) Cell 62, 579-589. [DOI] [PubMed] [Google Scholar]
- 28.McNally, K. P., Bazirgan, O. A. & McNally, F. J. (2000) J. Cell Sci. 113, 1623-1633. [DOI] [PubMed] [Google Scholar]
- 29.Vandre, D. D. & Borisy, G. G. (1989) J. Cell Sci. 94, 245-258. [DOI] [PubMed] [Google Scholar]
- 30.Buendia, B., Draetta, G. & Karsenti, E. (1992) J. Cell Biol. 116, 1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohta, K., Shiina, N., Okumura, E., Hisanaga, S., Kishimoto, T., Endo, S., Gotoh, Y., Nishida, E. & Sakai, H. (1993) J. Cell Sci. 104, 125-137. [DOI] [PubMed] [Google Scholar]
- 32.Fry, A. M., Mayor, T. & Nigg, N. E. (2000) Curr. Top. Dev. Biol. 49, 291-312. [DOI] [PubMed] [Google Scholar]
- 33.Carazo-Salas, R. E., Gruss, O. J., Mattaj, I. W. & Karsenti, E. (2001) Nat. Cell Biol. 3, 228-234. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, E. E. & Askham, J. M. (2001) Eur. J. Cell Biol. 80, 749-753. [DOI] [PubMed] [Google Scholar]
- 35.O'Connell, C. B. & Wang, Y. L. (2000) Mol. Biol. Cell 11, 1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, H., Oliferenko, S. & Balasubramanian, M. K. (2003) Curr. Opin. Cell Biol. 15, 82-87. [DOI] [PubMed] [Google Scholar]
- 37.Tran, P. T., Marsh, L., Doye, V., Inoue, S. & Chang, F. (2001) J. Cell Biol. 153, 397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.