Abstract
Structural studies on T cell receptors (TCRs) specific for foreign antigens demonstrated a remarkably similar topology characterized by a central, diagonal TCR binding mode that maximizes interactions with the MHC bound peptide. However, three recent structures involving autoimmune TCRs demonstrated unusual interactions with self-peptide/MHC complexes. Two TCRs from multiple sclerosis patients bind with unconventional topologies, and both TCRs are shifted toward the peptide N terminus and the MHC class II β chain helix. A TCR from the experimental autoimmune encephalomyelitis (EAE) model binds in a conventional orientation, but the structure is unusual because the self-peptide only partially fills the binding site. For all three TCRs, interaction with the MHC bound self-peptide is suboptimal, and only two or three TCR loops contact the peptide. Optimal TCR binding modes confer a competitive advantage for antimicrobial T cells during an infection, whereas altered binding properties may permit survival of a subset of autoreactive T cells during thymic selection.
Escape from Negative Selection by Autoreactive T Cells
Autoimmune diseases are caused by self-reactive T cells that have escaped negative selection during T cell development in the thymus (Goodnow et al., 2005). In many autoimmune diseases, pathogenic T cells recognize “tissue- specific” antigens, and it was previously thought that the presence of such autoreactive T cells is due to a lack of self-antigen expression in the thymus. However, more recent work has convincingly demonstrated that most tissue-specific self-antigens are in fact expressed in the thymus by an unusual subpopulation of medullary thymic epithelial cells (mTECs) that express a wide range of tissue-specific genes in a promiscuous manner (Derbinski et al., 2001). The expressed genes encode autoantigens relevant in human autoimmune diseases; such autoantigens include myelin basic protein (MBP) and proteolipid protein (PLP), which are considered to be target antigens in multiple sclerosis (MS) (Pribyl et al., 1996; Derbinski et al., 2001). The promiscuous expression of these genes in mTECs is in part controlled by the transcription factor AIRE, and both humans (The Finnish-German APECED Consortium, 1997) and mice with a defective AIRE gene (Anderson et al., 2002) develop autoimmunity in multiple organs.
Several explanations for a failure of negative selection have been proposed on the basis of experiments in animal models of autoimmunity. Particularly instructive has been a comparison of the T cell response to a self-antigen between wild-type and knockout mice that lack expression of the self-antigen in question. Collectively, these experiments have demonstrated that expression of the self-antigen has striking effects on the T cell repertoire and that T cells directed against certain epitopes are largely deleted, whereas T cells that recognize other epitopes escape negative selection (Harrington et al., 1998; Klein et al., 2000). Comparison of the CD4 T cell response in MBP-deficient and wild-type H-2u mice showed that the response to MBP is far more vigorous in MBP-deficient mice and that the majority of responding T cells in MBP-deficient mice recognize the 121–150 region of MBP (Harrington et al., 1998). In contrast, the CD4 T cell response in wild-type H-2u mice is primarily focused on the N-terminal Ac1-11 epitope of MBP that binds with low affinity to I-Au, whereas a T cell response to the high-affinity 121–150 peptide is barely detectable (Zamvil et al., 1986; Harrington et al., 1998). Alternative splicing has been implicated as another mechanism for a failure of negative selection in another EAE model in which the T cell response is primarily focused on an epitope of PLP. The PLP splice variant present in the thymus lacks the critical T cell epitope, resulting in a selective defect of thymic negative selection to the PLP 139–151 peptide (Anderson et al., 2000; Klein et al., 2000).
These two mechanisms—low-affinity peptide binding and alternative splicing—do not account for the majority of cases in which autoreactive T cells are present in the mature T cell repertoire. A substantial number of peptides that are recognized by self-reactive T cells bind with an intermediate or high affinity to the relevant MHC molecule (Wall et al., 1992; Valli et al., 1993; Wucherpfennig et al., 1994a), and in most cases the relevant antigen and the epitope in question are expressed in the thymus (Pribyl et al., 1996; Derbinski et al., 2001). Two recent studies have indicated that general alterations in T cell signaling thresholds can profoundly affect the outcome of thymic selection events. The molecular defects can affect either early TCR signaling events or more distal signaling pathways that initiate apoptosis in thymocytes (Sakaguchi et al., 2003; Liston et al., 2004). In a spontaneously occurring mouse model of rheumatoid arthritis (SKG mice), the genetic defect was pinpointed to the SH2 domain of ZAP-70, a tyrosine kinase that associates with the ζ chain of the TCR-CD3 complex (Sakaguchi et al., 2003). Diminished TCR signaling dramatically affected thymic selection and resulted in positive selection of otherwise negatively selected autoreactive T cells (Sakaguchi et al., 2003). It is possible that alterations in TCR binding to peptide/MHC, which modulate the strength of TCR signaling in the thymus, can lead to a similar outcome.
The Conventional Topology of Peptide/MHC Class I Binding by TCRs
The first crystal structures of TCR/peptide/MHC class I complexes showed remarkable similarities in the overall topology of TCR binding to peptide/MHC, even though the TCRs originated from two different species and had been isolated from distinct biological settings, a chronic infectious disease (human A6 TCR), and an alloimmune response (murine 2C TCR) (Garboczi et al., 1996; Garcia et al., 1996, 1998). In both structures, the TCR is positioned diagonally across the compound surface created by the peptide and the long MHC helices that flank the peptide binding site such that the TCR covers most of the MHC bound peptide (illustrated for A6 TCR in Figures 1A and 1B). The most diverse TCR loops, the CDR3 loops of TCRα and β chains, are located over the central peptide residue and form a pocket that accommodates this peptide side chain (P5 tyrosine of the nine amino acid Tax peptide in the A6 structure, Figures 1B–1D). This TCR position permits extensive interactions between the most diverse TCR loops and the central segment of the MHC bound peptide.
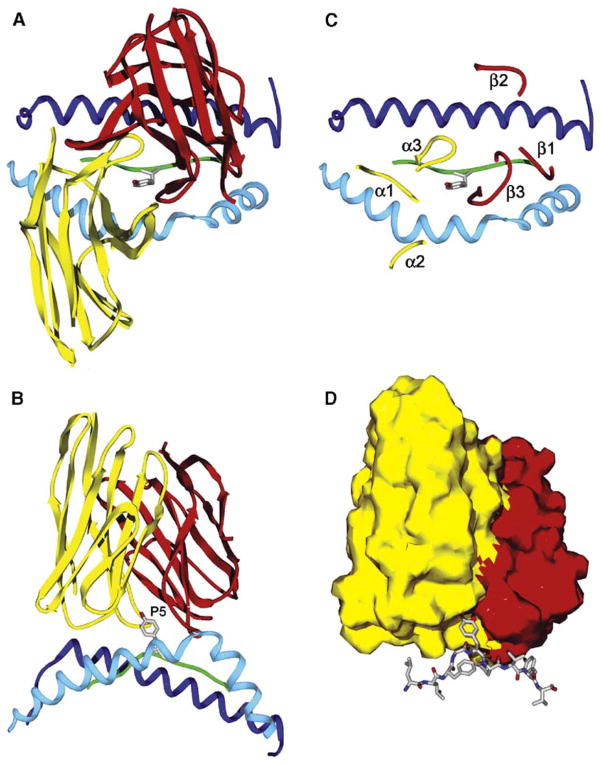
Figure 1. The Conventional Topology of TCR Binding to Peptide/MHC Complexes.
(A and B) The conventional TCR binding mode is illustrated with the human A6 TCR as an example (Protein Data Bank [PDB] accession number 1AO7). This TCR recognizes the HTLV-1 Tax 11–19 peptide bound to HLA-A2 (Garboczi et al., 1996). The A6 TCR binds in a diagonal orientation to the peptide/MHC class I complex and is centered over the P5 tyrosine residue of the nine amino acid Tax peptide. Top view from the T cell surface (A) and side view (B) are shown; for clarity, only the TCR variable domains, the peptide backbone, and the MHC helices are shown. The side view shows the high points of the MHC helices that are avoided by the diagonal orientation of the TCR across the peptide/MHC surface. The P5 tyrosine side chain of the peptide is shown, and the backbone of the peptide is colored green. The MHC helices are colored blue (α1 helix in dark blue and α2 helix in light blue, respectively). The Vα and Vβ domains of TCR are colored yellow and red, respectively. The same color code is used throughout the figures for TCR V domains, MHC molecules, and peptides.
(C) Location of the TCR loops on the peptide/MHC surface. The CDR3 loops of both TCR chains interact directly with the peptide and form a pocket for P5 tyrosine. The CDR1α and CDR1β loops are located over the N-terminal and C-terminal segments of the peptide, respectively. In the conventional diagonal orientation, the CDR2 loops do not contact the peptide, but make contacts to the MHC helices. The TCRα and TCRβ chain loops are colored yellow and red, respectively. The CDR1, CDR2, and CDR3 loops are labeled as α1, α2, and α3 for TCRα and as β1, β2, and β3 for TCRβ.
(D) Space-filling model of the TCR variable domains (Vα yellow, Vβ red), illustrating the TCR pocket for the P5 tyrosine side chain of the peptide located in the center of the TCR interaction surface with peptide/MHC. The Tax peptide is shown as a stick-and-ball model. The figure was prepared with Deep View/Swiss-PDB Viewer (Guex and Peitsch, 1997).
The other TCR loops (CDR1 and CDR2) are encoded by the V gene segments and are less diverse in sequence among different TCRs. The diagonal orientation places the CDR1 loops of TCRα and TCRβ over the N-terminal and C-terminal segments of MHC class I bound peptides (Figure 1C), such that a total of four TCR loops (CDR1 and CDR3 loops of both chains) can participate in peptide recognition. The CDR1 loops can also contact the MHC helices. In contrast, the CDR2 loops of both chains are positioned over the MHC helices and do not participate in peptide recognition in these structures (Figure 1C) (Garcia et al., 1999; Hennecke and Wiley, 2001; Rudolph and Wilson, 2002).
The TCR surface that contacts peptide/MHC was found to be rather flat, with the exception of the central cavity described above. In contrast, the MHC helices form two high points at opposite ends, and the TCR avoids these high points by a diagonal orientation (Figure 1B). Subsequent MHC class I/peptide/TCR structures enforced the view that this binding topology may be general and led to the hypothesis that the germline-encoded CDR1 and CDR2 loops have evolved to recognize structural features of MHC molecules (Ding et al., 1998; Garcia et al., 1999; Reiser et al., 2000, 2002, 2003; Hennecke and Wiley, 2001; Rudolph and Wilson, 2002; Stewart-Jones et al., 2003). A contribution of the CDR1 or CDR2 loop of TCRα to MHC binding was shown by experiments in which single amino acid substitutions in these TCR loops affected T cell differentiation to the CD4 or CD8 lineage (Sim et al., 1996). However, mutagenesis experiments failed to identify conserved MHC side chains required for recognition by all TCRs (Sun et al., 1995; Baker et al., 2001).
MHC Class II Restricted TCRs that Recognize Foreign Peptides Also Bind with Conventional Topology
Subsequently, the structures of two MHC class II restricted TCRs were determined that recognize foreign peptides: the mouse D10 TCR specific for a conalbumin peptide bound to I-Ak (Reinherz et al., 1999), and the human HA1.7 TCR specific for an influenza hemagglutinin (HA, residues 306–318) peptide bound to HLA-DR1 (DRA, DRB1*0101) (Figure 2A; Table 1) (Hennecke et al., 2000). In contrast to MHC class I molecules, the peptide binding site of MHC class II molecules is open at both ends, permitting binding of longer peptides (Brown et al., 1993). Despite these differences between MHC class I and class II molecules, both of these MHC class II restricted TCRs bind with a very similar topology as the MHC class I restricted TCRs described above. The only substantial difference between MHC class I and class II restricted TCRs appeared to be the crossing angle (defined by a line drawn through the peptide and a line through the centers of mass of the TCR variable domains) because the first structure of an MHC class II restricted TCR (D10 TCR) (Reinherz et al., 1999) suggested a more orthogonal position (80°) than previously reported structures for MHC class I restricted T cells (45°–70°) (Garcia et al., 1999). However, subsequent studies demonstrated that there are no global differences in the binding angle between MHC class I and class II restricted TCRs (Hennecke and Wiley, 2001; Rudolph and Wilson, 2002; Stewart-Jones et al., 2003). The overall similarities between these initial structures of MHC class I and class II TCR complexes further enforced the notion that all TCRs bind to peptide/MHC complexes in a similar fashion. All human TCRs analyzed in these studies recognized viral peptides, and the T cell clones from which they had been isolated represented predominant T cell populations in the immune response to the virus. In vivo competition (Kedl et al., 2003) may therefore have resulted in the expansion of T cells whose TCRs have optimal binding properties for these viral peptide/MHC ligands.
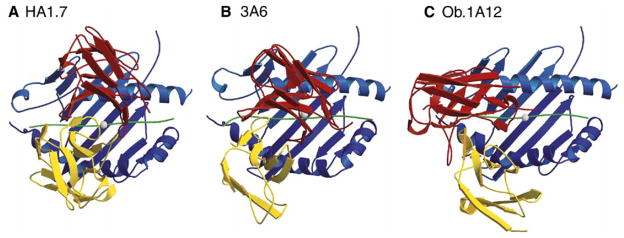
Figure 2. Altered Topology of Self-Peptide/MHC Binding by Two Human Autoimmune TCRs.
(A) The HA1.7 TCR (PDB accession number 1FYT) is specific for the influenza HA 306–318 peptide bound to DRA, DRB1*0101 and binds with a conventional topology over the center of the peptide/MHC surface. The white sphere marks the position of the P5 peptide residue in the center of the binding site over which the CDR3 loops of the TCRα and β chains converge.
(B) The 3A6 TCR was isolated from a patient with MS and recognizes the MBP 89–101 peptide bound to DRA, DRB5*0101 (PDB accession number 1ZGL). Compared to HA1.7 TCR, the position of this TCR is shifted toward the peptide N terminus and the DRβ chain helix (the lower helix in all three DR molecules in this figure).
(C) The Ob.1A12 TCR was isolated from another MS patient and recognizes a different epitope of MBP (residues 85–99) bound to another HLA-DR molecule (DRA, DRB1*1501) (PDB accession number 1YMM). This TCR is also shifted toward the peptide N terminus and tilted toward the DRβ chain helix. In addition, it is rotated counterclockwise relative to HA1.7 TCR. This counterclockwise rotation distinguishes it from 3A6 TCR. The TCR Vα and Vβ domains are colored in yellow and red, respectively, the MHC molecule in blue, and the bound peptide in green. The figure was prepared with Molscript (http://www.avatar.se/molscript/) and rendered with Raster3d (Merritt and Murphy, 1994).
Table 1.
MHC Class II Restricted TCRs for which the Structure of TCR/Peptide/MHC Complex Has Been Determined
| TCR | Peptide | MHC Class II | Species |
|---|---|---|---|
| TCRs Specific for Foreign Peptides | |||
|
| |||
| HA1.7 | Influenza hemagglutinin, 306–318 | DRA, DRB1*0101 | Human |
| D10 | Conalbumin, 131–144 | I-Ak | Mouse |
|
| |||
| TCRs Specific for Self Peptides | |||
|
| |||
| Ob.1A12 | Human myelin basic protein, 85–99 | DRA, DRB1*1501 | Human |
| 3A6 | Human myelin basic protein, 89–101 | DRA, DRB5*0101 | Human |
| 172.10 | Mouse myelin basic protein, Ac1–11 | I-Au | Mouse |
The HA1.7 and D10 TCRs are specific for foreign peptides, whereas the human Ob.1A12 and 3A6 TCRs as well as the murine 172.10 TCR recognize different epitopes of the self-antigen myelin basic protein.
Unconventional Topology of Self-Peptide/MHC Binding by a TCR from a Multiple Sclerosis Patient
The first crystal structure of a human autoimmune TCR (Ob.1A12 TCR) bound to its self-peptide/MHC ligand showed a strikingly different topology (Figure 2C) (Hahn et al., 2005), which was surprising given the strong similarities between all of the previously reported TCR/peptide/MHC structures. The Ob.1A12 TCR originated from a patient with relapsing-remitting MS and is specific for a major epitope of human MBP (residues 85– 99) bound to a MS-associated MHC class II molecule (DRA, DRB1*1501) (Ota et al., 1990; Wucherpfennig et al., 1994a). Transgenic mice that express this human TCR and the MHC class II molecule develop a spontaneous inflammatory disease in the CNS that has similarities with the human disease, demonstrating that this TCR has the potential to be pathogenic in vivo (Madsen et al., 1999; Ellmerich et al., 2004). The structure showed that the Ob.1A12 TCR is not centered over the peptide/MHC surface and that it only contacts the N-terminal segment of the peptide (Figure 2C). In addition, the TCR does not make symmetrical interactions with the MHC helices, but is shifted and tilted toward the DRβ chain helix. Another unusual feature is the counterclockwise rotation of Ob.1A12 TCR relative to the MHC molecule when compared to HA1.7 TCR. The crossing angle is 70° for HA1.7 TCR, within the 45°–80° range observed for other TCRs, but 110° for Ob.1A12 TCR. This altered position of the TCR dramatically affects peptide recognition. This is particularly evident in the position of the two CDR3 loops, which create a large cavity over the P2 side chain, rather than the P5 side chain (Figures 3C, 4, and 5C, left panel). The lateral shift in the location of the CDR3 loops is substantial and corresponds to three peptide residues. The cavity created by the CDR3 loops not only accommodates a peptide side chain (P2 histidine, Figure 3C), but also a side chain from a MHC helix (DRβ81 histidine, Figure 5C, left panel).
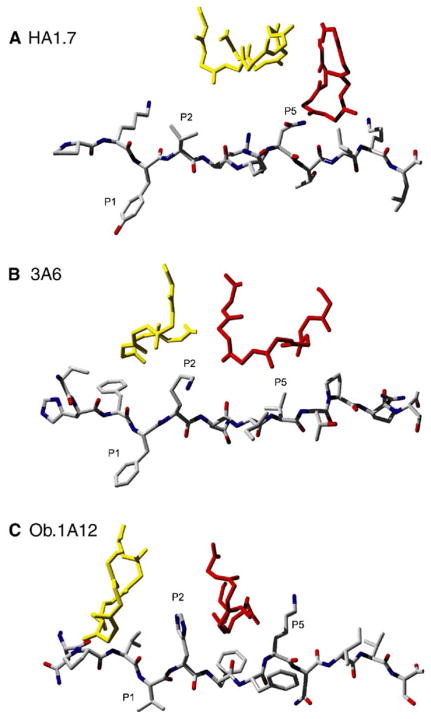
Figure 3. Unusual Location of the TCR CDR3α and CDR3β Loops over the MHC Bound Peptide in the 3A6 and Ob.1A12 Structures.
(A) The influenza HA peptide is shown as a stick model with the peptide backbone and side chain carbons in gray and the nitrogen and oxygen atoms in blue and red, respectively. The two CDR3 loops meet over the P5 side chain of the HA peptide. Only the backbone of the CDR3 loops (yellow, CDR3α; red, CDR3β) is shown to avoid crowding.
(B and C) In the 3A6 (B) and Ob.1A12 (C) structures, the CDR3 loops are instead positioned over the P2 peptide residue, a lysine in the 3A6 structure, and a histidine in the Ob.1A12 structure. The CDR3β loop of Ob.1A12 TCR forms lateral contacts with the P5 lysine of the MBP peptide. The figure was prepared with Deep View/Swiss-PDB Viewer (Guex and Peitsch, 1997) and POV-Ray (http://www.povray.org/).
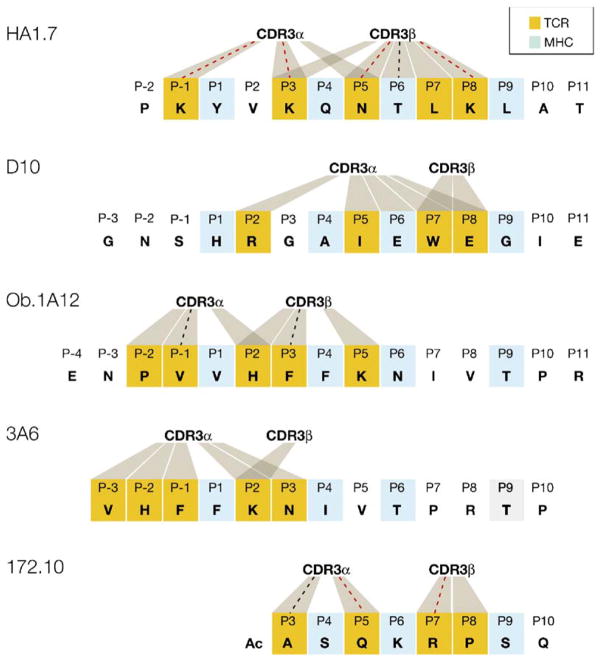
Figure 4. Peptide Contacts Established by TCR CDR3 Loops in Human and Murine TCR/Peptide/MHC Class II Structures.
The peptide residues that occupy the P1, P4, P6, and P9 pockets of the MHC class II peptide binding site are colored light blue in all five peptide sequences. Peptide residues contacted by CDR3 loops are colored orange, and contacts to CDR3α or CDR3β are indicated by shaded areas. Contacts that represent hydrogen bonds are marked with a dotted line, which is colored red when the contact involves a side chain of the peptide. In the HA1.7 (human) and D10 (mouse) structures in which the TCR recognizes a foreign peptide, both CDR3 loops are located over the center of the peptide. In contrast, both human autoimmune TCRs (Ob.1A12 and 3A6) are characterized by a shift of the CDR3 loops toward the N terminus of the peptide. The mouse 172.10 TCR recognizes the N-terminal MBP Ac1-11 peptide that only partially fills the peptide binding site. This structure also contains a peptide extension by the insect leader peptide, which is not part of the native MBP peptide and thus not included in this figure. The CDR3 loops of D10 TCR do not form hydrogen bonds to the peptide, but a hydrogen bond is present between the CDR1α loop and P2 arginine of the peptide.
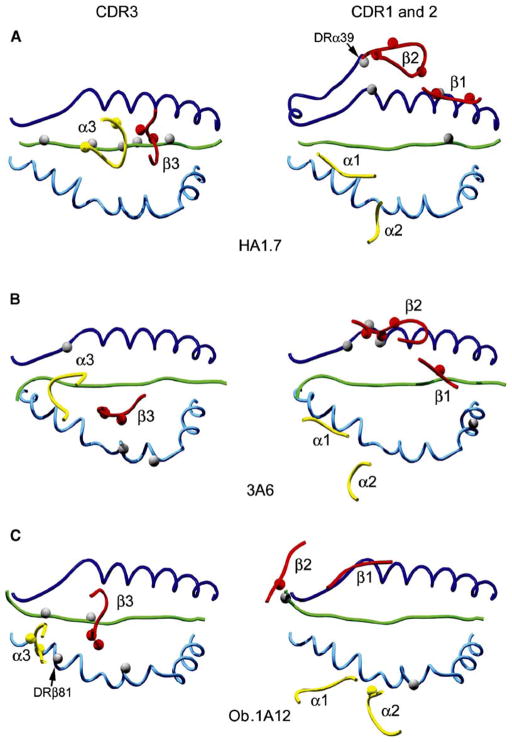
Figure 5. Location of the TCR Loops on the Peptide/MHC Surface in the Three Human TCR/Peptide/MHC Class II Structures.
In the left panel, the CDR3 loops are located over the center of the peptide/MHC surface in the HA1.7 structure, but are shifted toward the peptide N terminus in the 3A6 and Ob.1A12 structures. The CDR3α (yellow) and CDR3β (red) loops are labeled as α3 and β3, respectively. Residues involved in hydrogen bonds are represented as small spheres.
In the right panel, the CDR1 and CDR2 loops of TCRβ chains are located in different positions in each of the three structures, and their position is most extreme in the Ob.1A12 structure. The position of the CDR1 and CDR2 loops of TCRα chains is more similar among the three structures, but the CDR2α loop of 3A6 TCR fails to contact peptide/MHC. The CDR1 and CDR2 loops of TCRα and β are labeled as α1, α2, β1, and β2. The figure was prepared with Deep View/Swiss-PDB Viewer (Guex and Peitsch, 1997) and POV-Ray (http://www2.povray.org/).
Several other rules thought to be universally applicable to TCR recognition do not apply to this TCR. The majority of contacts to the MHC helices are made by the hypervariable CDR3 loops rather than by the germline- encoded CDR1 and CDR2 loops (Figure 5C). Particularly prominent is the contribution of the TCRβ CDR3 loop, which spans across the peptide binding site and contacts both MHC helices (Figure 5C, left panel). The majority of residues in this TCR loop that contact the MHC helices (three out of four residues) are encoded by randomly inserted N-region nucleotides at the V-D-J junction, indicating that the unique sequence elements of this TCR make a significant contribution to MHC binding. It therefore appears that unconventional topologies are possible because of the sequence diversity of TCRs in the CDR3 loops.
The functional relevance of the topology observed in this structure is supported by a large body of experimental data. Experiments with peptide analogs had demonstrated that P2 histidine and P3 phenylalanine represent important TCR contact residues (Wucherpfennig et al., 1994a; Hausmann et al., 1999), and the structure shows that the CDR3 loops are centered over P2 histidine (Figure 3C) and make a number of interactions with both P2 histidine and P3 phenylalanine (Figure 4). The structure also explains why any substitution in the C-terminal segment of the peptide that is not contacted by the TCR in this structure had no effect on TCR recognition as long as it did not reduce MHC binding (Wucherpfennig et al., 1994a; Hausmann et al., 1999). Furthermore, it accounts for the observation that an N-terminal extension of the peptide up to the P −4 residue was required for optimal T cell stimulation (Wucherpfennig et al., 1994a). In the structure, the backbone of the P −4 residue is contacted by the TCRβ CDR2 loop, which is located in a very unusual position in this complex (Figure 5C, right panel).
These results raised the question of whether this TCR represents an isolated case (Wilson and Stanfield, 2005). The T cell response to MBP in this MS patient was largely focused on this epitope because 60 of 75 T cell lines reactive with MBP responded to the MBP (84–102) peptide (Wucherpfennig et al., 1994b). A second independent T cell clone (Ob.2F3) from this patient had the same Vα-Jα and Vβ-Jβ rearrangements, and the TCR protein sequences differed only at one position in CDR3α and two positions in CDR3β to Ob.1A12 TCR (Wucherpfennig et al., 1994b). This T cell clone had a remarkably similar fine specificity to Ob.1A12 when assayed on a large panel of peptide analogs. The only difference among these clones was observed with analogs of P5 lysine (Hausmann et al., 1999), a peptide side chain that is contacted in the Ob.1A12 structure by a CDR3β residue that differs between the two TCRs (Figure 3C). The Ob.1A12 and Ob.2F3 TCRs therefore make very similar interactions with the MBP peptide, strongly suggesting that the overall topology is almost identical.
The Structure of a Second Human Autoreactive TCR Also Shows an Unconventional Topology
The recent structure of a second TCR from a different MS patient (3A6 TCR) provides another example of an unusual interaction of an autoimmune TCR with a self-peptide/MHC complex (Figure 2B) (Li et al., 2005). This TCR also recognizes MBP (residues 89–101), but the peptide is shifted by three residues in the HLA-DR binding site such that a different set of peptide residues is available for TCR recognition (Figure 4). As a consequence, peptide residues VxHFxK are solvent exposed in the P −1 to P5 segment in the Ob.1A12 complex, compared to FxKNxV in the 3A6 complex. In addition, two different MHC class II molecules are involved: DRB1*1501 (Ob.1A12) and DRB5*0101 (3A6), both of which are associated with susceptibility to MS (Hillert, 1994). Previous studies had shown that this MBP peptide binds in different registers to these MHC molecules (Vogt et al., 1994; Wucherpfennig et al., 1994a). Thus, both the MHC and the peptide epitope differ between these trimolecular complexes.
Compared to TCRs with conventional topology such as HA1.7 (Figure 2A), the position of the 3A6 TCR (Figure 2B) is shifted toward the N terminus of the peptide and the DRβ chain helix (the lower helix in all MHC structures in Figure 2). This shift is thus a common feature between 3A6 and Ob.1A12 TCRs (Figure 2B and 2C, respectively), and it substantially alters recognition of both peptide and MHC. Whereas the CDR3 loops of HA1.7 TCR meet over the central peptide residue P5 (Figure 3A, CDR3α colored yellow and CDR3β colored red), the CDR3 loops of both 3A6 and Ob.1A12 TCRs create a pocket that accommodates the P2 side chain of the respective peptide (Figure 3B and 3C, respectively). For both 3A6 and Ob.1A12 TCRs, the P2 peptide side chain is critical for recognition because substitution of P2 lysine (3A6 TCR) or P2 histidine (Ob.1A12 TCR) by any other residue abrogates T cell activation (Hausmann et al., 1999; Hemmer et al., 2000). The shift toward the peptide N terminus is also evident on a map of the peptide residues contacted by the CDR3α and CDR3β loops (Figure 4, peptide residues contacted by CDR3 loops colored orange). Whereas the CDR3 loops of HA1.7 TCR establish contacts with the peptide over the central peptide segment spanning from P −1 to P8, the footprint of the CDR3 loops of Ob.1A12 and 3A6 TCRs is located over the N-terminal peptide segment, spanning from P −2 to P5 for Ob.1A12 TCR and P −3 to P3 for 3A6 TCR. The binding angle of 3A6 TCR is 47° and thus falls within the range observed for the majority of TCRs (45°–80°) (Rudolph and Wilson, 2002). The two human autoimmune TCRs thus differ substantially in the crossing angle (47° for 3A6 TCR and 110° for Ob.1A12 TCR) even though they share a shift toward the N terminus of the peptide. Both human TCRs that recognize self-peptide/MHC complexes therefore have unusual binding properties that distinguish them from TCRs specific for peptides from infectious agents.
Substantial Differences in Both Peptide and MHC Binding by Three Human MHC Class II Restricted TCRs
Comparison of the three human MHC class II restricted TCRs (HA1.7, 3A6 and Ob.1A12) demonstrates large differences in the location of several TCR loops (Figure 5). In these three structures, the CDR3 loops of TCRα (colored yellow) and TCRβ (colored red) assume a different conformation and position: They are centered over the P5 peptide position in the HA1.7 structure but located over the P2 peptide residue in the 3A6 and Ob.1A12 structures (Figure 5, left panels). Also, the CDR3 loops of Ob.1A12 TCR make a larger number of contacts to the MHC helices than the CDR1 and CDR2 loops. Particularly unusual is the interaction between the CDR3α loop of Ob.1A12 TCR and a histidine residue of the MHC class II molecule (DRβ81 histidine, Figure 5C, left panel); this histidine residue is located in the central cavity between the two CDR3 loops in the Ob.1A12 structure.
Large differences are also observed in the position of the germline-encoded CDR1 and CDR2 loops of the TCRβ chain in these three structures (Figure 5, right panels). These loops are located in an entirely different position in the Ob.1A12 structure compared to the HA1.7 structure, and the CDR2β loop of Ob.1A12 TCR establishes a highly unusual interaction with the N-terminal residue of the MBP peptide (Figure 5C, right panel). The unusual location of these TCRβ chain loops is in part caused by a 40° counter-clockwise rotation of the variable domains of Ob.1A12 TCR relative to HA1.7 TCR. Significant differences in the location of the CDR1 and CDR2 loops of TCRβ are also observed between the HA1.7 and 3A6 structures (Figures 5A and 5B, right panels). In the 3A6 structure, the CDR2β loop is positioned directly over the DRα helix, and the CDR1β loop is located closer to the peptide (Figures 5A and 5B, right panels) because of the shift of the TCR toward the peptide N terminus and the DRβ chain helix (the lower MHC helix in all pictures in Figure 5).
Nevertheless, the CDR1 and CDR2 loops of TCRα are in a similar overall location in all three structures (Figure 5, right panels). In the 3A6 structure, the CDR2α loop does not contact the MHC molecule (Figure 5B, right panel), and only a limited number of MHC contact residues are shared between the CDR1α loops of these TCRs (contacts to DRβ 77 by all three TCRs, contacts to DRβ 81 by 3A6 and HA1.7 TCRs, and contacts to DRβ 76 by 3A6 and Ob.1A12 TCRs). None of the contacts made by the CDR1α loops of these TCRs to the MHC molecule represent hydrogen bonds. The similarity in the location of the CDR1 and CDR2 loops of TCRα is the only feature that the Ob.1A12 TCR has in common with TCRs that bind with a conventional topology to peptide/MHC.
Structural Characteristics of a TCR that Causes EAE
The structure of the TCR/peptide/MHC complex has also been recently determined for the TCR from a T cell clone that causes EAE (172.10 TCR) (Maynard et al., 2005). This clone recognizes the acetylated N-terminal peptide of MBP (Ac1-11) bound to the mouse MHC class II molecule I-Au (Zamvil et al., 1986; Goverman et al., 1993). As discussed in the introduction, this MBP peptide binds with low affinity to I-Au and is immunodominant in wild-type, but not MBP-deficient, mice (Fairchild et al., 1993; Harrington et al., 1998). TCR binding to this peptide is unusual because the peptide only partially fills the MHC class II binding site, as shown in Figure 4 where the peptides are aligned on the basis of their MHC anchor residues (He et al., 2002). 172.10 TCR only establishes a limited number of contacts to the MBP-peptide segment that occupies part of the binding site, and only two TCR loops are involved in contacts to the MBP peptide (CDR3α and CDR3β) (Figure 4), compared to four TCR loops in the HA1.7 structure (CDR1 and CDR3 loops of both chains) (Figure 4; Figure 5A, left and right panels). As a consequence, 172.10 TCR recognizes a substantially shorter span of the peptide (six MBP peptide residues from P3 to P8, Figure 4) compared to HA1.7 and D10 TCRs (nine HA peptide residues, from P −1 to P8). These results, combined with the low affinity of Ac1-11 to I-Au (Fairchild et al., 1993), provide an explanation for the finding that another region of MBP (the 121–150 segment) is immunodominant in MBP-deficient mice and that T cell reactivity to Ac1-11 only becomes prominent when T cells that recognize the 121–150 region are deleted in the thymus (Harrington et al., 1998). The interaction with the MHC bound peptide is thus highly unusual in the 172.10 structure, even though this TCR binds in a conventional diagonal orientation to the Ac1-11/I-Au complex. In the Ob.1A12 TCR structure, the interaction surface with the peptide is also reduced by the altered position of the TCR on the peptide/MHC surface, and again only two TCR loops contact peptide side chains. All three autoimmune TCRs for which the structure has been determined thus differ substantially in the interaction with peptide/MHC compared to TCRs specific for foreign antigens.
Distinct Selection Pressures Exerted on Self-Reactive and Antimicrobial T Cells
Foreign antigens are normally not present during T cell development in the thymus, and as a result, T cells that express TCRs with an optimal fit for a given microbial peptide/MHC complex are not deleted from the repertoire. During an infection, there is intense competition among T cells that recognize peptides derived from the infectious agent, leading to the outgrowth of those clones that express TCRs with the highest affinity for the most abundant peptide/MHC complexes (Busch and Pamer, 1999; Kedl et al., 2000). When the TCR repertoire against a peptide from an infectious agent is analyzed, this clonal competition is reflected by the predominance of clones with particular TCR rearrangements. The HA1.7 TCR is a representative of such a predominant T cell population because its Vβ segment is highly over-represented among HA 306–318-specific T cells. The Vβ segments commonly used by HA 306–318-specific T cells share acidic residues in the CDR1 loop that form a salt bridge to the P8 lysine of the HA peptide in the HA1.7 structure (Wedderburn et al., 1995; Hennecke et al., 2000). The central diagonal position of the HA1.7 and D10 TCRs that recognize foreign peptides bound to MHC class II molecules appears to represent the optimal binding mode that maximizes the interaction surface with the bound peptide.
Autoreactive T cells face different selection pressures. Comparison of the T cell repertoire between mice that lack expression of a particular autoantigen and wild-type mice has shown that a substantial number of T cells are deleted in the thymus in the presence of the autoantigen and that T cells with certain peptide specificities are more severely affected (Harrington et al., 1998; Klein et al., 2000). The structures of the three autoimmune TCRs that have been determined reveal in each case unusual TCR binding properties that appear to result in suboptimal recognition of the MHC bound self-peptide. For the two human TCRs, it results from an altered TCR position over the peptide/MHC surface, whereas for the murine TCR, it is the consequence of partial filling of the peptide binding site by the N-terminal MBP Ac1-11 peptide. It is likely that T cells with an optimal conventional binding mode to a self-peptide/MHC complex are present in animals rendered deficient in expression of the self-antigen, but when the autoantigen is present, such T cells may be more susceptible to negative selection than T cells with altered binding properties.
Low-Affinity TCR Binding and Crossreactivity
Binding studies have demonstrated low-affinity interactions of both Ob.1A12 and 3A6 TCRs with their peptide/MHC ligands. In Biacore experiments, binding could only be detected when either the TCR or the peptide/MHC was made multivalent (Appel et al., 2000; Li et al., 2005). The affinity of the two human MBP-reactive TCRs thus appears to be substantially lower than for other crystallized TCRs for which such measurements have been made (Li et al., 2005). The affinity of TCR interaction with peptide/MHC is critical in setting the thresholds for positive and negative selection in the thymus; low-affinity ligands have been shown to promote positive selection, whereas higher-affinity ligands induce negative selection (Alam et al., 1996). The HA1.7 TCR binds with higher avidity to its peptide/MHC ligand than Ob.1A12 TCR because the HA1.7 T cell clone is brightly labeled by dimeric or multimeric peptide/MHC (Cochran et al., 2000) whereas the Ob.1A12 T cell clone is only weakly stained (Appel et al., 2000). However, the affinity of HA1.7 binding to DR1/HA peptide has not been measured by Biacore. The 172.10 TCR has been reported to have an affinity within the normal range of interactions between TCR and peptide/MHC, but it has to be kept in mind that these experiments used a recombinant peptide/MHC ligand in which the low-affinity interaction of the MBP peptide with the MHC molecule was stabilized by covalent attachment to the MHC β chain (Garcia et al., 2001; Maynard et al., 2005).
For both human TCRs, peptide recognition appears to be suboptimal: In the case of Ob.1A12 TCR, only two TCR loops contact peptide side chains, whereas four HA1.7 TCR loops are involved in direct recognition of the viral peptide. In the case of 3A6 TCR, no hydrogen bonds or salt bridges are observed between TCR loops and the MHC bound peptide. The shift toward the peptide N terminus may constitute an important component of suboptimal peptide recognition for both TCRs. Overall, the autoimmune TCRs establish a limited number of contacts to the peptide, and only a small number of these contacts represent hydrogen bonds (Figure 4). Particularly striking is the observation that only two TCR loops contact the MHC bound peptide in both the murine 172.10 and the human Ob.1A12 structures.
It is possible that autoreactive T cells that express such TCRs are inherently more peptide crossreactive because microbial peptides that create a better interface with these TCRs can represent ligands that are more potent. Such crossreactivity may be relevant for the activation of autoreactive T cells by microbial peptides during infections (Fujinami and Oldstone, 1985; Wucherpfennig and Strominger, 1995; Hemmer et al., 1997). Analogs of the relevant self-peptides have been identified that significantly improve T cell stimulation, with the most striking example being a substitution at P −1 from phenylalanine to tryptophan that augmented the stimulatory capacity of the peptide for the 3A6 T cell clone by several orders of magnitude, presumably by increasing the affinity of TCR interaction with peptide/MHC (Hemmer et al., 2000). The 172.10 TCR appears to be less crossreactive than the two human MBP-reactive TCRs because sequence identity with the self-peptide is required at three peptide positions (Maynard et al., 2005), but a substantial number of microbial peptides that are recognized by this TCR have nevertheless been identified (Grogan et al., 1999).
Contribution of TCR/MHC Coevolution and Thymic Selection to TCR Topology
It has been suggested that the conventional topology represents the product of coevolution of TCR and MHC genes (Garcia and Adams, 2005). The new structures demonstrate that alternative binding modes are possible, indicating that coevolution may not be the only explanation for the predominant conventional topology. It is possible that unconventional binding topologies are fairly common in the TCR repertoire before positive and negative selection have occurred and that one of the functions of thymic selection is to eliminate T cells whose TCRs bind to peptide/MHC with a geometry outside of the functionally useful range.
A common feature of TCR binding to both MHC class I and class II molecules is the general location of the CDR1 and CDR2 loops of TCRα (Figure 5, right panels), but it is not known whether essential contacts are made to the corresponding MHC helix or whether a particular geometry is required, for example for coreceptor function. A substantial effort has been made to identify MHC residues essential for TCR binding (Baker et al., 2001), but these experiments have failed to identify universal MHC contact residues. The new structures demonstrate that most of the TCR loops can bind to peptide/MHC in alternative locations. Nevertheless, certain MHC residues are contacted by TCRs in several structures and may thus be significant for a substantial subset of receptors. For example, three of the five MHC class II restricted TCRs for which the structure is known (HA1.7, D10, 172.10) form a salt bridge between glutamic acid 56 in the CDR2 loop of TCRβ and lysine 39 on the MHC class II α chain (DRα39 in the HA1.7 complex, Figure 5A, right panel). This contact is not made by the Ob.1A12 and 3A6 TCRs because of the altered topology of TCR binding to peptide/MHC.
The structures of the two human autoimmune TCRs have shown that not all αβ TCRs bind with the same overall topology to peptide/MHC complexes. At present, it is not known whether the Ob.1A12 TCR represents the most extreme case of an alternative topology and what range of alternative topologies are compatible with TCR function. It is even possible that a single TCR can bind with different topologies to distinct peptide/MHC complexes and that alternative topologies are thus extreme cases of TCR crossreactivity.
An interesting question is whether a shift, observed for both Ob.1A12 and 3A6 TCRs, toward the peptide N terminus and the MHC class II β chain helix is a common alternative topology, a finding that would suggest that this arrangement alters the assembly of higher-order signaling complexes in a way that is favorable for autoreactive T cells, for example during thymic selection. Alterations in the biochemical interaction between TCR and peptide/MHC may modify TCR signaling thresholds during thymic selection, analogous to the changes in TCR signaling thresholds described in the SKG model of spontaneous arthritis, in which a point mutation in ZAP-70 leads to positive selection of autoreactive T cells that are otherwise negatively selected (Sakaguchi et al., 2003). Additional structures, in particular of TCRs from other autoimmune diseases, are needed to determine what fraction of autoreactive TCRs have unusual binding properties. Such an effort will require a global analysis of TCR binding properties in terms of affinity, overall topology, and interaction surface with peptide. The three available structures show that each autoimmune TCR has individual features that distinguish it from antimicrobial TCRs. Escape from negative selection may for individual TCRs be due to a distinct combination of structural and functional alterations, such as reduced affinity, suboptimal interface with peptide, and altered topology.
Acknowledgments
This work was supported by grants from the National Institutes of Health to K.W.W. (RO1 AI064177 and PO1 AI45757).
References
- Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. T-cell-receptor affinity and thymocyte positive selection. Nature. 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- Anderson AC, Nicholson LB, Legge KL, Turchin V, Zaghouani H, Kuchroo VK. High frequency of autoreactive myelin proteolipid protein-specific T cells in the periphery of naive mice: Mechanisms of selection of the self-reactive repertoire. J Exp Med. 2000;191:761–770. doi: 10.1084/jem.191.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Appel H, Gauthier L, Pyrdol J, Wucherpfennig KW. Kinetics of T-cell receptor binding by bivalent HLA-DR. Peptide complexes that activate antigen-specific human T-cells. J Biol Chem. 2000;275:312–321. doi: 10.1074/jbc.275.1.312. [DOI] [PubMed] [Google Scholar]
- Baker BM, Turner RV, Gagnon SJ, Wiley DC, Biddison WE. Identification of a crucial energetic footprint on the alpha1 helix of human histocompatibility leukocyte antigen (HLA)-A2 that provides functional interactions for recognition by tax peptide/HLA-A2-specific T cell receptors. J Exp Med. 2001;193:551–562. doi: 10.1084/jem.193.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG, Strominger JL, Wiley DC. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Busch DH, Pamer EG. T cell affinity maturation by selective expansion during infection. J Exp Med. 1999;189:701–710. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran JR, Cameron TO, Stern LJ. The relationship of MHC-peptide binding and T cell activation probed using chemically defined MHC class II oligomers. Immunity. 2000;12:241–250. doi: 10.1016/s1074-7613(00)80177-6. [DOI] [PubMed] [Google Scholar]
- Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- Ding YH, Smith KJ, Garboczi DN, Utz U, Biddison WE, Wiley DC. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 1998;8:403–411. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- Ellmerich S, Takacs K, Mycko M, Waldner H, Wahid F, Boyton RJ, Smith PA, Amor S, Baker D, Hafler DA, et al. Disease- related epitope spread in a humanized T cell receptor transgenic model of multiple sclerosis. Eur J Immunol. 2004;34:1839–1848. doi: 10.1002/eji.200324044. [DOI] [PubMed] [Google Scholar]
- Fairchild PJ, Wildgoose R, Atherton E, Webb S, Wraith DC. An autoantigenic T cell epitope forms unstable complexes with class II MHC: A novel route for escape from tolerance induction. Int Immunol. 1993;5:1151–1158. doi: 10.1093/intimm/5.9.1151. [DOI] [PubMed] [Google Scholar]
- The Finnish-German APECED Consortium. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- Fujinami RS, Oldstone MB. Amino acid homology between the encephalitogenic site of myelin basic protein and virus: Mechanism for autoimmunity. Science. 1985;230:1043–1045. doi: 10.1126/science.2414848. [DOI] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Adams EJ. How the T cell receptor sees antigen—a structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An αβ T cell receptor structure at 2.5 Å and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Radu CG, Ho J, Ober RJ, Ward ES. Kinetics and thermodynamics of T cell receptor- autoantigen interactions in murine experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2001;98:6818–6823. doi: 10.1073/pnas.111161198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow CC, Sprent J, de St Groth BF, Vinuesa CG. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature. 2005;435:590–597. doi: 10.1038/nature03724. [DOI] [PubMed] [Google Scholar]
- Goverman J, Woods A, Larson L, Weiner LP, Hood L, Zaller DM. Transgenic mice that express a myelin basic protein-specific T cell receptor develop spontaneous autoimmunity. Cell. 1993;72:551–560. doi: 10.1016/0092-8674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- Grogan JL, Kramer A, Nogai A, Dong L, Ohde M, Schneider-Mergener J, Kamradt T. Cross-reactivity of myelin basic protein-specific T cells with multiple microbial peptides: Experimental autoimmune encephalomyelitis induction in TCR transgenic mice. J Immunol. 1999;163:3764–3770. [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss- PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington CJ, Paez A, Hunkapiller T, Mannikko V, Brabb T, Ahearn M, Beeson C, Goverman J. Differential tolerance is induced in T cells recognizing distinct epitopes of myelin basic protein. Immunity. 1998;8:571–580. doi: 10.1016/s1074-7613(00)80562-2. [DOI] [PubMed] [Google Scholar]
- Hausmann S, Martin M, Gauthier L, Wucherpfennig KW. Structural features of autoreactive TCR that determine the degree of degeneracy in peptide recognition. J Immunol. 1999;162:338–344. [PubMed] [Google Scholar]
- He XL, Radu C, Sidney J, Sette A, Ward ES, Garcia KC. Structural snapshot of aberrant antigen presentation linked to autoimmunity: The immunodominant epitope of MBP complexed with I-Au. Immunity. 2002;17:83–94. doi: 10.1016/s1074-7613(02)00340-0. [DOI] [PubMed] [Google Scholar]
- Hemmer B, Fleckenstein BT, Vergelli M, Jung G, McFarland H, Martin R, Wiesmuller KH. Identification of high potency microbial and self ligands for a human autoreactive class II-restricted T cell clone. J Exp Med. 1997;185:1651–1659. doi: 10.1084/jem.185.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmer B, Pinilla C, Gran B, Vergelli M, Ling N, Conlon P, McFarland HF, Houghten R, Martin R. Contribution of individual amino acids within MHC molecule or antigenic peptide to TCR ligand potency. J Immunol. 2000;164:861–871. doi: 10.4049/jimmunol.164.2.861. [DOI] [PubMed] [Google Scholar]
- Hennecke J, Wiley DC. T cell receptor-MHC interactions up close. Cell. 2001;104:1–4. doi: 10.1016/s0092-8674(01)00185-4. [DOI] [PubMed] [Google Scholar]
- Hennecke J, Carfi A, Wiley DC. Structure of a covalently stabilized complex of a human αβ T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J. 2000;19:5611–5624. doi: 10.1093/emboj/19.21.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillert J. Human leukocyte antigen studies in multiple sclerosis. Ann Neurol. 1994;36:S15–S17. doi: 10.1002/ana.410360706. [DOI] [PubMed] [Google Scholar]
- Kedl RM, Kappler JW, Marrack P. Epitope dominance, competition and T cell affinity maturation. Curr Opin Immunol. 2003;15:120–127. doi: 10.1016/s0952-7915(02)00009-2. [DOI] [PubMed] [Google Scholar]
- Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, Marrack P. T cells compete for access to antigen-bearing antigen-presenting cells. J Exp Med. 2000;192:1105– 1113. doi: 10.1084/jem.192.8.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L, Klugmann M, Nave KA, Tuohy VK, Kyewski B. Shaping of the autoreactive T-cell repertoire by a splice variant of self protein expressed in thymic epithelial cells. Nat Med. 2000;6:56–61. doi: 10.1038/71540. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Lue J, Quandt JA, Martin R, Mariuzza RA. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lesage S, Gray DH, O’Reilly LA, Strasser A, Fahrer AM, Boyd RL, Wilson J, Baxter AG, Gallo EM, et al. Generalized resistance to thymic deletion in the NOD mouse: A polygenic trait characterized by defective induction of Bim. Immunity. 2004;21:817–830. doi: 10.1016/j.immuni.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Madsen LS, Andersson EC, Jansson L, Krogsgaard M, Andersen CB, Engberg J, Strominger JL, Svejgaard A, Hjorth JP, Holmdahl R, et al. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat Genet. 1999;23:343–347. doi: 10.1038/15525. [DOI] [PubMed] [Google Scholar]
- Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC. Structure of an autoimmune T cell receptor complexed with class II peptide- MHC: Insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Murphy ME. Raster-3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D Biol Crystallogr. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Ota K, Matsui M, Milford EL, Mackin GA, Weiner HL, Hafler DA. T-cell recognition of an immunodominant myelin basic protein epitope in multiple sclerosis. Nature. 1990;346:183–187. doi: 10.1038/346183a0. [DOI] [PubMed] [Google Scholar]
- Pribyl TM, Campagnoni C, Kampf K, Handley VW, Campagnoni AT. The major myelin protein genes are expressed in the human thymus. J Neurosci Res. 1996;45:812–819. doi: 10.1002/(SICI)1097-4547(19960915)45:6<812::AID-JNR18>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, Hussey RE, Smolyar A, Hare B, Zhang R, et al. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- Reiser JB, Darnault C, Gregoire C, Mosser T, Mazza G, Kearney A, van der Merwe PA, Fontecilla-Camps JC, Housset D, Malissen B. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat Immunol. 2003;4:241–247. doi: 10.1038/ni891. [DOI] [PubMed] [Google Scholar]
- Reiser JB, Darnault C, Guimezanes A, Gregoire C, Mosser T, Schmitt-Verhulst AM, Fontecilla-Camps JC, Malissen B, Housset D, Mazza G. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat Immunol. 2000;1:291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- Reiser JB, Gregoire C, Darnault C, Mosser T, Guimezanes A, Schmitt-Verhulst AM, Fontecilla-Camps JC, Mazza G, Malissen B, Housset D. A T cell receptor CDR3β loop undergoes conformational changes of unprecedented magnitude upon binding to a peptide/MHC class I complex. Immunity. 2002;16:345–354. doi: 10.1016/s1074-7613(02)00288-1. [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Wilson IA. The specificity of TCR/pMHC interaction. Curr Opin Immunol. 2002;14:52–65. doi: 10.1016/s0952-7915(01)00298-9. [DOI] [PubMed] [Google Scholar]
- Sakaguchi N, Takahashi T, Hata H, Nomura T, Tagami T, Yamazaki S, Sakihama T, Matsutani T, Negishi I, Nakatsuru S, Sakaguchi S. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature. 2003;426:454–460. doi: 10.1038/nature02119. [DOI] [PubMed] [Google Scholar]
- Sim BC, Zerva L, Greene MI, Gascoigne NR. Control of MHC restriction by TCR Vα CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol. 2003;4:657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- Sun R, Shepherd SE, Geier SS, Thomson CT, Sheil JM, Nathenson SG. Evidence that the antigen receptors of cytotoxic T lymphocytes interact with a common recognition pattern on the H-2Kb molecule. Immunity. 1995;3:573–582. doi: 10.1016/1074-7613(95)90128-0. [DOI] [PubMed] [Google Scholar]
- Valli A, Sette A, Kappos L, Oseroff C, Sidney J, Miescher G, Hochberger M, Albert ED, Adorini L. Binding of myelin basic protein peptides to human histocompatibility leukocyte antigen class II molecules and their recognition by T cells from multiple sclerosis patients. J Clin Invest. 1993;91:616–628. doi: 10.1172/JCI116242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt AB, Kropshofer H, Kalbacher H, Kalbus M, Rammensee HG, Coligan JE, Martin R. Ligand motifs of HLA-DRB5* 0101 and DRB1*1501 molecules delineated from self-peptides. J Immunol. 1994;153:1665–1673. [PubMed] [Google Scholar]
- Wall M, Southwood S, Sidney J, Oseroff C, del Guericio MF, Lamont AG, Colon SM, Arrhenius T, Gaeta FC, Sette A. High affinity for class II molecules as a necessary but not sufficient characteristic of encephalitogenic determinants. Int Immunol. 1992;4:773–777. doi: 10.1093/intimm/4.7.773. [DOI] [PubMed] [Google Scholar]
- Wedderburn LR, Searle SJ, Rees AR, Lamb JR, Owen MJ. Mapping T cell recognition: The identification of a T cell receptor residue critical to the specific interaction with an influenza hemagglutinin peptide. Eur J Immunol. 1995;25:1654–1662. doi: 10.1002/eji.1830250627. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Stanfield RL. MHC restriction: Slip-sliding away. Nat Immunol. 2005;6:434–435. doi: 10.1038/ni0505-434. [DOI] [PubMed] [Google Scholar]
- Wucherpfennig KW, Strominger JL. Molecular mimicry in T cell-mediated autoimmunity: Viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig KW, Sette A, Southwood S, Oseroff C, Matsui M, Strominger JL, Hafler DA. Structural requirements for binding of an immunodominant myelin basic protein peptide to DR2 isotypes and for its recognition by human T cell clones. J Exp Med. 1994a;179:279–290. doi: 10.1084/jem.179.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig KW, Zhang J, Witek C, Matsui M, Modabber Y, Ota K, Hafler DA. Clonal expansion and persistence of human T cells specific for an immunodominant myelin basic protein peptide. J Immunol. 1994b;152:5581–5592. [PubMed] [Google Scholar]
- Zamvil SS, Mitchell DJ, Moore AC, Kitamura K, Steinman L, Rothbard JB. T-cell epitope of the autoantigen myelin basic protein that induces encephalomyelitis. Nature. 1986;324:258–260. doi: 10.1038/324258a0. [DOI] [PubMed] [Google Scholar]