Abstract
BACKGROUND AND AIMS
The intestinal microbiomes of healthy children and pediatric patients with irritable bowel syndrome (IBS) are not well defined. Studies in adults have indicated that the gastrointestinal microbiota could be involved in IBS.
METHODS
We analyzed 71 samples from 22 children with IBS (pediatric Rome III criteria) and 22 healthy children, ages 7–12 years, by 16S rRNA gene sequencing, with an average of 54,287 reads/stool sample (average 454 read length = 503 bases). Data were analyzed using phylogenetic-based clustering (Unifrac), or an operational taxonomic unit (OTU) approach using a supervised machine learning tool (randomForest). Most samples were also hybridized to a microarray that can detect 8,741 bacterial taxa (16S rRNA PhyloChip).
RESULTS
Microbiomes associated with pediatric IBS were characterized by a significantly greater percentage of the class Gammaproteobacteria (0.07% vs 0.89% of total bacteria; P <.05); one prominent component of this group was Haemophilus parainfluenzae. Differences highlighted by 454 sequencing were confirmed by high-resolution PhyloChip analysis. Using supervised learning techniques, we were able to classify different subtypes of IBS with a success rate of 98.5%, using limited sets of discriminant bacterial species. A novel Ruminococcus-like microbe was associated with IBS, indicating the potential utility of microbe discovery for gastrointestinal disorders. A greater frequency of pain correlated with an increased abundance of several bacterial taxa from the genus Alistipes.
CONCLUSIONS
Using16S metagenomics by Phylochip DNA hybridization and deep 454 pyrosequencing, we associated specific microbiome signatures with pediatric IBS. These findings indicate the important association between gastrointestinal microbes and IBS in children; these approaches might be used in diagnosis of functional bowel disorders in pediatric patients.
INTRODUCTION
Abdominal pain is a common complaint in childhood accounting for a large percentage (approximately 30%) of health care visits for children ages 4 to 16 years1,2. The defining criteria for recurrent abdominal pain (RAP) are based on the pioneering work of Apley and Naish in England1. Abdominal pain accounts for at least 5% of all pediatric office visits and, approximately 25% of office visits to a pediatric gastroenterologist3. A minority of children have an identifiable organic reason for their pain4. Community-based studies from different geographic regions demonstrate that 10% – 46% of children (4–16 years of age) meet the criteria for RAP5,6.
As reflected in the Pediatric Rome III Criteria, RAP can be considered an umbrella term that includes several subtypes of abdominal pain described in children and adults [e.g., functional abdominal pain (FAP), irritable bowel syndrome, (IBS)]7,8. The Pediatric Rome III criteria for IBS include episodic or continuous abdominal pain that occurs ≥ 1 time/week for at least two months associated with 2 or more of the following at least 25% of the time: pain improved with defecation, onset associated with a change in frequency of stool, and/or onset associated with a change in form (appearance) of stool, and no evidence of an inflammatory, anatomic, metabolic, or neoplastic process that explains the subject's symptoms7. IBS comprises a large proportion of RAP9. Follow-up studies indicate that 30%–66% of children with RAP will experience similar pain as adults and meet the adult Rome criteria for IBS3,10. IBS in children and adults may be the same syndrome at different developmental stages11,12.
Components or deficiencies of the pediatric intestinal microbiome may contribute to symptoms and point to new therapies that ameliorate GI symptoms in childhood and adulthood. Studies suggest that fundamental differences in the composition of the human microbiome may be associated with the IBS disease phenotype in adults13. In the context of IBS with diarrhea, previous studies yielded evidence of diminished quantities of Lactobacillus spp. in adult patients, whereas patients with the phenotype of IBS with constipation had increased proportions of Veillonella spp.14. Studies with probiotics suggest that manipulation of intestinal microbial communities may modulate visceral hypersensitivity15 and alter the disease course in IBS in both adults8,16–18 and children19. However, more information is needed about how the human microbiome contributes to the constellation of symptoms in IBS, particularly in children.
Although common practice in adults with IBS, no prospective studies in the pediatric population have been undertaken in a parallel effort to identify subgroups of children with IBS (i.e., IBS-D, IBS-C, IBS-M, or IBS-U). Using next generation sequencing technology and a bacterial DNA microarray coupled with feature selection and supervised learning algorithms, we identified specific microbial signatures in healthy children and children with different IBS subtypes. With the aid of random forests20 as a supervised learning strategy, the relative abundance of specific bacterial taxa correlated with the phenotype of increased frequency of recurrent abdominal pain.
MATERIALS AND METHODS
Pediatric Subject Evaluation and Enrollment
School-age children (7–12 years of age) participated in the study and were recruited from a large healthcare network in the Houston metropolitan area. All recruitment and study procedures were approved by the Baylor College of Medicine Institutional Review Board, and informed consent was obtained from the parents and assent from the children. Children with IBS were identified from physician practices by screening medical charts for inclusion and exclusion criteria. Parents were further screened by phone to establish frequency, duration, and intensity of the child's complaints and ensure that symptoms were current prior to enrollment. Participants met Pediatric Rome III criteria for IBS7 (Table 1). Subtyping of IBS was based on previous recommendations for IBS in adults because no Pediatric Rome subtype criteria exist for children8.
TABLE 1.
Clinical features of the children enrolled in this study.
| Characteristics | Healthy | IBS | |||
|---|---|---|---|---|---|
| IBS-C• | IBS-D• | IBS-U• | Other IBS subjects* | ||
| Number of subjects | 22 | 13 | 1 | 7 | 1 |
| Age (year ± SD) | 9.32 ± 1.52 | 9.38 ± 1.19 | 10 | 9.26 ± 1.89 | 9 |
| Female/male | 11/11 | 5/8 | 0/1 | 3/4 | 0/1 |
| Recollection (times) | 6 (3×) | 1 (2×), 4 (3×) | 1 (2×) | 2 (3×) | 1 (2×) |
| Mean pain rating a | 0.01 ± 0.04 | 0.70 ± 0.57 | 0.21 | 0.30 ± 0.36 | NA |
| Maximum pain rating a | 0.51 ± 1.19 | 5.69 ± 2.53 | 4.00 | 4.43 ± 3.26 | NA |
IBS-C: IBS with constipation; IBS-D: IBS with diarrhea; IBS-U: unsubtyped IBS.
Not classifiable by IBS subtype (absence of diary)
Initial collection only (diary A complete). For IBS-U, data for mean and maximum pain values were obtained from 6 subjects only
NA: data not available
Specifically, children kept a 2-week pain and stool diary as we have described previously9, 21. Abdominal pain ratings were made 3 times per day (awakening, after lunch, and evening) during the 2-week period, and pain ratings were recorded in a database linked to a dedicated telephone line. The child rated the pain using a 0 to 10 scale with 0 being “no pain at all” and 10 representing the “worst pain you can imagine”. This pain scale has been validated for measuring abdominal pain in children22. The maximum level of pain was defined as the greatest intensity of pain recorded during the 2-week period. Pain frequency was recorded during the same 2-week period and stratified into two groups to provide statistical power. The high-medium (HM) group reported > 3 pain episodes and the low (LO) group reported ≤ 3 pain episodes in the 2-week period. Stools were compared to the Bristol stool form chart by the children and their parents23. Stool type 1 or 2 (hard) was considered to be consistent with constipation, and stool type 6 or 7 (loose) was considered to be consistent with diarrhea8. Patients were subtyped as having IBS-C (hard ≥25% and loose <25% of the time), IBS-D (hard <25% and loose ≥25%), IBS-M (hard or loose ≥25%), or IBS-U (does not meet criteria for IBS-C, IBS-D, or IBS-M8. IBS-U refers to unsubtyped IBS or recurrent abdominal pain associated with specific changes in bowel habits, but lacking sufficient constipation or diarrhea (hard or loose stool <25% of the time).
A total of 48 children were enrolled in the study. Stool samples (n=71) were obtained from 22 children with IBS and from 22 healthy controls. Exclusion criteria included use of antibiotics, probiotics, or steroids (oral or nasal (inhaled)) within 6 months of sampling. Four children withdrew from the study (2 children declined after the home visit; one child consumed an antibiotic; one child took a probiotic). Detailed inclusion and exclusion criteria, as well as metadata for these subjects, are available on dbGaP (http://www.ncbi.nlm.nih.gov/gap), under the study accession phs000265.v2.p1.
Stool samples were collected in a stool “hat” placed in the toilet, immediately transferred into a sterile cup, and stored frozen at −80°C. At the time the children collected the stool sample, they kept a two-week diary of any episodes of abdominal pain as we described previously9. In the diary, the children recorded the time of defecation and rated the stool consistency against a validated stool form scale21, 23. In a subset of children, stools were re-collected 2–3 times, and each serial collection occurred after a 2-month interval.
Fecal Microbial DNA Extraction
DNA extraction, bacterial 16S rRNA gene amplification and 454 16S metagenomic (microbial DNA) sequencing were performed as previously described for the JumpStart HMP study (http://www.hmpdacc.org/doc/HMP_MDG_454_16S_Protocol_V4_2_102109.pdf). In brief, DNA was extracted from stool samples using a commercial manual DNA extraction kit (MO-BIO PowerSoil® DNA Isolation Kit, MO-BIO Laboratories, Carlsbad, CA, USA), with modifications.
16S Metagenomic 454 Sequencing Data Generation
The V1–V3 and V3–V5 regions of the 16S rRNA gene were amplified by PCR using bar-coded universal primers 27F and 534R, or 357F and 936R respectively. Primers containing the A and B sequencing adaptors (454 Life Sciences, Branford, CT) were obtained from Eurofins MWG Operon (Huntsville, AL). Sequencing was performed using the 454/Roche B sequencing primer kit in the Roche Genome Sequencer GS-FLX Titanium platform. Samples were combined in a single region of the picotiter plate such that approximately 20,000 to 40,000 sequences were obtained from each group with each primer set. Samples were isolated and quality-filtered from each multiplexed Standard Flowgram Format (SFF) file. Raw sequences were deposited in the Short Read Archive Database (http://www.ncbi.nlm.nih.gov/sra, project number SRP002457).
16S Metagenomic 454 Sequencing Analyses
Quality filtered sequences were analyzed using three standard microbiome analysis techniques: operational taxonomic unit (OTU) generation, phylogenetic tree construction, and taxonomic binning of classified sequences. Sequences were taxonomically binned based on the output of a local copy of Ribosomal Database Project (RDP) Classifier24, and normalized data were produced from the relative abundance of taxa present in each sample. After filtering, 3,854,377 reads were obtained in total with an average read length of 503 nucleotides. This result corresponds to an average of 54,287 reads per pediatric sample. QIIME was utilized to produce OTU tables from the quality filtered sequences25. OTU tables were analyzed to study the relatedness of clinical metadata attributes for two types of diversity measurements: alpha (biodiversity within a group of samples) and beta diversity (biodiversity between groups of samples)26.
For the analysis and comparison of the sequencing data, the Mann-Whitney test was applied to normalized RDP counts to compare healthy pediatric samples and pediatric IBS samples using Genespring GX v 11.0 (Agilent Technologies, Santa Clara, CA). Machine learning algorithms (randomForest27) were used to determine the robustness of metadata clustering. These algorithms and the Boruta package28 were deployed to identify the most important variables involved in discriminating multiple groups of samples. Methods used for dataset sequence analysis are described in greater detail in the supplementary methods.
Bacterial DNA Microarray (PhyloChip) Hybridizations and Analyses
PhyloChip (version G2) hybridizations were performed with purified 16S rRNA gene amplicons from aliquots of DNA extracted from 27 samples of healthy children and 28 samples of children with IBS, using universal primers 27F (and 1492R as previously described29. Amplification, purification of the amplicons and microarray hybridizations were performed as reported elsewhere30–32. For microarray analyses, data were normalized, and taxa deemed present if they exhibited pf (positive fraction) value ≥0.95 (95% of all probes in a given probe set for an individual taxon report fluorescence). Taxonomy for PhyloChip studies is based on the Hugenholtz phylogenetic classification33. The PhyloChip contains probe sets additional details regarding the microarray analysis are provided in the supplementary methods.
Biostatistical Considerations
Sample size and power calculations were performed using NQuery Advisor 7.0. A sample size of 24 in each group will have 85% power to detect a probability of 0.750 that an observation in Group 1 (IBS) is less than an observation in Group 2 (Healthy) using a Wilcoxon (Mann-Whitney) rank-sum test with a 0.050 two-sided significance level. Retrospective analysis using mean values and standard deviations from genus-level data indicate that the sample size of 22 children per group (IBS and healthy) was sufficient to draw conclusions (average power of 88%) in the bacterial genera of interest.
RESULTS
Comparisons of pediatric gut microbiomes in healthy children and children with IBS
Although microbial dysbiosis has been described in adults with IBS, a paucity of data is available for the pediatric population. No evidence of bacterial overgrowth in pediatric IBS was found in this study by molecular methods. In support of this statement, no significant differences in total bacterial load by qPCR were observed in stool specimens from healthy children and children with IBS (Suppl. Fig. 1). PhyloChip-based taxonomic analyses enabled high-resolution investigations of global metrics such as bacterial richness, evenness or diversity. In addition to the total amount of bacterial DNA, the total number of bacterial taxa, otherwise known as bacterial community richness, and evenness did not significantly differ between IBS and healthy microbiomes (Suppl. Fig. 2). Greater overall phylogenetic diversity was evident among the bacteria in the IBS group (Suppl. Fig. 3).
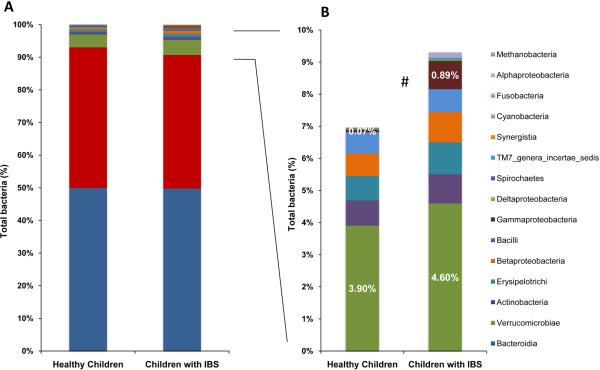
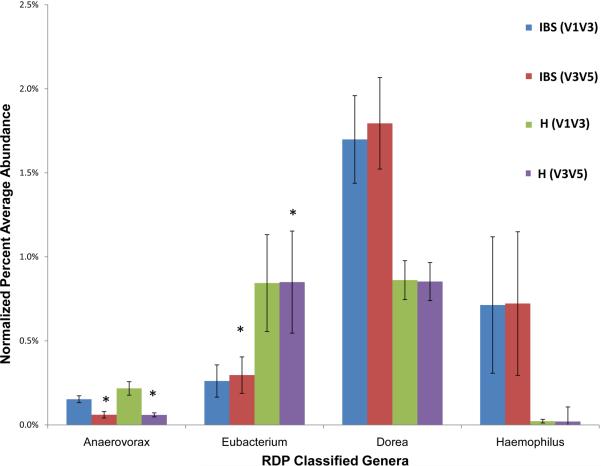
By 454 pyrosequencing and 16S metagenomics, we observed that microbial disease signatures exist in children with IBS (Fig. 1). Using the supervised learning algorithm, randomForest27, along with Boruta feature selection for individual taxa, children with IBS were classified correctly and separately from groups of 22 healthy children with a success rate of more than 96%. The microbiomes in children with IBS were characterized by a significantly greater percentage of the class Gammaproteobacteria compared with the distal gut microbiomes of healthy children (0.89% vs 0.07% of total bacteria; P <.05) (Figs. 1, 2). At the genus level, IBS microbiomes were characterized by greater percentages of Haemophilus (in class Gammaproteobacteria) and Dorea (P <.05) (Fig. 3). The species Haemophilus parainfluenzae was identified as a prominent component within the group of Haemophilus sequences, and taxa of the genus Veillonella were more abundant in children with IBS (data not shown). Based on the V1–V3 region, the IBS microbiomes were characterized by corresponding reductions in Eubacterium and Anaerovorax (P <.05) (Fig. 3).
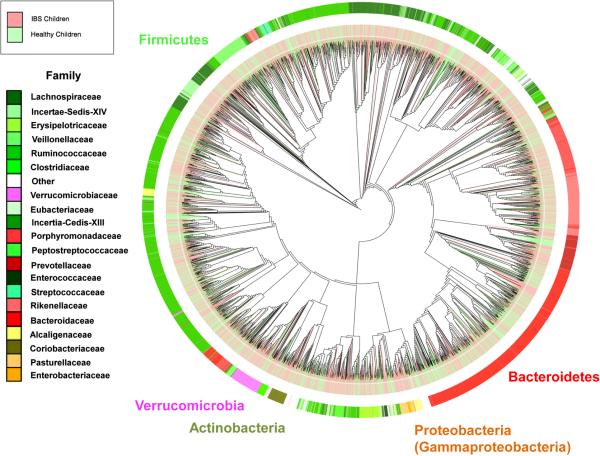
Figure 1. Global phylogenetic tree comparing the intestinal microbiomes of healthy children and children with IBS.
Phylogenetic tree was generated using QIIME and drawn with iTOL 54, including data from 22 healthy children (69 samples) and 22 children with IBS (71 samples). Map colored by phyla (exterior text), patient status (IBS - light red; Healthy - light green) and family (inset). Data were generated by 454 pyrosequencing (V1–V3 region).
Figure 2. The pediatric gut microbiomes of children with IBS are characterized by greater abundance of Gammaproteobacteria.
A) Percentage of all bacterial classes represented. B) Percentage of bacterial taxa found in lower abundance (< 5% of total bacteria). Healthy children: 29 samples from 22 subjects, IBS: 42 samples from 22 patients. #: Significantly different between IBS and healthy children (P <.05). Data were generated by 454 pyrosequencing (V1–V3 region).
Figure 3. Relative abundance of bacterial genera differentiates the distal intestinal microbiomes of healthy children and children with IBS.
Healthy (H) = 29 samples from 22 subjects, IBS (IBS): 42 samples from 22 patients (V1–V3 region or V3–V5). The data were generated by 454 pyrosequencing, and relative amounts were significantly different between IBS and healthy children (P <.05) except when labeled with *.
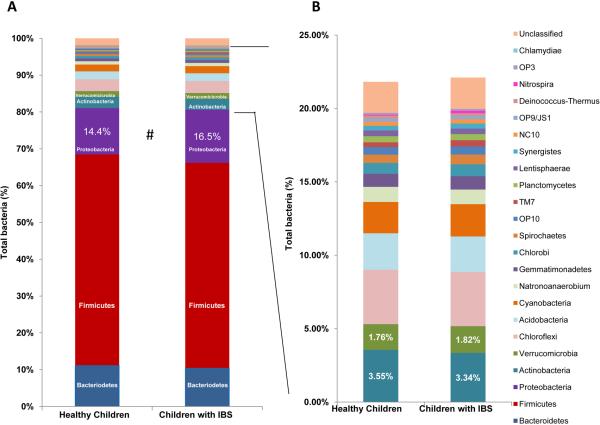
PhyloChip hybridization yielded similar results when compared to 454 pyrosequencing, in terms of commonly identified bacterial groups present in greater abundance in IBS microbiomes. Notably, Proteobacteria represented more than 16.5% of the total bacteria in IBS patients compared to 14.5% in healthy subjects (Fig. 4). The class Gammaproteobacteria mostly accounted for the relative difference in abundance of Proteobacteria in these two groups (6.14% in IBS patients compared with 4.53% in healthy subjects). In other words, the majority of other taxa found to be more abundant in IBS by PhyloChip analysis belonged to the class Gammaproteobacteria. Interestingly, the genus Haemophilus and more specifically, H. parainfluenzae, are common taxa detected within the class Gammaproteobacteria. Comparisons between the IBS and healthy groups demonstrated that 44 taxa were more abundant (>2 fold) in the IBS group (Suppl. Table 1A). One taxon not detected by 454 pyrosequencing and exhibiting the greatest fold change in relative abundance between children with IBS and healthy subjects (>19-fold) belonged to the phylum Nitrospira (otu_0984; G2 taxonomy). This taxon was present in 10 of 28 IBS samples (9 of 17 IBS subjects), but only in 1 of 27 samples from healthy subjects. However, attempts to amplify Nitrospira DNA using several sets of primers based on the PhyloChip probes and previously validated primer sets failed to confirm the presence of Nitrospira. Instead these sequences appear to be novel taxa related to the genus Ruminococcus.
Figure 4. The pediatric gut microbiomes in children with IBS are enriched in Proteobacteria (Gammaproteobacteria).
Healthy: 27 samples from 21 subjects IBS: 28 samples from 17 patients. A) Percentage of bacterial phyla represented in healthy children and children with IBS. B) Bacterial phyla representing less than 5% of total bacteria in healthy children and children with IBS. Data were generated by PhyloChip hybridization.
Bacterial taxa may also be associated with microbiomes from healthy children, and some microbes may be protective with respect to the recurrent abdominal pain phenotype. PhyloChip characterization of IBS microbiomes detected 12 taxa that were less abundant when compared to microbiomes of healthy children (> 2-fold, P <.05). These taxa included several Bacteroides species, including Bacteroides vulgatus, and other unclassified Bacteroides, present in much less abundance (by 15-30-fold) in IBS microbiomes when compared to healthy microbiomes, Suppl. Table 1B).
Bacterial taxa associated with increased frequency of abdominal pain in IBS subjects
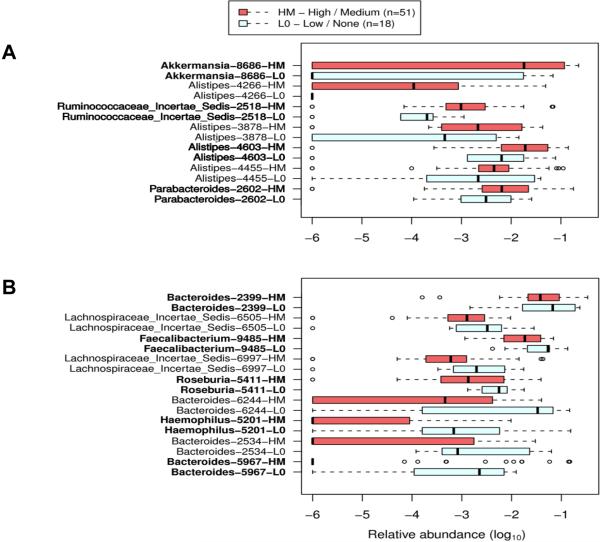
A primary aim of this study was to investigate the potential associations of differences in gut microbiome composition with the phenotype of recurrent abdominal pain in children. The clinical metadata obtained by history with 2-week diaries was analyzed together with 16S metagenomic 454 sequencing data by supervised learning (randomForest). Patients were distinguished by history as having a high-medium (HM) or low (LO) pain phenotype. A HM pain phenotype was characterized by greater than 3 pain episodes per 2-week period, and a LO pain phenotype was described by ≤3 pain episodes per 2-week period. Children with a HM pain phenotype (increased pain frequency) contained a relatively greater abundance of four different taxa belonging to the genus Alistipes (Fig. 5A). Additionally, individual taxa belonging to the genera Akkermansia and Parabacteroides, and a member of the family Ruminococcaceae were found in children with an increased number of abdominal pain episodes (Fig. 5A). Children with a LO pain phenotype had a greater preponderance of commensal taxa including Bacteroides, Faecalibacterium, Haemophilus and Roseburia (Fig. 5B).
Figure 5. Differential distribution of bacterial taxa in patients with recurrent abdominal pain was correlated with the relative frequency of abdominal pain.
Bacterial taxa (specified in leftmost column) were defined by randomForest and confirmed by feature selection using Boruta. The list is sorted first by Mann-Whitney U score followed by the largest disparity in medians for each group. Taxa represent the lowest taxonomic depth (Genus) that are labeled by RDP Classifier. Red rectangles display the HM recurrent abdominal pain phenotype. Light blue rectangles display the L0 recurrent abdominal pain phenotype. Boxes represent the first quartile, median, and third quartile of the OTU distributions for each pain group. Empty circles represent outliers that are 1.5× greater than the respective interquartile ranges. A) OTUs with greater abundance in patients with HM versus L0 recurrent abdominal pain phenotypes. B) OTUs with reduced abundance in patients with HM versus L0 recurrent abdominal pain phenotypes.
The recurrent abdominal pain phenotype was not associated with evidence of intestinal inflammation. Fecal calprotectin quantities were measured by ELISA in healthy children and children with IBS as a human biomarker of intestinal inflammation. However no significant differences were detected between the two groups (Suppl. Fig. 4).
Pediatric IBS subtypes can be distinguished by the microbial composition of distal gut microbiomes
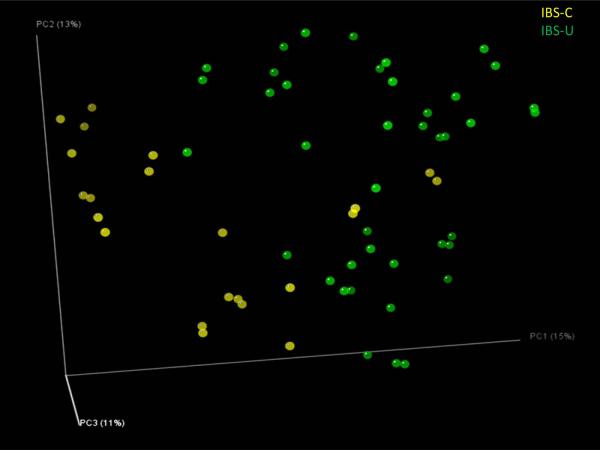
No prospective studies in pediatric populations have been undertaken to identify subgroups of children with IBS, as routinely done in adults. Therefore, we used the adult Rome III criteria to categorize different pediatric IBS subtypes using our recently published schema for analyzing pediatric IBS stooling diary data34. We attempted to refine the characterization of IBS subtypes in children by exploring the relative abundance of specific bacterial species. The IBS-C and IBS-U microbiomes (IBS-D was omitted in this analysis as only one subject was included into this category) were well separated by OTU composition (Fig. 6; Suppl. Table 2). IBS subtypes could be distinguished from each other, and children with IBS could be distinguished from healthy children by intestinal microbiome composition.
Figure 6. Distal gut microbiomes of children segregate the IBS-C and IBS-U subtypes.
 IBS-C: IBS with constipation (n=41 samples),
IBS-C: IBS with constipation (n=41 samples),  IBS-U: unsubtyped IBS (n=22 samples). Bray-Curtis analysis was used to generate a matrix of pairwise sample dissimilarities between communities. The scatterplot was generated from the matrix of distances using principal components analysis. Data were generated by 454 pyrosequencing (V1–V3 region only, 2 replicates per sample).
IBS-U: unsubtyped IBS (n=22 samples). Bray-Curtis analysis was used to generate a matrix of pairwise sample dissimilarities between communities. The scatterplot was generated from the matrix of distances using principal components analysis. Data were generated by 454 pyrosequencing (V1–V3 region only, 2 replicates per sample).
Classification of IBS-C and IBS-U based on distal gut microbiome composition of IBS subjects only was fully achieved using the supervised (machine) learning algorithm named randomForest27 with up to 98.5% accuracy (Fig. 6; Suppl. Table 2 for a complete list). The IBS-C subtype was characterized by the relative abundance of 54 OTUs, and the IBS-U group was distinguished by the relative abundance of 70 OTUs. Most of the OTUs that facilitated the classifications of these two IBS subtypes belong to the genera or groups such as Bacteroides, Ruminococcus, Lachnospiraceae Incertae Sedis, Veillonella, and Erysipelotrichaceae species. However no trend (lower or greater abundance) was observed for any individual bacterial genus. The data obtained by machine learning suggest that the composition of different microbial communities, as aggregate collections of species or strains rather than individual taxa, may facilitate the classification of each IBS subtype.
DISCUSSION
Our study has provided new insights about the composition of the distal gut microbiome in healthy, pre-adolescent children and children with IBS. We identified specific microbial signatures associated with recurrent abdominal pain in children and, specifically, pediatric IBS. Children with IBS yielded greater proportions of the phylum Proteobacteria, the class Gammaproteobacteria, and genera such as Dorea (member of Firmicutes) and Haemophilus (member of Gammaproteobacteria). Species such as H. parainfluenzae and a novel Ruminococcus-like organism were enriched in children with IBS. By contrast, taxa such as the genus Eubacterium and the species Bacteroides vulgatus were enriched in healthy children. Supervised learning and feature selection algorithms in combination with 16S metagenomics resulted in the successful classification and stratification of children with IBS and specific IBS subtypes with accuracies exceeding 95 percent. Finally, several taxa of the genus Alistipes were associated with the phenotype of frequently recurrent abdominal pain. One example from this genus, Alistipes putredinis, resembles members of the Bacteroides fragilis group35, 36, and is considered to be a commonly found gut commensal microbe of the Bacteroidetes phylum37. This species has been isolated from inflamed and non-inflamed intestinal tissues in studies of the bacteriology of appendicitis in children35,38.
Although conflicting results exist from studies in adults with IBS, enrichment in Proteobacteria has been described in adult studies39. In this study, the class Gammaproteobacteria accounted for most of this enrichment of the phylum Proteobacteria in children with IBS, and one prominent member of this class of organisms was H. parainfluenzae. H. parainfluenzae was cultured from the jejunal fluid of a substantial fraction of children in a prior Danish study40, suggesting that this bacterium is a gut commensal in children that may fluctuate quantitatively in disease states. In this study, increased abundance of the genus Veillonella was associated with pediatric IBS, although this genus was not enriched in a particular IBS subtype (data not shown). The gram-negative organism Veillonella (phylum Firmicutes) was more abundant in adults with IBS-C, and associated with increased quantities of fecal organic acids (acetic and propionic acids), and severity of pain41. A greater proportion of Haemophilus and acid-tolerant bacteria such as Veillonella, were also found to be more abundant in microbiomes associated with esophagitis42. Sequences amplified with Nitrospira primers were most similar to sequences of several unclassified Ruminococcus clones. Nonetheless, clones having 94% identity with Ruminococcus torques were present in greater abundance in IBS patients, and were correlated with greater pain severity43. Bacterial genera, such as Dorea, that were not previously linked with IBS were found to be associated with IBS in children. Dorea, a genus that includes species capable of formic acid production, has also been found in greater abundance in patients with ulcerative colitis44. In contrast, the PhyloChip could quantitatively examine different Bacteroides species, and B. vulgatus was found to be less abundant in children with IBS when compared to healthy subjects. These results corroborate previous data obtained in studies of adult patients with IBS-C45. Microbial community studies in other GI disorders may yield useful insights regarding the etiology and pathogenesis of IBS.
Two IBS subtypes dominated our patient population, and gastroenterologists are just beginning to appreciate the distribution of IBS subtypes in children. In our study, 59.1% of children presented with IBS-C, and 31.8% of children presented with IBS-U. The IBS-C and IBS-U subtypes were associated with differences in gut microbial composition encompassing at least 50–75 different taxa. Due to the paucity of clinical data, it remains unclear if the IBS subtype distribution in our study is typical for this age group in the USA, and this distribution is different from that seen in adults with IBS. Using supervised machine learning, specific bacterial communities were postulated to play a role in the pathogenesis of IBS. Considerations regarding statistical power in this study (22 subjects per group) were addressed by retrospective power/sample size calculations and review of data at the level of bacterial genera. Given that the Bristol chart reflects GI transit time in adults, it implies that the IBS-C group had a longer transit time than the IBS-U group. Further investigations are required to determine if colorectal transit influences the gut microbiota or vice-versa, and previous studies clearly show that the composition of the gut microbiota (i.e., methane producers) can affect transit time1, 2.
Our findings point to qualitative and quantitative differences in specific bacterial components of the gut microbiome as important features of IBS and its subtypes in children. The role of generic bacterial overgrowth46,47, and more specifically small intestinal bacterial overgrowth (SIBO), as a mechanism in the pathophysiology of IBS should be critically evaluated in light of these findings, detailed clinical assessment of SIBO48, and the recently published limitations of lactulose hydrogen breath testing49. In addition to SIBO, intestinal inflammation has been considered to contribute to disease in a subset of patients with IBS. Fecal calprotectin has been used as a biomarker of intestinal inflammation, and previous studies reported evidence of increased fecal calprotectin in some adults and children with IBS and FAP50. We speculate that abdominal pain in IBS may be associated with alterations in visceral sensitivity or pain perception induced by the composition of the gut microbiome, rather than immune activation5–7.
The present study yielded the most comprehensive dataset to date regarding the gut microbiome in children with IBS and highlighted specific differences in the gut microbiome related to IBS and its disease subtypes. Further analyses of clinical features, dietary and medication history, and human genetics may yield additional insights into the interplay of genetic, metagenomic, and environmental factors in the pathophysiology of IBS. This study also emphasizes the potential importance of microbial manipulation strategies in disease prevention and management. Rational manipulation of the microbiome by antibiotics or probiotics may ultimately result in the alleviation of symptoms. Treatment of IBS with the antibiotic rifaximin relieved symptom scores in patients51 and underscored the potential ability to manage IBS by eliminating “bad actors” in the gut microbiomes of some patients. In addition to antibiotics, probiotics including yet-to-be-defined beneficial microbes may yield beneficial effects in adults and children with IBS17, 52. Recent studies described the impact of diet on the human gut microbiome53, and the coupling of specific nutritional interventions with carefully selected antibiotics or probiotics may provide efficacious microbial manipulation strategies for the prevention and treatment of IBS.
CONCLUSIONS
Differences between the gut microbiomes of healthy children and children with IBS were identified in terms of relative abundance of specific bacterial phyla and genera. IBS subtypes (IBS-C and IBS-U) were effectively distinguished by global microbiome analyses. Our data suggest that gut microbiome studies may help clarify important associations between commensal microbes, microbe-derived molecular signals, and phenotypes of recurrent abdominal pain in children. Candidate bacterial genera have been identified that may serve as the basis for next-generation probiotics. Metagenomics data appear to be useful for stratification of patients with different IBS subtypes. We anticipate that compositional and functional discoveries related to the intestinal microbiome will translate into new biological therapeutics in neurogastroenterology.
Supplementary Material
ACKNOWLEDGEMENTS
This project is supported by the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK) and the National Human Genome Research Institute (NHGRI) from the National Institutes of Health (NIH) (grant UH2 DK083990-01 and UH3 DK083990-02). The work in JV's laboratory is supported by several awards from the NIH including NIDDK (R01 DK065075 and P30 DK56338), National Center for Complementary and Alternative Medicine (R01 AT004326 and R21 AT003102), and NHGRI (HMP sampling Jumpstart 5U54 HG003273-08, with Dr RG as PI). SVL is supported by the Rainin Foundation. We thank Jun Ma and Kjersti Aagaard for their assistance with phylogenetic tree construction. We are grateful to David Ottarson for his help with the PhyloChip validation.
Grant Support: This project is supported by the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK) and the National Center for Complementary and Alternative Medicine (NCCAM) from the National Institute of Health (NIH) (grant UH2 DK083990-01 and UH3 DK083990-02). The work in JV's laboratory is supported by NIDDK (R01 DK065075 and P30 DK56338), NIH National Center for Complementary and Alternative Medicine (R01 AT004326 and R21 AT003102), and NHGRI (HMP sampling Jumpstart 5U54 HG003273-08, with Dr RG as PI).
Abbreviations
- DGGE
denaturing gradient gel electrophoresis
- FAP
functional abdominal pain
- GI
gastrointestinal
- HMP
Human Microbiome Project
- IBS
irritable bowel syndrome
- OTU
operational taxonomic unit
- RAP
recurrent abdominal pain
- IBS-D
IBS with diarrhea
- IBS-C
IBS with constipation
- IBS-U
unsubtyped IBS
- RDP
Ribosomal Database Project
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: J. Versalovic serves on an advisory board for Danone and he receives unrestricted research support from Biogaia AB.
REFERENCES
- 1.Apley J. The Child with Abdominal Pains. Blackwell Scientific. 1975 [Google Scholar]
- 2.Zuckerman B, Stevenson J, Bailey V. Stomachaches and headaches in a community sample of preschool children. Pediatrics. 1987;79:677–82. [PubMed] [Google Scholar]
- 3.Miele E, Simeone D, Marino A, et al. Functional gastrointestinal disorders in children: an Italian prospective survey. Pediatrics. 2004;114:73–8. doi: 10.1542/peds.114.1.73. [DOI] [PubMed] [Google Scholar]
- 4.Thakkar K, Gilger MA, Shulman RJ, et al. EGD in children with abdominal pain: a systematic review. Am J Gastroenterol. 2007;102:654–61. doi: 10.1111/j.1572-0241.2007.01051.x. [DOI] [PubMed] [Google Scholar]
- 5.Saps M, Sztainberg M, Di Lorenzo C. A prospective community-based study of gastroenterological symptoms in school-age children. J Pediatr Gastroenterol Nutr. 2006;43:477–82. doi: 10.1097/01.mpg.0000235979.41947.f6. [DOI] [PubMed] [Google Scholar]
- 6.Uc A, Hyman PE, Walker LS. Functional gastrointestinal disorders in African American children in primary care. J Pediatr Gastroenterol Nutr. 2006;42:270–4. doi: 10.1097/01.mpg.0000189371.29911.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasquin A, Di Lorenzo C, Forbes D, et al. Childhood functional gastrointestinal disorders: child/adolescent. Gastroenterology. 2006;130:1527–37. doi: 10.1053/j.gastro.2005.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longstreth GF, Thompson WG, Chey WD, et al. Functional bowel disorders. Gastroenterology. 2006;130:1480–91. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 9.Shulman RJ, Eakin MN, Jarrett M, et al. Characteristics of pain and stooling in children with recurrent abdominal pain. J Pediatr Gastroenterol Nutr. 2007;44:203–8. doi: 10.1097/01.mpg.0000243437.39710.c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker LS, Guite JW, Duke M, et al. Recurrent abdominal pain: a potential precursor of irritable bowel syndrome in adolescents and young adults. J Pediatr. 1998;132:1010–5. doi: 10.1016/s0022-3476(98)70400-7. [DOI] [PubMed] [Google Scholar]
- 11.Jarrett M, Heitkemper M, Czyzewski DI, et al. Recurrent abdominal pain in children: forerunner to adult irritable bowel syndrome? J Spec Pediatr Nurs. 2003;8:81–9. doi: 10.1111/j.1088-145x.2003.00081.x. [DOI] [PubMed] [Google Scholar]
- 12.Scharff L. Recurrent abdominal pain in children: a review of psychological factors and treatment. Clin Psychol Rev. 1997;17:145–66. doi: 10.1016/s0272-7358(96)00001-3. [DOI] [PubMed] [Google Scholar]
- 13.Salonen A, de Vos WM, Palva A. Gastrointestinal microbiota in irritable bowel syndrome: present state and perspectives. Microbiology. 2010;156:3205–15. doi: 10.1099/mic.0.043257-0. [DOI] [PubMed] [Google Scholar]
- 14.Malinen E, Rinttila T, Kajander K, et al. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–82. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 15.Verdu EF, Bercik P, Verma-Gandhu M, et al. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut. 2006;55:182–90. doi: 10.1136/gut.2005.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyra A, Krogius-Kurikka L, Nikkila J, et al. Effect of a multispecies probiotic supplement on quantity of irritable bowel syndrome-related intestinal microbial phylotypes. BMC Gastroenterol. 2010;10:110. doi: 10.1186/1471-230X-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moayyedi P, Ford AC, Talley NJ, et al. The efficacy of probiotics in the treatment of irritable bowel syndrome: a systematic review. Gut. 2010;59:325–32. doi: 10.1136/gut.2008.167270. [DOI] [PubMed] [Google Scholar]
- 18.Spiller P. Review article: probiotics and prebiotics in irritable bowel syndrome (IBS) Aliment Pharmacol Ther. 2008;28:385–396. doi: 10.1111/j.1365-2036.2008.03750.x. [DOI] [PubMed] [Google Scholar]
- 19.Francavilla R, Miniello V, Magista AM, et al. A randomized controlled trial of lactobacillus GG in children with functional abdominal pain. Pediatrics. 2010;126:e1445–52. doi: 10.1542/peds.2010-0467. [DOI] [PubMed] [Google Scholar]
- 20.Breiman L. Random Forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 21.Chumpitazi BP, Lane MM, Czyzewski DI, et al. Creation and initial evaluation of a Stool Form Scale for children. J Pediatr. 2010;157:594–7. doi: 10.1016/j.jpeds.2010.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Baeyer CL, Spagrud LJ, McCormick JC, et al. Three new datasets supporting use of the Numerical Rating Scale (NRS-11) for children's self-reports of pain intensity. Pain. 2009;143:223–7. doi: 10.1016/j.pain.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 1997;32:920–4. doi: 10.3109/00365529709011203. [DOI] [PubMed] [Google Scholar]
- 24.Cole JR, Wang Q, Cardenas E, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittaker RH. Evolution and measurement of species diversity. Taxon. 1972;21:213–51. [Google Scholar]
- 27.Liaw A, Wiener M. Classification and regression by randomForest. R news. 2002;2:18–22. [Google Scholar]
- 28.Kursa MB, Rudnicki WR. Feature selection with the Boruta package. Journal of statistical software. 2010;36:1–13. [Google Scholar]
- 29.Cox MJ, Huang YJ, Fujimura KE, et al. Lactobacillus casei abundance is associated with profound shifts in the infant gut microbiome. PLoS One. 2010;5:e8745. doi: 10.1371/journal.pone.0008745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yergeau E, Schoondermark-Stolk SA, Brodie EL, et al. Environmental microarray analyses of Antarctic soil microbial communities. ISME J. 2009;3:340–51. doi: 10.1038/ismej.2008.111. [DOI] [PubMed] [Google Scholar]
- 31.Sunagawa S, DeSantis TZ, Piceno YM, et al. Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 2009;3:512–21. doi: 10.1038/ismej.2008.131. [DOI] [PubMed] [Google Scholar]
- 32.Brodie EL, DeSantis TZ, Parker JP, et al. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci U S A. 2007;104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Czyzewski DI, Lane MM, Weidler EM, et al. The interpretation of Rome III criteria and method of assessment affect the irritable bowel syndrome classification of children. Aliment Pharmacol Ther. 2010;33:403–11. doi: 10.1111/j.1365-2036.2010.04535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rautio M, Eerola E, Vaisanen-Tunkelrott ML, et al. Reclassification of Bacteroides putredinis (Weinberg et al., 1937) in a new genus Alistipes gen. nov., as Alistipes putredinis comb. nov., and description of Alistipes finegoldii sp. nov., from human sources. Syst Appl Microbiol. 2003;26:182–8. doi: 10.1078/072320203322346029. [DOI] [PubMed] [Google Scholar]
- 36.Rautio M, Lonnroth M, Saxen H, et al. Characteristics of an unusual anaerobic pigmented gram-negative rod isolated from normal and inflamed appendices. Clin Infect Dis. 1997;25(Suppl 2):S107–10. doi: 10.1086/516210. [DOI] [PubMed] [Google Scholar]
- 37.Rigottier-Gois L, Rochet V, Garrec N, et al. Enumeration of Bacteroides species in human faeces by fluorescent in situ hybridisation combined with flow cytometry using 16S rRNA probes. Syst Appl Microbiol. 2003;26:110–8. doi: 10.1078/072320203322337399. [DOI] [PubMed] [Google Scholar]
- 38.Rautio M, Saxen H, Siitonen A, et al. Bacteriology of histopathologically defined appendicitis in children. Pediatr Infect Dis J. 2000;19:1078–83. doi: 10.1097/00006454-200011000-00010. [DOI] [PubMed] [Google Scholar]
- 39.Krogius-Kurikka L, Lyra A, Malinen E, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Justesen T, Nielsen OH, Hjelt K, et al. Normal cultivable microflora in upper jejunal fluid in children without gastrointestinal disorders. J Pediatr Gastroenterol Nutr. 1984;3:683–6. doi: 10.1097/00005176-198411000-00007. [DOI] [PubMed] [Google Scholar]
- 41.Tana C, Umesaki Y, Imaoka A, et al. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–e115. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 42.Yang L, Lu X, Nossa CW, et al. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology. 2009;137:588–97. doi: 10.1053/j.gastro.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malinen E, Krogius-Kurikka L, Lyra A, et al. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2010;16:4532–40. doi: 10.3748/wjg.v16.i36.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nomura T, Ohkusa T, Okayasu I, et al. Mucosa-associated bacteria in ulcerative colitis before and after antibiotic combination therapy. Aliment Pharmacol Ther. 2005;21:1017–27. doi: 10.1111/j.1365-2036.2005.02428.x. [DOI] [PubMed] [Google Scholar]
- 45.Rajilić-Stojanović M. Diversity of the Human Gastrointestinal Microbiota_Novel erspectives from High Troughput Analyses. Wageningen University; Wageningen: 2007. [Google Scholar]
- 46.Pimentel M. Evaluating a bacterial hypothesis in IBS using a modification of Koch's postulates: part 1. Am J Gastroenterol. 2010;105:718–21. doi: 10.1038/ajg.2009.678. [DOI] [PubMed] [Google Scholar]
- 47.Spiegel BM. Questioning the Bacterial Overgrowth Hypothesis of Irritable Bowel Syndrome: An Epidemiologic and Evolutionary Perspective. Clin Gastroenterol Hepatol. 2011;9:461–469. doi: 10.1016/j.cgh.2011.02.030. [DOI] [PubMed] [Google Scholar]
- 48.Posserud I, Stotzer PO, Bjornsson ES, et al. Small intestinal bacterial overgrowth in patients with irritable bowel syndrome. Gut. 2007;56:802–8. doi: 10.1136/gut.2006.108712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu D, Cheeseman F, Vanner S. Combined oro-caecal scintigraphy and lactulose hydrogen breath testing demonstrate that breath testing detects oro-caecal transit, not small intestinal bacterial overgrowth in patients with IBS. Gut. 2011;60:334–40. doi: 10.1136/gut.2009.205476. [DOI] [PubMed] [Google Scholar]
- 50.Shulman RJ, Eakin MN, Czyzewski DI, et al. Increased gastrointestinal permeability and gut inflammation in children with functional abdominal pain and irritable bowel syndrome. J Pediatr. 2008;153:646–50. doi: 10.1016/j.jpeds.2008.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pimentel M, Lembo A, Chey WD, et al. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med. 2011;364:22–32. doi: 10.1056/NEJMoa1004409. [DOI] [PubMed] [Google Scholar]
- 52.Hoveyda N, Heneghan C, Mahtani KR, et al. A systematic review and meta-analysis: probiotics in the treatment of irritable bowel syndrome. BMC Gastroenterol. 2009;9:15. doi: 10.1186/1471-230X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ciccarelli FD, Doerks T, von Mering C, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–7. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.