Abstract
Human metastatic lymph node 64 (MLN64) is a transmembrane protein that shares homology with the cholesterol-binding vertebrate steroid acute regulatory protein (StAR)-related lipid transfer domain (START) and is involved in cholesterol traffic and steroid synthesis. We identified a Drosophila melanogaster gene whose putative protein product shows extensive homology with MLN64 and that we name Start1 (FlyBase CG3522). The putative Start1 protein, derived from Start1 cDNA sequences, contains an additional 122 aa of unknown function within the StAR-related lipid transfer domain. Similar inserts seem to exist in the Start1 homologues of Drosophila pseudoobscura and Anopheles gambiae, but not in the homologous protein of the urochordate Ciona intestinalis. Immunostaining using an insert-specific antibody confirms the presence of the insert in the cytoplasm. Whereas RT-PCR data indicate that Start1 is expressed ubiquitously, RNA in situ hybridizations demonstrate its overexpression in prothoracic gland cells, where ecdysteroids are synthesized from cholesterol. Transcripts of Start1 are detectable in embryonic ring gland progenitor cells and are abundant in prothoracic glands of larvae showing wave-like expression during larval stages. In adults, Start1 is expressed in nurse cells of the ovary. These observations are consistent with the assumption that Start1 plays a key role in the regulation of ecdysteroid synthesis. Vice versa, the expression of Start1 itself seems to depend on ecdysone, as in the ecdysone-deficient mutant ecd-1, Start1 expression is severely reduced.
In insects, pulses of ecdysteroid hormones induce larval molting and metamorphosis. Changes of puffing patterns in the polytene chromosomes in the course of the salivary gland development of Drosophila melanogaster had led Becker (1) to propose in 1962 a role for ecdysone in the regulation of gene expression. Since than, numerous reports have demonstrated that ecdysone is the dominant player in regulating cascades of gene activities, thereby controlling the major developmental events of insects. In Drosophila, we are beginning to understand at the molecular level how ecdysone induces gene expression (2).
Much less is known about the regulation of the synthesis of ecdysone itself and the generation of the wave-like diurnal change of the ecdysone titer during development. In the postembryonic developmental stages of insects, ecdysone is synthesized in the endocrine prothoracic gland (PG). In cyclorrhaphus diptera like Drosophila, the PG, the corpora allata, and the corpora cardiaca form the ring gland, which is innervated by the CNS (3). In Lepidoptera, prothoracicotropic hormones (PTTH) are synthesized by neurosecretory cells of the CNS and act on the PG, thereby initiating a transductory cascade, which results in the up-regulation of ecdysteroid synthesis from its precursor cholesterol (4). PTTH activity has been found in brain extracts of several insect species like the lepidoptera Bombyx mori (5) and Manduca sexta (6); however, we are only beginning to identify the components of the transductory cascade.
In vertebrates, the regulation of steroid hormone synthesis is much better understood. Steroidogenic hormones like adrenocorticotrophic hormone are released from the pituitary, recognized by their target cell receptors, and regulate the expression of genes involved in the steroid hormone pathway. The rate-limiting step in this pathway seems to be the induction of the synthesis of the steroid acute regulatory protein (StAR). StAR mediates, by means of the StAR-related lipid transfer (START) domain, the transfer of cholesterol across the mitochondrial membrane to the side-chain cleavage P450 enzyme that converts cholesterol to pregnenolon (7). A similar function has been assigned to the vertebrate cholesterol transporter MLN64 (metastatic lymph node 64), which shares the START domain with the StAR protein but, in addition, has a transmembrane domain region (8).
Here, we describe a gene of D. melanogaster, Start1, as the likely homologue of MLN64. Start1 seems to be the only gene in Drosophila that codes for a START domain protein of the StAR-type START subfamily (9). It is expressed in a spatial and temporal pattern consistent with it being the rate-limiting factor in ecdysteroidogenesis. On the other hand, our data indicate that the expression of Start1 itself depends on ecdysone, implying that in Drosophila ecdysone might be involved in the regulation of its own synthesis. This possibility seems especially attractive to us, because for Drosophila a PTTH similar to Bombyx PTTH has not yet been identified, and the role of a Drosophila protein with PTTH-like activity (10) remains open.
Materials and Methods
Computer Analyses. blast searches were performed at the Berkeley Drosophila Genome Project (BDGP) server (www.fruitfly.org/blast/). blastp, matrix blosum 52, the Predicted Proteins database, and human StAR (GenBank accession no. P49675) as a query sequence led to CG3522 (P = 10-15).
Start1 homologues in the genomes of Drosophila pseudoobscura, Anopheles gambiae, and Ciona intestinalis were detected by tblastn, and the D. melanogaster Start1 cDNA or protein sequence as a query. The amino acid sequence for D. pseudoobscura is derived from contig 5050 by analyzing nucleotides 16.001 to 19.000 by GeneMark.hmm (htpp://opal.biology.gatech.edu/GeneMark/hmmchoice.html). The A. gambiae genomic Start1 sequence (GenBank accession no. EAA03945) contained a gap and was completed (see below). C. intestinalis Start1 homologous protein is contained in the Scaffold 100 sequence (www.jgi.doe.gov.) within nucleotides 221032 and 228458.
clustal w (version 1.8.1) was used for protein alignment; toppred2 was used for transmembrane prediction.
Stocks. D. melanogaster strains Kochi-R and ecd-1 (obtained from C. Thummel, University of Utah, Salt Lake City) were kept on cornmeal medium at 25°C. For age-staging of third instar larvae, the medium was supplemented by bromophenol blue (11). Kc cells were provided by H. Saumweber, Humboldt-Universitæt, Berlin.
Cloning and RT-PCR. cDNA clones LD23890, RE28156, and RE40430 were obtained from BDGP. Plasmid pStart1179 was constructed by amplifying a 1,179-bp fragment by RT-PCR using total RNA from D. melanogaster third instar larvae and primers Start4 (5′-GCCTGGTTCCTGGACTGTAG) and Start5 (5′-CAGTTCGTTGACATGCTTGC). The fragment was cloned into pGEM-T (Promega) and sequenced.
A 332-bp DNA fragment coding for part of the putative Start1 protein insert was amplified from pStart1179, using primers Start10 (5′-ccaagaattcCAATGGTCAAATCTGCGATG) containing an EcoRI site (lowercase letters), and Start11 (5′-ccaactcgagTCGTCATGTTATTCGCTTCG) containing an XhoI site, cloned into pBlueScript II SK(-) and sequenced. A 1,204-bp sequence covering the transmembrane region of Start1 was cloned into pGEM-T by using primers Start25 (5′-GCATTCGGTGTTCCTTTTGT) and Start26 (5′-CCCTACAGTCCAGGAACCAG).
To fill a gap within the Start1 gene of the A. gambiae genome sequence, we amplified and sequenced this region (A. gambiae DNA was provided by F. Kafatos, European Molecular Biology Laboratory, Heidelberg). This replaces C11 N20 at position 7457741 of the National Center for Biotechnology Information sequence AAAB01008807.1 by 5′-C7ACAGGCCTACACA, thereby producing an acceptor splice site (BDGP splice site prediction, P = 0.95), and leads to the given amino acid sequence.
RNA was isolated by TRIzol reagent (Invitrogen). For RT-PCR, the OneStep RT-PCR kit (Qiagen, Valencia, CA) and primers Start4 and Start5 were used.
D. melanogaster cDNA libraries were constructed from RNA of third instar larvae (Clontech) or brain/ring glands (12).
Northern Analysis. Poly(A) RNA was prepared from brain/ring gland complexes of third instar larvae by using message maker (GIBCO), separated in a 1% agarose/formaldehyde gel and transferred to a Nytran Super Charge membrane (Schleicher and Schuell). A [32P]UTP-labeled Start1 antisense RNA probe was synthesized from a subclone of cDNA RE28156. By primers Start13 (5′-ccaactcgagCTGAACTCGCTCTCCTGCTT) and Start14 (5′-ccaagaattcAATATAACCGCTCGCCATGT), a 1,882-bp fragment containing exons 1-10 was amplified and cloned into pBlueScript SK(-) as an EcoRI/XhoI (lowercase letters in the primers) fragment. To reduce the length of the transcribed polylinker, it was again subcloned into pGEM-9Zf(-) (Promega) as a SalI/XbaI fragment. Hybridization was carried out in a roll oven. X-Omat AR (Kodak) and Biomax MS screens (Kodak) were used for exposure.
Whole-Mount in Situ Hybridization. Digoxygenin-labeled sense and antisense RNA probes were synthesized by using the DIG RNA labeling kit (Roche Molecular Biochemicals). An EcoRI/SalI fragment containing the 5′ sequence up to nucleotide 1,736 of cDNA LD23890 was excised from pOT2, subcloned in pBlueScript SK(-), and used for all experiments, except for ovaries where the transmembrane domain clone was used. For hybridization of embryos we used formaldehyde for fixation (13). Hybridization of larval and adult tissue was done as given in ref. 14, except that tissue was digested with Pepsin. Probe was detected by an anti-digoxigenin-alkaline phosphatase antibody (Roche Molecular Biochemicals), nitroblue tetrazolium, and 5-bromo-4-chloro-3-indolyl phosphate.
Antibodies and Immunostaining. For polyclonal antibody production, two regions of Start1 were expressed in Escherichia coli. Region S comprises only the START domain with insert (amino acids 228-572). It was amplified by primers Start17 (5′-ggggggcatATGGACACGGCTCGTCAT) containing an NdeI site and Start16 (5′-ccccccctcgagCTTCTGCCTCAGTTCGTT) containing an XhoI site. Region I comprises the insert only (amino acids 363-465), amplified by primers Start10 (see above) and Start19 (5′-ccccccctcgagGTCGACCTTGTCCTTAGC) containing an XhoI site. Fragments were cloned into pET21b (Invitrogen) and sequenced. Start1 protein was purified by His-tag columns. Rabbits were immunized by Eurogentec (Brussels). Whole-mount immunostaining was done as described (15) by using goat anti-rabbit Cy3-labeled secondary antibody (Jackson ImmunoResearch). Preparations were examined with a Zeiss Axiophot fluorescence microscope and a Quantix (Photometrics, Tucson, AZ) video camera. Images were processed by an Apple computer and photoshop 5 (Adobe Systems, Mountain View, CA).
Results
Insect START Domain Proteins Related to Vertebrate MLN64 Contain an Unusual Insert. A computer search of the D. melanogaster genome for the existence of cholesterol transport proteins, using the START domain of the human StAR protein as the query sequence, leads to only one significant hit, a gene annotated by the BDGP as CG3522 and that we name Start1. Further comparisons revealed that the putative Start1 protein is more closely related to the vertebrate cholesterol transporter MLN64 than to StAR by having four putative transmembrane helices in addition to the START domain. The predicted START domain of the D. melanogaster Start1 protein differs from the vertebrate START domains by containing an insert of 122 aa (Fig. 1). The bipartite structure of the START domain is indicated by a National Center for Biotechnology Information conserved domain search, which places part I to amino acids 262-362 and part II to amino acids 487-574 (Pfam E value 10-7 for both parts). According to the BDGP prediction, Start1 consists of 12 exons, with the sequence coding for the 122-aa insert (insert coding sequence, ICS) contained within exon 10. Because the ICS is flanked by weak splice sites, we verified its existence in mRNA and protein. (i) Sequence analysis of the BDGP reference cDNA for CG5322, LD23890, revealed that it contains a splice artifact at the exon 10/exon11 junction, being not suited to clarify the problem. Therefore, we cloned by RT-PCR a DNA fragment (pStart1179) covering exon 10. The sequence of this fragment and its fusion with the 5′ and 3′ adjacent fragments of LD23890 leads to a virtual cDNA with a conceptual translation product containing the insert in accordance with the BDGP prediction. (ii) We sequenced other BDGP cDNAs, RE28156 and RE40430. Both contain the ICS. (iii) By PCR analysis of different cDNA libraries and RT-PCR on RNA from different tissues (see below), we did not obtain fragments without ICS. (iv) RNA in situ hybridization and immunostaining indicate the existence of the insert (see below). (v) In the genome databases of D. pseudoobscura, A. gambiae, and the urochordate C. intestinalis, we identified their Start1 homologues (Fig. 1). Both insect genes, but not the one of Ciona, contain an insert related to that of D. melanogaster. In both cases, we did not detect putative splice sites adjacent to the insert. In sum, we conclude that the amino acid insert is a native component of Start1 protein.
Fig. 1.
Insect Start1 proteins differ from their chordate homologues by an insert within the START domain. Alignment of Start1 proteins from D. melanogaster (Dm-Start1; AAF47232), D. pseudoobscura (Dp-Start1), A. gambiae (Ag-Start1; EAA03945, see Materials and Methods), C. intestinalis (Ci-Start1), and MLN64 from Homo sapiens (Hs-MLN64; Q14849). Positions with at least one insect residue identical to at least one chordate residue are shown in gray (52%). Identical insert residues are boxed. Putative transmembrane helices for Hs-MLN64 are underlined in black; the START domain for MLN64 (19) is in gray. Triangles 1-6, positions of splice sites conserved between insect-Start1 and Hs-MLN64. Triangles 1′-7′, splice sites common to the chordate proteins. Sites 1 and 6 are common to the five species. ♦, residues playing a role in forming the tunnel of Hs-MLN64 (20), 29% of which are identical and 63% are chemically similar between the species. * indicate the dileucine motif and the tyrosine residue, shown to be necessary for endosomal location of MLN64 (16).
Sequence Comparison. Amino acid sequence comparison of the proteins aligned as in Fig. 1 leads to 22% identity and 52% similarity between the five species. The degree of conservation is highest for the putative transmembrane helices-containing domain (55% identity for transmembrane 1) and the START domain. The transmembrane domain has been shown to be necessary for endosomal location of MLN64, but does not seem to contain conventional endosomal targeting signals (16). Our comparison suggests that this domain represents an evolutionary conserved endosomal localization sequence. Residues shown to be necessary for endosomal location of MLN64 are conserved between the five distantly related species. The insert is the least conserved of all domains. A comparison of splice sites between the genes shows that six of nine splice sites of MLN64 are maintained in Start1 of the insects, and two sites are common to the five genes (Fig. 1). As each of the genes seems to be single copy, we conclude that they are homologous.
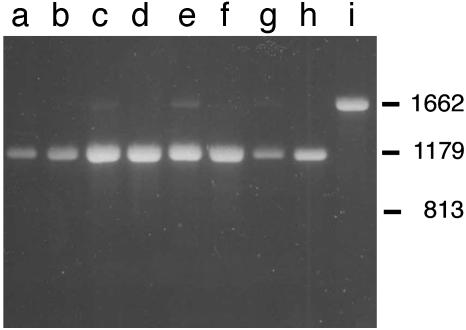
Start1 Is Expressed Ubiquitously. To analyze the expression pattern of Start1 we used RT-PCR and whole-mount RNA in situ hybridization. RT-PCR analyses indicate that Start1 is expressed ubiquitously (Fig. 2). Three larval tissues (brain/ring glands, imaginal disks, salivary glands), three adult tissues (heads, ovaries, testes), and Kc cells contain transcripts of Start1. The fragment size obtained indicates that the mRNA contains the ICS. For the six introns covered by the primers, there is no indication that Start1 is processed in a tissue-specific way.
Fig. 2.
Start1 mRNA exists in many cell types and contains the insert. RT-PCR on RNA from different larval and adult tissue and from cultured cells using primers spanning six introns. Lane a, total third larva (3L); lane b, 3L brain/ring gland complexes; lane c, 3L imaginal discs; lane d, 3L salivary glands; lane e, adult heads; lane f, ovaries; lane g, testes; lane h, Kc-cells; lane i, genomic DNA control. Approximate positions of bands with the expected size for genomic DNA (1,662 bp), cDNA with insert (1,179 bp), and without (813 bp) are derived from marker lanes. The data are not meant to be quantitative.
Start1 Is Expressed Predominantly in Steroidogenic Tissues. A differential expression pattern of Start1 emerges from whole-mount RNA in situ hybridization (Fig. 3).
Fig. 3.
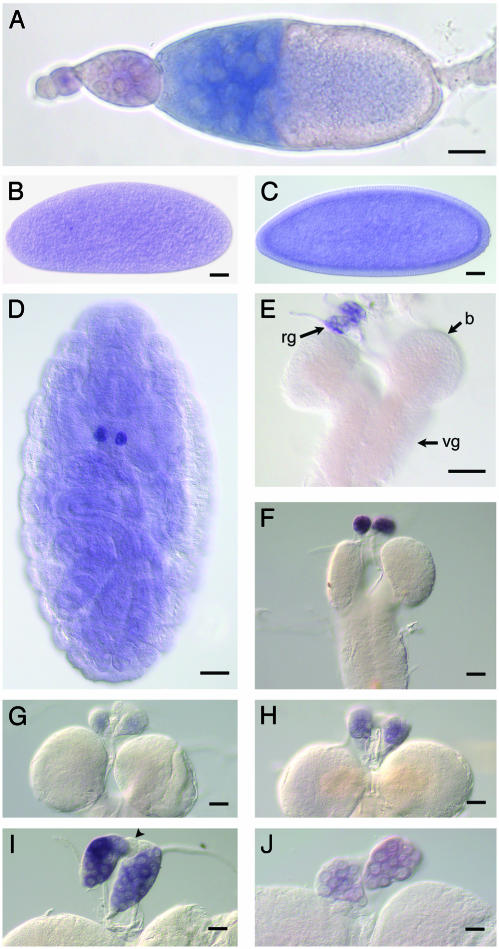
Start1 is expressed in nurse cells of ovaries and the prothoracic cells of the ring gland. Whole-mount RNA in situ hybridization using digoxygenin-labeled Start1 antisense RNA as a probe is shown. (A) Ovariol with egg chambers; nurse cells of stage 10 egg chambers show staining. (B) Praeblastoderm. (C) Blastoderm. (D) Embryo, stage 16; the strongly stained cells are the presumed precursor cells of the PG. (E-J) Brain/ring gland complexes from first larval instar (E) and second larval instar (F). (G-I) third larval instar of different stages: freshly hatched larva (G), feeding larva (H), nonfeeding/crawling larva (I), and white prepupa (J). The difference in expression between the second larva (F) and the young third larva (G) indicates that Start1 transcription is shut off during hatching. Staining conditions were kept constant for all larval stages. No staining was obtained for the sense RNA probes (data not shown). rg, ring gland; b, brain; vg, ventral ganglion. Arrowhead in I indicates unstained corpus allatum cells. (Bar, 40 μm.)
Ovaries from 2-day-old adults show Start1 expression in nurse cells of stage 10 egg chambers. In some egg chambers, we observed an apparent influx of staining into the oocyte, which could represent maternal Start1 RNA. We did not detect Start1 expression in adult brains, the only other adult organ we analyzed, indicating differential expression of Start1 in adults.
During development, Start1 RNA is found in praeblastoderm and blastoderm embryos, reflecting maternal mRNA. Start1 is specifically expressed in the PG cells of the ring gland of the three larval instars and its presumed embryonic precursor cells. A specific staining of the embryo is observed earliest at stage 16. A precise analysis of the level of expression of Start1 throughout larval development reveals that Start1 RNA is abundant at the end of both the second and third instar, but is almost undetectable in freshly hatched third instar larvae and declines in white prepupae, exhibiting a development-related wave-like diurnal pattern of expression (Fig. 3). Besides the PG cells, no other larval cells show significant Start1 RNA signals.
The PG cells of the ring gland are the predominant source of ecdysteroids in Drosophila postembryonic development, synthesized in a wave-like manner. In addition, there is growing evidence that the ovary is the source of ecdysteroids in adults (4, 17). Therefore, the observed spatial and temporal pattern of expression of Start1 and its probable involvement in the transport of cholesterol infer that Start1 is involved in the synthesis of ecdysteroids.
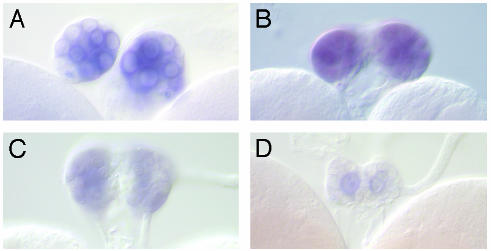
Start1 Protein Can Be Detected in PG Cells. The presence of the ICS in cDNAs and RT-PCR products had indicated that the insert exists in the Start1 protein. Further experiments support this conclusion. First, in situ hybridization using an ICS-specific antisense RNA probe resulted in hybridization signals in the cytoplasm of PG cells (data not shown). Second, Northern analysis using RNA from brain/ring gland complexes leads to a band, the size of which equals the expected size for Start1 mRNA containing the ICS (Fig. 4). Third, an antibody, directed specifically against the amino acid sequence of the insert, stains PG cells (Fig. 5) in the same way as an antibody against the START domain including the insert (data not shown), also indicating that Start1 mRNA is readily translated into protein.
Fig. 4.
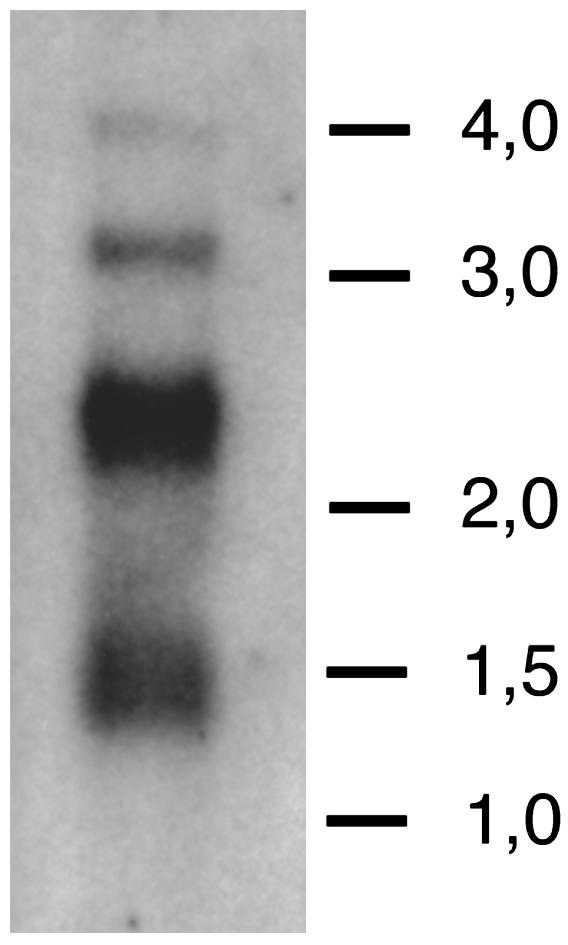
Northern analysis of Start1. Two micrograms of poly(A) RNA from brain/ring gland complexes was size-fractionated by gel electrophoresis, transferred to a membrane, and hybridized with a 32P-labeled RNA probe complementary to Start1. Exposure time was 14 h. Lines show the position of the high range RNA ladder marker bands (Fermentas; sizes are in kb). The bulk of the signal appears at ≈2.4 kb, as expected for Start1 transcripts, including 100 nt of poly(A) tail. No signal is detected at 2.1 kb, the approximate size for transcripts without the 366-nt ICS. As yet, we have no conclusive explanation for the origin of the other bands.
Fig. 5.
Immunofluorescence detection of Start1 protein in the PG cells. As primary antibody we used a polyclonal antibody risen against the insert amino acid sequence. The secondary antibody was labeled with Cy3. There was no noticeable difference in staining with antibodies against the complete START domain, without the transmembrane domains (data not shown). Besides the PG cells, no other tissue showed prominent staining.
Start1 Expression Seems to Depend on Ecdysone. The temperature-sensitive mutant ecd-1 has a reduced titer of ecdysone after a shift from 20°Cto29°C early in the third larval instar stage. Such larvae have only 5% of the WT ecdysone titer at pupariation and do not pupariate (18). Analysis of Start1 expression of WT and ecd-1 larvae by in situ hybridization demonstrates a severely reduced amount of Start1 transcripts in the mutant at the restrictive temperature (Fig. 6). The level of expression in the WT at 29°C seems to be normal; however, compared to 20°C, there is a change in the subcellular distribution of Start1 transcripts. In the mutant, even at 20°C the hybridization signals are different from those in the WT, concerning both intensity and subcellular distribution. At 29°C in the mutant, most of the PG cells do not show hybridization signals at all (Fig. 6D).
Fig. 6.
Start1 expression in the PG cells is reduced in the temperature-sensitive ecdysone-deficient mutant ecd-1. Animals were kept at 20°C, the permissive temperature for ecd-1, up to the late second instar. Then one group was allowed to continue development at 20°C, whereas the other group was transferred to 29°C, the restrictive temperature for ecd-1. RNA in situ hybridization was performed on late third instar larvae. WT strain Kochi-R is shown at 20°C(A) and 29°C(B). At this temperature nuclei also show staining. ecd-1 kept at 20°C(C) and 29°C(D) is shown. In ecd-1 at 29°C ring glands are smaller compared to 20°C as observed (35). Only some of the ring glands in the preparation showed staining, and in those only a few cells are stained.
Discussion
The family of START domain proteins is believed to function in the intracellular transport of lipids, lipid metabolism, and cell signaling (19). The START protein characterized best is the mammalian StAR protein, which transfers cholesterol, the precursor for steroid hormones, to mitochondria in steroidogenic cells (7). In the human and mouse genomes, 15 genes have been identified that encode START domain proteins. Some of these contain only START; in others, START is combined with other domains. A phylogenetic analysis divides this protein family into six subfamilies (9). According to this computer analysis, the Drosophila genome has members of only three subfamilies. Our own data indicate and are in agreement with ref. 9 that Start1 is the only START domain-coding gene in the Drosophila genome that belongs to the StAR subfamily. However, Start1 is not homologous to StAR itself, which contains only the START domain, but is homologous to MLN64, which in addition to the START domain carries a domain with four putative transmembrane helices.
To test our negative computer search for a homologue of StAR, we performed nonstringent genomic Southern hybridizations, using Start1 as a probe, but did not detect DNA fragments, indicating the existence of a related gene (data not shown). This result is especially critical because of the finding that the START domain of Start1 of D. melanogaster is different from all other START domains known so far by containing a 122-aa insert. The structure of the Start1 homologues of D. pseudoobscura and A. gambiae are similar (Fig. 1). Both StAR and MLN64 have been shown to bind cholesterol by means of their START domain. X-ray crystal structures imply that the START domain forms a cavity, accommodating a single cholesterol molecule (20).
We show that the insert in D. melanogaster exists in the translation product of Start1. Analysis of cDNAs, RT-PCR, whole cDNA libraries, Northern analysis, insert-specific in situ hybridization, and finally immunostaining with insert-specific antibodies indicate its presence in the Start1 protein. So far, we do not know whether the insert influences the function of Start1 with respect to the formation of the cavity.
MLN64, which was originally isolated by its overexpression and coamplification with the ERBB2 oncogene in human breast cancers, is expressed in all tissues and probably plays a role in intracellular sterol trafficking (8, 21). In addition, it is supposed to be involved in the synthesis of steroid hormones in the placenta (22). The expression pattern, which we observe for Start1, is similar to that of MLN64 in mammals. It is found expressed ubiquitously, obviously also in nonsteroidogenic tissues, when analyzed by RT-PCR (Fig. 2), but is found heavily expressed in the steroidogenic PG when analyzed by in situ hybridization (Fig. 3). Possibly the level of expression in the other tissues is too low to be detected by in situ hybridization.
MLN64 has been found localized on late endosomes and lysosomes, which are involved in cholesterol trafficking. The N-terminal region containing the transmembrane helices is necessary for this location (16). The degree of conservation of this region between the proteins is striking. Residues found to be essential for the endosomal location are conserved in Start1 (Fig. 1), indicating an identical location and similar function in Drosophila cells.
It was supposed earlier that in steroidogenic placental tissue MLN64 might deliver cholesterol to mitochondria. This view was supported by the finding of fragments of MLN64 containing the START domain in placental mitochondria, implying proteolytic cleavage of MLN64 (23). In addition, it was demonstrated in an in vitro system that the MLN64 START domain transfers cholesterol from liposomes to mitochondrial membranes (24). For insects, there is evidence that, like in mammals, cholesterol has to get into mitochondria for the synthesis of ecdysteroids. Recently, two mitochondrial cytochrome P450 monooxygenases required for the production of ecdysone have been described for Drosophila (25).
Given these data on MLN64 and in view of the similarities between Start1 and MLN64 in sequence and expression pattern, we are convinced that Start1 plays a role in insects analogous to that of MLN64 in mammals.
There is evidence for the involvement of human peripheral benzodiazepine receptor (PBR) in steroidogenesis (26), and there are indications that StAR protein and PBR interact at mitochondrial membranes (27). This finding led to speculation about the possible involvement of an insect PBR in ecdysteroidogenesis (4). The Drosophila protein most closely related to human PBR is the predicted protein product of gene CG2789 (DmPBR) showing 47% identity. We amplified a fragment of CG2789 and used this for RNA in situ hybridization with embryos, with brain/ring gland complexes of third instar larvae, and with adult ovaries. None of these experiments yielded a significant hybridization signal (data not shown). However, RT-PCR is positive with RNA, both from third larval instar brain/ring gland complexes and the carcass (data not shown). These data do not support a model in which Start1 and DmPBR would interact in insect steroidogenic tissue, as one would expect DmPBR to be hyperexpressed in the PG.
The expression pattern of Start1, in combination with its possible dependence on ecdysone, has some implications on the view of how the synthesis of ecdysone might be regulated during Drosophila development. There is evidence that in adult Drosophila females ecdysteroids are synthesized in the ovary (17), and it was suggested that nurse cells are the site of ecdysteroid synthesis, as the activation of ecdysteroid-dependent genes was observed in these cells (28). In consistency, we observe expression of Start1 in nurse cells. This observation agrees well with the nurse cell expression of dare, which codes for a mitochondrial protein transporting electrons to P450 enzymes and was shown to be necessary for ecdysteroidogenesis (29). However, the genes shadow (sad) (25) and disembodied (dib) (30), which encode hydroxylases involved in the conversion of cholesterol to ecdysteroids, are expressed in follicle cells. In conclusion, it seems likely that the mRNAs of the four genes are transported into the oocyte where ecdysteroids could be synthesized to be used in embryogenesis. The existence of such maternal ecdysteroids has been proposed (31). It remains to be analyzed if the activation of Start1 in nurse cells depends on ecdysteroids.
Besides this function for Start1, we think that part of the ovarian Start1 mRNA is stored in the oocyte as maternal mRNA. This is inferred from the observation of considerable staining of preblastoderm and blastoderm embryos after in situ hybridization (Fig. 3). The existence of maternal transcripts has also been reported for sad (25) and dib (30) and was postulated for dare (29). Maternal mRNA was also found from the Drosophila gene woc encoding a transcription factor involved in ecdysone biosynthesis (32). Translation of these maternal mRNAs would explain the surge in ecdysteroids, which has its peak at stages 11-12, well before the formation of the embryonic ring gland at stage 15, where at this stage sad and dib and, as shown here, Start1 are activated. So far, there are no data on the embryonic activation of dare. We suppose that the coordinated activation of these genes of the steroidogenic pathway initiates the next wave of ecdysteroids. How are the pulses of ecdysteroids generated? We observe a cycling of expression of Start1 in the second and third larval instar. The same cycling was found for sad and dib (25). Given the seeming dependency of Start1 on ecdysteroids, it is tempting to speculate that the two cycles might drive each other. A PTTH-like activity could connect these cycles to the circadian rhythm.
Presently, we cannot rule out that the weak expression of Start1 in the ecd-1 mutant has other reasons but the reduced ecdysone level. However, additional observations support our idea that Start1 regulation depends on ecdysone. One of the transcription factors regulating the StAR gene is steroidogenic factor-1 (SF-1) (33). The D. melanogaster homologue of SF-1 is the transcription factor βFTZ-F1. As (i) Start1 contains two possible binding sites for βFTZ-F1, (ii) βFTZ-F1 is expressed in PG cells (G.E.R., unpublished results) and (iii) βFTZ-F1 is controlled by ecdysone (34), we think that ecdysone is indeed a good candidate to regulate Start1.
Acknowledgments
We thank two unknown reviewers for helpful comments on the manuscript. This work was financially supported as a Pilot Project by the Freie Universitaet Berlin (to G.E.R.).
Abbreviations: PG, prothoracic gland; BDGP, Berkeley Drosophila Genome Project; StAR, steroid acute regulatory protein; START, StAR-related lipid transfer; ICS, insert coding sequence; PTTH, prothoracicotropic hormones; PBR, peripheral benzodiazepine receptor; MLN64, metastatic lymph node 64.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY455866).
References
- 1.Becker, H. J. (1962) Chromosoma 13, 341-384. [Google Scholar]
- 2.Thummel, C. S. (2002) Insect Biochem. Mol. Biol. 32, 113-120. [DOI] [PubMed] [Google Scholar]
- 3.Siegmund, T. & Korge, G. (2001) J. Comp. Neurol. 431, 481-491. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert, L. I., Rybczynski, R. & Warren, J. T. (2002) Annu. Rev. Entomol. 47, 883-916. [DOI] [PubMed] [Google Scholar]
- 5.Nagasawa, H., Guo, F., Zhong, X. C., Xia, B. Y., Wang, Z. S., Qui, X. J., Wei, D. Y., Chen, E. I., Wang, J. Z., Suzuki, A., et al. (1980) Sci. Sin. Ser. A 23, 1053-1060. [PubMed] [Google Scholar]
- 6.Bollenbacher, W. E., Katahira, E. J., O'Brien, M., Gilbert, L. I., Thomas, M. K., Agui, N. & Baumhover, A. H. (1984) Science 224, 1243-1245. [DOI] [PubMed] [Google Scholar]
- 7.Stocco, D. M. (2001) Annu. Rev. Physiol. 63, 193-213. [DOI] [PubMed] [Google Scholar]
- 8.Moog-Lutz, C., Tomasetto, C., Regnier, C. H., Wendling, C., Lutz, Y., Muller, D., Chenard, M. P., Basset, P. & Rio, M. C. (1997) Int. J. Cancer 71, 183-191. [DOI] [PubMed] [Google Scholar]
- 9.Soccio, R. E. & Breslow, J. L. (2003) J. Biol. Chem. 278, 22183-22186. [DOI] [PubMed] [Google Scholar]
- 10.Kim, A. J., Cha, G. H., Kim, K., Gilbert, L. I. & Lee, C. C. (1997) Proc. Natl. Acad. Sci. USA 94, 1130-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andres, A. J. & Thummel, C. S. (1994) Methods Cell Biol. 44, 565-573. [DOI] [PubMed] [Google Scholar]
- 12.Grams, R. & Korge, G. (1998) Gene 215, 191-201. [DOI] [PubMed] [Google Scholar]
- 13.Tautz, D. & Pfeifle, C. (1989) Chromosoma 98, 81-85. [DOI] [PubMed] [Google Scholar]
- 14.Wolf, T. (2000) in Drosophila Protocols, eds. Sullivan, W., Ashburner, M. & Hawley, R. S. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 201-227.
- 15.Ito, K., Urban, J. & Technau, G. M. (1995) Roux's Arch. Dev. Biol. 204, 284-307. [DOI] [PubMed] [Google Scholar]
- 16.Alpy, F., Stoeckel, M. E., Dierich, A., Escola, J. M., Wendling, C., Chenard, M. P., Vanier, M. T., Gruenberg, J., Tomasetto, C. & Rio, M. C. (2001) J. Biol. Chem. 276, 4261-4269. [DOI] [PubMed] [Google Scholar]
- 17.Riddiford, L. M. (1993) in The Development of Drosophila melanogaster, eds. Bate, M. & Martinez-Arias, A. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. 2, pp. 899-939. [Google Scholar]
- 18.Garen, A., Kauvar, L. & Lepesant, J. A. (1977) Proc. Natl. Acad. Sci. USA 74, 5099-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponting, C. P. & Aravind, L. (1999) Trends Biochem. Sci. 24, 130-132. [DOI] [PubMed] [Google Scholar]
- 20.Tsujishita, Y. & Hurley, J. H. (2000) Nat. Struct. Biol. 7, 408-414. [DOI] [PubMed] [Google Scholar]
- 21.Tomasetto, C., Regnier, C., Moog-Lutz, C., Mattei, M. G., Chenard, M. P., Lidereau, R., Basset, P. & Rio, M. C. (1995) Genomics 28, 367-376. [DOI] [PubMed] [Google Scholar]
- 22.Strauss, J. F., III, Kishida, T., Christenson, L. K., Fujimoto, T. & Hiroi, H. (2003) Mol. Cell. Endocrinol. 202, 59-65. [DOI] [PubMed] [Google Scholar]
- 23.Watari, H., Arakane, F., Moog-Lutz, C., Kallen, C. B., Tomasetto, C., Gerton, G. L., Rio, M. C., Baker, M. E. & Strauss, J. F., III (1997) Proc. Natl. Acad. Sci. USA 94, 8462-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, M., Liu, P., Dwyer, N. K., Christenson, L. K., Fujimoto, T., Martinez, F., Comly, M., Hanover, J. A., Blanchette-Mackie, E. J. & Strauss, J. F., III (2002) J. Biol. Chem. 277, 33300-33310. [DOI] [PubMed] [Google Scholar]
- 25.Warren, J. T., Petryk, A., Marques, G., Jarcho, M., Parvy, J. P., Dauphin-Villemant, C., O'Connor, M. B. & Gilbert, L. I. (2002) Proc. Natl. Acad. Sci. USA 99, 11043-11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beurdeley-Thomas, A., Miccoli, L., Oudard, S., Dutrillaux, B. & Poupon, M. F. (2000) J. Neurooncol. 46, 45-56. [DOI] [PubMed] [Google Scholar]
- 27.West, L. A., Horvat, R. D., Roess, D. A., Barisas, B. G., Juengel, J. L. & Niswender, G. D. (2001) Endocrinology 142, 502-505. [DOI] [PubMed] [Google Scholar]
- 28.Kozlova, T. & Thummel, C. S. (2000) Trends Endocrinol. Metab. 11, 276-280. [DOI] [PubMed] [Google Scholar]
- 29.Freeman, M. R., Dobritsa, A., Gaines, P., Segraves, W. A. & Carlson, J. R. (1999) Development (Cambridge, U.K.) 126, 4591-4602. [DOI] [PubMed] [Google Scholar]
- 30.Chavez, V. M., Marques, G., Delbecque, J. P., Kobayashi, K., Hollingsworth, M., Burr, J., Natzle, J. E. & O'Connor, M. B. (2000) Development (Cambridge, U.K.) 127, 4115-4126. [DOI] [PubMed] [Google Scholar]
- 31.Bownes, M., Shirras, A., Blair, M., Collins, J. & Coulson, A. (1988) Proc. Natl. Acad. Sci. USA 85, 1554-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren, J. T., Wismar, J., Subrahmanyam, B. & Gilbert, L. I. (2001) Mol. Cell. Endocrinol. 181, 1-14. [DOI] [PubMed] [Google Scholar]
- 33.Stocco, D. M., Clark, B. J., Reinhart, A. J., Williams, S. C., Dyson, M., Dassi, B., Walsh, L. P., Manna, P. R., Wang, X. J., Zeleznik, A. J. & Orly, J. (2001) Mol. Cell. Endocrinol. 177, 55-59. [DOI] [PubMed] [Google Scholar]
- 34.Woodard, C. T., Baehrecke, E. H. & Thummel, C. S. (1994) Cell 79, 607-615. [DOI] [PubMed] [Google Scholar]
- 35.Dai, J. D., Henrich, V. C. & Gilbert, L. I. (1991) Cell Tissue Res. 265, 435-445. [DOI] [PubMed] [Google Scholar]