Abstract
Rotifers of the asexual class Bdelloidea are unusual in possessing two or more divergent copies of every gene that has been examined. Phylogenetic analysis of the heat-shock gene hsp82 and the TATA-box-binding protein gene tbp in multiple bdelloid species suggested that for each gene, each copy belonged to one of two lineages that began to diverge before the bdelloid radiation. Such gene trees are consistent with the two lineages having descended from former alleles that began to diverge after meiotic segregation ceased or from subgenomes of an alloploid ancestor of the bdelloids. However, the original analyses of bdelloid gene-copy divergence used only a single outgroup species and were based on parsimony and neighbor joining. We have now used maximum likelihood and Bayesian inference methods and, for hsp82, multiple outgroups in an attempt to produce more robust gene trees. Here we report that the available data do not unambiguously discriminate between gene trees that root the origin of hsp82 and tbp copy divergence before the bdelloid radiation and those which indicate that the gene copies began to diverge within bdelloid families. The remarkable presence of multiple diverged gene copies in individual genomes is nevertheless consistent with the loss of sex in an ancient ancestor of bdelloids.
If a diploid lineage abandons the cycle of meiosis and syngamy that characterizes sexual reproduction and engages in no other form of lateral gene transfer, former alleles will no longer segregate and, because of the cessation of genetic drift, will no longer be kept nearly identical. Conversion, mitotic crossing over, and nondisjunction then will be the only processes opposing the mutational accumulation of neutral divergence between sequences at loci of former alleles in individual genomes. In such an asexual system, divergence between such sequences will reflect the time since they last experienced one of these processes. If there are particular cases in which descendants of former alleles have escaped these processes, their divergence will reflect the time since sexual reproduction ceased.
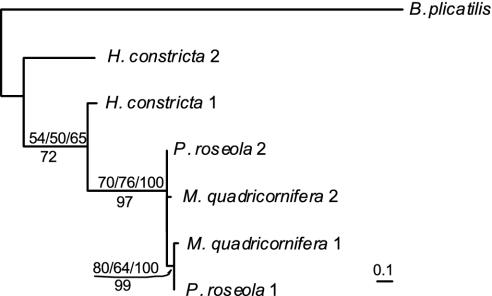
Genomes of rotifers of the asexual class Bdelloidea are unusual in that they possess two or more divergent copies of every gene that has been examined and lack closely similar copies (ref. 1 and D.B.M.W. and M.M., unpublished data). Previously (1), two of us presented gene trees of the heat-shock gene hsp82 and the TATA-box-binding protein gene tbp that were consistent with the separation of the most divergent copies of both genes before the bdelloid radiation and after the separation of bdelloids from their sister taxon, the facultatively sexual monogonont rotifers (for hsp82, see Fig. 1A). Although the possibility was cited that these copies could represent ancient duplications or components of an ancient polyploid, the time of their apparent separation was seen as consistent with their descent from a pair of former alleles.
Fig. 1.
Three gene trees of hsp82 in Rotifera. Monogononts, acanthocephalans, and the seisonid are described in ref. 1; bdelloids and the notation of bdelloid gene copies are described in ref. 2. (A) The tree presented in ref. 1. Branch lengths are proportional to the number of changes by maximum parsimony, with support for clades shown as a percent of 1,000 bootstrap replicates by maximum parsimony (above) and neighbor joining of a Kimura two-parameter distance matrix (below).(B) The tree found by a heuristic ML search using a nucleotide model of all positions. Branch lengths are proportional to the inferred number of substitutions, with support for clades shown as percent of bootstrap support (above) and posterior probability (below). Support for Philodinidae ranged from 71% to 76% of bootstrap replicates and from 91% to 97% posterior probability using various models of nucleotide substitution; the same rooting of bdelloid sequences is obtained when only codon third positions are used. (C) The tree found by a heuristic ML search using a nucleotide model of codon first and second positions. Branch lengths are proportional to the inferred number of substitutions, with support for clades shown as percent of bootstrap support (above) and posterior probability (below). The same rooting of bdelloid sequences is found when a codon-based model is used with ML.
The original trees were generated by parsimony and neighbor joining based on 4-fold degenerate codon positions and used a single outgroup to root the bdelloid gene copies. Because multiple outgroups, more sophisticated evolutionary models, and maximum likelihood (ML) and Bayesian inference methods can produce a more robust phylogeny, we conducted new analyses (initiated by D.M.H.). We find that phylogenetic analysis of the available data cannot discriminate between gene trees that differ in the placement of the root of hsp82 and of tbp bdelloid gene divergence, thereby leaving it unclear whether the most divergent gene copies separated before the bdelloid radiation or within bdelloid families.
Materials and Methods
Sequences Examined. Sequences and alignments have been reported (1, 2) and are available from D.B.M.W. or the rotifer biology database Wheelbase (http://jbpc.mbl.edu/wheelbase). GenBank accession numbers are AF143849-AF143858, AF249985-AF250004, AF375825, and AF375826. The species examined were Seisonida: Seison nebaliae Grube 1859; Acanthocephala: Moniliformis moniliformis (Bremser 1811), Oligacanthorhynchus tortuosa (Leidy 1850), and Oncicola sp. (Archiacanthocephala); Monogononta: Brachionus plicatilis Mueller 1786 strain AUS, Brachionus calyciflorus Pallas 1766 (Ploima, Brachionidae), Eosphora ehrenbergi Weber 1918 (Ploima, Notommatidae), and Sinantherina socialis (Linnaeus 1758) (Flosculariacea, Flosculariidae); and Bdelloidea: Philodina roseola Ehrenberg 1832, Macrotrachela quadricornifera (Milne 1886) (Philodinida, Philodinidae), Habrotrocha constricta (Dujardin 1841) (Philodinida, Habrotrochidae), and Adineta vaga (Davis 1873) (Adinetida, Adinetidae).
ML Analysis of hsp82. modeltest (3) was used to determine appropriate nucleotide-based evolutionary models. Three partitions of the data were considered: all nucleotides, codon first and second positions, and codon third positions. For each partition, the best model by the Akaike Information Criterion was General Time Reversible with a gamma-shape parameter and proportion of invariant sites to estimate rate heterogeneity. The alpha-shape parameter of the gamma distribution and the percent of invariant sites for the three partitions of the data set were 1.27 and 0.35, 0.32 and 0, and 2.51 and 0, respectively. Likelihood ratio tests showed a simpler model (Tajima-Nei) to be sufficient for all nucleotides and for codon third positions; this model produced the same topology with slightly lower bootstrap support (data not shown).
paup* 4.0b10 (4) was used to find the best ML tree by using a heuristic search with tree bisection-reconnection and 1,000 random-addition-sequence replications. Bootstrap values were generated in heuristic searches with 1,000 bootstrapped data sets and 10 random-addition-sequence replications for each bootstrap replicate.
For codon-based ML, the codeml program in paml 3.13a (5) was used to determine the ML score for the tree in Fig. 1B under a variety of models. The most appropriate model, as determined by likelihood ratio tests and Akaike Information Criterion, was codon equilibrium frequencies estimated as free parameters from the existing codon frequencies (60 parameters) and nonsynonymous/synonymous (dN/dS) ratios estimated for each branch of the tree (39 parameters). The transition-transversion frequency was estimated (1 parameter). The ML trees were found by nearest-neighbor interchange using kappa = 1.5, found when exploring models. When comparing hsp82 trees, the parameter estimations did not always converge; therefore, the tree-comparison program was run 10 times. The standard deviations of the average ML scores for the best and second-best trees were 0.005% and 0.076%, respectively. The best model that did not include the 60-parameter estimate of equilibrium codon frequencies was identical to the one above except that codon equilibrium frequencies were assumed to be equal.
ML trees were evaluated with consel (6), which uses the site likelihoods of each position to determine the significance of a tree by the bootstrap and posterior probability criteria as well as Kishino-Hasegawa and Shimodaira-Hasegawa tests.
Bayesian Analysis of hsp82. Bayesian analyses were performed with mrbayes 3.0b4 (7). The nucleotide-substitution model was General Time Reversible with a gamma-shape parameter and proportion of invariant sites to estimate rate heterogeneity. For analysis of all nucleotide positions, substitution rates and rate-heterogeneity parameters were estimated for codon first and second positions and for codon third positions independently by using the “unlink” option. Similar posterior probability support for the clade of Philodinidae was obtained without this option or by using a site-specific rate model (data not shown). Markovchain Monte Carlo chain length for all analyses was 2 × 106 generations with trees sampled every 100 generations; the first 104 trees (representing 106 generations) were discarded as burn-in. Additional runs with the same conditions produced the same topology with insignificant differences in posterior probability of any node.
Phylogenetic Analysis of tbp. The likelihood ratio tests implemented in modeltest evaluated the Hasegawa-Kishino-Yano model (empirical base frequencies, one transition-transversion rate) with gamma-shape parameters of 0.25 and 0.36 and no invariant sites to be a sufficient explanation of the total nucleotide and codon-only data, respectively. The program dnaml in phylip 3.6 was used to find the best ML tree by using the “jumble” option and global rearrangements. Bootstrap values were generated in an identical manner from 1,000 bootstrapped data sets; for the codons-only analysis, the bootstrap replicates were made at the codon rather than nucleotide level by using a perl script written by D.B.M.W. (available on request). Codon-based ML analysis was conducted as for hsp82 except that likelihood ratio tests and Akaike Information Criterion indicated that determining codon equilibrium frequencies from nucleotide frequencies (3 parameters) was sufficient. Because of the small number of sequences, analysis of bootstrap replicates was practical; 200 bootstrap replicates were each used in nearest-neighbor interchange searches for the best ML tree. Bayesian analysis using the Hasegawa-Kishino-Yano model with a gamma-shape parameter was conducted as described for hsp82 except that the data set included an additional unlinked partition containing the two introns. For analyses of codon first and second positions, the simpler model of Jukes-Cantor with a gamma-shape parameter of 0.12 was selected; more complex models produced similar results (data not shown).
Results and Discussion
By using multiple outgroups and a nucleotide-substitution model of all codon positions, neither ML nor Bayesian trees show hsp82 lineages that diverged before the bdelloid radiation. Instead, the bdelloid gene copies are rooted such that the highly divergent copies of hsp82 in P. roseola and in M. quadricornifera separate within the bdelloid family Philodinidae (Fig. 1B). This family-specific clade is well supported by bootstrap and posterior probability. Under ML this tree receives significantly better bootstrap and posterior probability support than one in which bdelloid gene copies diverge before the bdelloid radiation, although the ML scores of the two trees are not significantly different by Kishino-Hasegawa or Shimodaira-Hasegawa tests (Table 1). In contrast, the lineages of hsp82 diverge before the bdelloid radiation in both ML and Bayesian trees when codon third positions are excluded (Fig. 1C). However, the rooting of the bdelloid gene copies in the tree of Fig. 1C is not significantly supported by bootstrap or posterior probability, nor is the likelihood score of the tree significantly different from one in which the gene copies separate within families. Divergence of the two lineages before the bdelloid radiation is also seen in the best ML tree by using a codon-substitution model. This tree is strongly supported by bootstrap and posterior probability criteria and by Kishino-Hasegawa and Shimodaira-Hasegawa tests when compared with one that differs from it only in having the outgroups root the bdelloid gene copies as in Fig. 1B. The codon model used, although favored by likelihood ratio tests and the Akaike Information Criterion, has 60 free parameters estimating codon equilibrium frequencies, which may be inadequately specified by the available data. A codon-based model with the best likelihood score that does not include these parameters supports the same rooting of the bdelloid clade, but under this model the tree is not significantly better than one in which the B clade is repositioned as in Fig. 1B.
Table 1. Comparison of tree alternatives for hsp82.
| Criteria
|
|||||
|---|---|---|---|---|---|
| Model | Tree | BP | PP | KH | SH |
| Nucleotide model, all positions | B | 0.732 (0.004) | 0.900 (0.000) | 0.729 (0.004) | 0.729 (0.004) |
| C | 0.268 (0.004) | 0.100 (0.000) | 0.271 (0.004) | 0.271 (0.004) | |
| Nucleotide model, codon positions 1 and 2 | B | 0.408 (0.005) | 0.313 (0.000) | 0.362 (0.005) | 0.362 (0.005) |
| C | 0.592 (0.005) | 0.687 (0.000) | 0.638 (0.005) | 0.638 (0.005) | |
| Nucleotide model, codon position 3 | B | 0.923 (0.003) | 0.998 (0.000) | 0.912 (0.003) | 0.994 (0.001) |
| C | 0.076 (0.003) | 0.002 (0.000) | 0.088 (0.003) | 0.168 (0.004) | |
| Codon model, estimated frequency | B | 0.021 (0.001) | 5 × 10-11 (0.000) | 0.016 (0.001) | 0.016 (0.001) |
| C | 0.979 (0.001) | 1.000 (0.000) | 0.984 (0.001) | 0.984 (0.001) | |
| Codon model, equal frequency | B | 0.362 (0.005) | 0.011 (0.000) | 0.349 (0.005) | 0.349 (0.005) |
| C | 0.638 (0.005) | 0.989 (0.000) | 0.651 (0.005) | 0.651 (0.005) | |
Tree B has all gene copies diverging after the separation of bdelloid families as in Fig. 1B; in tree C the most divergent copies separate before the bdelloid radiation as in Fig. 1C. Scores are shown for the bootstrap (BP), posterior probability (PP), Kishino-Hasegawa (KH), and Shimodaira-Hasegawa (SH) tests, with standard error in parentheses.
No new outgroup sequences are available for tbp. Gene trees produced by ML and Bayesian analysis of all nucleotides and by ML using codon models are identical to the tree presented previously (1), as are ML and Bayesian trees when codon third positions and introns are excluded (Fig. 2). However, this tree topology is not well supported and is not significantly better than one in which the outgroup is positioned such that highly divergent copies separate only within bdelloid families. When codon third positions are included but introns, which are not present in the outgroup, are excluded, ML and Bayesian analyses produce several different trees depending on small changes in model parameters, and none of the trees are well supported (data not shown). Under any method of analysis, the very long branch length to the single outgroup probably precludes accurate rooting of the bdelloid gene copies.
Fig. 2.
Phylogenetic analyses of tbp. Numbers are percentage support of bootstrap replicates under nucleotide models of all positions, codon first and second positions only, and under a codon model, respectively (above the line), and of posterior probability with separate models for codon first and second positions, codon third positions, and introns (below the line). (Scale bar, changes per nucleotide according to ML using all positions.)
Thus, the available hsp82 and tbp data (alignments of 304 codons for four bdelloid species and of 109 codons plus 136 intron positions for three bdelloid species, respectively) are insufficient to discriminate between gene trees in which the highly divergent copies of hsp82 and of tbp within individual bdelloid genomes separated before the bdelloid radiation or within individual bdelloid families. The existing evidence for the persistence of the descendants of former allele pairs or homeologs in individual bdelloid genomes is therefore equivocal. Nevertheless, the presence in individual bdelloid genomes of divergent copies of each gene examined, the presence of chromosomes without morphological homologs, the lack of long interspersed nuclear element-like and gypsy-like retrotransposons, and the long-standing failure to find meiosis, males, or hermaphrodites in so large and diverse a taxon continues to argue for the ancient asexuality of bdelloid rotifers (1, 7-16).
Acknowledgments
We thank Irina Arkhipova, Bill Birky, Jim Bull, Jessica Mark Welch, Benjamin Normark, and Tom Wilcox for discussion and helpful comments on the manuscript. This work was supported by the U.S. National Science Foundation.
Abbreviation: ML, maximum likelihood.
References
- 1.Mark Welch, D. B. & Meselson, M. (2000) Science 288, 2111-1215. [DOI] [PubMed] [Google Scholar]
- 2.Mark Welch, D. B. & Meselson, M. (2001) Proc. Natl. Acad. Sci. USA 98, 6720-6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 917-818. [DOI] [PubMed] [Google Scholar]
- 4.Swofford, D. L. (2002) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.
- 5.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 6.Shimodaira, H. & Hasegawa, M. (2001) Bioinformatics 17, 1246-1247. [DOI] [PubMed] [Google Scholar]
- 7.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 8.Arkhipova, I. & Meselson, M. (2001) Proc. Natl. Acad. Sci. USA 97, 14473-14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donner, J. (1956) Ordnung Bdelloidea (Akademie, Berlin).
- 10.Hsu, W. (1956) Cellule 57, 283-296. [Google Scholar]
- 11.Hsu, W. (1956) Biol. Bull. 111, 364-374. [Google Scholar]
- 12.Hudson, C. T. & Gosse, P. H. (1886) The Rotifera, or Wheel-Animalcules (Longmans, Green, London).
- 13.Mark Welch, J. L. & Meselson, M. (1998) Hydrobiologia 387/388, 403-407. [Google Scholar]
- 14.Ricci, C. (1987) Hydrobiologia 147, 117-127. [Google Scholar]
- 15.White, M. J. D. (1978) Modes of Speciation (Freeman, San Francisco).
- 16.Mark Welch, J. L., Mark Welch, D. B. & Meselson, M. (2004) Proc. Natl. Acad. Sci. USA 101, 1618-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]