Abstract
We know little about several important properties of beneficial mutations, including their mutational origin, their phenotypic effects (e.g., protein structure changes vs. regulatory changes), and the frequency and rapidity with which they become fixed in a population. One signature of the spread of beneficial mutations is the reduction of heterozygosity at linked sites. Here, we present population genetic data from several loci across chromosome arm 2R in Drosophila simulans. A 100-kb segment from a freely recombining region of this chromosome shows extremely reduced heterozygosity in a California population sample, yet typical levels of divergence between species, suggesting that at least one episode of strong directional selection has occurred in the region. The 5′ flanking sequence of one gene in this region, Cyp6g1 (a cytochrome P450), is nearly fixed for a Doc transposable element insertion. Presence of the insertion is correlated with increased transcript abundance of Cyp6g1, a phenotype previously shown to be associated with insecticide resistance in Drosophila melanogaster. Surveys of nucleotide variation in the same genomic region in an African D. simulans population revealed no evidence for a high-frequency Doc element and no evidence for reduced polymorphism. These data are consistent with the notion that the Doc element is a geographically restricted beneficial mutation. Data from D. simulans Cyp6g1 are paralleled in many respects by data from its sister species D. melanogaster.
The spread of beneficial mutations is expected to reduce variation at linked sites, a phenomenon known as genetic hitchhiking (1, 2). All else being equal, the size of the swept region depends on the selection coefficient of the beneficial mutant and the local recombination rate. Theoretical results show that for regions of normal recombination in Drosophila, hitchhiking effects associated with moderately strong selection should result in localized regions of reduced heterozygosity (3). Thus, in principle, the frequency and locations of selective sweeps can be determined by scanning chromosomes for “valleys” of reduced variation (4). The paucity of large genomic regions of severely reduced heterozygosity from recombining regions in Drosophila and other organisms (4-7) suggests that novel mutations with large positive selection coefficients are rare, although it does not rule out the evolutionary importance of such mutations.
In this study, we document the existence of a 100-kb chromosomal region that has extremely reduced heterozygosity in a Drosophila simulans population sample from California, but not in a sample from Africa, indicating the recent and geographically restricted sweep of a unique, beneficial mutation. Furthermore, we report the unusual observation of an intact transposable element in this region, which occurs at very high frequency in the California, but not Africa, sample. The transposon insertion is associated with increased transcript abundance of the downstream cytochrome P450 gene Cyp6g1. These data are consistent with the notion that the transposable element insertion is the beneficial mutation responsible for the selective sweep.
Materials and Methods
Drosophila Stocks. California D. simulans and Drosophila melanogaster sequence data are from sets of eight highly inbred lines made from field-caught inseminated females collected in the Wolfskill Orchard, Winters, CA. PCR assays for the detection of transposon insertions were conducted on independent collections of male flies of both species from this same locality. African D. simulans and D. melanogaster sequence data are from sets of 10 isofemale lines collected in Zimbabwe and Malawi, respectively. As African lines retain some heterozygosity, PCR products from these stocks were generated by using a high-fidelity polymerase and then cloned before sequencing. A Cyp6g1 allele was also sequenced from Drosophila yakuba, a close outgroup to the sister species D. simulans and D. melanogaster. Survey sequence data from Table 1, Cyp6g1 sequences, and the single D. simulans Cyp6g1 Doc sequence are deposited in GenBank under accession nos. AY349854-AY349861, AY508487, AY521635-AY521652, AY521673, and AY523077-AY523383.
Table 1. Summary of population sequence data.
| Position, kb | No. sites | S | Theta CS | Theta AS | Theta CM | Divergence | Hd | ZnS | D |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 764 | 18 | 0.0087 | 0.0293 | 0.750 | 0.3574 | -0.0368 | ||
| 490 | 1021 | 9 | 0.0035 | 0.0341 | 0.750 | 0.4520 | -0.6648 | ||
| 914 | 818 | 10 | 0.0048 | 0.0282 | 0.464* | 0.7116 | -1.5123 | ||
| 1,210 | 660 | 31 | 0.0171 | 0.0353 | 0.893 | 0.2736 | 0.4064 | ||
| 1,391 | 842 | 11 | 0.0046 | 0.0211 | 0.607* | 0.5636 | 0.5134 | ||
| 1,476 | 929 | 9 | 0.0037 | 0.0035 | 0.0241 | 0.679 | 0.5429 | -0.5136 | |
| 1,554 | 778 | 12 | 0.0060 | 0.0411 | 0.464* | 0.8367* | -1.7700* | ||
| 1,649 | 739 | 7 | 0.0037 | 0.0342 | 0.250* | 1.0000* | -1.6741* | ||
| 1,699 | 739 | 15 | 0.0074 | 0.0514 | 0.429* | 1.0000* | 0.5691 | ||
| 1,729 | 910 | 0 | 0.0000 | 0.0386 | 0.000 | na | na | ||
| 1,744 | 887 | 0 | 0.0000 | 0.0496 | 0.000 | na | na | ||
| 1,772 | 774 | 0 | 0.0000 | 0.0130 | 0.0054 | 0.0492 | 0.000 | na | na |
| 1,798 | 937 | 0 | 0.0000 | 0.0523 | 0.000 | na | na | ||
| 1,807 | 1092 | 0 | 0.0000 | 0.0370 | 0.000 | na | na | ||
| 1,814 | 789 | 0 | 0.0000 | 0.0240 | 0.0015 | 0.0874 | 0.000 | na | na |
| 1,821 | 984 | 0 | 0.0000 | 0.0004 | 0.0601 | 0.000 | na | na | |
| 1,825 | 719 | 0 | 0.0000 | 0.0157 | 0.0000 | 0.1101 | 0.000 | na | na |
| 1,830 | 899 | 14 | 0.0060 | 0.0041 | 0.0441 | 0.250* | 1.0000* | -1.7912* | |
| 1,839 | 799 | 10 | 0.0048 | 0.0374 | 0.250* | 1.0000* | -1.7415* | ||
| 1,849 | 1613 | 71 | 0.0150 | 0.0073 | 0.0526 | 0.464* | 0.5496 | -0.8811 | |
| 1,858 | 971 | 43 | 0.0177 | 0.0526 | 0.464* | 0.5759 | -0.8057 | ||
| 1,873 | 1044 | 32 | 0.0119 | 0.0613 | 0.643* | 0.3519 | -0.4979 | ||
| 1,897 | 951 | 18 | 0.0067 | 0.0565 | 0.607* | 0.5107 | 0.3893 | ||
| 1,999 | 740 | 15 | 0.0074 | 0.0364 | 0.429* | 1.0000* | 0.5691 | ||
| 2,117 | 827 | 45 | 0.0204 | 0.0067 | 0.0532 | 0.821* | 0.3284 | 0.5527 | |
| 2,374 | 1072 | 20 | 0.0070 | 0.0237 | 0.643* | 0.3685 | -0.6262 | ||
| 2,737 | 826 | 21 | 0.0095 | 0.0375 | 0.821 | 0.3520 | 0.8797 | ||
| 3,219 | 957 | 75 | 0.0306 | 0.0522 | 0.893 | 0.2555 | -0.3747 |
Position is relative to the first locus. S is the number of segregating sites in the California D. simulans sample. Theta CS is nuclear diversity in the California D. simulans sample, AS is African D. simulans, and CM is California D. melanogaster. Hd is haplotype diversity, ZnS is linkage disequilibrium, and D is Tajima's D. * indicates significance at 0.05 level (see Materials and Methods). na, not available.
Calculation of P Values. Significance of Hd (haplotype diversity), ZnS (linkage disequilibrium), and Tajima's D statistics (Table 1) were calculated by comparing the observed values to those obtained from neutral coalescence simulations. Simulated data were generated by using the observed number of segregating sites in the sample (S) and under the conservative assumption of no recombination.
Estimation of Selection Coefficient. A maximum-likelihood estimate for the magnitude of the selection coefficient required to cause the reduction in heterozygosity observed in our data (3) was calculated assuming standard estimates of Drosophila population parameters: a population size of 106 (8), a per-site recombination rate of 10-8 (9), and a per-site heterozygosity (θ) of 0.008 (10, 11).
Dot Blots. cDNA was isolated from 20 adult flies (10 males, 10 females) from each stock. Aliquots of cDNA were then bound to nylon membranes and hybridized with 32P-labeled probes generated from species-specific Cyp6g1 PCR products. Signal was measured by using a PhosphorImager. Dot blots were then stripped and hybridized with 32P-labeled species-specific Gapdh1 PCR products as a control. Hybridizations carried out in the reverse order yielded the same results.
DDT Resistance Bioassay. D. simulans and D. melanogaster lines were tested for resistance to the insecticide DDT by using a contact assay (12). Glass scintillation vials were coated with DDT by rolling 200 μl of acetone containing 20 μg of DDT inside the vials until the acetone evaporated. The vials were plugged with cotton soaked in 5% sucrose. For three replicates of each line, 20 female flies 2-5 days posteclosion were placed in the vials, and percentage mortality was measured the next morning, 18 h after initiation. Percentage mortality for each line was calculated as the average of the three replicates.
Results
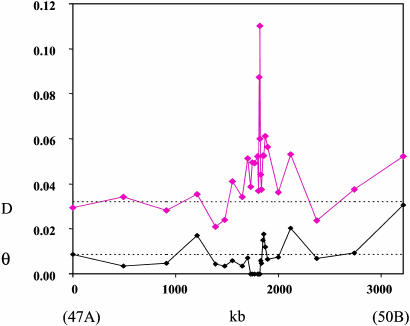
Hitchhiking Effects. We collected DNA sequence data from a California population of D. simulans at various intervals across 3 Mb of chromosome 2R (cytological position 47-50), a freely recombining region of the genome (13). (This project was originally begun to assess haplotype structure near Sr-CII, a scavenger receptor gene located at position 1849 in Table 1.) The data consist of 28 primarily noncoding loci averaging 900 bp, which were sequenced from each of eight inbred California D. simulans lines. Although levels of autosomal heterozygosity for most of the loci were relatively typical of both the species (0.0074, ref. 10) and other loci sampled from these particular D. simulans lines (0.0086, ref. 11), a set of eight consecutive ≈900-bp segments encompassing 100 kb of the genome were completely invariant (Table 1 and Fig. 1).
Fig. 1.
Heterozygosity (θ) and divergence (D) across chromosome 2R, cytological position 47A to 50B, in California D. simulans. Position in kb is relative to the first locus. Dashed lines represent an independent estimate of average values for the species (11).
The probability of observing one 900-bp locus devoid of polymorphism given an expected heterozygosity (θ) value of 0.0070 (the average of all 28 loci) was generated by comparison with simulated neutral coalescence trees assuming mutations along the lineages are Poisson distributed. This probability was extremely small (P = 0.0004), indicating that the probability of observing eight consecutive invariant 900-bp loci caused by stochastic variation in the average mutation rate alone is nil. A historically low mutation rate in this region cannot explain the reduced heterozygosity either, as levels of interspecific divergence between D. simulans and D. melanogaster in the invariant region are above average (Fig. 1). Instead, the data from this unusual region of the D. simulans genome are probably best explained by the recent fixation of a single haplotype by directional selection. A maximum-likelihood estimate for the magnitude of the selection coefficient required to cause such a reduction in heterozygosity is 0.022 (3). Evidence for significantly reduced haplotype diversity (for loci spanning >800 kb), significantly high levels of linkage disequilibrium, and significantly large negative Tajima's D values immediately flanking the region lacking polymorphism (Table 1) all support the selective sweep hypothesis (14-16).
The lack of even low-frequency mutations over >7,000 bp of surveyed DNA spanning a 100-kb region suggests that the presumed selective sweep occurred very recently (17). D. simulans, like D. melanogaster, is thought to have originated in Africa and colonized the rest of the world as a human commensal in evolutionarily recent times (18). A survey of three of the eight loci located in the 100-kb invariant region in a population sample from Harare, Zimbabwe revealed high levels of polymorphism (Table 1). The contrast between the distribution of variation in Zimbabwe and California suggests that the selective sweep occurred outside of the ancestral range of the species, perhaps associated with novel selection pressures in recently established D. simulans populations.
Further evidence in support of the sweep hypothesis comes from recent simulation studies of the probability of observing a “valley” of reduced heterozygosity by chance under the neutral model (3). Results using parameter values of population size, mutation rate, and recombination rate that are plausible for Drosophila suggest that it is extremely unlikely that drift could have fixed a haplotype sufficiently rapidly to eliminate variation over a 100-kb region. Local heterogeneity in heterozygosity may be common under the neutral model, but the physical scale is only on the order of 1-2 kb (3). We have no theoretical guidance on the effects of nonequilibrium population histories on the likelihood of observing a large window of reduced heterozygosity under neutrality. However, the fact that the African D. simulans samples from this region have typically high levels of nucleotide variation and low levels of linkage disequilibrium make it unlikely that sampling error during the establishment of North American populations from an ancestral African population (19, 20) could explain the data. Furthermore, large, freely recombining invariant regions in D. melanogaster have not been found, despite evidence that non-African D. melanogaster underwent a stronger bottleneck than non-African D. simulans (21).
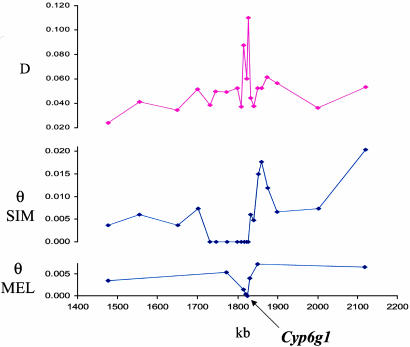
Given that D. simulans and its sister species D. melanogaster are sympatric and have similar demographic histories (18), we decided to investigate the distribution of polymorphism in the homologous chromosomal region of California D. melanogaster. Of the 28 loci sampled from California D. simulans, a representative subset of eight was surveyed for variation in eight inbred California D. melanogaster lines (Table 1). One of these eight loci completely lacked polymorphism in the California D. melanogaster sample. Although both species were invariant at this locus (Fig. 2), it was the most highly diverged of the 28 loci sampled in this region, a very unlikely result under a neutral model of evolution (22). Reduced heterozygosity at the same locus in D. simulans and D. melanogaster is consistent with the hypothesis that both species experienced recent selective sweeps in this region (although the breadth of the swept region is much greater in D. simulans), whereas the unusually high divergence suggests that recurrent directional selection has occurred at this locus in the past.
Fig. 2.
Heterozygosity (θ) and divergence (D) within and between California populations of D. simulans (SIM) and D. melanogaster (MEL) near the Cyp6g1 locus.
Candidate Sites. The hypothesis of geographically restricted strong selection in D. simulans suggests that the target of such selection should be fixed in our California population sample but absent or at extremely low frequency in the African population sample. The 22 known or predicted genes located in the invariant region (Table 3, which is published as supporting information on the PNAS web site) include a large number of candidate sites to sample. However, as noted above, only one of the surveyed loci was invariant from California population samples of both D. simulans and D. melanogaster. This locus was comprised primarily of the first intron of the gene Cyp6g1, located at the 3′ edge of the 100-kb region of reduced heterozygosity in California D. simulans. Cyp6g1 encodes a cytochrome P450 protein, a class of proteins that detoxifies xenobiotic compounds (23). Given that data from D. melanogaster implicate Cyp6g1 as an insecticide resistance gene (12), we decided to investigate whether the distribution of variation at Cyp6g1 provided any evidence for a candidate site of selection in D. simulans. Note, that although a selected site is expected to occur at the center of a swept region, results from simulated selective sweeps suggest that the small number of recombination events sampled during a rapid selective sweep may often cause the selected site to be positioned asymmetrically within the associated region of reduced heterozygosity (figure 3 b and d of ref. 3). Variation in recombination rates along chromosomes (24, 25) would presumably further inflate the variance of the location of the selected site.
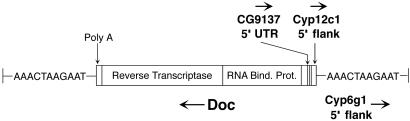
Fig. 3.
The structure of the D. simulans Doc insertion. The genomic sequence duplicated by Doc is shown to either side. Horizontal arrows represent the direction of transcription or, for noncoding sequence, the direction of transcription of the associated gene.
DNA encompassing the complete transcript for Cyp6g1 (2,776 bp) and 200 bp of 5′ flanking sequence was surveyed in eight Californian and 10 African D. simulans lines. As expected, the African sample harbors considerable variation (θ = 0.196; Table 4, which is published as supporting information on the PNAS web site), whereas the Californian sample does not. Interestingly, three singleton mutations were observed within the coding region of one California D. simulans line, suggesting that the 3′ boundary of the invariant region in the California sample may occur within Cyp6g1. The only mutation meeting our criteria for a candidate selected site was a 4,803-bp Doc non-LTR retrotransposable element found in all Californian lines (n = 8), but absent from the African lines (n = 10). The transposon is inserted ≈200 bp upstream of the putative transcription start site of Cyp6g1 (≈1 kb upstream of the coding start site, within the 3′ UTR of the upstream predicted gene CG8447; Table 4). This Doc element is inserted in reverse orientation and is associated with an 11-bp duplication of genomic sequence that causes a direct flanking repeat (Fig. 3). Sequence data from one California D. simulans line show that the inserted Doc element is full length and has no mutational lesions, indicating that it is functional and has a recent origin. Allele-specific PCR was used to determine the diploid genotypes of 26 freshly caught D. simulans males from the same California population used for the original survey; the frequency of the Doc insertion was 0.98. This is one of the few examples of a complete transposable element insertion at high frequency in natural Drosophila populations (26).
With the exception of its 5′ end, our D. simulans Doc sequence differed from the canonical D. melanogaster Doc element (27) at only three bases (all C-T transitions) over the 4,697 bp compared, suggesting that Doc may have recently invaded some melanogaster subgroup species through horizontal transfer (28). Conversely, the most 5′ 106 bp of the D. simulans Doc element was homologous neither to any known euchromatic D. melanogaster Doc sequence (29) nor to the Cyp6g1 5′ flanking sequence from a second outgroup, D. yakuba. blast comparisons of these 106 bp to the D. melanogaster genome yielded two matches to unique sequence. The first 72 bp were homologous (identity at 71 of 72 bases) to the 5′ flanking region of the D. melanogaster mitochondrial cytochrome P450 gene Cyp12c1 (≈2 kb upstream of the translation start site). Our sequence data from the Cyp12c1 5′ flanking region of one California D. simulans line showed that it was a perfect match (72 of 72 bases) to the Doc element-associated Cyp12c1 sequence. Unlike Cyp6g1, on chromosome 2, Cyp12c1 is located at polytene band 75D on chromosome arm 3L. The last 23 bp of the 5′ noncanonical Doc sequence were identical to DNA from an intron in the 5′ UTR of the D. melanogaster predicted gene CG9137, a protein inferred from sequence similarity to have an esterase/lipase/thioesterase active site. This gene is located on chromosome arm 3L at cytological position 61F. Thus, the 5′ end of the D. simulans Doc element located upstream of Cyp6g1 contains DNA from two different genomic regions (Fig. 3), one of which is the 5′ flanking region of a different cytochrome P450 gene. Although similar patterns were not observed in the 35 Doc elements located in the D. melanogaster euchromatic genome sequence (29), inclusion of genomic sequence during transposition has been observed for other transposable elements (30-32).
We also surveyed Cyp6g1 sequence variation in flanking and protein-coding regions in eight Californian and 10 African D. melanogaster lines. During the course of this study, an independent study reported the presence of an Accord (Gypsy-like) LTR retrotransposable element insertion in the 5′ regulatory region of Cyp6g1 in several D. melanogaster lab stocks from around the world (33). Our data support these results. The Accord insertion occurs ≈300 bp upstream of the transcription start site (1.1 kb upstream of the coding start site, Supporting Text and Fig. 5, which are published as supporting information on the PNAS web site) and is present in 7 of 8 D. melanogaster lines from California and in 2 of 10 D. melanogaster lines from Malawi, Africa. The Cyp6g1 Accord element is inserted in reverse orientation and causes a 4-bp duplication of genomic sequence (CGTG). Although the canonical length for Accord elements is 7,404 bp, there is variation in Accord insert PCR product lengths across lines, suggesting that the Cyp6g1 Accord element has accumulated indels since inserting and thus may not be as young as the D. simulans Doc insertion. Allele-specific PCR was used to determine the diploid genotypes of 30 freshly caught D. melanogaster males from the California population; the Accord insertion was present at a frequency of 0.98.
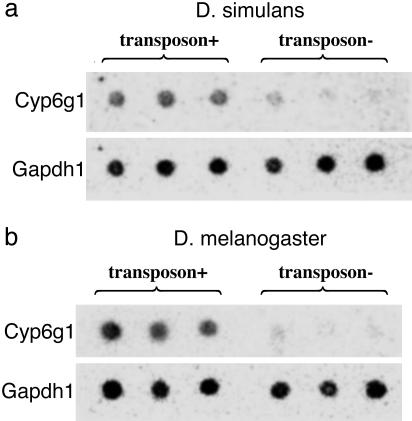
Cyp6g1 Transcription. Some D. melanogaster lines resistant to DDT have substantially higher levels of Cyp6g1 transcript compared to susceptible lines (12, 33), suggesting that mutations increasing transcript abundance of Cyp6g1 might be favored in certain environments. We compared Cyp6g1 mRNA transcript levels in adult flies from Californian D. simulans lines homozygous for the Doc insertion (n = 3) and African D. simulans lines homozygous for absence of the insertion (n = 3) (Fig. 4). Dilution series (data not shown) demonstrate that the D. simulans Doc insertion lines have at least 2-fold higher levels of Cyp6g1 transcript. We also compared Cyp6g1 mRNA transcript abundance in adult D. melanogaster homozygous for the Accord insertion (n = 3, two Californian lines and one African line) or homozygous for the absence of the Accord insertion (n = 3, one Californian line and two African lines) (Fig. 4). As expected (12, 33), all Accord insertion lines had substantially higher levels of Cyp6g1 transcript. These data are consistent with the hypothesis that both the D. simulans Doc insertion and the D. melanogaster Accord insertion cause constitutive Cyp6g1 up-regulation.
Fig. 4.
Dot blots comparing levels of Cyp6g1 transcript in adult flies by using the control Gapdh1 (see Materials and Methods). (a) The first three D. simulans dots are from the Californian lines CS1, CS2, and CS3, whereas the last three are from the African lines AS1, AS2, and AS3. (b) The first two D. melanogaster dots and the fourth dot are from the Californian lines CM1, CM2, and CM3, whereas the third dot and the fifth and sixth dots are from the African lines AM3, AM2, and AM5.
We cannot rule out the possibility that mutations other than the transposon insertions are responsible for Cyp6g1 up-regulation in these lines. In our D. simulans population samples Doc presence/absence is conflated with the California/Africa D. simulans genomes. Nevertheless, the hypothesis that the transposons cause Cyp6g1 up-regulation is plausible given previous reports of transposon insertions disrupting existing repressor elements (34), or cases in which regulatory elements within transposons up-regulate transcription of the nearby gene (31, 32, 35). To investigate these possibilities we used the matinspector tool (version 2.2) of the Transcription Factor Database (36) to identify potential transcription factor binding sites for xenobiotic response elements near the Doc and Accord insertion sites, and within the genomic sequences that the Doc element acquired. Although results from this bioinformatics approach must be considered speculative until tested by functional experiments, such transcription factor binding sites were found within the D. simulans Doc-associated Cyp12c1 sequence as well as in close proximity to the D. melanogaster Accord insertion site (Supporting Text and Fig. 5). Identification of currently unannotated transcription factor binding sites in the Cyp6g1 regulatory region may be possible with the sequencing of additional Drosophila species and the use of the phylogenetic shadowing approach (37). Of course, any one of the large number of potential promoter/enhancer sequences that occur within the Doc or Accord elements themselves may also be the underlying cause of Cyp6g1 up-regulation. It is also possible that the Doc and Accord transposon insertions may influence regulation of Cyp6g1 by altering the physical distance between regulatory elements and the transcriptional start site.
DDT Resistance. The similarities between the population genetic data and gene expression data from Cyp6g1 in D. simulans and D. melanogaster, along with the evidence that Cyp6g1 can confer DDT resistance in D. melanogaster (12), led us to investigate whether the Doc insertion is associated with DDT resistance in D. simulans. We compared DDT resistance between Californian D. simulans lines homozygous for the Doc transposon insertion (n = 8) and African D. simulans lines homozygous for the absence of the Doc insertion (n = 10). We also measured resistance in D. melanogaster lines homozygous for the Accord transposon insertion (n = 9, seven Californian lines and two African lines) and D. melanogaster lines homozygous for the absence of the Accord insertion (n = 9, one Californian line and eight African lines). In our DDT resistance assay the average percent mortality for the D. simulans Doc insertion lines (0.72) was marginally significantly lower than the average percent mortality for the non-Doc insertion lines (0.94) (Table 2). However, much of the difference in mean resistance between population samples is attributable to one line, CS8, a highly resistant line from California. Larger population samples will be required for more powerful association studies of the Doc insertion and the DDT-resistance phenotype. In D. melanogaster, lines harboring the Accord insertion had a significantly lower percent mortality (0.47) than lines without the Accord insertion (0.97), consistent with previous results from this species (33). However, we observed abundant variation in DDT resistance within each class of D. melanogaster Cyp6g1 alleles. In fact, for both species, some transposon insertion lines faired worse than nontransposon insertion lines (Table 2). Thus, for both species, it seems probable that DDT resistance is not determined solely by transposon insertions at Cyp6g1, but instead is multifactorial.
Table 2. Percent mortality per line for DDT bioassay.
| Transposon + | Transposon - | ||
|---|---|---|---|
| D. simulans | |||
| CS1 | 0.84 | AS1 | 0.69 |
| CS2 | 0.60 | AS2 | 0.82 |
| CS3 | 0.77 | AS3 | 1.00 |
| CS4 | 1.00 | AS4 | 0.95 |
| CS5 | 0.95 | AS5 | 0.97 |
| CS6 | 0.57 | AS7 | 1.00 |
| CS7 | 1.00 | AS9 | 0.98 |
| CS8 | 0.04 | AS10 | 1.00 |
| Average | 0.72 | AS11 | 0.98 |
| AS12 | 1.00 | ||
| Average | 0.94 | ||
| D. melanogaster | |||
| CM1 | 0.00 | CM3 | 0.87 |
| CM2 | 1.00 | AM1 | 0.95 |
| CM4 | 0.97 | AM2 | 1.00 |
| CM5 | 0.11 | AM5 | 1.00 |
| CM6 | 0.02 | AM6 | 0.94 |
| CM7 | 0.00 | AM7 | 1.00 |
| CM9 | 0.28 | AM8 | 1.00 |
| AM3 | 0.82 | AM9 | 0.97 |
| AM4 | 1.00 | AM10 | 0.97 |
| Average | 0.47 | Average | 0.97 |
For D. simulans, ANOVA P = 0.057. For D. melanogaster, ANOVA P = 0.006.
Discussion
We have documented the existence of a large, freely recombining region showing severely reduced heterozygosity in a California population of D. simulans, a phenomena best explained by the selective sweep of a new beneficial mutation. Analysis of a simple hitchhiking model yields a maximum-likelihood estimate of the selection coefficient of 2%. The rarity of such “heterozygosity valleys,” despite the growing amount of population sequence data (including data from 61 other genes from these same D. simulans lines; refs. 11 and 38), suggests that beneficial mutations with selection coefficients of this magnitude occur infrequently or that selection coefficients change on a faster scale than the substitution rate. Furthermore, this mutation must have occurred at low frequency in the California population when it became favored and must have swept relatively recently to explain the apparent lack of even low-frequency mutations in the swept region. The fact that the Doc insertion has not yet accumulated indels and is limited to the California sample is also consistent with a recent origin.
Although the data do not allow us to rule out the possibility that the Doc element hitchhiked to high frequency as a result of its linkage with some unsampled selected mutation, the Doc insertion is a good candidate mutation for the Cyp6g1 up-regulation phenotype and thus is a plausible candidate as the target of selection. Until recently, the role of transposable elements in adaptive evolution of Drosophila was thought to be minimal because surveys of particular element insertions suggested that they occur only at low frequency within species and are never fixed between species (39). However, recent studies have shown that transposon insertions play a large role in transcriptional regulation of hsp70 in Drosophila (40, 41) and in the evolution of regulatory and coding sequences of genomes in general (42-44). Our data are consistent with the notion that transposable element insertions occasionally act as beneficial mutations, particularly with respect to transcriptional regulation. The genomic bias toward deletion of DNA in Drosophila (45) may tend to obscure most transposon-mediated adaptive mutations by causing rapid loss of transposon-derived DNA not associated with novel function.
The Cyp12c1 genomic sequence associated with the D. simulans Cyp6g1 Doc insertion raises the intriguing possibility that this Doc element has moved transcriptional information between functionally related genes on different chromosomes (31, 46). Transcription of non-LTR retroposon insertions poses somewhat of a paradox because transcripts from which such insertions originate are not expected to contain upstream promoter elements (LTR retroposons, on the other hand, duplicate the 3′ terminal repeats, which are generally thought to contain promoters, to the 5′ end upon each insertion). This finding has led to the hypothesis that non-LTR retroposons use internal promoters downstream of the transcription start site (47). The use of internal promoters may bias non-LTR retroposons toward occasionally acquiring new transcription start sites in genomic sequence further to their 5′ flanks, allowing them a potentially larger role in the mobilization of genomic information than other transposable elements.
The widespread, largely indiscriminate use of DDT for insect pest eradication in the United States from 1945 until 1972 would seem to make it a candidate for the selective agent affecting the unusual genomic region of California D. simulans. Our bioassay data provide weak support for the hypothesis that selection for DDT resistance favored the Cyp6g1 Doc insertion haplotype. However, some data argue against DDT as the agent responsible for the D. simulans selective sweep. For example, mutations conferring resistance to pesticides are often disfavored and decline in frequency in the absence of the pesticide (48, 49). Although agricultural use of DDT in Zimbabwe was not banned until 1982, and continues to be used in tse-tse fly control programs there (50), the Doc insertion haplotype occurs at negligible frequency in Zimbabwe, yet persists at 98% frequency in the California D. simulans population sample where DDT has been banned for >30 years. Because the Cyp6g1 protein has broad insecticide detoxification activity (12, 33), and is also up-regulated in D. melanogaster lines selected for increased resistance to caffeine (G. Passador-Gurgel, personal communication), other types of selection pressures at Cyp6g1 remain highly plausible. Selection at Cyp6g1 could have been caused by an insecticide, a natural toxin, or an environmental contaminant found in California but not in Zimbabwe, with the beneficial mutation only tangentially conferring weak cross-resistance to DDT.
In summary, the localized reduction in heterozygosity around Cyp6g1 in California populations of both D. simulans and D. melanogaster, the existence of different transposable element insertions at high frequency in the 5′ regulatory region of Cyp6g1 in these species, and the associated transcriptional up-regulation of Cyp6g1 in both species provide a striking example of parallel evolution. The evidence that nucleotide variation linked to Cyp6g1 may be influenced by positive selection in D. simulans and D. melanogaster suggests that cytochrome P450s and other detoxification proteins may be hotspots for recent adaptive evolution in many insects.
Supplementary Material
Acknowledgments
We thank C. Bergman for helpful advice regarding Doc and Accord sequences and Y. Kim for the use of his program for estimating selection coefficients associated with heterozygosity valleys. This work was supported by the National Institutes of Health and the National Science Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY349854 -AY349861, AY508487, AY521635-AY521652, AY521673, and AY523077-AY523383).
References
- 1.Maynard-Smith, J. & Haigh, J. (1974) Genet. Res. 23, 23-35. [PubMed] [Google Scholar]
- 2.Kaplan, N. L., Hudson, R. R. & Langley, C. H. (1989) Genetics 123, 887-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim, Y. & Stephan, W. (2002) Genetics 160, 765-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harr, B., Kauer, M. & Schlotterer, C. (2002) Proc. Natl. Acad. Sci. USA 99, 12949-12954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, R. L., Stec, A., Hey, J., Lukens, L. & Doebley, J. (1999) Nature 398, 236-239. [DOI] [PubMed] [Google Scholar]
- 6.Fullerton, S. M., Clark, A. G., Weiss, K. M., Nickerson, D. A., Taylor, S. L., Stengard, J. H., Salomaa, V., Vartiainen, E., Perola, M., Boerwinkle, E., et al. (2000) Am. J. Hum. Genet. 67, 881-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nurminsky, D., DeAguiar, D., Bustamante, C. D. & Hartl, D. L. (2001) Science 291, 128-130. [DOI] [PubMed] [Google Scholar]
- 8.Przeworski, M., Wall, J. D. & Andolfatto, P. (2001) Mol. Biol. Evol. 18, 291-298. [DOI] [PubMed] [Google Scholar]
- 9.Comeron, J. M., Kreitman, M. & Aguade, M. (1999) Genetics 151, 239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriyama, E. N. & Powell, J. R. (1996) Mol. Biol. Evol. 13, 261-277. [DOI] [PubMed] [Google Scholar]
- 11.Begun, D. J. & Whitley, P. (2000) Proc. Natl. Acad. Sci. USA 97, 5960-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daborn, P., Boundy, S., Yen, J., Pittendrigh, B. & ffrench-Constant, R. (2001) Mol. Genet. Genomics 266, 556-563. [DOI] [PubMed] [Google Scholar]
- 13.True, J. R., Mercer, J. M. & Laurie, C. C. (1996) Genetics 142, 507-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depaulis, F. & Veuille, M. (1998) Mol. Biol. Evol. 15, 1788-1790. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, J. K. (1997) Genetics 146, 1197-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braverman, J. M., Hudson, R. R., Kaplan, N. L., Langley, C. H. & Stephan, W. (1995) Genetics 140, 783-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonsen K. L., Churchill, G. A. & Aquadro, C. F. (1995) Genetics 141, 413-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachaise, D., Cariou, M. L., David, J. R., Lemeunier, F., Tsacas, L. & Ashburner, M. (1988) Evol. Biol. 22, 159-225. [Google Scholar]
- 19.Hamblin, M. T. & Veuille, M. (1999) Genetics 153, 305-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andolfatto, P. (2001) Mol. Biol. Genet. 18, 279-290. [DOI] [PubMed] [Google Scholar]
- 21.Aquadro, C. F., Lado, K. M. & Noon, W. A. (1988) Genetics 119, 875-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudson, R. R., Kreitman, M. & Aguade, M. (1987) Genetics 116, 153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feyereisen, R. (1999) Annu. Rev. Entomol. 44, 507-533. [DOI] [PubMed] [Google Scholar]
- 24.Jeffreys, A. J., Kauppi, L. & Neumann, R. (2001) Nat. Genet. 29, 217-222. [DOI] [PubMed] [Google Scholar]
- 25.Petes, T. D. (2001) Nat. Rev. Genet. 2, 360-369. [DOI] [PubMed] [Google Scholar]
- 26.Petrov, D. A., Aminetzach, Y. T., Davis, J. C., Bensasson, D. & Hirsh, A. E. (2003) Mol. Biol. Evol. 20, 880-892. [DOI] [PubMed] [Google Scholar]
- 27.O'Hare, K., Alley, M. R., Cullingford, T. E., Driver, A. & Sanderson, M. J. (1991) Mol. Gen. Genet. 225, 17-24. [DOI] [PubMed] [Google Scholar]
- 28.Biemont, C. & Cizeron, G. (1999) Genetica 105, 43-62. [DOI] [PubMed] [Google Scholar]
- 29.Kaminker, J. S., Bergman, C. M., Kronmiller, B., Carlson, J., Svirskas, R., Patel, S., Frise, E., Wheeler, D. A., Lewis, S., Rubin, G. M., et al. (2002) Genome Biol. 3, research0084.1-research0084.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozmahel, R., Heng, H. H. Q., Duncan, A. M. V., Shi, X. M., Rommens, J. M. & Tsui, L. C. (1997) Genomics 45, 554-561. [DOI] [PubMed] [Google Scholar]
- 31.Ackerman, H., Udalova, I., Hull, J. & Kwiatkowski, D. (2002) Mol. Biol. Evol. 19, 884-890. [DOI] [PubMed] [Google Scholar]
- 32.Moran, J. V., DeBerardinis, R. J. & Kazazian, H. H., Jr. (1999) Science 283, 1530-1534. [DOI] [PubMed] [Google Scholar]
- 33.Daborn, P. J., Yen, J. L., Bogwitz, M. R., LeGoff, G., Feil, E., Jeffers, S., Tijet, N., Perry, T., Heckel, D., Batterham, P., et al. (2002) Science 297, 2253-2256. [DOI] [PubMed] [Google Scholar]
- 34.Wallace, M. R., Anderson, L. B., Saulino, A. M., Gregory, P. E., Glover, T. W. & Collins, F. S. (1991) Nature 353, 864-866. [DOI] [PubMed] [Google Scholar]
- 35.Willoughby, D. A., Vilalta, A. & Oshima, R. G. (2000) J. Biol. Chem. 275, 759-768. [DOI] [PubMed] [Google Scholar]
- 36.Quandt, K., Frech, K., Karas, H., Wingender, E. & Werner, T. (1995) Nucleic Acids Res. 23, 4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K. D., Ovcharenko, I., Pachter, L. & Rubin, E. M. (2003) Science 299, 1331-1333.12610290 [Google Scholar]
- 38.Schlenke, T. A. & Begun, D. J. (2003) Genetics 164, 1471-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charlesworth, B. & Langley, C. H. (1989) Annu. Rev. Genet. 23, 251-287. [DOI] [PubMed] [Google Scholar]
- 40.Maside, X., Bartolome, C. & Charlesworth, B. (2002) Curr. Biol. 12, 1686-1691. [DOI] [PubMed] [Google Scholar]
- 41.Lerman, D. N., Michalak, P., Helin, A. B., Bettencourt, B. R. & Feder, M. E. (2003) Mol. Biol. Evol. 20, 135-144. [DOI] [PubMed] [Google Scholar]
- 42.Brosius, J. (1999) Genetica 107, 209-238. [PubMed] [Google Scholar]
- 43.Nekrutenko, A. & Li, W.-H. (2001) Trends Genet. 17, 619-621. [DOI] [PubMed] [Google Scholar]
- 44.Jordan, I. K., Rogozin, I. B., Glazko, G. V. & Koonin, E. V. (2003) Trends Genet. 19, 68-72. [DOI] [PubMed] [Google Scholar]
- 45.Petrov, D. A. (2002) Genetica 115, 81-91. [DOI] [PubMed] [Google Scholar]
- 46.Britten, R. J. & Davidson, E. H. (1971) Q. Rev. Biol. 46, 111-138. [DOI] [PubMed] [Google Scholar]
- 47.Eickbush, T. H. (1992) New Biol. 4, 430-440. [PubMed] [Google Scholar]
- 48.Crow, J. F. (1957) Annu. Rev. Entomol. 2, 227-246. [Google Scholar]
- 49.Cochran, D. G. (1993) J. Econ. Entomol. 86, 1639-1644. [DOI] [PubMed] [Google Scholar]
- 50.Chikuni, O., Polder, A., Skaare, J. U. & Nhachi, C. F. B. (1997) Bull. Environ. Contam. Toxicol. 58, 776-778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.