Abstract
It has been suggested that fish have more genes than humans. Whether most of these additional genes originated through a complete (fish-specific) genome duplication or through many lineage-specific tandem gene or smaller block duplications and family expansions continues to be debated. We analyzed the complete genome of the pufferfish Takifugu rubripes (Fugu) and compared it with the paranome of humans. We show that most paralogous genes of Fugu are the result of three complete genome duplications. Both relative and absolute dating of the complete predicted set of protein-coding genes suggest that initial genome duplications, estimated to have occurred at least 600 million years ago, shaped the genome of all vertebrates. In addition, analysis of >150 block duplications in the Fugu genome clearly supports a fish-specific genome duplication (≈320 million years ago) that coincided with the vast radiation of most modern ray-finned fishes. Unlike the human genome, Fugu contains very few recently duplicated genes; hence, many human genes are much younger than fish genes. This lack of recent gene duplication, or, alternatively, the accelerated rate of gene loss, is possibly one reason for the drastic reduction of the genome size of Fugu observed during the past 100 million years or so, subsequent to the additional genome duplication that ray-finned fishes but not land vertebrates experienced.
Ohno believed that duplications of genes and genomes are more important in shaping the evolution of novelty and complexity than what he considered to be only modifying forces of natural selection (1). Although based on rather inaccurate indicators, such as genome size and isozyme complexity, he suggested that the genomes of vertebrates have been shaped by two complete genome duplications, one on the shared lineage leading to both cephalochordates and vertebrates and a second one at the “fish or amphibian” line. Later, important indications for two rounds of large-scale gene duplications in the early vertebrates came from the analysis of Hox genes and Hox gene clusters (2). The observation that protostome invertebrates and the deuterostome cephalochordate Amphioxus possess a single Hox cluster, whereas the lobe-finned fish, such as the coelacanth and lungfishes, amphibians, reptiles, birds, and mammals have four clusters (3, 4), supports the hypothesis of two rounds (2R) of entire-genome duplications early in vertebrate evolution, although Holland et al. (2) proposed that a first duplication occurred after the divergence of the cephalochordates, and a second one occurred after the divergence of the jawless vertebrates. Since then, evidence for and against the 2R hypothesis has been put forward and several modifications have been proposed, assuming a diversity of small- and large-scale gene duplication events. Based on quadruplicate paralogy between different genomic segments, some have strongly argued for 2R (5, 6), whereas others, often analyzing the same data but using different techniques, found only clear evidence for one genome-doubling event early in the evolution of vertebrates (7-9). Still others reject whole-genome duplications in vertebrates all together and only accept a continuous rate of gene duplication (10, 11). As a consequence, the 2R hypothesis of vertebrate genome evolution is still vividly debated, and opinions range from strong belief (12-14) to strong skepticism (15-17).
A decade ago, Brenner et al. (18) proposed to sequence the pufferfish genome as a cost-effective way to identify and characterize genes in the human genome. The pufferfish Takifugu rubripes (Fugu) genome is only about one-eighth the size of the human genome but was expected to contain a similar gene repertoire. However, the discovery of “extra” Hox gene clusters in other ray-finned fishes (19-21), together with mapping data (19, 22-25) and the inference of phylogenetic trees (26, 27), recently suggested that the genomes of ray-finned fishes might be considerably different from those of their sister group, the land vertebrates, because of an additional genome duplication in their evolutionary past (19, 28-30). Others have argued that an ancestral whole-genome duplication event might not be responsible for the abundance of duplicated fish genes. For example, Robinson-Rechavi and coworkers (31, 32) counted orthologous genes in fish and mice and, where extra genes were found in fish, compared the number of gene duplications occurring in a single fish lineage with the number of gene duplications shared by more than one lineage. They found that most mouse genes surveyed occurred only once in fish. Duplicated fish genes were detected, but most were the products of lineage-specific duplication events and not an ancient duplication event. Preliminary analysis of the draft sequence of the Fugu genome on its release also did not provide evidence for an entire genome duplication event in Fugu, although segmental block duplications were detected (30). However, the recent publication of this draft sequence of the genome of the pufferfish (30) now allows comparison of the paranomes (the complete set of duplicated genes in a genome) of ray-finned fishes and land vertebrates. We have studied the differences in genome evolution between fish and land vertebrates to address hypotheses about large-scale gene duplications (i.e., chromosomal block duplications and polyploidy events) in both early vertebrates and ray-finned fishes.
Materials and Methods
Gene Families in Fugu and Humans. We retrieved a total of 8,597 Fugu scaffolds (total of 319 megabases) containing 34,615 genes, from Ensembl (www.ensembl.org; Fugu release 13.2.1). If, for one gene, multiple transcripts were reported due to splice variants, only the longest transcript was used to represent that gene. The 24,847 predicted protein sequences from the human genome were downloaded from Ensembl (release 13.31.1).
To get a general overview of all duplications in the complete Fugu and human genomes, gene families of paralogous proteins had to be created. Therefore, we carried out a similarity search (blastp) from all Fugu and human proteins against the full set of Fugu and human proteins, respectively. Gene families were created based on sequence similarity (33). Only gene families with two to ten paralogous genes were used for further analysis. Larger gene families were ignored because they often pose problems in the reliable construction and automatic interpretation of phylogenetic trees. Furthermore, such large gene families represent only a small fraction (i.e., 3.4%) of the total number of gene families found. To enrich these gene families with sequences from other organisms, all proteins present in these families were used as query sequences in a blastp search carried out against a protein database containing all proteins available from Fugu (Ensembl release 13.2.1), all other fish species available in Swiss-Prot [all proteins from ray-finned fishes, excluding T. rubripes, downloaded from Swiss-Prot plus TrEMBL (release February 4, 2003; http://us.expasy.org/sprot/)], human (Ensembl, release 13.31.1), mouse (Ensembl, release 14.30.1), Ciona (release 1.0 from http://genome.jgips-f.org/ciona4/ciona4.home.html), and Drosophila (Ensembl release 13.3.1). Either Drosophila or Ciona sequences were used as outgroup sequences (see below). To avoid adding local domain hits to our subfamily classification, the alignable regions for every possible homolog had to cover at least 50% of the length of the query sequence. Because the focus of this study was to identify genes that were duplicated during vertebrate evolution, only blast hits giving a higher score than the sequence of Drosophila were retained. Finally, to date the duplication events before or after the divergence between ray-finned fishes and land vertebrates, gene families had to contain at least two Fugu genes, one land vertebrate sequence (a so-called calibration point), and one outgroup sequence. For the analysis of the human genome, gene families had to contain at least two human genes, one fish sequence (as calibration), and one outgroup sequence.
Once the different gene families were defined, alignments were created by using clustalw 1.82. Alignment columns containing gaps were removed when a gap was present in >10% of the sequences. To reduce the chance of including misaligned amino acids, all positions in the alignment left or right from the gap were also removed until a column in the sequence alignment was found where the residues were conserved in all genes included in our analyses. This was determined as follows: For every pair of residues in the column, the blosum62 value was retrieved. Next, the median value for all these values was calculated. If this median was ≥0, the column was considered as containing homologous amino acids. Finally, only alignments with >50 positions were retained for phylogenetic analysis.
Dating Duplication Events. Dating gene duplication events in the Fugu and humans was done by phylogenetic means, considering both relative and absolute dating. Relative dating was performed by construction of neighbor-joining trees by using lintree (34) for every gene family, based on Poisson corrected evolutionary distances. Empirical analyses on a subset of gene families showed that the use of an estimated α-value for γ-correction instead of Poisson correction gave the same results. All trees were bootstrapped 1,000 times.
Inferring the absolute duplication date of paralogous genes was based on the construction of linearized trees (34), which assumes equal rates of evolution in different lineages of the tree, i.e., a molecular clock. Therefore, all sequences in the tree were tested for their homogeneous rate of evolution. To create these linearized trees, the two-cluster test and branch-length test for rate heterogeneity were applied to these trees to test for deviations from the molecular clock at 1% significance (34). Faster or more slowly evolving sequences were removed and the procedure was repeated until the data set contained only sequences evolving at a similar rate. Only trees significantly supported by bootstrap analysis (BS ≥ 70%) for the relevant branches were retained. As a reference point for dating, the divergence time between ray-finned fishes and land vertebrates, i.e., 450 million years, was used, because this divergence date is well agreed on, both on the basis of fossil data (35-39) and on the basis of molecular clocks (40). By comparing the divergence of duplicated genes with this calibration point, the absolute date of origin of paralogous genes can be inferred. In some trees, we found more than one node representing the split between ray-finned fishes and land vertebrates. In these cases, the duplication date was calculated by using the mean evolutionary distance of all these calibration points. A flowchart, describing the whole automated procedure of building data sets and dating duplication events on the basis of constructed phylogenetic trees, is given in Fig. 1. All sequence alignments and nonlinearized and linearized phylogenetic trees can be found at our web site (www.psb.ugent.be/bioinformatics).
Fig. 1.
Flowchart describing the automatic procedure for data selection, tree construction, and relative and absolute dating of duplication events. Numbers in bold denote the number of retained data sets or trees including Fugu duplicates (left, all duplicated genes; right, only duplicated genes in block duplications) after different steps of the procedure. In some cases, the number of nodes that could be used for dating was larger than the number of trees, because some data sets contained more than two Fugu paralogs. As a result, the box denoted as “unambiguously dated duplications” refers to the number of nodes in trees that could be used to date duplication events. Supporting material, showing results at different stages of the process can be found at www.psb.ugent.be/bioinformatics.
Detection of Block Duplications. For the detection of duplicated segments, only scaffolds with five or more genes were retained, which reduced, for this part of the analysis, our Fugu data set from 6,788 to 2,029 scaffolds (225.3 megabases), representing 25,170 genes (≈71% of the genome). To identify colinear regions in the Fugu genome, pointing to block or segmental duplications, the ADHoRe algorithm (41) was used, with parameters Q = 0.9 and G = 25. Parameter G refers to the maximum distance (in number of intervening “unique” genes) between two pairs of homologous genes in a duplicated segment, whereas Q refers to how well the different pairs of homologous genes fit on a single diagonal line in a gene homology matrix, i.e., a dot plot for homologous genes (41). The Q value was set at 0.9 based on previous similar analyses of the Arabidopsis and rice genomes (42, 43). Only block duplications that had a probability to be generated by chance <0.1% (or a significance of 99.9%) were retained in the analysis. For the determination of the number of tandem duplications, only homologous genes (i.e., belonging to the same gene family; see above) with 25 or fewer nonhomologous intervening genes were considered.
Results and Discussion
Phylogenetic trees were constructed for all (i.e., 3,077) gene families containing two to ten duplicated Fugu genes. Larger gene families were ignored because the automatic interpretation of the tree topology often becomes too complicated. Furthermore, such large gene families represent only a small fraction (i.e., 3.4%) of the total number of gene families found. For each gene family in Fugu, first, relative dating of duplication events was performed to test whether gene duplications occurred before or after the divergence of the lineages that led to ray-finned fishes and land vertebrates, subsequently referred to as 1R/2R and 3R, respectively (Fig. 2). To this end, neighbor-joining trees were created for each of the Fugu gene families with homologous sequences from mice and humans (as representatives for the land vertebrates), different genes of several species of ray-finned fishes, which were available in the databases, Ciona, and Drosophila (see Materials and Methods). After elimination of trees that were not supported by a significant bootstrap value (BP ≥ 70), 752 gene families were available for relative dating (Fig. 1).
Fig. 2.
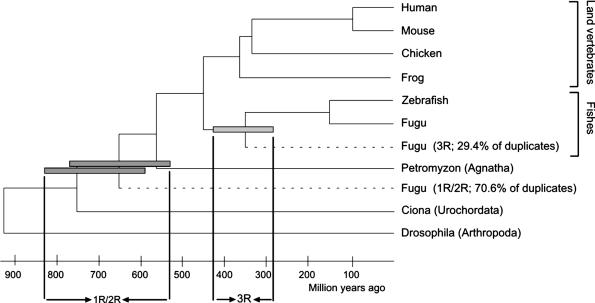
Phylogenetic tree of major vertebrate groups and superimposed Fugu gene duplication events. Dark and light gray bars denote large-scale gene duplication events observed in the Fugu genome based on absolute and relative dating and the detection of segmental duplications (see text for details). The time of divergence for Petromyzon, as a representative of the Agnatha, was taken from ref. 51.
Absolute dating of duplication events was performed through the inference of linearized trees (ref. 34; see Materials and Methods). In these linearized trees, where branch length is directly proportional to time, the split between ray-finned fishes and land vertebrates, dated at 450 million years ago (mya) (35-37), was used as a calibration point for the dating of gene duplication events. After removing trees with bootstrap values of <70%, for the relevant branches, an absolute date could be inferred for 595 nodes, based on the analyses of 488 gene families. Combining the results of relative and absolute dating, we then subdivided 565 duplication events for which an absolute date could be inferred into 166 3R and 399 1R/2R duplications (Figs. 1, 2, 3).
Fig. 3.
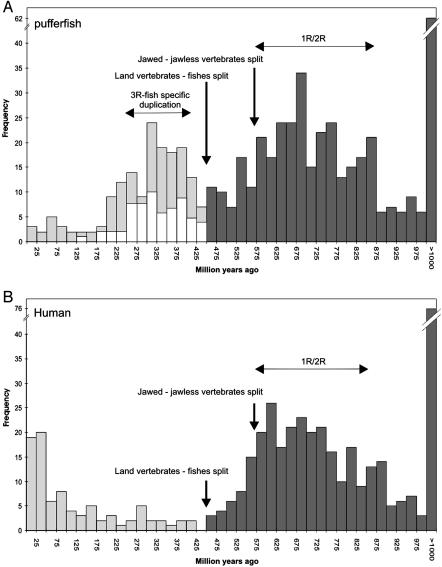
Age distribution of duplicated genes in the Fugu (A) and human (B) genome. Black bars correspond to duplications before the divergence of ray-finned fishes and land vertebrates; gray bars correspond to duplications after the split between ray-finned fishes and land vertebrates. White bars (A) refer to duplicated Fugu genes that are part of block (chromosomal block) duplications (see text for details).
A major fraction of the paralogs (i.e., 30%) is younger than the split between ray-finned fishes and land vertebrates and seems to have arisen between 225 and 425 mya. The most plausible and parsimonious explanation for this observation would be a large-scale gene or entire-genome duplication. To test whether the sudden increase in the number of duplicated genes in the Fugu genome (Fig. 3A) is the result of an entire-genome duplication rather than an increased rate of independent tandem duplications events, we investigated whether these duplicated genes appear in duplicated blocks on chromosomes. To this end, we identified statistically significant regions of microcolinearity (showing the same gene content and gene order) within the complete Fugu genome. All genes within such a region are supposed to be duplicated at the same time and hence of identical age, because it is unlikely that these colinear regions are created independently on different chromosomes. By applying the ADHoRe algorithm to scaffolds of the available pufferfish genome sequence (see Materials and Methods), 159 statistically significant duplicated blocks (i.e., those that contained at least three homologous gene pairs with a significant density) were identified (41). The complete set of block duplications, without tandem duplications, contains 544 paralogous gene pairs (so-called anchor points), which is an average of 3.4 anchor points per block and includes 8.0% (2,024) of all Fugu proteins used in this analysis (or 9.2% of all of the base pairs used). To date the origin of all these 159 chromosomal blocks, a similar phylogenetic approach was applied to the set of Fugu anchor points (paralogous genes within duplicated blocks) as before on all Fugu paralogs (see Materials and Methods). Four hundred and six subfamilies, this time involving two Fugu anchor points, at least one land vertebrate sequence (human or mouse), and one outgroup sequence (Drosophila or Ciona), were created. Regarding Fugu anchor points, the topologies of 148 neighbor-joining trees could be used for relative dating. An absolute duplication date for these anchor points was obtained by constructing linearized trees for 99 gene families (Fig. 1). Considering all 59 3R-dated anchor points (representing 50 duplicated chromosomal blocks), for which an absolute date could be inferred and no conflict is observed among different relative dates between genes within the same duplicated block, we find that the 3R blocks of duplicated genes all arose at approximately the same time, i.e., at ≈320 mya with a standard deviation of 67 million years.
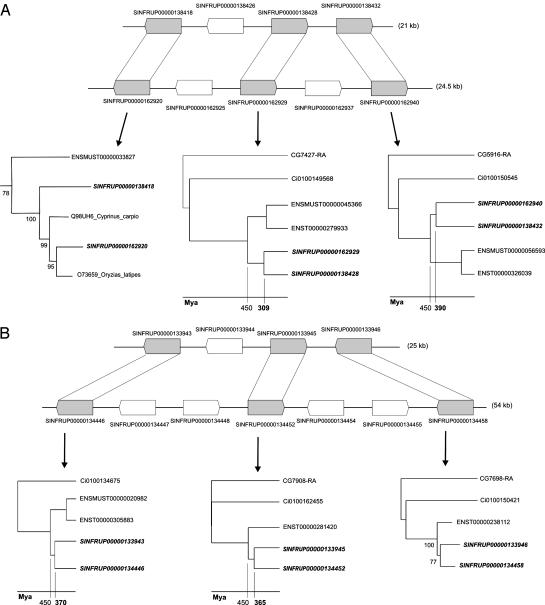
One might argue that a standard deviation of 67 million years is rather large and could point to independent block duplications, rather than all block duplications having originated from the same genome duplication event. However, when using an absolute dating approach, such as applied in this and other studies (7, 8), such a variance on estimated duplication times is expected. This finding is clarified by Fig. 4, which shows two duplicated blocks in the Fugu genome. Both block duplications contain homologous genes that have been duplicated at the same time, yet, at least for one block, a considerable difference exists in the estimated duplication date for different genes, namely 309 and 390 million years (Fig. 4A). The main reason for such differences is deviation of the molecular clock, by which some genes evolve slower or faster and have different rates in different evolutionary lineages, although we have taken into account the latter phenomena through the construction of linearized trees. For example, in both Fig. 4, for one anchor point, an absolute date could not be inferred, because (at least) one of the Fugu or land vertebrate sequences in the data set did not pass the molecular clock test as implemented in the linearized tree approach. Nevertheless, smaller deviations of the molecular clock, accepted by linearized tree construction, in 300 or so million years of evolution are to be expected and can result in considerably different inferred duplication times (as shown in Fig. 4A). Fig. 4B shows a counterexample, where both duplication date estimates are congruent, even almost identical, i.e., 370 and 365 million years. However, in general, for many duplicated blocks, quite a large difference exists in the estimated duplication date, although we can show, with statistical significance, that all duplicated genes in such a block have been created simultaneously. We have plotted the number of duplicated genes in blocks against their estimated age by which a distribution is obtained that is very similar to the distribution based on all duplicated Fugu genes (Fig. 3A). Forty-five percent of all paralogs dated between 225 and 425 mya are located in blocks such as the ones shown in Fig. 4. Given the very fragmented nature of the Fugu scaffold data set (see Materials and Methods), the number of duplicated blocks is probably much higher and is expected to rise considerably, once better assemblies of the Fugu genome become available. This finding suggests that the observation of the rather wide distribution of duplicated Fugu genes as observed is in perfect agreement with the hypothesis of a single, complete genome duplication event. Therefore, we believe that the observed peak in our age distribution of duplicated fish genes provides very strong support for a complete genome duplication event in the early stages of fish evolution, predating the origin of most modern ray-finned fish species that are believed to have (started to) diverge(d) from each other >200 mya (44).
Fig. 4.
Example of two duplicated blocks in the Fugu genome. Genes in gray represent anchor points (retained homologs). To infer the (relative) duplication date for every anchor point, phylogenetic trees are constructed. Neighbor-joining trees show that these blocks are the result of a fish-specific duplication event. For absolute dating, if possible, linearized trees are constructed, with branch lengths directly proportional to time. Although all genes in a duplicated region originated at the same time, a considerable deviation of the inferred duplication times can be observed between different anchor points (A). Genes represented in bold are Fugu genes. ENSMUS, ENST, Ci, and CG represent mouse, human, Ciona, and Drosophila genes, respectively. Other genes correspond to different fish species.
The age distribution of duplicated genes also supports the hypothesis of a second wave of duplication events between 525 and 875 mya. Several authors have suggested that, in early vertebrate evolution, one or two complete genome duplications may have occurred (1, 2, 5-9, 12-14, 45). The sharp increase in the number of duplicates seems to point to the remnants of at least one and maybe even two large-scale genome duplication events early in the evolution of chordates (see below). To compare the paranome of Fugu with that of a land vertebrate species, a similar approach to date duplication events by phylogenetic means was applied to the human genome. For the human genome, 2,873 gene families containing 2-10 human family members could be identified based on 8,138 proteins. As with the Fugu genome, neighbor-joining trees were created for each of these gene families and after bootstrap validation (BS ≥ 70%), the topologies of 707 human trees were used for relative dating. Absolute dating of duplicated genes using linearized trees resulted in 447 duplication events, of which 87 are specific to humans and 360 are attributed to 1R/2R (see Fig. 3B). The distribution of inferred ages of duplicated genes shows a similar increase in the number of duplication events ≈675 mya, as observed in Fugu. As expected, the fish-specific genome duplication event cannot be detected in the human genome (Fig. 3B). On the other hand, the human genome clearly contains a higher number of recent duplicates than the Fugu genome. If (tandem) duplications are a continuous process during evolution (8, 46) and occur with an average frequency of 0.01 per gene per million years, and an exponential decay of the number of retained gene duplicates over time can be expected, as observed by Lynch and Conery (46), one would expect a high number of recent duplicates in both the human and the Fugu genomes. Although this trend can be observed in the human genome (Fig. 3B), it is absent in the Fugu genome (Fig. 3A). The fact that a smaller-than-expected number of recently duplicated genes occur in the Fugu genome might be due to its extreme tendency for compaction of its genome (30). Neafsey and Palumbi (47) recently demonstrated that the genomes within the family Tetraodontideae, including the smooth pufferfish T. rubripes used in this study, have undergone a major contraction in the past 50-70 million years. They explain this drop in genome size by a reduction of large insertions and by a higher rate of deletions. Alternatively, the latter phenomenon might also have been responsible for the fast removal of redundant copies of duplicated genes in the Fugu genome. In this respect, it is interesting to note that the number of tandem duplications in the human genome is ∼7-fold higher than the number of tandem duplications in the Fugu genome, namely 1,248 tandem duplications involving 3,927 genes in human versus 268 tandem duplications covering 546 genes in Fugu. The recent peak of duplication activity observed in the human genome is also consistent with the results of Eichler (48), who described an increased rate of segmental and tandem duplications in primate genomes.
The relative and absolute dating of almost 500 different gene families together with the detection of many duplicated blocks that originated at about the same time provides strong support for the hypothesis of a fish-specific genome duplication ≈320 mya that was not experienced in the lineage of vertebrates leading to humans. We showed that this genome duplication event (3R) accounts for the large majority of retained Fugu gene duplicates, contrary to the situation in the human genome where many more recent tandem and segmental duplication events (49) account for the majority of our genes (Fig. 3). Most other Fugu paralogs seem to have been created by one or two much older large-scale duplication events, predating the split between ray-finned fishes and land vertebrates. By using the fish-specific genome duplication as a benchmark, and assuming equal rates of gene loss throughout vertebrate evolution, two genome duplications rather than one seem to have occurred, as originally proposed by Ohno (1) in 1970, and later corroborated by the observation of quadruplicate paralogy between different parts of vertebrate genomes (5-7). Indeed, ≈70.6% of the Fugu duplicates are dated between 500 and 900 million years, whereas Fugu duplicates that originated between 250 and 450 million years only account for 29.4%. However, one would expect that, if two genome duplications had taken place in the early evolution of vertebrates, the distribution of ancient duplicates should show two peaks instead of one (Fig. 3). This assumption is not necessarily true. Some advocates of the 2R hypothesis believe that the two rounds of genome duplications occurred in very short succession (6). This finding would also explain why it is generally hard to infer phylogenetic trees of the form [(A,B)(C,D)], which are to be expected if two tetraploidy events had happened (11, 15, 50). If both genome doublings indeed took place almost contemporaneously, it is not surprising that we cannot discriminate, based on age differences between genes or the topology of gene family trees, between two genome duplication events early in vertebrate evolution. It would be nearly impossible to discriminate between two almost contemporaneous events that happened such a long time ago. Furthermore, because the two polyploidy events occurred maybe only within 10 million years, as suggested by some (6), and these events took place >600 mya, the dating of duplications events in the early vertebrate evolution will show an even larger variance (as observed in Fig. 3A) than is the case with duplicates that are 300 million years old and the result of one fish-specific genome duplication.
Acknowledgments
We thank Cedric Simillion for technical assistance and Jeff Boore and Michael Lynch for discussion and comments on an earlier version of the manuscript. K.V. and W.D.V. thank the Vlaams Instituut voor de Bevordering van het Wetenschappelijk-Technologisch Onderzoek in de Industrie for a predoctoral fellowship. J.S.T. was supported by a Natural Sciences and Engineering Research Council (Canada) Discovery Grant, and A.M. was supported by the Deutsche Forschungsgemeinschaft.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: mya, million years ago; Fugu, pufferfish Takifugu rubripes.
References
- 1.Ohno, S. (1970) Evolution by Gene Duplication (Springer, New York).
- 2.Holland, P. W., Garcia-Fernandez, J., Williams, N. A. & Sidow, A. (1994) Development (Cambridge, U.K.) 1994, Suppl., 125-133. [PubMed] [Google Scholar]
- 3.Garcia-Fernandez, J. & Holland, P. W. (1996) Int. J. Dev. Biol. 1996, Suppl. 1, 71S-72S. [PubMed] [Google Scholar]
- 4.Holland, P. W. (1997) Curr. Biol. 7, R570-R572. [DOI] [PubMed] [Google Scholar]
- 5.Abi-Rached, L., Gilles, A., Shiina, T., Pontarotti, P. & Inoko, H. (2002) Nat. Genet. 31, 100-105. [DOI] [PubMed] [Google Scholar]
- 6.Lundin, L. G., Larhammar, D. & Hallbook, F. (2003) J. Struct. Funct. Genomics 3, 53-63. [PubMed] [Google Scholar]
- 7.McLysaght, A., Hokamp, K. & Wolfe, K. H. (2002) Nat. Genet. 31, 200-204. [DOI] [PubMed] [Google Scholar]
- 8.Gu, X., Wang, Y. & Gu, J. (2002) Nat. Genet. 31, 205-209. [DOI] [PubMed] [Google Scholar]
- 9.Panopoulou, G., Hennig, S., Groth, D., Krause, A., Poustka, A. J., Herwig, R., Vingron, M. & Lehrach, H. (2003) Genome Res. 13, 1056-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman, R. & Hughes, A. L. (2001) Genome Res. 11, 1842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes, A. L. (1999) J. Mol. Evol. 48, 565-576. [DOI] [PubMed] [Google Scholar]
- 12.Furlong, R. F. & Holland, P. W. (2002) Philos. Trans. R. Soc. London B 357, 531-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larhammar, D., Lundin, L. G. & Hallbook, F. (2002) Genome Res. 12, 1910-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spring, J. (2003) J. Struct. Funct. Genomics 3, 19-25. [PubMed] [Google Scholar]
- 15.Martin, A. (2001) Mol. Biol. Evol. 18, 89-93. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, A. L., da Silva, J. & Friedman, R. (2001) Genome Res. 11, 771-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman, R. & Hughes, A. L. (2003) Mol. Biol. Evol. 20, 154-161. [DOI] [PubMed] [Google Scholar]
- 18.Brenner, S., Elgar, G., Sandford, R., Macrae, A., Venkatesh, B. & Aparicio, S. (1993) Nature 366, 265-268. [DOI] [PubMed] [Google Scholar]
- 19.Amores, A., Force, A., Yan, Y. L., Joly, L., Amemiya, C., Fritz, A., Ho, R. K., Langeland, J., Prince, V., Wang, Y. L., et al. (1998) Science 282, 1711-1714. [DOI] [PubMed] [Google Scholar]
- 20.Naruse, K., Fukamachi, S., Mitani, H., Kondo, M., Matsuoka, T., Kondo, S., Hanamura, N., Morita, Y., Hasegawa, K., Nishigaki, R., et al. (2000) Genetics 154, 1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Málaga-Trillo, E. & Meyer, A (2001) Am. Zool. 41, 676-686. [Google Scholar]
- 22.Postlethwait, J. H., Woods, I. G., Ngo-Hazelett, P., Yan, Y. L., Kelly, P. D., Chu, F., Huang, H., Hill-Force, A. & Talbot, W. S. (2000) Genome Res. 10, 1890-1902. [DOI] [PubMed] [Google Scholar]
- 23.Woods, I. G., Kelly, P. D., Chu, F., Ngo-Hazelett, P., Yan, Y. L., Huang, H., Postlethwait, J. H. & Talbot, W. S (2000) Genome Res. 10, 1903-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, S. F., Snell, P., Gruetzner, F., Bench, A. J., Haaf, T., Metcalfe, J. A., Green, A. R. & Elgar, G. (2002) Genome Res. 12, 776-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elgar, G., Clark, M. S., Meek, S., Smith, S., Warner, S., Edwards, Y. J., Bouchireb, N., Cottage, A., Yeo, G. S., Umrania, Y., et al. (1999) Genome Res. 9, 960-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor, J. S., Braasch, I., Frickey, T., Meyer, A. & Van de Peer, Y. (2003) Genome Res. 13, 382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor, J. S., Van de Peer, Y., Braasch, I. & Meyer, A. (2001) Philos. Trans. R. Soc. London B 356, 1661-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wittbrodt, J., Meyer, A. & Schartl, M. (1998) BioEssays 20, 511-512. [Google Scholar]
- 29.Meyer, A. & Schartl, M. (1999) Curr. Opin. Cell Biol. 11, 699-704. [DOI] [PubMed] [Google Scholar]
- 30.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J. M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. (2002) Science 297, 1301-1310. [DOI] [PubMed] [Google Scholar]
- 31.Robinson-Rechavi, M., Marchand, O., Escriva, H. & Laudet, V. (2001) Curr. Biol. 11, R458-R459. [DOI] [PubMed] [Google Scholar]
- 32.Robinson-Rechavi, M., Marchand, O., Escriva, H., Bardet, P. L., Zelus, D., Hughes, S. & Laudet, V. (2001) Genome Res. 11, 781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, W. H., Gu, Z., Wang, H. & Nekrutenko, A. (2001) Nature 409, 847-849. [DOI] [PubMed] [Google Scholar]
- 34.Takezaki, N., Rzhetsky, A. & Nei, M. (1995) Mol. Biol. Evol. 12, 823-833. [DOI] [PubMed] [Google Scholar]
- 35.Carroll, R. L. (1988) Vertebrate Paleontology and Evolution (Freeman, New York).
- 36.Benton, M. J. (1990) J. Mol. Evol. 30, 409-424. [DOI] [PubMed] [Google Scholar]
- 37.Zhu, M., Yu, X. & Janvier, P. (1999) Nature 397, 607-610. [Google Scholar]
- 38.Schultze, H.-P. & Cumbaa, S. L. (2001) in Major Events in Early Vertebrate Evolution, ed. Ahlberg, P. E. (Taylor and Francis, London), pp. 315-332.
- 39.Zhu, M. & Yu, X. (2002) Nature 418, 767-770. [DOI] [PubMed] [Google Scholar]
- 40.Hedges, S. B. & Kumar, S. (2003) Trends Genet. 19, 200-206. [DOI] [PubMed] [Google Scholar]
- 41.Vandepoele, K., Saeys, Y., Simillion, C., Raes, J. & Van de Peer, Y. (2002) Genome Res. 12, 1792-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simillion, C., Vandepoele, K., Van Montagu, M. C., Zabeau, M. & Van De Peer, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 13627-13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vandepoele, K., Simillion, C. & Van de Peer, Y. (2003) Plant Cell 15, 2192-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrol, R. L. (1997) Patterns and Processes of Vertebrate Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 45.Spring, J. (1997) FEBS Lett. 400, 2-8. [DOI] [PubMed] [Google Scholar]
- 46.Lynch, M. & Conery, J. S. (2000) Science 290, 1151-1155. [DOI] [PubMed] [Google Scholar]
- 47.Neafsey, D. E. & Palumbi, S. R. (2003) Genome Res. 13, 821-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eichler, E. E. (2001) Trends Genet. 17, 661-669. [DOI] [PubMed] [Google Scholar]
- 49.Bailey, J. A., Gu, Z., Clark, R. A., Reinert, K., Samonte, R. V., Schwartz, S., Adams, M. D., Myers, E. W., Li, P. W. & Eichler, E. E. (2002) Science 297, 1003-1007. [DOI] [PubMed] [Google Scholar]
- 50.Skrabanek, L. & Wolfe, K. H. (1998) Curr. Opin. Genet. Dev. 8, 694-700. [DOI] [PubMed] [Google Scholar]
- 51.Shu, D.-G., Luo, H.-L., Conway Morris, S., Zhang, X. L., Hu, S.-X., Chen, L., Han, J., Zhu, M., Li, Y. & Chen, L. Z. (1999) Nature 402, 42-46. [Google Scholar]