Abstract
There are two main pathways in eukaryotic cells for the repair of DNA double-strand breaks: homologous recombination and nonhomologous end joining. Because eukaryotic genomes are packaged in chromatin, these pathways are likely to require the modulation of chromatin structure. One way to achieve this is by the acetylation of lysine residues on the N-terminal tails of histones. Here we demonstrate that Sin3p and Rpd3p, components of one of the predominant histone deacetylase complexes of Saccharomyces cerevisiae, are required for efficient nonhomologous end joining. We also show that lysine 16 of histone H4 becomes deacetylated in the proximity of a chromosomal DNA double-strand break in a Sin3p-dependent manner. Taken together, these results define a role for the Sin3p/Rpd3p complex in the modulation of DNA repair.
In eukaryotic cells, the genome is compacted into chromatin. The basic unit of chromatin is the nucleosome, which is composed of the conserved core histones H2A, H2B, H3, and H4. The structure of nucleosomal DNA is further compacted by a range of other factors, including proteins such as the linker histone H1 that binds to DNA between the nucleosome repeats (1). Various DNA-templated processes such as transcription, replication, and DNA repair take place in the context of chromatin and so must involve the manipulation of nucleosomes. One way to achieve this is through the reversible acetylation, phosphorylation, ubiquitination, or ADP-ribosylation of the histone tails that extend to the accessible surface of the nucleosome core particle (2). It has been proposed that combinations of these modifications create a “histone code” that is recognized by other proteins to bring about specific downstream events (3).
The most extensive body of data has been accumulated for the effect of nucleosome modifications in transcription (4). However, there is growing evidence that there is cross-talk between chromatin and DNA damage response and repair pathways. This is particularly well established for nucleotide excision repair (NER), in which several chromatin remodeling and modification factors have been implicated (ref. 5 and references therein). As discussed further below, there is also evidence for the importance of chromatin modification in the repair of DNA double-strand breaks (DSBs), which are generated by ionizing radiation and a range of chemotherapeutic agents. Given the highly recombinogenic and cytotoxic nature of such lesions (6, 7), the impact of chromatin on DSB repair is likely to be of considerable biological importance.
There are two principle mechanisms for the repair of DNA DSBs: homologous recombination (HR) and nonhomologous end joining (NHEJ). The former requires genes in the RAD52 epistasis group and relies on extensive homology between the damaged DNA and an undamaged partner [sister chromatid or homologous chromosome (8)]. By contrast, NHEJ requires little or no homology between the recombining molecules (9). Key components of the NHEJ machinery in mammals include the Ku70/Ku80 heterodimer and the Ligase IV/XRCC4 complex, all of which have functional counterparts in yeast [Yku70p, Yku80p, Dnl4p, and Lif1p, respectively (10)].
The role of chromatin modification in DNA DSB repair is probably best demonstrated by the rapid phosphorylation of the histone variant H2AX in mammalian cells after the formation of DNA DSBs (11, 12). A similar response has been documented in budding yeast, in which the C terminus of the histone H2A undergoes DNA damage-dependent phosphorylation and is required for efficient NHEJ (13). Although no other histone modification has yet been strongly implicated in DNA DSB repair, there are indications that proteins that mediate histone acetylation affect the NHEJ pathway. For example, it was recently demonstrated that the budding yeast histone acetyltransferase (HAT), Esa1p, is required for normal NHEJ (14). Significantly, the mammalian Esa1p homolog, TIP60, is required for apoptosis and DNA DSB repair (15). However, it is not yet clear whether TIP60 impinges on NHEJ or HR, or both. There is also some indirect evidence that the human histone acetyltransferase, hGCN5, might function in NHEJ, because the mammalian NHEJ protein DNA-PKcs (DNA-dependent protein kinase catalytic subunit) (16) interacts with and phosphorylates hGCN5, leading to reduction of hGCN5 HAT activity (17). Histone deacetylases (HDACs) have also been implicated in DNA DSB repair. For example, Sir2p, an NAD-dependent HDAC, and its interacting partners, Sir3p and Sir4p, influence NHEJ. It was shown that cells lacking these factors exhibit a defect in NHEJ, and furthermore, chromatin immunoprecipitation (ChIP) studies showed that these factors relocalize from telomeres to the site of DNA DSBs (18-21). Despite this localization, it is now clear that the defect of cells lacking these factors in NHEJ is mainly indirect because deletion of the SIR genes leads to pseudodiploidy in yeast as a result of loss of silencing at the silent mating type loci (22, 23). This results in expression of the a/α transcriptional regulator that suppresses a number of haploid-specific genes, including NEJ1, which encodes for a protein required for NHEJ (24, 25). Therefore, the main defect in NHEJ in sir mutants is due to the loss of silencing at the silent mating type loci. The exact function of the Sir protein localization to DNA DSBs is yet to be elucidated.
In light of the above, we decided to establish whether the Saccharomyces cerevisiae Sin3p/Rpd3p HDAC complex influences NHEJ. Previous work has shown that these factors interact with other proteins to form a heterogeneous multiprotein complex of >1 MDa that is recruited to the promoters of target genes by the DNA-binding protein Ume6p (26-28). The deacetylase activity of Rpd3p then results in the formation of a localized domain of hypoacetylated chromatin that is thought to inhibit transcription by preventing the recruitment of the transcriptional machinery (29-32). Significantly, genomewide localization studies have revealed that, apart from residing on the promoter regions of its target genes, the Sin3p/Rpd3p complex also binds to nonpromoter sequences independently of Ume6p. This finding indicates that the Sin3p/Rpd3p complex is capable of binding to the substrate histones directly or to other nucleosome binding proteins (28, 33) and raises the possibility that the complex might control other events in addition to transcription. Here we demonstrate that Sin3p and Rpd3p are required for normal levels of survival in response to DNA DSBs, and specifically influence the repair of these lesions by NHEJ. Furthermore, we establish that Sin3p is required for the efficient deacetylation of histone H4 lysine 16 in the vicinity of a chromosomal DNA DSB.
Materials and Methods
Yeast Strains. Standard genetic techniques were used for manipulating yeast strains. Media and growth of yeast strains were as described (34, 35). Yeast strains used are all in the W303 background. sin3Δ, yku80Δ, rad52Δ, and double mutants were constructed by one-step gene deletion method. Strains JKM179 and MK203 have been described (36, 37).
Phleomycin, Hydroxyurea (HU), UV, and Gal-HO Sensitivity Assays. Overnight cultures were diluted to A600 of 0.3, and 7 μl of 5-fold serial dilutions were spotted on yeast extract/peptone/dextrose plus adenine (YPAD) plates containing phleomycin (2.5 or 5 μg/ml) (Melford Laboratories, Chelsworth, U.K.) or HU (20 or 200 mM) (Sigma). For the UV-sensitivity experiment, the same amounts of cells were plated on YPAD plates that subsequently were placed under a UV lamp and irradiated at 254 nm and a delivery rate of 2.5 J/m2/sec. For galactose-inducible HO sensitivity experiments, cells were grown in the preinduction medium (YP plus 3% glycerol) first, then cultures were diluted to A600 of 0.7, and 5-fold serial dilution were plated either on YPA plates containing glucose or galactose. Plates were incubated at 30°C for 4-5 days.
Plasmid Repair Assay. These experiments were performed essentially as described (38). Briefly, the yeast-Escherichia coli shuttle plasmid pBTM116, which contains TRP1 for selection in yeast, was digested with the appropriate restriction enzyme to completion, and the enzyme was inactivated by treatment at 65°C for 20 min. One microgram of linearized DNA was used to transform each strain with the lithium acetate method (34). In parallel, the same amount of cells was transformed with the same amount of uncut plasmid to normalize for differences in transformation efficiency. Diluted samples were plated on minimal media lacking the appropriate amino acids, and colonies were counted after incubation at 30°C for 4-5 days. To test the accuracy of the repair DNA from single yeast transformants was isolated as described (34), and this was used to transform E. coli XL1-Blue cells (Stratagene) to ampicillin resistance. Plasmid DNA was then isolated and analyzed by restriction enzyme digestion.
ChIP. Cultures of WT and sin3Δ cells in the JKM179 background were grown to mid-log in rich medium containing 3% glycerol. Cells were then resuspended in either glucose or galactose as indicated. After 4 h of induction, cross-linking and chromatin preparations were done exactly as described (39). The ChIP with the anti-acetyl histone H4 lysine 16 (Upstate Cell Signaling Solutions, Waltham, MA) was performed as described (40). The same procedure was used with 5 μl of anti-H2A antibody (Upstate Cell Signaling Solutions). After DNA extraction and purification, PCR was performed in the presence of 0.8 μCi/μl (1 Ci = 37 GBq) [α-32P]dATP in the linear range. The PCR products were analyzed by electrophoresis on 6% nondenaturing TBE gel (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) followed by quantification with an Image Reader FLA-5000 (Fujifilm) phosphoimager. To analyze the DNA around the HO site, we used primers ≈2 kb upstream of the HO recognition site (HO2-1 5′-TTGTATTAGACGAGGGACGGAGTG-3′ and HO2-2 5′-ACAGAGGGTCACAGCACTAATACAG-3′). The internal control TEL band was amplified by using primers specific for a region ≈500 bp away from the end of chromosome VI-R (TEL1 5′-GCGTAACAAAGCCATAATGCCTCC-3′ and TEL2 5′-CTCGTTAGGTACACGTTCGAATCC-3′).
Results
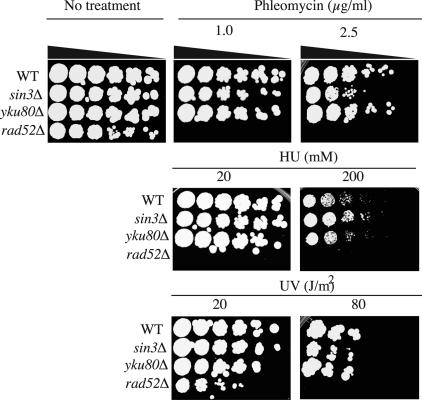
Disruption of SIN3 Results in Phleomycin Hypersensitivity. To address whether Sin3p modulates DNA repair, we analyzed the viability of cells containing or lacking Sin3p for their sensitivity to various DNA damaging agents. Thus, yeast strains deleted for SIN3, along with WT strains and control strains defective in HR (rad52Δ) or NHEJ (yku80Δ), were serially 5-fold diluted and spotted onto plates containing low or high concentrations of phleomycin or HU. Phleomycin causes DNA DSBs (41), whereas HU leads to depletion of the deoxyribonucleotide pool that in turn slows down S-phase progression and activates the DNA replication checkpoint (42). Furthermore, high levels of HU (e.g., 200 mM) cause the formation of S phase-specific DNA DSBs (43). In addition, we treated cells with UV light, which causes cyclobutane pyrimidine dimers and 6-4 photoproducts that are repaired primarily by nucleotide excision repair (44).
As anticipated from previous studies, the rad52Δ mutant but not the yku80Δ mutant displayed marked hypersensitivity toward phleomycin, HU, and UV (Fig. 1). Disruption of SIN3 led to a marked (≈25-fold) decrease in viability in the presence of phleomycin, as compared to WT cells or sin3Δ cells complemented by a low copy number vector bearing full-length SIN3 (Fig. 1 and data not shown). By contrast, in these assays sin3Δ cells did not exhibit significant hypersensitivity toward HU or UV. These results therefore suggest that deletion of SIN3 specifically confers sensitivity to DNA DSBs, although it does not markedly affect sensitivity to S phase-specific DNA DSBs that are caused by exposure to high concentrations of HU. Consistently, we also observed no increased hypersensitivity of sin3Δ cells to camptothecin, which also causes S phase-specific DSBs (data not shown).
Fig. 1.
SIN3 deletion results in hypersensitivity to phleomycin. Five-fold serial dilutions of WT, sin3Δ, yku80Δ, and rad52Δ strains were plated on YPAD medium containing the indicated doses of hydroxyurea or phleomycin. Cells were exposed to the indicated doses of UV after plating. Plates were incubated for 3-4 days at 30°C.
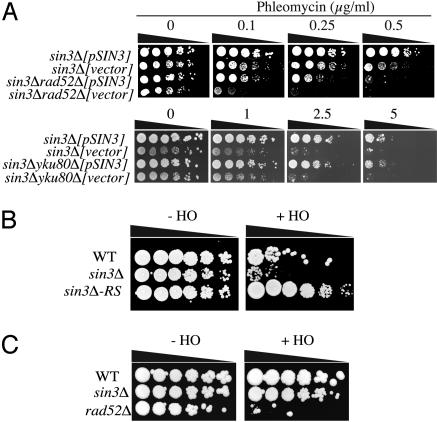
Deletion of SIN3 Causes a Defect in NHEJ. The above results prompted us to further investigate the potential role of Sin3p in DNA DSB repair. It has previously been demonstrated that deletion of NHEJ genes, such as YKU80, further hypersensitizes rad52Δ cells to agents that generate DNA DSBs (45, 38). To see whether this was also the case for SIN3 deletion, we compared the phleomycin sensitivity of a sin3Δrad52Δ double-mutant strain to that of a sin3Δ single-mutant strain, and to derivatives of these strains that were complemented by a low copy number plasmid containing full-length SIN3. As shown in Fig. 2A, the sin3Δrad52Δ double mutant was significantly more sensitive to phleomycin than the sin3Δ or rad52Δ single mutants (compare sin3Δrad52Δ[vector] with sin3Δ[vector] and sin3Δrad52Δ[pSIN3]). Notably, when we subjected a sin3Δyku80Δ double-mutant strain to a similar analysis, we found that it was no more sensitive than the sin3Δ single mutant (Fig. 2 A; compare sin3Δyku80Δ[vector] with sin3Δ[vector] and sin3Δyku80Δ[pSIN3]). Taken together, these results indicate that Sin3p influences phleomycin sensitivity by a pathway that is distinct from Rad52p-dependent HR and, furthermore, strongly suggest that Sin3p influences NHEJ.
Fig. 2.
Sin3p functions in NHEJ. (A) Phleomycin sensitivity analysis of sin3Δrad52Δ and sin3Δyku80Δ mutants. Assays were done as in Fig. 1. Each strain was complemented with the full-length SIN3 under control of its own promoter on low-copy plasmid pRS416. Plates were incubated at 30°C for 4-5 days. (B) NHEJ-dependent repair of a HO endonuclease-induced DSB. In the parental strain (WT), the HMR and HML loci are deleted, and HO endonuclease expression is controlled by a galactose inducible promoter. Strain sin3Δ-RS is a sin3Δ strain with a mutated HO recognition site. Five-fold serial dilutions of each strain were plated on either YPA with glucose or YPA with galactose plates. (C) HR-dependent repair of HO endonuclease-induced DSB. In the parental strain (WT), the HO recognition site placed within the URA3 gene on chromosome V, with homology on chromosome II, and HO endonuclease expression is controlled by a galactose inducible promoter. Five-fold serial dilutions of WT, sin3Δ, and rad52Δ strains were plated on either YPA with glucose or YPA with galactose plates.
To confirm the above conclusions, we took advantage of a galactose-inducible HO endonuclease system that generates a single DNA DSB at the chromosomal MAT locus. First, we carried out assays in a strain background that contains a deletion of the HML and HMR loci, meaning that the HO-induced DSB can only be repaired by NHEJ (36). As shown in Fig. 2B, in this background sin3Δ mutant cells were markedly impaired in their ability to grow on galactose medium as compared to WT control cells. This was due to a specific defect in DSB repair rather than an inability to metabolize galactose, because a control sin3Δ mutant strain (sin3Δ-RS) that contained a mutated, uncleavable, HO target site was able to grow efficiently on the galactose medium (Fig. 2B). In a complementary set of experiments, we analyzed the ability of sin3Δ mutants to repair a HO endonuclease-induced DSB by HR. To this end, we took advantage of a strain in which the HO recognition site is inserted in the URA3 gene on chromosome V and there is 1.2 kb of homologous sequence on chromosome II that serves as a template for HR repair (37). In contrast to the rad52Δ strain, which displays a severe loss of viability in this background (46), the sin3Δ mutant grew as well as the WT control strain (Fig. 2C). These results provide further evidence that Sin3p facilitates NHEJ but not HR. These conclusions are thus consistent with sin3Δ mutants being hypersensitive to phleomycin, which induces DSBs throughout the cell cycle, but not to high levels of HU, which produces DSBs mainly in S phase, where they are thought to be repaired mainly by HR (43).
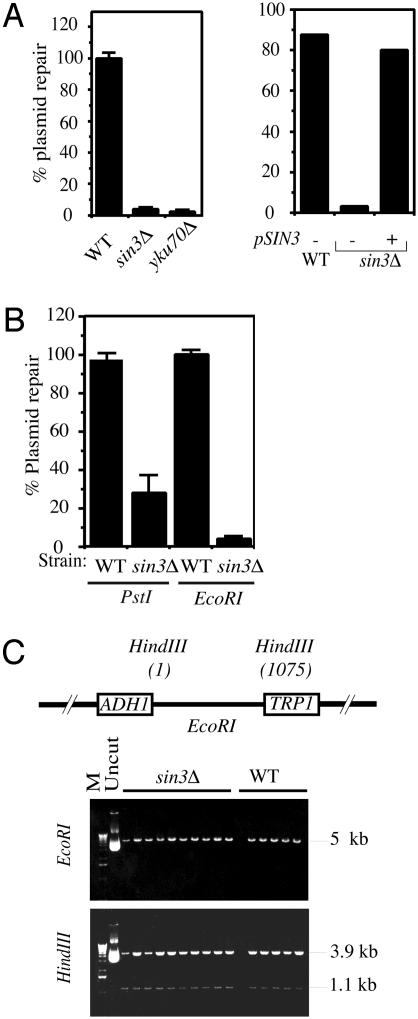
Deletion of SIN3 Confers a Plasmid Repair Defect. NHEJ mutants exhibit severe defects in the in vivo plasmid repair assay (38, 47). In this assay, a plasmid is linearized in a region with no significant homology to the yeast genome and is then transformed into yeast cells. Because plasmid maintenance requires its recircularization, the number of colonies formed provides a readout of the NHEJ capacity of the yeast strain being tested. To normalize for possible differences in transformation efficiency, each strain is transformed in parallel with supercoiled plasmid, and the number of colonies formed from the linearized plasmid transformation is normalized to that from the supercoiled transformation. Compared to the WT strain, the sin3Δ mutant displayed a severe defect in plasmid repair, and this defect was complemented by a low-copy plasmid bearing full-length SIN3 but not by the parental vector (Fig. 3A). As shown in Fig. 3B, the reduction in plasmid repair efficiency in the sin3Δ strain was more marked with a plasmid bearing 5′ overhanging termini produced by EcoRI digestion than with 3′ overhangs produced by PstI digestion, a pattern that has been observed previously for yku70Δ strains (47). These results confirm that disruption of SIN3 leads to markedly impaired NHEJ.
Fig. 3.
SIN3 deletion causes a plasmid repair defect. (A) Defect in plasmid repair assay. WT, sin3Δ, and yku70Δ strains were transformed, in parallel, with EcoRI-linearized or supercoiled pBTM116, then plated and incubated at 30°C for 4-5 days. (Right) sin3Δ mutant cells were complemented with either full-length SIN3 or parental vector. Values plotted are the numbers of transformants obtained with EcoRI-linearized vector expressed as a percentage of the numbers obtained with supercoiled vector. (B) Plasmid repair assay with different types of DNA ends. pBTM116 linearized with PstI (3′ overhang) or EcoRI (5′ overhang) was used as described above. Values plotted are the means of values from three experiments; error bars represent the SD. (C) Accuracy of repair. The repaired plasmids from WT or sin3Δ cells were retrieved into E. coli, and after their amplification and purification, plasmids were examined for the accuracy of the repair by digesting the plasmids with EcoRI or HindIII. The schematic diagram demonstrates the positions of the cut-sites for the above enzymes in pBTM116.
If a yeast cell performs accurate NHEJ of an EcoRI-linearized pBTM116 plasmid, then the repaired plasmid should be linearized by EcoRI treatment and should produce fragments of ≈4 and ≈1 kb when digested with HindIII. When we retrieved repaired plasmids from sin3Δ and WT cells and analyzed them in this way, we found that in all cases they had been repaired accurately (Fig. 3C). This is in marked contrast to high levels of inaccurate repair that occur in the absence of Yku70p, Yku80p, or Dnl4p (47, 48) but is similar to the accurate residual repair observed in strains deleted for RAD50, MRE11, or XRS2 (18). This raised the possibility that SIN3 disruption might affect NHEJ indirectly by impairing transcription of one or other of these latter genes. Arguing against this idea, however, is the finding that SIN3 disruption does not reduce the transcription levels for any of the known NHEJ genes (49). Moreover, unlike strains lacking Rad50p, Mre11p, Xrs2p, Yku70p, or Yku80p (18, 50, 51), strains disrupted for SIN3 do not exhibit shortened telomeres as ascertained by Southern blot analysis (data not shown). Taken together, these results imply that SIN3 disruption does not impair NHEJ through affecting the transcription or protein levels of any of the known NHEJ factors. Instead, they suggest that Sin3p acts in a more direct way to modulate NHEJ.
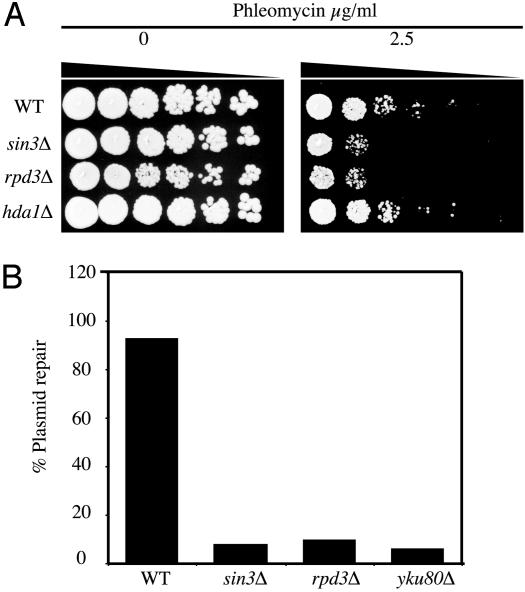
rpd3Δ Mutants Are also Defective for NHEJ. Previous work has shown that Sin3p functions in a complex with the HDAC Rpd3p (27, 29). Significantly, we found that, like sin3Δ cells, strains disrupted for RPD3 were markedly hypersensitive to phleomycin but not to HU or UV (Fig. 4A and data not shown). By contrast, deletion of HDA1, which encodes the other predominant deacetylase activity in budding yeast, did not result in phleomycin hypersensitivity (Fig. 4A). This suggests that the DNA damage hypersensitivity of rpd3Δ and sin3Δ strains is unlikely to be due to a general and nonspecific change in histone acetylation levels. In addition, we found that rpd3Δ mutant cells exhibited a similar plasmid repair defect to that of sin3Δ cells (Fig. 4B), suggesting that the HDAC activity of the Sin3p/Rpd3p complex is required for efficient NHEJ.
Fig. 4.
Deletion of RPD3 phenocopies the repair defects of a sin3Δ strain. (A) Phleomycin sensitivity. The experiment was performed as described in Fig. 1, with WT, sin3Δ, and rpd3Δ cells. (B) Plasmid repair assay. This assay was performed as described in Fig. 3A, with EcoRI-linearized pBTM116.
DSB Induction Triggers Sin3p-Dependent Deacetylation of Histone H4 Lysine 16. To study the potential consequences of the genetic data described above, we used ChIP to examine whether there is any change in the levels of acetylation of histone H4 around the site of a single HO-induced chromosomal DNA DSB in a strain in which such a DSB can only be repaired by NHEJ (see above). For these studies, we concentrated on histone H4 lysine 16. One reason for this was that deacetylation of this residue is generally thought to be less important than deacetylation of lysines 5, 8, and 12 for the transcriptional regulation of Sin3p/Rpd3p target genes (52, 40), raising the possibility that it might, instead, control other events such as DNA repair. In addition, we reasoned that it would be difficult to establish whether any alteration in the acetylation of lysines 5 and 12 after DNA DSB induction genetically depended on the Sin3p/Rpd3p complex, because of the large increase in the total levels of acetylation of these residues in the absence of this complex (52).
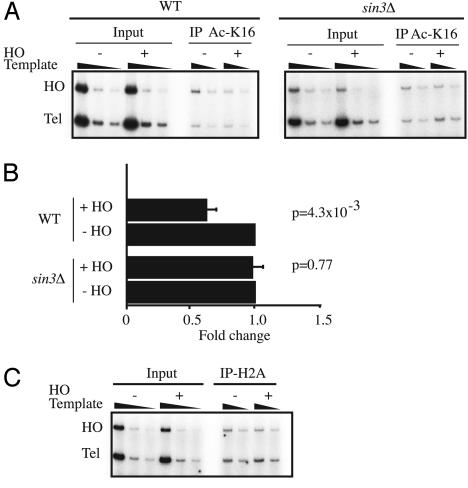
After the induction of HO endonuclease, acetylated histone H4 lysine 16 was immunoprecipitated from cross-linked and sheared chromatin preparations, then DNA from the precipitates was analyzed by PCR in the linear range. For the HO site we designed primers to a region that is ≈2 kb upstream of the HO recognition site on chromosome III, and as an internal control, we used primers to a region ≈500 bp away from the end of chromosome VI. In multiple experiments we consistently observed a statistically significant 40-50% reduction in the levels of acetylation of histone H4 lysine 16 at the HO target locus after DSB induction (Fig. 5 A and B). Moreover, this decrease required Sin3p, because it no longer occurred in sin3Δ mutant cells (Fig. 5 A and B). Importantly, Southern blot analysis did not reveal any difference in the extent of DSB induction between the WT and sin3Δ strains (data not shown). Thus, the absence of deacetylation in the sin3Δ strain presumably reflects the absence of the deacetylase activity rather than reduced DSB formation. It is important to note that Southern blot analyses carried in parallel with the above studies consistently revealed <100% DSB formation (data not shown). The significant amount of uncut DNA (generally 30-40%) in these experiments therefore presumably led to a significant underestimation of the level of histone H4 lysine 16 deacetylation measured after HO induction in cells containing functional Sin3p.
Fig. 5.
Sin3p-dependent hypoacetylation of histone H4 lysine 16. (A) Level of acetylation on the histone H4 lysine residue after induction of a HO-induced DSB. Using an antibody specific to the acetylated form of histone H4 lysine 16 (Ac-K16), ChIP was performed with extracts from cells with or without HO-induced DSB in the presence or absence of Sin3p (WT or sin3Δ). PCR was done with primers specific to a region ≈2 kb upstream of the HO recognition site. For internal control, primers specific to a region ≈0.5 kb away from the end of chromosome VI-R (Tel) were used. Input represents 10% of the total extract used for immunoprecipitation. (B) Quantification of the histone H4 lysine 16 acetylation after HO DSB induction. For quantification, the ratio of the control signal to the HO signal in the immunoprecipitation samples was calculated, then this ratio was normalized to the same ratio derived from the input samples. Signals from the samples before HO endonuclease induction were assigned as one, and the change in the signal after HO induction was calculated for each experiment. The graph represents the average of at least three experiments; error bars represent the SD, and the P value was calculated by using Student's t test. Quantification was done by using the phosphoimager system and macbas 2.5 software (Fujifilm). (C) Levels of the H2A around the site of HO-induced DSB. ChIP was done with an antibody against histone H2A from cells with or without the HO-induced DSB. PCR was performed with primers described above.
To ensure that reduction in acetylation did not result from the loss of nucleosomes after the processing of the HO-induced DSB, we performed the same ChIP experiments with an antibody that recognizes histone H2A. As shown in Fig. 5C, DSB induction resulted in no detectable loss of this signal from the HO target locus, indicating that the amount of the H2A protein around the site of the DSB is not markedly altered under these conditions. Taken together, these data establish that, after the formation of a chromosomal DSB, there is a Sin3p-dependent reduction of the levels of lysine 16 acetylation of histone H4 in the vicinity of the DNA damage. It is therefore likely that the role of Sin3p in NHEJ reflects this histone deacetylation function, although it is formally possible that Sin3p might also influence NHEJ via it targeting additional, nonhistone proteins.
Discussion
We have described a previously uncharacterized role for Sin3p and Rpd3p in DNA NHEJ and have shown that Sin3p-dependent hypoacetylation of lysine 16 of histone H4 occurs in the vicinity of DNA DSBs. These results are consistent with the recent finding that the Caenorhabditis elegans homolog of Rpd3p, Hda-3, is required for efficient repair of x-ray-induced damage in the nematode (53). It is also notable that C. elegans RPD3 and SIN3 homologs were also recently identified in an RNAi-based screen to be important for the protection of the genome against mutations (54).
Although our data suggest that deletion of SIN3 or RPD3 affects NHEJ, we found that sin3Δ cells are hypersensitive to phleomycin and yku80Δ mutant is not (Fig. 1). This is consistent with previously published reports showing that yku70Δ, yku80Δ, or dnl4Δ mutants are not sensitive to ionizing radiation (38, 47, 48). Furthermore, we have found that the sin3Δyku80Δrad52Δ triple mutant is more sensitive to phleomycin than the corresponding double mutants (data not shown). Taken together, these results suggest that Sin3p/Rpd3p may have additional roles in response to DNA damage other than NHEJ. We do not think that Sin3p functions in HR because we detected no defect in this pathway in sin3Δ cells (Fig. 2C). Also, SIN3 deletion did not result in any detectable hypersensitivity to S phase-induced DSBs that are repaired mainly by HR (Fig. 1, HU sensitivity). It is therefore possible that Sin3p has a role in DNA damage signaling. In this regard it is noteworthy that there is some evidence from other model systems that this might indeed be the case. For example, human HDAC1 (the homolog of Rpd3p) interacts with the PCNA-like checkpoint proteins Hus1 and Rad9 (55). Furthermore, Hda-3, the C. elegans homolog of Rpd3p, interacts with the C. elegans Hus-1 (53). In addition, human HDAC2 (another homolog of Rpd3p) interacts with the checkpoint kinase ATR (the homolog of yeast Mec1p) (56). Although we have so far been unable to detect a checkpoint defect in the absence of SIN3, it is possible that deletion of SIN3 results in a subtle defect in these events or affects these pathways only in a particular phase of the cell cycle.
The fidelity of residual repair in the absence of Sin3p is in contrast to the inaccurate residual repair in strains lacking the core NHEJ component (Yku70p/80p and Dnl4p; 38, 47, 48). Although there are other interpretations, these data suggest that the core NHEJ machinery can function without Sin3p but that the efficiency is reduced. It is possible that this reduction in repair efficiency is caused by the global hyperacetylation of histones that occurs in the absence of Sin3p/Rpd3p making the chromatin a suboptimal substrate for NHEJ. However, arguing against such a general effect is the demonstration that there is a damage-specific hypoacetylation of lysine 16 of histone H4 in the proximity of the site of damage that is genetically dependent on Sin3p. This finding provides evidence for the direct involvement of Sin3p (and by extension Rpd3p) in NHEJ events. It is presumably the inability of the sin3Δ cells to create the hypoacetylated region that results in defective NHEJ. In combination with the genetic data, this argues that, in the absence of Sin3p, the chromatin structure around the site of damage is not hypoacetylated and thus does not constitute a good substrate for the NHEJ machinery. This model is consistent with the recent demonstration that mutating the four acetylable lysine residues to glutamine, which mimics hyperacetylated chromatin (as would be the case in the absence of the Sin3p or Rpd3p), results in a significant plasmid repair defect (14).
Our finding that efficient NHEJ requires a HDAC are perhaps surprising in light of the recent report that NHEJ also requires a HAT activity (14). How can these results be reconciled? One possibility is that NHEJ requires the combination of both HDAC and HAT activities, presumably for different stages of the end-joining process. For example, it is possible that a HAT is initially used to relax the chromatin structure before the initiation of the repair and that later a HDAC is recruited to create a local region of hypoacetylation, which might help the stabilization and juxtaposition of the two broken ends. Alternatively, it is possible that the HDAC is first recruited to create a hypoacetylated region to allow efficient stabilization and juxtaposition of the two broken ends and that once the repair process is completed the HAT is recruited to help reestablish the correct histone code to allow efficient gene activity. An attractive model is that these two activities might even be recruited to the site of damage as parts of one complex. In this regard, it is noteworthy that HDAC1 and hGCN5 have been found to be in the same complex (57). Interestingly, a conserved common motif has recently been found in Esa1p and Rpd3p, and this motif is essential for the HDAC and HAT activities of Rpd3p and Esa1p, respectively (58). Although the functional role of this motif is currently unclear, an intriguing possibility is that it defines part of a regulatory interaction domain with the proteins in DNA processes that require both HAT and HDAC activities, such as transcription or possibly repair. Modification of this dual activity by other proteins might allow the cell to ensure that the correct histone modification is established for each step of the DNA repair process. It will be interesting to see whether Rpd3p/Sin3p interact with Esa1p as part of one complex that is required for efficient NHEJ.
Acknowledgments
We are extremely grateful to Drs. Martin Kupiec, David Stillman, and James Haber for strains and reagents. We thank members of the Jackson laboratory for helpful discussions. We are especially thankful to Andy Bannister, Jane Bradbury, Jessica Downs, Kate Dry, and Muriel Grenon for their critical reading of the manuscript. This study was funded by Cancer Research UK.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; DSB, double-strand break; HDAC, histone deacetylase; HR, homologous recombination; NHEJ, nonhomologous end joining.
See Commentary on page 1427.
References
- 1.Kornberg, R. D. & Lorch, Y. L. (1999) Cell 98, 285-294. [DOI] [PubMed] [Google Scholar]
- 2.Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251-260. [DOI] [PubMed] [Google Scholar]
- 3.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41-45. [DOI] [PubMed] [Google Scholar]
- 4.Narlikar, G. J., Fan, H. Y. & Kingston, R. E. (2002) Cell 108, 475-487. [DOI] [PubMed] [Google Scholar]
- 5.Green, C. M. & Almouzni, G. (2002) EMBO Rep. 3, 28-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Gent, D. C., Hoeijmakers, J. H. J. & Kanaar, R. (2001) Nat. Rev. Genet. 2, 196-206. [DOI] [PubMed] [Google Scholar]
- 7.Khanna, K. K. & Jackson, S. P. (2001) Nat. Genet. 27, 247-254. [DOI] [PubMed] [Google Scholar]
- 8.Paques, F. & Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Featherstone, C. & Jackson, S. P. (1999) Curr. Biol. 9, R759-R761. [DOI] [PubMed] [Google Scholar]
- 10.Critchlow, S. E. & Jackson, S. P. (1998) Trends Biochem. Sci. 23, 394-398. [DOI] [PubMed] [Google Scholar]
- 11.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. (1998) J. Biol. Chem. 273, 5858-5868. [DOI] [PubMed] [Google Scholar]
- 12.Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. (1999) J. Cell Biol. 146, 905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Downs, J. A., Lowndes, N. F. & Jackson, S. P. (2000) Nature 408, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 14.Bird, A. W., Yu, D. Y., Pray-Grant, M. G., Qiu, Q., Harmon, K. E., Megee, P. C., Grant, P. A., Smith, M. M. & Christman, M. F. (2002) Nature 419, 411-415. [DOI] [PubMed] [Google Scholar]
- 15.Ikura, T., Ogryzko, V. V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J. & Nakatani, Y. (2000) Cell 102, 463-473. [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb, T. M. & Jackson, S. P. (1993) Cell 72, 131-142. [DOI] [PubMed] [Google Scholar]
- 17.Barlev, N. A., Poltoratsky, V., OwenHughes, T., Ying, C., Liu, L., Workman, J. L. & Berger, S. L. (1998) Mol. Cell. Biol. 18, 1349-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulton, S. J. & Jackson, S. P. (1998) EMBO J. 17, 1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukamoto, Y., Kato, J. & Ikeda, H. (1997) Nature 388, 900-903. [DOI] [PubMed] [Google Scholar]
- 20.Martin, S. G., Laroche, T., Suka, N., Grunstein, M. & Gasser, S. M. (1999) Cell 97, 621-633. [DOI] [PubMed] [Google Scholar]
- 21.Mills, K. D., Sinclair, D. A. & Guarente, L. (1999) Cell 97, 609-620. [DOI] [PubMed] [Google Scholar]
- 22.Astrom, S. U., Okamura, S. M. & Rine, J. (1999) Nature 397, 310. [DOI] [PubMed] [Google Scholar]
- 23.Lee, S. E., Paques, F., Sylvan, J. & Haber, J. E. (1999) Curr. Biol. 9, 767-770. [DOI] [PubMed] [Google Scholar]
- 24.Kegel, A., Sjostrand, J. O. O. & Astrom, S. U. (2001) Curr. Biol. 11, 1611-1617. [DOI] [PubMed] [Google Scholar]
- 25.Valencia, M., Bentele, M., Vaze, M. B., Herrmann, G., Kraus, E., Lee, S. E., Schar, P. & Haber, J. E. (2001) Nature 414, 666-669. [DOI] [PubMed] [Google Scholar]
- 26.Lechner, T., Carrozza, M. J., Yu, Y., Grant, P. A., Eberharter, A., Vannier, D., Brosch, G., Stillman, D. J., Shore, D. & Workman, J. L. (2000) J. Biol. Chem. 275, 40961-40966. [DOI] [PubMed] [Google Scholar]
- 27.Kasten, M. M., Dorland, S. & Stillman, D. J. (1997) Mol. Cell. Biol. 17, 4852-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurdistani, S. K., Robyr, D., Tavazoie, S. & Grunstein, M. (2002) Nat. Genet. 31, 248-254. [DOI] [PubMed] [Google Scholar]
- 29.Kadosh, D. & Struhl, K. (1997) Cell 89, 365-371. [DOI] [PubMed] [Google Scholar]
- 30.Kadosh, D. & Struhl, K. (1998) Mol. Cell. Biol. 18, 5121-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadosh, D. & Struhl, K. (1998) Genes Dev. 12, 797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deckert, J. & Struhl, K. (2002) Mol. Cell. Biol. 22, 6458-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurdistani, S. K. & Grunstein, M. (2003) Nat. Rev. Mol. Cell Biol. 4, 276-284. [DOI] [PubMed] [Google Scholar]
- 34.Adams, A., Gottschling, D. E., Kaiser, C. A. & Stearns, T. (1997) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 35.Rose, M. D., Meluh, P. B. & Hieter, P. (1990) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 36.Moore, J. K. & Haber, J. E. (1996) Mol. Cell. Biol. 16, 2164-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inbar, O., Liefshitz, B., Bitan, G. & Kupiec, M. (2000) J. Biol. Chem. 275, 30833-30838. [DOI] [PubMed] [Google Scholar]
- 38.Boulton, S. J. & Jackson, S. P. (1996) Nucleic Acids Res. 24, 4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rouse, J. & Jackson, S. P. (2002) Mol. Cell 9, 857-869. [DOI] [PubMed] [Google Scholar]
- 40.Suka, N., Suka, Y., Carmen, A. A., Wu, J. S. & Grunstein, M. (2001) Mol. Cell 8, 473-479. [DOI] [PubMed] [Google Scholar]
- 41.Moore, C. W. (1988) Cancer Res. 23, 6837-6843. [PubMed] [Google Scholar]
- 42.Desany, B. A., Alcasabas, A. A., Bachant, J. B. & Elledge, S. J. (1998) Genes Dev. 12, 2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merrill, B. J. & Holm, C. (1999) Genetics 153, 595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC).
- 45.Milne, G. T., Jin, S., Shannon, K. B. & Weaver, D. T. (1996) Mol. Cell. Biol. 16, 4189-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aylon, Y., Liefshitz, B., Bitan-Banin, G. & Kupiec, M. (2003) Mol. Cell. Biol. 23, 1403-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulton, S. J. & Jackson, S. P. (1996) EMBO J. 15, 5093-5103. [PMC free article] [PubMed] [Google Scholar]
- 48.Teo, S.-H. & Jackson, S. P. (1997) EMBO J. 16, 4788-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernstein, B. E., Tong, J. K. & Schreiber, S. L. (2000) Proc. Natl. Acad. Sci. USA 97, 13708-13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gravel, S., Larrivee, M., Labrecque, P. & Wellinger, R. J. (1998) Science 280, 741-744. [DOI] [PubMed] [Google Scholar]
- 51.Laroche, T., Martin, S. G., Gotta, M., Gorham, H. C., Pryde, F. E., Louis, E. J. & Gasser, S. M. (1998) Curr. Biol. 8, 653-656. [DOI] [PubMed] [Google Scholar]
- 52.Rundlett, S. E., Carmen, A. A., Kobayashi, R., Bavykin, S., Turner, B. M. & Grunstein, M. (1996) Proc. Natl Acad. Sci. USA 93, 14503-14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulton, S. J., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D. E. & Vidal, M. (2002) Science 295, 127-131. [DOI] [PubMed] [Google Scholar]
- 54.Pothof, J., van Haaften, G., Thijssen, K., Kamath, R. S., Fraser, A. G., Ahringer, J., Plasterk, R. H. & Tijsterman, M. (2003) Genes Dev. 17, 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai, R. L., Yan-Neale, Y., Cueto, M. A., Xu, H. & Cohen, D. (2000) J. Biol. Chem. 275, 27909-27916. [DOI] [PubMed] [Google Scholar]
- 56.Schmidt, D. R. & Schreiber, S. L. (1999) Biochemistry 38, 14711-14717. [DOI] [PubMed] [Google Scholar]
- 57.Yamagoe, S., Kanno, T., Kanno, Y., Sasaki, S., Siegel, R. M., Lenardo, M. J., Humphrey, G., Wang, Y., Nakatani, Y., Howard, B. H. & Ozato, K. (2003) Mol. Cell. Biol. 23, 1025-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adachi, N., Kimura, A. & Horikoshi, M. (2002) J. Biol. Chem. 277, 35688-35695. [DOI] [PubMed] [Google Scholar]