Abstract
We demonstrate a label-free peptide-coated carbon nanotube based immunosensor for the direct assay of human serum. A rheumatoid arthritis (RA) specific (cyclic citrulline-containing) peptide, was immobilized to functionalized single-walled carbon nanotubes deposited on a quartz crystal microbalance (QCM) sensing crystal. Serum from RA patients was used to probe these nanotube-based sensors, and antibody binding was detected by QCM sensing. Specific antibody binding was also determined by comparing the assay of two serum control groups (normal and diseased sera), and the native unmodified peptide. The sensitivity of the nanotube-based sensor (detection in the femtomol range) was higher than that of the established ELISA and recently described microarray assay systems, detecting 34.4% and 37.5% more RA patients with anti-citrullinated peptide antibodies than those found by ELISA and microarray respectively. As well as 18.4% and 19.6% greater chance of a negative test being a true indicator of a person not having RA than by either ELISA or microarray respectively. The performance of our label-free biosensor constitutes its application in the direct assay of sera in research and diagnostics.
1. Introduction
In the study of proteins, traditional and microtechnological methods to date require at least one protein component be covalently labeled with a marker molecule, such as a fluorophore, to enable the detection of specific protein-protein binding. However, this modification may disrupt binding sites involved in specific protein recognition. In some studies (such as drug discovery) molecule-labeling is unacceptable, and labeling procedures require excess material for an acceptable yield. Some protein assays require a secondary molecule for the detection of protein-protein binding, which may result in decreased assay sensitivity. Novel nano-materials for bioassay applications are being developed to overcome these problems of current technologies, resulting in a rapidly progressing field of nanobiotechnology.
Following their discovery (Kroto et al., 1985, Ebbesen and Ajayan, 1992, Iijima, 1991, Hamada et al., 1992), the electronic, mechanical and optical properties of single-walled carbon nanotubes (SWNTs)(Bethune et al., 1993, Iijima and Ichihashi, 1993) have made them popular as nanoscale probes and sensors in not only electronic (An et al., 2001, Niu et al., 1997, Baughman et al., 1999) but also biological devices (Mattson et al., 2000, Williams et al., 2002). Their solubility, functionalization and chemical modification enable their use as membrane channels (Hummer et al., 2001, Park et al., 2003, Zhu and Schulten, 2003), molecular tweezers (Kim and Lieber, 1999), probes for imaging biomolecules (Wong et al., 1998, Woolley et al., 2000), and biosensors (Kong et al., 2000, Ng et al., 2001, Sotiropoulos et al., 2003, Chen et al., 2003). Their three-dimensional structure enables higher sample loading hence increased antigen density. Their simple chemistry for protein immobilization makes nanotubes versatile as biosensor substrates. Protein immobilization on nanotubes is not only simple and noninvasive but also bypasses the need for the synthesis of peptides with specific chemical linkages as needed in other nanostructure sensing methods such as nanowires (Zheng et al., 2005) and self-assembled monolayers (Chou et al., 2002, Shen et al., 2005) which could alter peptide conformation and functionality. We, and others, demonstrated that nanotubes could be used to immobilize antigens, preserving protein bioactivity (Chen et al., 2003, Fu et al., 2002), and these antigen-coated nanotubes can be used to detect antibodies in simple solutions such as salt buffers. Our current study overcomes a critical barrier to using nanotubes for clinical assays by demonstrating that antigen-coated nanotubes can be used as biosensors to detect specific antibodies in a highly complex mixture of proteins such as serum.
We describe a novel label-free detection method using SWNTs as platforms to immobilize peptides for the detection of specific autoantibodies in serum from patients with the autoimmune disease rheumatoid arthritis (RA). Current diagnosis of RA is based on the presentation of clinical features, supported by X-rays and a set of molecular markers including autoantibodies (Arnett et al., 1988). Rheumatoid factor (RF), immunoglobulins directed against the Fc portion of IgG and IgM, is the only molecular marker included in the current criteria. Recent studies show a relevance of autoantibodies to citrullinated peptides (citrulline-containing peptides) in RA (Schellekens et al., 1998, Girbal-Neuhauser et al., 1999, Nakamura, 2000), specifically for citrullinated regions of pro-filaggrin (Girbal-Neuhauser et al., 1999, Schellekens et al., 1998) and fibrin (Masson-Bessière et al., 2001); a family of enzymes (peptidyl arginine deiminases) post-translationally convert arginine to citrulline by deimination. Measurement of these autoantibodies is traditionally by reliable yet laborious Western-blotting, and recently ELISA, using various synthetic cyclic citrullinated peptides (CCPs) or modified proteins as antigens (van Jaarsveld et al., 1999, Schellekens et al., 1998, Schellekens et al., 2000, Nogueira et al., 2001).
Using a quartz crystal microbalance (QCM) sensing device, we demonstrate for the first time a peptide-coated nanotube technique that can assay human serum, with assay performance superior to ELISA, the diagnostic gold-standard assay, and the recently described microarray methodology (Hueber et al., 2005). Our nanotube-based sensor overcomes limitations of other nanostructure-based sensors, without requiring the pretreatment of serum for analysis. Our study provides the basis for using nanotube-based sensors for the direct assay of sera in diagnostics, research, and therapeutics.
2. Materials and Methods
2.1. Patient samples
The diseased patient group consisted of 35 serum samples from RA patients (all rheumatoid factor positive) diagnosed according to ACR criteria (Arnett et al., 1988) with a diagnosis of RA for less than one year. Two control groups were used: the normal control group consisting of 13 serum samples from healthy individuals, and the diseased control group consisting of 11 serum samples from patients with osteoarthritis (a non-autoimmune arthritis). All sera were collected under internal review board (of Stanford) (IRB) approved protocols and with informed consent.
2.2. Peptides
A 14-mer cyclic filaggrin peptide derived from the amino acid sequence deduced from cDNA of human filaggrin (Gan et al., 1990, Gan et al., 1991) was synthesized as previously described (Schellekens et al., 2000). A citrulline-modified version of this peptide was used as the antigen, where arginine deimination results in citrulline. The control peptide was the non-citrullinated peptide (arginine substituted for citrulline).
2.3. Anti-peptide ELISA
Based on a previously described method (Schellekens et al., 2000), wells of 96-well ELISA microtiter plates (Nunc, USA) were coated with 0.125 µg/well of citrullinated or non-citrullinated peptide in carbonated buffer, or buffer alone to determine background binding, overnight at 4 °C. The wells were washed with phosphate buffered saline/0.1%Tween20 (PBS/Tween), and blocked with 3% fetal calf serum (FCS) in PBS/Tween, at room temperature for 1 h. The wells were then washed with PBS/Tween, and 100 µl of serum sample diluted at 1/200 in 3%FCS/PBS/Tween (optimal serum dilution was determined by assaying a number of RA and control serum samples for anti-citrullinated peptide reactivity (see Supplementary Fig. 1 online)) was added and incubated for 1.5 h at room temperature. The wells were washed, and 100 µl/well of 1/5000 HRP-conjugated donkey anti-human immunoglobulin IgG/M (Jackson Immunoresearch Laboratories, West Grove, PA.) was added. After a further 1 h incubation the wells were washed and developed by the addition of 3,3´,5,5´ tetramethylbenzidine (TMB) substrate mixture (Pierce, USA) and the absorbance read at 450 nm. The specific antibody-peptide binding was corrected for the absorbance of wells coated with buffer alone. Each assay contained a positive and negative control serum. The optimal normal cut-off was established by receiver operating characteristic (ROC) curve analysis.
2.4. Production of antigen-coated nanotubes
Carbon nanotube film formation and antigen immobilization procedures were modified from previously described methods (Chen et al., 2003). Specifically a carbon nanotube film was formed on a QCM sensing crystal (5 MHz, AT cut, Au coated; Q-sense, Newport Beach, CA) by depositing a total of 100 µl of 50 µg/ml SWNT (Carbon Nanotechnologies, Houston) suspension in chloroform dropwise, followed by a 1 h bake at 60 °C. The antigen was immobilized on top of this film via a layer of carboxy-terminated Tween20 (Tween-COOH) containing polyethylene glycol units for reduced nonspecific binding and carboxylic acid groups for subsequent attachment. This layer of polyethylene glycol chains was not only used for linking the antigen to the nanotube substrate, but also served as a spacer, separating the antigen from the denaturing effect of the hydrophobic nanotube surface. Tween-COOH was synthesized by oxidation of its hydroxyl groups according to a previously described procedure (Hermanson, 1996). After coating the film with 1% Tween-COOH in water for 1 h, the carboxylic acid groups on the Tween-COOH were activated with 100 mM 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride and 250 mM N-hydroxysulfosuccinimide in 0.1 M phosphate buffer (pH 6.0) for 15 min. The surface was then rinsed quickly to remove excess reagents, before incubating with antigen (citrullinated or noncitrullinated peptide) in 0.1 M phosphate buffer (pH 7.5) overnight at 4 °C. Background binding was decreased with 3% FCS/0.1% Tween20 in 10 mM phosphate buffer (pH 7.0) for 2 h at room temperature. The QCM crystal was then rinsed and loaded into the QCM chamber (D300; Q-sense, Newport Beach, CA) with 10 mM phosphate buffer (pH 7.0) for serum analysis. The antigen was never allowed to dry prior to and during QCM analysis.
2.4.1. Probing of antigen-coated nanotubes
QCM measurements for the detection of serum antibodies binding to the antigen-functionalized carbon nanotubes were performed at the third harmonic resonance of the sensing crystal. Antibodies binding to immobilized antigen on the nanotube surface resulted in a mass uptake that decreased the natural level of vibration of the tubes, and was measured as a change in frequency of the quartz crystal. Following an initial equilibration period, serum diluted 1/300 in 10 mM phosphate buffer (pH 7.0) (total volume of 800 µl) was serially injected into the Q-sense instrument; each serum was allowed to interact with the tubes for 15 min, then flowed out, and after a 15 min re-equilibration period, the change in the resonance frequency was recorded. Dissipation was monitored during the entire QCM analysis, and at any significant change in dissipation, the analysis was discontinued. The optimal normal cut-off was established by ROC curve analysis.
At the completion of an experimental run, the carbon nanotube film on a QCM crystal was removed by oxygen plasma, and the crystal was reused with a newly deposited nanotube film.
2.5. Antigen microarray
Antigen arrays containing peptides and proteins representing autoantigen candidates in RA were fabricated as previously described (Robinson et al., 2002, Hueber et al., 2005). These microarrays were blocked overnight at 4 °C in PBS/0.5% Tween20/3% FCS, and then washed and incubated with 300 µl of serum diluted 1/150 in PBS/3% FCS for 1 h at 4 °C. Arrays were washed twice for 30 min and 20 min, respectively, at room temperature in blocking buffer, and incubated with 300 µl of 1/4000 Cy3-conjugated goat-anti-human IgG/M secondary antibody (Jackson Immunoresearch, West Grove, PA) for 1 h at room temperature. The arrays were then washed twice for 30 min with blocking buffer, twice for 20 min with PBS, and rinsed twice for 15 sec in water before being spun dry and scanned using a GenePix4000 Scanner. GenePix Pro 3.0 software (Molecular Devices, Union City, CA) was used to determine median pixel intensities of features and background. Data was normalized to anti-IgG/M, and threshold values were set based on the negative control (buffer alone) (Hueber et al., 2005). The optimal normal cut-off was established by ROC analysis.
2.6. Peptide Modeling
The non-citrullinated peptide and its citrullinated analog were modeled using SYBYL® 7.0 (Tripos Inc., MO). Fold prediction was done using GeneFold (Jaroszewski et al., 1998). A disulfide link was inserted to create an internal cyclization of each peptide, and the predicted structures were further minimized using Amber95 force field before superimposing.
2.7. Statistical Analysis
Statistical analysis of the data was performed using SPSS® software to obtain ROC to determine the optimal normal cut-off values for each test system and antigen, and to perform correlation tests. Statview® was used for contingency table analysis. A p value < 0.05 was considered statistically significant.
3. Results
3.1. Antigenicity of the citrullinated peptide
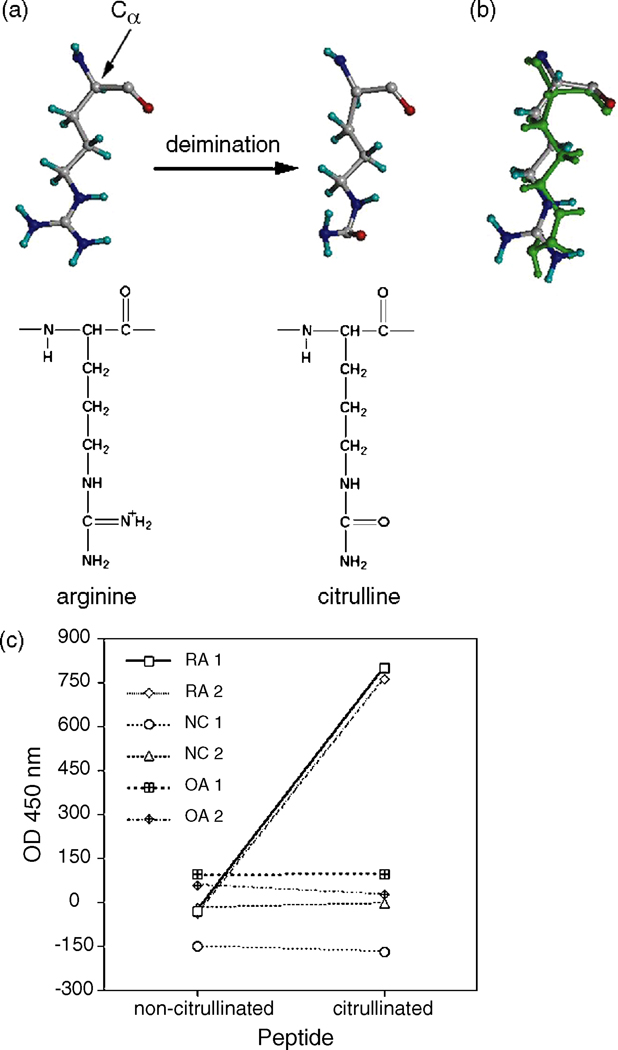
The deimination of arginine to citrulline results in a slight decrease in residue size, as well as a change in charge of +1 to 0 at physiological pH (Fig. 1a and b). This conformational change to the residue results in the presentation of different epitopes and contributes to the ‘antigenicity’ of the peptide. This is evident by ELISA where there are markedly elevated levels of antibodies in the RA sera to the citrulline-modified peptide compared to its non-citrullinated form (Fig. 1c). Control sera (normal persons and osteoarthritic patients) did not exhibit reactivity against this candidate autoantigen, the citrullinated peptide, nor its non-citrullinated form.
Figure 1.
Arginine deimination and RA autoantibody specificity. (a) Deimination of arginine in the peptide to form citrulline; amino acid orientation with respect to the rest of the peptide is denoted by the alpha carbon. (b) Superimposed model of arginine and citrulline (on top, in green). (c) Graph of RA, normal control (NC) and the diseased control osteoarthritis (OA) sera tested for antibodies to the citrullinated peptide and non-citrullinated peptide by ELISA, showing markedly elevated serum levels of anti-citrullinated peptide antibodies in the RA sera.
3.2. Design of the nanotube bioassay
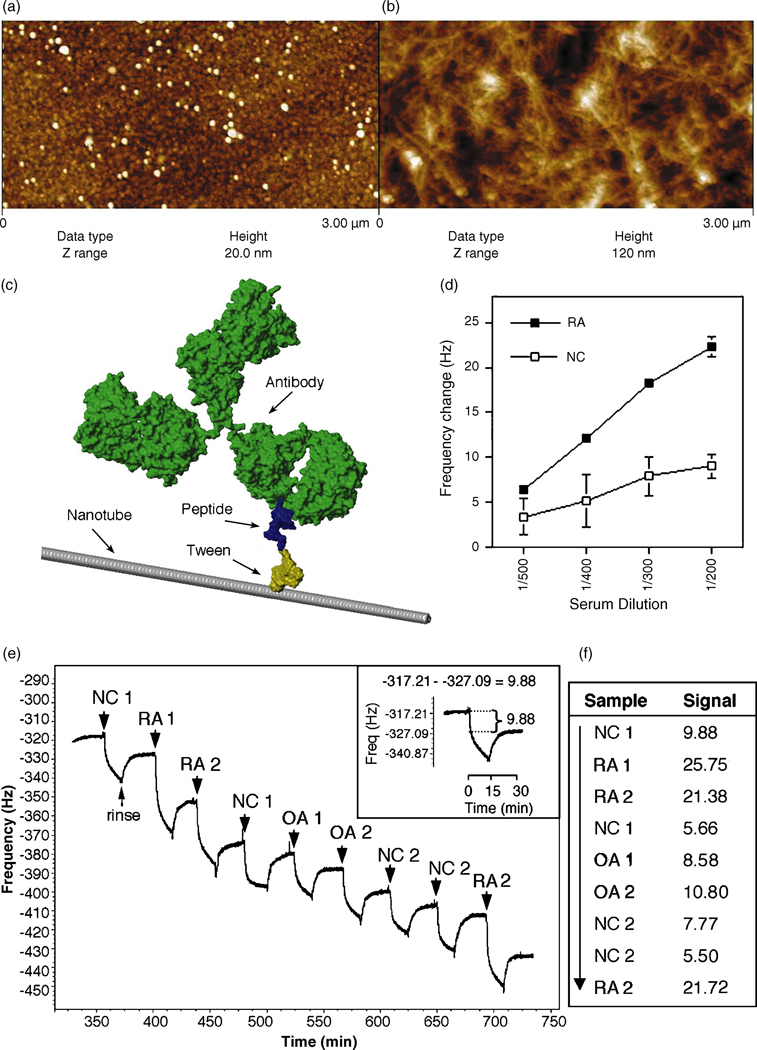
Due to its high sensitivity in direct, label-free detection and the ability of real time data collection (Decker et al., 2000, Chou et al., 2002, Shen et al., 2005, Janshoff and Steinem, 2005), QCM was used to measure serum autoantibody binding to peptide-coated nanotubes. Carbon nanotubes deposited on QCM sensing crystals, as shown in figure 2a and b (for SEM image see Supplementary Fig. 2 online), were co-functionalized with Tween-COOH and either the test antigen, the citrullinated peptide, or the same peptide with arginines substituted for citrullines. After blocking unbound portions of nanotubes, the crystals were loaded into the QCM chamber. These peptide-nanotube platforms were tested for reactivity with specific autoantibodies by assaying serum from RA patients, using serum from healthy individuals as controls; a hypothetical space-filled molecular model of an antibody bound to a Tween-functionalized peptide-nanotube is shown in figure 2c. The natural resonance of the QCM crystal decreases upon a mass uptake on the surface, so a QCM measurement of serum antibodies that bind to the peptide-coated nanotubes was expressed as a net change of this frequency. An optimal serum dilution of 1/300 was determined in preliminary experiments by assaying a number of RA and control serum samples for anti-citrullinated peptide reactivity; a representative example of each is shown in figure 2d. Assay reproducibility is evident by the very low standard error of means (s.e.m.) (Fig. 2d). Inter-assay variability was less than 14%, and intra-assay variability was less than 15% (data not shown).
Figure 2.
Nanotube film topography by AFM imaging of (a) a bare Au surface on a QCM crystal and (b) the carbon nanotube film deposited on top; the z range (height) is much higher for the nanotube network, illustrating its high surface area and three-dimensional structure. (c) A visual representation of an antibody bound to a peptide-Tween-nanotube complex that comprises the nanotube biosensor. (d) A sample representation of a dilution curve of an RA and normal control (NC) serum in the nanotube assay measuring antibody binding to citrullinated peptide-coated nanotubes as a change in frequency (Hz). Presented is the mean ± s.e.m. (e) Raw QCM data measuring a mass uptake of antibodies in RA, normal control (NC) and osteoarthritic (OA) sera binding to citrullinated peptide-coated nanotubes seen as a decrease in frequency (Hz); each serum sample is allowed to interact with the peptide-coated nanotubes for 15 mins, followed by a 15 min rinse between sample runs. The net change in frequency post rinse is recorded as the signal of relative antibody binding (inset). (f) The signal for each sample tested in (e) is given; the arrow indicates consecutive order of samples tested in one experimental run.
Each serum sample was injected into the QCM chamber and allowed to incubate with the peptide-coated nanotubes for 15 mins before rinsing the tubes for a further 15 mins to remove unbound material and to establish a baseline between sample testing (Fig. 2e). The signal (a change in frequency post rinse) is calculated as initial frequency – frequency post rinse (Fig. 2e inset and f). Serum samples having higher titers of antibodies to the citrullinated peptide result in a greater change in frequency as more antibodies bind to citrullinated peptide-coated nanotubes, thus the RA sera display a greater signal than do the control sera (Fig. 2f). Assay reproducibility is evident by the repeat serum testing in this assay (Fig. 2f).
Antigen loading as estimated by saturation experiments for each peptide using RA versus control serum was approximately 3 ng (data not shown). In a single experimental run, approximately 22 RA and 7 control sera at a 1/300 dilution could be tested on such a platform (peptide-coated nanotubes on a QCM crystal) before reaching saturation. Detection levels were as low as 89 fmol of specific antibody.
3.3. Preservation of antigenicity in the nanotube system
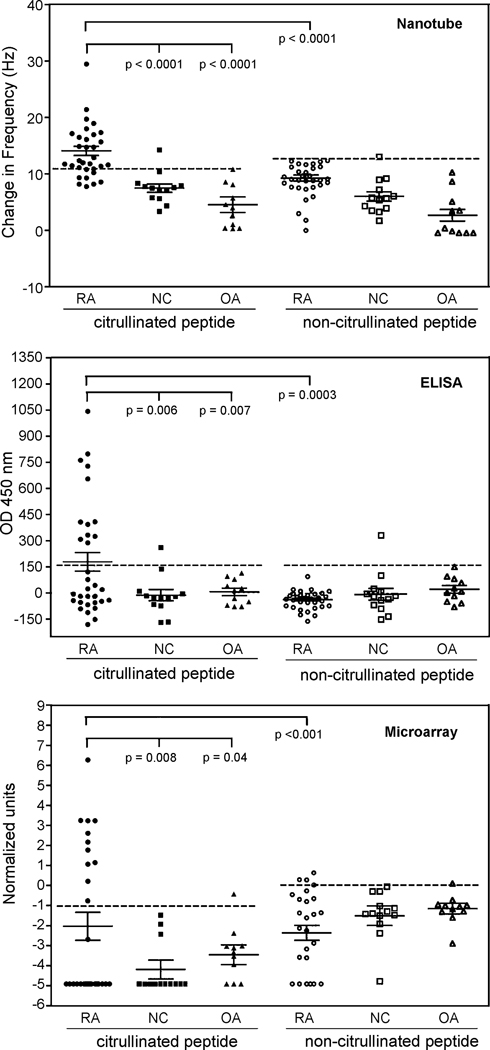
Immobilization of proteins may cause a conformational change that could result in the loss of important binding sites. Preservation of antibody-recognition sites is a major component of any immunological assay for the reliable detection of antigen and antibody binding, hence reliable measurements of antibody levels. The antibody reactive epitope of the citrullinated peptide was conserved upon immobilization to nanotubes as seen by the significantly greater reactivity of RA sera to the citrullinated peptide than to the non-citrullinated peptide (p < 0.0001) (Fig. 3, top panel). Reactivity against the citrullinated peptide when compared to the non-citrullinated peptide separated the RA patient group from the control groups; i.e., there were significantly higher titers of antibodies to the citrullinated peptide in RA sera when compared with the control sera (p < 0.0001) (Fig. 3). The RA patient group had a higher mean serum level of anti-citrullinated peptide antibodies when compared to the control groups (p < 0.05) (Fig. 3). There was no significant difference in the mean serum antibody levels to the non-citrullinated peptide between the three patient study groups in the nanotube system (p > 0.05). These results confirm that the reactivity to the citrullinated peptide is specific to the citrulline moiety, and this reactive conformation is preserved upon immobilization of the peptide on nanotubes. This preservation (seen as antigenic function) is due in part to the Tween20 functionalization of the nanotubes, thereby separating the peptide from the denaturing effect of the hydrophobic nanotube surface.
Figure 3.
Scatter plots of the level of antibodies to the citrullinated peptide or non-citrullinated peptide in the RA, normal control (NC) and osteoarthritic (OA) patient groups as measured by the three assay systems (nanotube, ELISA and microarray). The normal cut-off is indicated below the horizontal dashed line, and presented is also the mean ± s.e.m. for each serum sample group in each assay.
The preservation of antibody-recognition epitopes of the peptide in the nanotube system is also seen by the significantly greater number of RA patients with antibodies to the citrullinated peptide (71.8%) when compared to the healthy (7.7%) and OA (0%) control groups by this nanotube assay (p < 0.05) (Table 1). The 7.7% of healthy controls with antibodies to the citrullinated peptide was due to 1 of the 13 control sera testing positive for antibodies to the citrullinated peptide, however at a specificity level of 100% none of the healthy controls were anti-citrullinated peptide positive. With a specificity level of < 100%, it is expected to see RA and control sera positive for antibodies to the native non-citrullinated peptide. It is well known that autoantibodies may be found in up to 15% of healthy individuals (Smolen, 1996). Less than 1% of healthy individuals possess antibodies to citrullinated antigens (Schellekens et al., 2000).
Table 1.
The number (and percentage in that sample group) of serum samples with antibodies to the citrullinated peptide (CitPept) or the negative control peptide (nonCitPept), by the three different assays (nanotube, ELISA and microarray). The serum groups are rheumatoid arthritis (RA), normal control (NC) and osteoarthritis (OA).
| RA (n = 32) |
NC (n = 13) |
OA (n = 11) |
|||||
|---|---|---|---|---|---|---|---|
| Nanotube | CitPept | 23* | (71.8%) | 1# | (7.7%) | 0 | 0 |
| nonCitPept | 0 | 1 | 0 | ||||
| ELISA | CitPept | 12* | (37.5%) | 1 | 0 | ||
| nonCitPept | 0 | 1# | 0 | ||||
| Microarray | CitPept | 11* | (34.3%) | 0 | 1† | (9.1%) | |
| nonCitPept | 3¥ | (9.3%) | 0 | 1† | |||
P values for anti-CitPept positive RA serum samples by ELISA versus nanotube p = 0.002, ELISA versus microarray p = 0.797, and nanotube versus microarray p = 0.0003.
Two of the three sera also have reactivity to the citrullinated peptide by ELISA, nanotube and microarray testing.
This is the same NC serum sample that has antibodies to the citrullinated peptide by the nanotube assay, as well as antibodies to the non-citrullinated peptide by ELISA.
This is the same serum sample in the OA group that has antibodies to both the citrullinated and non-citrullinated peptide by microarray testing.
3.4. Performance of the nanotube assay compared to ELISA and microarray for each serum sample
In all three assay systems, nanotubes, ELISA and microarray, RA sera had greater reactivity to the citrullinated peptide in comparison to the non-citrullinated peptide (p < 0.001), and higher mean serum levels of anti-citrullinated peptide antibodies when compared to the control groups (p < 0.05) (Fig. 3). There was no significant difference in the mean serum antibody levels to the non-citrullinated peptide between the three patient study groups in any of the three assay systems (p > 0.05).
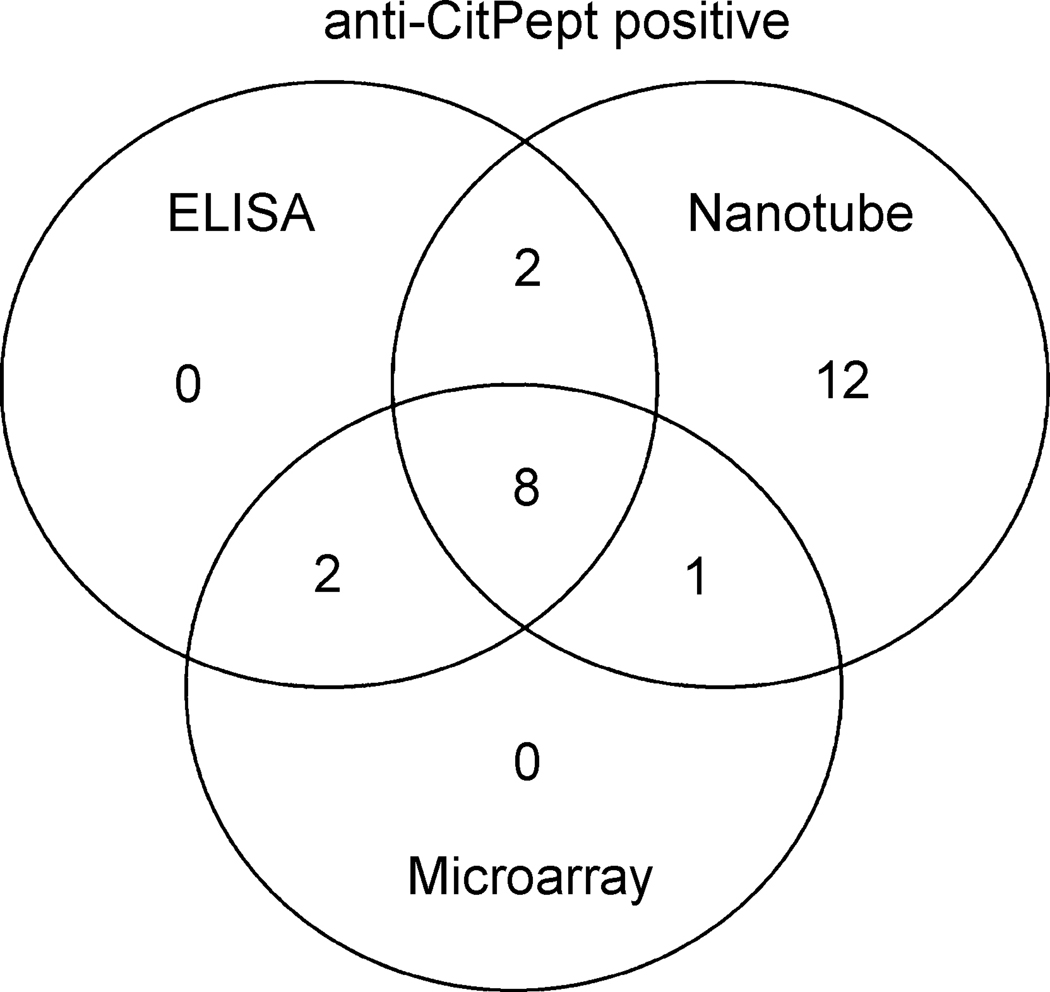
The direct performance of the nanotube system compared to the ELISA and microarray systems was assessed by comparing the total number of anti-citrullinated peptide positive patients in each assay system. Of the 32 RA patients tested 12 (37.5%) were positive for antibodies to the citrullinated peptide by ELISA and 11 (34.3%) by microarray assay. However, 23 (71.8%) RA patients were positive for antibodies to the citrullinated peptide by the nanotube assay, significantly more than by either ELISA or microarray (p < 0.01) (Table 1). Of these 12 RA patients positive for anti-citrullinated peptide antibodies by ELISA, 10 were also positive by nanotube testing, and 8 of these 10 were positive by microarray testing (Fig. 4). Also 9/10 positive by microarray testing were positive in the nanotube system (Fig. 4). However, 12 RA sera found to have antibodies to the citrullinated peptide by the nanotube system were not detected by either ELISA or microarray testing (Fig. 4). Thus the anti-citrullinated peptide nanotube assay detected 34.4% more RA patients with antibodies to the citrullinated peptide than those found by ELISA, and 37.5% more than by microarray testing. This greater number of RA patients found to be positive for anti-citrullinated peptide antibodies by the nanotube method than by ELISA or microarray suggests a greater level of sensitivity for antibody detection by the nanotube method.
Figure 4.
Venn diagram of the number of RA serum samples with antibodies to the citrullinated peptide (CitPept) as measured by ELISA, nanotubes and microarray.
Although by ELISA and nanotube testing none of the RA sera were positive for antibodies to the non-citrullinated peptide, the microarray assay identified 3 RA sera containing antibodies specific for this peptide (Table 1). However, 2 of these 3 RA serum samples also had reactivity to the citrullinated peptide by microarray, ELISA and nanotube testing. Antibodies to non-citrullinated filaggrin sequences have recently been reported in the occasional RA patient (Vittecoq et al., 2004).
One of 13 normal serum samples had reactivity to the control peptide and citrullinated peptide by ELISA, as well as antibodies to the citrullinated peptide by nanotube testing (Table 1). The nanotube assay did not detect antibodies to the native nor the antigenic peptide in the OA sera group but one of these sera did have reactivity to both the citrullinated peptide and the non-citrullinated peptide in the microarray system. So, there was no significant difference in the frequency of patients with antibodies to the citrullinated peptide between the two control serum groups in all of the three assay systems (p > 0.05); therefore within the control serum groups the nanotube assay performed similarly to ELISA and microarray.
3.5. Diagnostic values
To assess the validity of the nanotube assay as a potential diagnostic tool the assay characteristics of sensitivity, specificity, and positive and negative predictive values for disease (RA) were derived and compared to those from the more common assay method ELISA, and to recently described microarray technology (Hueber et al., 2005). Sensitivity describes the proportion of patients with RA who have a positive anti-citrullinated peptide test result, and specificity defines the proportion of patients without RA who have a negative test result. The nanotube assay resulted in a significantly higher test sensitivity of 71.9% for the detection of anti-citrullinated peptide antibodies in RA patients compared to 37.5% and 34.4% for the ELISA and microarray systems respectively (p < 0.03) (Table 2). This high sensitivity of the nanotube test for RA is at the upper end of the 41% to 88% values reported in the literature, depending on the cohort of patients studied (Schellekens et al., 2000, Berthelot et al., 1998, Goldbach-Mansky et al., 2000, Pruijn et al., 2005). There was no significant difference in the test specificity between nanotubes, ELISA and microarrays (all at 95.8%). The 95.8% specificity of the nanotube assay is close to the reported 96% to 100% range. Based on the test characteristics of sensitivity and specificity, this nanotube-based assay is highly sensitive and specific for the detection of this RA-specific autoantibody.
Table 2.
The diagnostic values of the three tests for RA based on the detection of antibodies to the citrullinated peptide.
| Test | Sensitivity | Specificity | PPV | NPV | AccuracyΔ |
|---|---|---|---|---|---|
| Nanotube | 71.9% * | 95.8% | 95.8% | 71.9% ¥ | 0.94 |
| ELISA | 37.5% * | 95.8% | 92.3% | 53.5% ¥ | 0.64 |
| Microarray | 34.4% * | 95.8% | 91.6% | 52.3% ¥ | 0.54 |
P < 0.03 for nanotube versus ELISA, and nanotube versus microarray.
P < 0.03 for nanotube versus ELISA, and nanotube versus microarray.
Maximum accuracy of 1.0.
As a clinical characteristic, all three testing systems had a good positive predictive value (PPV), that is the probability of a person with a positive anti-citrullinated peptide test also having RA (Table 2). With a PPV of 95.8% for the nanotube assay, slightly higher than 92.3% by ELISA, there is only a 4.2% chance of a false positive by the nanotube detection method. This translates to a greater number of persons with disease actually being identified by a positive anti-citrullinated peptide test in the nanotube assay in comparison with ELISA. The nanotube assay has an NPV of 71.9% which is a significantly greater chance of a negative test result being a true indicator of the absence of RA when compared to only a 53.5% chance of a true negative by ELISA and 52.3% by microarray (p < 0.03) (Table 2). Thus by the nanotube system there is a greater probability of a person with a negative anti-citrullinated peptide test actually not having RA. These predictive values of the nanotube test are comparable to those reported in the literature, depending on the cohort of patients studied (Schellekens et al., 2000, Pruijn et al., 2005).
The area under the ROC for the three test methods illustrates the accuracy of the test to separate the group being tested into those with and without the disease (i.e., RA). The nanotube based detection method exhibited greater observed accuracy than the ELISA or microarray detection systems (Table 2).
4. DISCUSSION
We have developed a label-free carbon nanotube-based method for the detection of disease-specific autoantibodies in human serum. A RA-specific peptide immobilized on nanotubes maintained its functionality to bind specific autoantibodies in RA patient serum. This is the first time a complex mixture of proteins in a biological sample (serum) has been used to successfully probe antigens immobilized on nanotubes. This method bypasses the need of a secondary molecule, or labeling of sera or a protein component to detect antibody-antigen binding as do ELISA and microarray methods, hence is a more time-efficient and cost-effective assay. The nanotube system had greater sensitivity without sacrificing specificity than either the traditional gold-standard method ELISA or recently described microarray (Hueber et al., 2005) for these autoantibodies. The increased assay accuracy could be explained by the direct detection of protein binding. The enhanced sensitivity of this system is useful for the detection of even the lowest amount of autoantibody in patient sera. The protein detection level (in the femtomolar range) of the nanotube-based sensor exceeds those of existing technologies for these autoantibodies.
Other nanostructures used for protein detection devices, such as nanowire-based sensors (Zheng et al., 2005), may exhibit better detection levels but require the pretreatment of serum (required to remove salts in serum that interfere with electrical sensing), or are not as sensitive as our nanotube-based sensor (Chou et al., 2002, Shen et al., 2005, Wang et al., 2005). Desalting can fundamentally change protein structure, denature a protein of interest, result in substantial sample loss, and is time consuming. Our nanotube-based sensor overcomes the requirement of serum preprocessing, and overcomes limitations of other nanostructure-based sensors. Self-assembled monolayers of proteins on QCM crystals by direct linkage chemistry (Chou et al., 2002, Shen et al., 2005) requires chemical modification of the peptide or designing peptides with specific chemical linkages resulting in altered conformation and functionality of bound peptide; in our case direct chemical linkage to QCM crystals was not feasible due to the cyclic nature of our peptide. Nanotubes also provide spatial separation of the peptide from the gold-coated QCM crystal, thereby avoiding potential oxidation of the disulphide bond that gives our peptide its cyclic-feature.
The microarray system requires the least effort with a higher sample throughput than the nanotube-based assay but data analysis can be time consuming. The present nanotube-based assay serially tests sera against one antigen, however, the system can be multiplexed into an array, enabling label-free protein profiling with high throughput, minimal sample volume, assay time, and post-assay data analysis. The nanotube assay is also performed in liquid phase, essentially eliminating potential problems of drying and steric hinderance seen with protein microarrays.
5. Conclusions
We demonstrate for the first time the use of a nanotube-based label-free immunosensor for the direct detection of disease-related autoantibodies in human serum. Serum antibody detection by peptide-coated carbon nanotubes, deposited on QCM as the sensing device, exhibited greater sensitivity and diagnostic accuracy than traditional ELISA and microarray assaying. The nanotube-based biosensor (i) validates the use of peptides immobilized to nanotubes for the direct assay of human sera in diagnostics; (ii) demonstrates diagnostic speed and accuracy; (iii) bypasses problems associated with serum sample preparation inherent with other nanostructure sensors; (iv) advances research for molecular markers of disease; (v) could be applied to develop patient-specific therapeutics based on their autoantibody and protein profile; (vi) can be used as an efficient drug screening tool for the pharmaceutical industry; (vii) and can be modified into a multiplexed sensor. A key aspect of our nanotube-based analytical system is its potential for the assay of other complex biological materials (such as synovial or cerebrospinal fluids) with many practical applications.
Supplementary Material
Figure 1. Serum dilution curve of a RA and normal control (NC) serum by ELISA for antibody binding to citrullinated peptide. Presented is the mean ± s.e.m.
Figure 2. Nanotube film topography. Scanning electron microscope (SEM) image of the carbon nanotube coated QCM crystal.
Acknowledgements
We are grateful to Professor Curtis Frank for the use of QCM equipment. This work was supported by a grant to H.D. and P.J.U. from the Stanford Program in Molecular and Genetic Medicine (PMGM). P.J.U. is the recipient of a Donald E. and Delia B. Baxter Foundation Career Development Award, a gift from the Floren Family Foundation, and is supported by the Dana Foundation, the Northern California Chapter of the Arthritis Foundation, DK61934, AI50854, AI50865, AR49328, and NHLBI Proteomics Contract N01-HV-28183 during the time period when this work was performed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An KH, Kim WS, Park YS, Moon JM, Bae DJ, Lim SC, Lee YS, Lee YH. Adv Funct Mater. 2001;11:387–392. [Google Scholar]
- Arnett FC, Edworthy SM, Block DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Baughman RH, Cui C, Zakhidov AA, Iqbal Z, Barisci JN, Spinks GM, Wallace GG, Mazzoldi A, de Rossi D, Rinzler AG, Jaschinski O, Roth S, Kertesz M. Science. 1999;284:1340–1344. doi: 10.1126/science.284.5418.1340. [DOI] [PubMed] [Google Scholar]
- Berthelot JM, Garnier P, Glémarec J, Flipo RM. Rev Rhum Engl Ed. 1998;65:9–14. [PubMed] [Google Scholar]
- Bethune DS, Klang CH, de Vires MS, Gorman G, Savoy R, Vazquez J, Beyers R. Nature. 1993;363:605–607. [Google Scholar]
- Chen RJ, Bangsaruntip S, Drouvalakis KA, Kam NW, Shim M, Li Y, Kim W, Utz PJ, Dai H. Proc Natl Acad Sci U S A. 2003;100:4984–4989. doi: 10.1073/pnas.0837064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S-F, Hsu W-L, Hwang J-M, Chen C-Y. Clin Chem. 2002;48:913–918. [PubMed] [Google Scholar]
- Decker J, Weinberger K, Prohaska E, Hauck S, Kosslinger C, Wolf H, Hengerer A. J Immunol Methods. 2000;233:159–165. doi: 10.1016/s0022-1759(99)00187-8. [DOI] [PubMed] [Google Scholar]
- Ebbesen TW, Ajayan PM. Nature. 1992;358:220–222. [Google Scholar]
- Fu K, Huang W, Lin Y, Zhang D, Hanks TW, Rao AM, Sun Y-P. J Nanosci Nanotechnol. 2002;2:457–461. doi: 10.1166/153348802760393981. [DOI] [PubMed] [Google Scholar]
- Gan S-Q, McBride OW, Idler WW, Markova N, Steinert PM. Biochem. 1990;29:9432–9440. doi: 10.1021/bi00492a018. [DOI] [PubMed] [Google Scholar]
- Gan S-Q, McBride OW, Idler WW, Markova N, Steinert PM. Biochem. 1991;30:5814. [PubMed] [Google Scholar]
- Girbal-Neuhauser E, Durieux J-J, Arnaud M, Dalbon P, Sebbag M, Vincent C, Simon M, Senshu T, Masson-Bessière C, Jolivet-Reynaud C, Jolivet M, Serre G. J Immunol. 1999;162:585. [PubMed] [Google Scholar]
- Goldbach-Mansky R, Lee J, McCoy A, Hoxworth J, Yarboro C, Smolen JS, Steiner G, Rosen A, Zhang C, Menard HA, Zhou ZJ, Palosuo T, Van Venrooij WJ, Wilder RL, Klippel JH, Schumacher HRJ, El-Gabalawy HS. Arthritis Res. 2000;2:236–243. doi: 10.1186/ar93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Sawada S, Oshiyama A. Phys Rev Lett. 1992;68:1579–1581. doi: 10.1103/PhysRevLett.68.1579. [DOI] [PubMed] [Google Scholar]
- Hermanson G. Bioconjugate Techniques. San Diego: Academic Press; 1996. [Google Scholar]
- Hueber W, Kidd BA, Toomoka BH, Lee BJ, Bruce B, Fries JF, Sonderstrup G, Monach P, Drijfhout J-W, van Venrooij WJ, Utz PJ, Genovese MC, Robinson WH. Arthritis Rheum. 2005;52:2645. doi: 10.1002/art.21269. [DOI] [PubMed] [Google Scholar]
- Hummer G, Rasaiah JC, Noworyta JP. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- Iijima S. Nature. 1991;354:56–58. [Google Scholar]
- Iijima S, Ichihashi T. Nature. 1993;363:603–605. [Google Scholar]
- Janshoff A, Steinem C. Methods Mol Biol. 2005;305:47–64. doi: 10.1385/1-59259-912-5:047. [DOI] [PubMed] [Google Scholar]
- Jaroszewski L, Rychlewski L, Zhang B, Godzik A. Protein Science. 1998;7:1431–1440. doi: 10.1002/pro.5560070620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P, Lieber CM. Science. 1999;286:2148–2150. doi: 10.1126/science.286.5447.2148. [DOI] [PubMed] [Google Scholar]
- Kong J, Franklin NR, Zhou C, Chapline MG, Peng S, Cho K, Dai H. Science. 2000;287:622–625. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- Kroto HW, Heath JR, O'Brien SC, Curl RF, Smalley RE. Nature. 1985;318:162–163. [Google Scholar]
- Masson-Bessière C, Sebbag M, Girbal-Neuhauser E, Nogueira L, Vincent C, Senshu T, Serre G. J Immunol. 2001;166:4177–4184. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Haddon RC, Rao AM. J Mol Neurosci. 2000;14:175–182. doi: 10.1385/JMN:14:3:175. [DOI] [PubMed] [Google Scholar]
- Nakamura RM. J Clin Lab Anal. 2000;14:305–313. doi: 10.1002/1098-2825(20001212)14:6<305::AID-JCLA10>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HT, Fang A, Li J, Li SF. J Nanosci Nanotechnol. 2001;1:375–379. doi: 10.1166/jnn.2001.056. [DOI] [PubMed] [Google Scholar]
- Niu C, Sichel EK, Hoch R, Moy D, Tennent H. Appl Phys Lett. 1997;70:1480–1482. [Google Scholar]
- Nogueira L, Nogueira L, Sebbag M, Vincent C, Arnaud M, Fournie B, Cantagrel A, Jolivet M, Serre G. Ann Rheum Dis. 2001;60:882–887. [PMC free article] [PubMed] [Google Scholar]
- Park KH, Chhowalla M, Iqbal Z, Sesti F. J Biol Chem. 2003;278:50212–50216. doi: 10.1074/jbc.M310216200. [DOI] [PubMed] [Google Scholar]
- Pruijn GJM, Vossenaar ER, Drijfhout JW, van Venrooij WJ, Zendman JW. Curr Rheum Revs. 2005;1:1–7. [Google Scholar]
- Robinson WH, DiGennaro C, Hueber W, Haab BB, Kamachi M, Dean EJ, Fournel S, Fong D, Genovese MC, de Vegvar HE, Skriner K, Hirschberg DL, Morris RI, Muller S, Pruijn GJ, van Venrooij WJ, Smolen JS, Brown PO, Steinman L, Utz PJ. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- Schellekens GA, de Jong BAW, van den Hoogen FHJ, van de Putte LBA, van Venrooij WJ. J Clin Invest. 1998;101:273–281. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellekens GA, Visser H, de Jong BAW, van de Hoogen FHJ, Hazes JMW, Breedveld FC, van Venrooij WJ. Arthritis Rheum. 2000;43:155–163. doi: 10.1002/1529-0131(200001)43:1<155::AID-ANR20>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Shen Z, Stryker GA, Mernaugh RL, Yu L, Yan H, Zeng X. Anal Chem. 2005;77:797–805. doi: 10.1021/ac048655w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen JS. In: Manual of biological markers of disease. van Venrooij WJ, Maini RN, editors. Dordrecht: Kluwer Academic Publishers; 1996. pp. C1.1/1–C1.1/18. [Google Scholar]
- Sotiropoulos S, Gavalas V, Vamvakaki V, Chaniotakis NA. Biosensors and Bioelectronics. 2003;18:211–215. doi: 10.1016/s0956-5663(02)00183-5. [DOI] [PubMed] [Google Scholar]
- van Jaarsveld CHM, ter Borg EJ, Jacobs JWG, Schellekens GA, Gmelig-Meyling FHJ, van Booma-Frankfort C, de Jong BAW, van Venrooj WJ, Bijlsma JWJ. Clin Exp Rheumatol. 1999;17:689–697. [PubMed] [Google Scholar]
- Vittecoq O, Inçaurgarat B, Jouen-Beades F, Legoedec J, Letourneur O, Rolland D, Gervasi G, Ménard JF, Gayet A, Fardellone P, Daragon A, Jolivet M, Loët LE, Tron F. Clin Exp Immunol. 2004;135:173–180. doi: 10.1111/j.1365-2249.2004.02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wu J, Li J, Ding Y, Shen G, Yu R. Biosens Bioelectron. 2005;20:2210–2217. doi: 10.1016/j.bios.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Williams KA, Veenhuizen PTM, de la Torre BG, Eritja R, Dekker C. Nature. 2002;420:761. doi: 10.1038/420761a. [DOI] [PubMed] [Google Scholar]
- Wong SS, Joselevich E, Woolley AT, Cheung CL, Lieber CM. Nature. 1998;394:52–55. doi: 10.1038/27873. [DOI] [PubMed] [Google Scholar]
- Woolley AT, Cheung CL, Hafner JH, Lieber CM. Chem Biol. 2000;7:193–204. doi: 10.1016/s1074-5521(00)00037-5. [DOI] [PubMed] [Google Scholar]
- Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- Zhu F, Schulten K. Biophys J. 2003;85:236–244. doi: 10.1016/S0006-3495(03)74469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1. Serum dilution curve of a RA and normal control (NC) serum by ELISA for antibody binding to citrullinated peptide. Presented is the mean ± s.e.m.
Figure 2. Nanotube film topography. Scanning electron microscope (SEM) image of the carbon nanotube coated QCM crystal.