SUMMARY
Upon host infection, the protozoan parasite Entamoeba histolytica is confronted with reactive oxygen and nitrogen species (ROS and RNS) and must survive these stresses in order to cause invasive disease. We analyzed the parasite’s response to oxidative and nitrosative stresses, probing the transcriptional changes of trophozoites of a pathogenic strain after a 60min exposure to H2O2 (1mM) or a NO donor (DPTA-NONOate, 200μM), using whole-genome DNA microarrays. Genes encoding ROS and RNS detoxification enzymes had high transcriptional levels under basal conditions and upon exposure to both stresses. On a whole genome level, there was significant modulation of gene expression by H2O2 (286 genes regulated) and DPTA-NONOate (1,036 genes regulated) with a significantly overlap of genes modulated under both conditions (164 genes). A number of transcriptionally regulated genes were in signaling/regulatory and repair/metabolic pathways. However, the majority of genes with altered transcription encode unknown proteins, suggesting as yet unraveled response pathways in E. histolytica. Trophozoites of a non-pathogenic E. histolytica strain had a significantly muted transcriptional response to H2O2 compared to the pathogenic strain hinting that differential response to oxidative stress may be one factor that contributes to the pathogenic potential of E. histolytica.
Keywords: parasite, virulence, microarray, gene expression, ROS, RNS
INTRODUCTION
Infection by Entamoeba histolytica, the causative agent of amoebiasis, is a global health problem, which affects 500 million people worldwide (Stanley, 2003). Most commonly, this pathogen causes hemorrhagic dysentery and liver abscesses. During tissue invasion, E. histolytica adapts to changing oxygen tensions as it goes from the anaerobic colonic lumen to an oxygen rich environment in the colonic tissue (Stanley, 2003). Additionally, the parasite must cope with cytotoxic reactive oxygen species (ROS) and reactive nitrogen species (RNS) that are produced and released by activated phagocytes that are attracted to the site of infection (Stanley, 2003; Bogdan et al., 2000; MacMicking et al., 1997). Therefore, a significant contribution to E. histolytica’s pathogenic potential is likely to be due to its ability to cope with oxidative and nitrosative stresses generated during tissue invasion.
The cellular components targeted by ROS and RNS include proteins (metal cofactors, thiolate side chains, tyrosine and methionine residues), nucleic acids and lipids (Halliwell and Gutteridge, 2007). Common defense strategies against oxidative and nitrosative stresses include detoxification enzymes and repair systems that enable cells to resist RNS and ROS (Vandenbroucke et al., 2008; Justino et al., 2005). Several microbial transcription factors and regulons, which are involved in the response to both oxidative and nitrosative stress as well as in the transition from anaerobic to aerobic metabolism, have redox sensitive active sites that are modified and/or damaged by both ROS and RNS. Not surprisingly, given the overlap in the types of damage caused by ROS and RNS, common mechanisms exist to deal with these stressors. These include the Crp-Fnr superfamily of transcriptional regulators, which respond both to nitrosative and oxidative stress (Korner et al., 2003), as do other transcriptional regulators, e.g., in Escherichia coli: NsrR, OxyR, SoxRS, MetR, ferric uptake regulator and NorR, regulating a wide range of cellular processes (Spiro, 2006). A survey of the genomes of the parasitic protists E. histolytica (Loftus et al., 2005), Giardia lamblia (Morrison et al., 2007) and Trichomonas vaginalis (Carlton et al., 2007) revealed the absence of homologues of any of the above mentioned transcriptional regulators. In contrast, genes coding for detoxification systems for ROS and RNS are present in the genomes of these anaerobic protists. Some of these genes may have been acquired by lateral gene transfer from prokaryotes (Andersson et al., 2006; Andersson et al., 2003). The E. histolytica genome has four genes encoding flavodiiron proteins (FDPs), enzymes endowed with oxygen and/or nitric oxide reductase activity which are widespread in prokaryotes (Kurtz, 2007; Saraiva et al., 2004), and have been studied in the protozoa T. vaginalis (Sarti et al., 2004) and G. lamblia (Di Matteo et al., 2008). E. histolytica’s genome also contains genes encoding other enzymes involved in the detoxification of ROS, including peroxiredoxin, rubrerythrin, hybrid-cluster protein, and superoxide dismutase (SOD). Peroxiredoxin constitutes a major defense against oxidative stress as it is induced by a high-oxygen environment (Akbar et al., 2004) and trichostatin A (Isakov et al., 2008), and contributes to E. histolytica’s virulence (Sen et al., 2007; Davis et al., 2006). Although peroxiredoxin and SOD are ubiquitous in all domains of life, FDPs, rubrerythrin and hybrid-cluster proteins have thus far been identified only in prokaryotes and in these anaerobic protists.
Whole-genome expression profiling has been used to assess the effects of oxidative and nitrosative stress in diverse eukaryotes and prokaryotes (Vandenbroucke et al., 2008; Thum and Bauersachs, 2007). A recent meta-analysis of microarray data performed to assess the common denominators in the oxidative stress response across different domains of life revealed that there are both strong species-specific responses, and common strategies for diverse organisms to cope with this challenge (Vandenbroucke et al., 2008). Microarray technology has been used in Entamoeba to investigate a wide variety of biological questions, including virulence (Davis et al., 2007; MacFarlane and Singh, 2006), host colonic and hepatic invasion (Santi-Rocca et al., 2008; Gilchrist et al., 2006) and development (Ehrenkaufer et al., 2007). In order to determine the molecular mechanisms by which E. histolytica trophozoites respond when challenged with oxidative and nitrosative stresses, we used whole genome expression profiling using a short oligonucleotide microarray containing 9,435 of the annotated 9,938 amebic genes. Our results demonstrated a significant transcriptional response of E. histolytica HM-1:IMSS, a canonical virulent strain, to H2O2 (286 genes regulated), NO (1,036 genes regulated), and a significant overlap among the genes responsive to both conditions (164 genes). To further identify which components of these response mechanisms may be correlated with E. histolytica’s virulence potential, the response to oxidative stress was assessed for a non-pathogenic strain, E. histolytica Rahman. In contrast to the observations for the virulent HM-1:IMSS strain, the Rahman strain had fewer transcriptional changes and the overall fold-changes for the regulated genes were significantly lower. Overall, our results provide insights into the molecular network regulating adaptation to oxidative and nitrosative stresses in E. histolytica and suggest that one important difference between virulent and non-virulent amebae is their ability to deal with the stresses encountered during host invasion.
RESULTS AND DISCUSSION
Sensitivity of E. histolytica trophozoites to oxidative and nitrosative stress
To determine the response of E. histolytica to nitrosative and oxidative stress, trophozoites from the HM-1:IMSS strain were exposed to dipropylenetriamine (DPTA)-NONOate (nitric oxide releaser) or hydrogen peroxide (H2O2). We analyzed the viability of E. histolytica trophozoites in varying concentrations of H2O2 and DPTA-NONOate in order to identify conditions in which E. histolytica trophozoites stressed but still ≥90% viable. A 1-hour exposure to 1 mM H2O2 or 200 μM DPTA-NONOate resulted in a significant fraction of the cells being stressed (as judged by rounded morphology) but only a few dead cells (≤5% and≤10%, respectively), based on Trypan blue staining (data not shown). We tested the sensitivity of both the virulent HM-1:IMSS and the non-virulent Rahman strains to 1 mM H2O2, and found a similar percentage of dead cells (data not shown). The two stress agents are differently released into the cultures: addition of hydrogen peroxide results in immediate exposure to the added concentration, whereas DPTA-NONOate is a slow releaser of nitric oxide. The tested concentration of H2O2 (1 mM) is within physiologically relevant concentrations found in the gastrointestinal lumen (Mayol et al., 2006). At the chosen concentration (200 μM), DPTA-NONOate releases NO at a rate of ~25 nM NO.s−1, which after 1 hour of exposure would accumulate to a maximum of ~82 μM of NO in the medium, a concentration above physiological concentrations (Halliwell and Gutteridge, 2007). It is however likely that the NO concentration experienced by E. histolytica under the tested conditions is lower than that released, due to the general reactivity of nitric oxide within biological milieu. Both NO and H2O2 may react with components of the TYI-S-33 medium, such as serum and cysteine, in which E. histolytica is cultured. However, no defined medium lacking serum or cysteine has been developed in which E. histolytica can be reliably grown (Diamond and Cunnick, 1991; Gillin and Diamond, 1980). Thus, the TYI-S-33 medium, in which parasite growth is well standardized and robust and in which many of the previous transcriptome analyses done previously have been performed, is the best option for our studies.
Expression analysis of E. histolytica strains challenged with oxidative or nitrosative stress
Affymetrix whole-genome microarrays were used to determine the global transcriptional changes in E. histolytica HM-1:IMSS trophozoites upon exposure to 1 mM hydrogen peroxide for 1 hour. Data from three independent H2O2-exposed cultures were compared with those from E. histolytica HM-1:IMSS trophozoites from standard axenic culture conditions. All sets of arrays displayed high correlation values (0.94–0.98) (Table 1). Genes with a ≥ 2-fold change and FDR < 0.05 were considered differentially expressed: 184 genes were up-regulated by H2O2 and 102 genes were down-regulated (Tables 2 and 3; Supplementary Table 2).
Table 1A.
Correlations between individual DNA microarrays for E. histolytica HM-1:IMSS and Rahman under each experimental condition.
| HM-1:IMSS | |||
|---|---|---|---|
| 1 | 2 | 3 | |
| 1 | 0.97 | 0.95 | |
| 2 | 0.94 | ||
| 3 | |||
| HM-1:IMSS + H2O2 | |||
| 1 | 2 | 3 | |
| 1 | 0.97 | 0.97 | |
| 2 | 0.98 | ||
| 3 | |||
| HM-1:IMSS + DPTA-NONOate | |||
| 1 | 2 | 3 | |
| 1 | 0.96 | 0.95 | |
| 2 | 0.97 | ||
| 3 | |||
| Rahman | |||
| 1 | 2 | 3 | |
| 1 | 0.99 | 0.97 | |
| 2 | 0.98 | ||
| 3 | |||
| Rahman + H2O2 | |||
| 1 | 2 | 3 | |
| 1 | 0.99 | 0.98 | |
| 2 | 0.99 | ||
| 3 | |||
Table 2.
Genes up-regulated by hydrogen peroxide in Entamoeba histolytica HM-1:IMSS. The probe ID, accession number, description, baseline expression level, fold-change, p-value, and regulation under DPTA or other conditions are shown. The 30 most highly regulated genes are listed. HS (heat shock) (adapted from Hackney et al); Cysts (E. histolytica cysts) (adapted from Ehrenkaufer et al).
| Probe ID | Accession number | Description | Baseline expression level | Fold-change | p-value | Regulated under DPTA | Regulated under other conditions |
|---|---|---|---|---|---|---|---|
| 879.m00008_at | XM_642785 | hypothetical protein | 0.07 | 189.6 | 0.000 | - | - |
| 194.m00102_s_at | XM_645780 | hypothetical protein | 0.06 | 127.1 | 0.001 | + | HS |
| 654.m00032_x_at | XM_642891 | hypothetical protein | 1.39 | 114.9 | 0.023 | + | - |
| 363.m00049_x_at | XM_643869 | hypothetical protein | 0.17 | 89.05 | 0.015 | + | HS |
| 256.m00084_x_at | XM_644865 | hypothetical protein | 0.12 | 86.64 | 0.014 | + | HS |
| 256.m00083_x_at | XM_644864 | hypothetical protein | 1.22 | 82.85 | 0.014 | + | - |
| 692.m00024_s_at | XM_642872 | hypothetical protein | 0.05 | 76.14 | 0.000 | + | - |
| 248.m00060_s_at | XM_644979 | hypothetical protein | 0.05 | 71.78 | 0.000 | + | - |
| 266.m00066_s_at | XM_644755 | hypothetical protein | 0.13 | 53.25 | 0.002 | + | - |
| 194.m00123_x_at | XM_645776 | dUTP diphosphatase, putative | 0.11 | 50.95 | 0.011 | + | - |
| 397.m00055_s_at | XM_643659 | hypothetical protein | 0.18 | 42.56 | 0.020 | + | - |
| 654.m00031_s_at | XM_642894 | hypothetical protein | 0.06 | 41.05 | 0.000 | + | HS |
| 397.m00062_x_at | XM_643664 | hypothetical protein | 1.35 | 37.88 | 0.034 | + | - |
| 692.m00026_s_at | XM_642873 | hypothetical protein | 0.06 | 35.78 | 0.001 | + | - |
| 711.m00021_x_at | XM_642861 | hypothetical protein | 0.10 | 33.68 | 0.018 | + | HS |
| 375.m00057_x_at | XM_643790 | deoxyuridine 5 -triphosphate nucleotidohydrolase | 0.05 | 21.75 | 0.001 | + | - |
| 397.m00054_x_at | XM_643658 | hypothetical protein | 0.06 | 15.74 | 0.001 | + | - |
| 344.m00044_at | XM_644000 | hypothetical protein | 0.24 | 15.1 | 0.025 | + | - |
| 245.m00039_x_at | XM_645025 | hypothetical protein | 0.47 | 14.39 | 0.026 | + | Cysts |
| 219.m00107_at | XM_645351 | cell division control protein 42, putative | 0.10 | 11.34 | 0.003 | - | HS |
| 267.m00069_at | XM_644746 | hypothetical protein | 0.10 | 11.2 | 0.034 | - | - |
| 358.m00033_at | XM_643905 | hypothetical protein | 0.64 | 11.05 | 0.012 | + | Cysts |
| 194.m00103_at | XM_645781 | hypothetical protein | 0.07 | 10.44 | 0.005 | + | - |
| 8.m00393_at | XM_651687 | late competence protein, putative | 0.25 | 8.85 | 0.027 | - | HS + Cysts |
| 356.m00029_s_at | XM_643911 | hypothetical protein | 0.18 | 7.68 | 0.021 | + | HS |
| 20.m00272_at | XM_650868 | conserved hypothetical protein | 0.13 | 6.83 | 0.046 | + | - |
| 36.m00211_at | XM_650002 | hypothetical protein | 0.09 | 6.78 | 0.009 | + | Cysts |
| 35.m00253_at | XM_650038 | iron-sulfur flavoprotein, putative | 2.16 | 6.7 | 0.006 | - | Cysts |
| 312.m00037_at | XM_644279 | iron-sulfur flavoprotein, putative | 0.99 | 6.62 | 0.005 | - | HS + Cysts |
| 1.m00705_at | XM_652401 | BspA-like leucine rich repeat protein, putative | 0.27 | 6.56 | 0.004 | - | HS |
Table 3.
Genes down-regulated by hydrogen peroxide in Entamoeba histolytica HM-1:IMSS. The probe ID, accession number, description, baseline expression level, fold-change, p-value, and regulation under DPTA or other conditions are shown. The 30 most highly regulated genes are listed. HS (heat shock) (adapted from Hackney et al); Colitis (ameba from a mouse model of amebic colitis) (adapted from Gilchrist et al); Trophs (E. histolytica trophozoites) (adapted from Ehrenkaufer et al).
| Probe ID | Accession number | Description | Baseline expression level | Fold-change | p-value | Regulated under DPTA | Regulated under other conditions? |
|---|---|---|---|---|---|---|---|
| 172.m00078_at | XM_646199 | hypothetical protein | 0.86 | −9.09 | 0.000 | + | HS |
| 537.m00017_x_at | XM_643063 | AIG1 family protein, putative | 4.58 | −7.41 | 0.039 | + | - |
| 2.m00528_at | XM_652349 | hypothetical protein | 0.35 | −6.8 | 0.019 | + | HS |
| 3.m00563_at | XM_652186 | hypothetical protein | 6.39 | −5.75 | 0.002 | + | - |
| 86.m00158_s_at | XM_648244 | hypothetical protein | 0.46 | −5.59 | 0.039 | + | HS |
| 226.m00092_at | XM_645246 | Rab family GTPase | 0.26 | −4.83 | 0.000 | + | - |
| 233.m00105_at | XM_645153 | hypothetical protein | 6.57 | −4.41 | 0.004 | + | - |
| 9.m00372_x_at | XM_651612 | hypothetical protein | 0.32 | −4.18 | 0.004 | + | HS |
| 87.m00165_at | XM_648195 | hypothetical protein | 0.21 | −4.13 | 0.009 | + | - |
| 87.m00154_at | XM_648212 | formate nitrite transporter family protein, putative | 0.78 | −4.02 | 0.001 | - | Colitis |
| 31.m00209_x_at | XM_650257 | conserved hypothetical protein | 0.25 | −3.79 | 0.019 | + | - |
| 66.m00150_at | XM_648887 | high mobility group protein, putative | 0.39 | −3.77 | 0.006 | + | HS |
| 343.m00064_x_at | XM_644015 | WD repeat protein | 0.21 | −3.76 | 0.039 | + | - |
| 125.m00091_at | XM_647175 | cyclin, putative | 0.71 | −3.69 | 0.006 | + | HS |
| 286.m00057_at | XM_644512 | mitotic inducer phosphatase, putative | 0.21 | −3.31 | 0.000 | + | HS |
| 37.m00216_at | XM_649962 | hypothetical protein | 0.79 | −3.26 | 0.018 | + | - |
| 56.m00175_at | XM_649238 | elongation factor 1 beta, putative | 12.26 | −3.15 | 0.038 | - | - |
| 46.m00250_at | XM_649627 | hypothetical protein | 3.53 | −3.07 | 0.005 | - | - |
| 19.m00301_at | XM_650878 | hypothetical protein | 2.32 | −3.03 | 0.015 | + | HS |
| 408.m00045_s_at | XM_643583 | putative GTPase | 2.43 | −3.02 | 0.006 | + | HS |
| 80.m00159_at | XM_648429 | hypothetical protein | 7.76 | −2.99 | 0.004 | + | - |
| 1.m00606_s_at | XM_652473 | hypothetical protein | 9.86 | −2.91 | 0.006 | + | HS |
| 40.m00246_s_at | XM_649855 | rRNA biogenesis protein, putative | 4.83 | −2.89 | 0.041 | + | HS |
| 95.m00147_at | XM_647953 | PfkB family carbohydrate kinase, putative | 6.85 | −2.89 | 0.005 | - | HS |
| 51.m00159_s_at | XM_649424 | hypothetical protein | 0.30 | −2.88 | 0.031 | + | - |
| 221.m00089_s_at | XM_645315 | hypothetical protein | 3.74 | −2.85 | 0.027 | + | - |
| 127.m00147_at | XM_647143 | predicted protein | 1.33 | −2.82 | 0.019 | + | HS |
| 4.m00641_at | XM_652060 | tRNA-specific adenosine deaminase, putative | 13.13 | −2.8 | 0.040 | + | - |
| 34.m00273_at | XM_650116 | Rab family GTPase | 4.84 | −2.77 | 0.048 | - | Trophs |
| 13.m00334_at | XM_651296 | conserved hypothetical protein | 2.85 | −2.67 | 0.027 | + | Trophs |
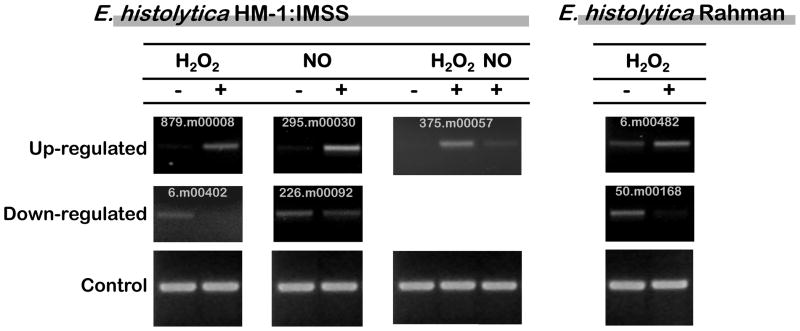
E. histolytica HM-1:IMSS trophozoites were also subjected to nitrosative stress by addition of the NO releaser DPTA-NONOate (200 μM, 1 hour) and resulting transcriptional changes assayed. Three arrays from independent parasite cultures challenged with DPTA-NONOate were performed; all arrays displayed good correlation values (0.94–0.97 (Table 1)). Although E. histolytica displayed similar percentages of cell death in H2O2 compared to NO, a substantially greater number of genes were transcriptionally regulated by nitrosative stress compared to oxidative stress. Using the same statistical criteria applied above (≥2-fold change and FDR < 0.05), 443 genes were up-regulated and 593 genes down-regulated by DPTA-NONOate (Tables 4 and 5, and Supplementary Table 3). To confirm the array results, seven genes were selected for semi-quantitative RT-PCR analysis. In every case the data from the microarray analyses were confirmed (Figure 1).
Table 4.
Genes up-regulated by DPTA-NONOate in Entamoeba histolytica HM-1:IMSS. The probe ID, accession number, description, baseline expression level, fold-change, p-value, and regulation under other conditions are shown. The 30 most highly regulated genes are listed. HS (heat shock) (adapted from Hackney et al); Colitis (ameba from a mouse model of amebic colitis) (adapted from Gilchrist et al); Cysts (E. histolytica cysts) (adapted from Ehrenkaufer et al).
| Probe ID | Accession number | Description | Baseline expression level | Fold-change | p-value | Regulated under other conditions? |
|---|---|---|---|---|---|---|
| 194.m00102_s_at | XM_645780 | hypothetical protein | 0.06 | 380.2 | 0.009 | HS |
| 363.m00049_x_at | XM_643869 | hypothetical protein | 0.17 | 305.5 | 0.002 | - |
| 256.m00084_x_at | XM_644865 | hypothetical protein | 0.12 | 252.8 | 0.002 | - |
| 248.m00060_s_at | XM_644979 | hypothetical protein | 0.05 | 240.9 | 0.006 | - |
| 692.m00024_s_at | XM_642872 | hypothetical protein | 0.05 | 219.5 | 0.003 | - |
| 194.m00123_x_at | XM_645776 | dUTP diphosphatase, putative | 0.11 | 191.5 | 0.003 | - |
| 266.m00066_s_at | XM_644755 | hypothetical protein | 0.13 | 176.2 | 0.004 | - |
| 654.m00032_x_at | XM_642891 | hypothetical protein | 1.39 | 162.9 | 0.015 | - |
| 654.m00031_s_at | XM_642894 | hypothetical protein | 0.06 | 130.4 | 0.008 | - |
| 256.m00083_x_at | XM_644864 | hypothetical protein | 1.22 | 123.6 | 0.006 | - |
| 397.m00055_s_at | XM_643659 | hypothetical protein | 0.18 | 119.3 | 0.004 | - |
| 692.m00026_s_at | XM_642873 | hypothetical protein | 0.06 | 107.7 | 0.010 | - |
| 375.m00057_x_at | XM_643790 | deoxyuridine 5 -triphosphate nucleotidohydrolase | 0.05 | 82.54 | 0.020 | - |
| 711.m00021_x_at | XM_642861 | hypothetical protein | 0.10 | 81.62 | 0.006 | - |
| 397.m00062_x_at | XM_643664 | hypothetical protein | 1.35 | 58.47 | 0.019 | - |
| 397.m00054_x_at | XM_643658 | hypothetical protein | 0.06 | 57.66 | 0.020 | - |
| 36.m00211_at | XM_650002 | hypothetical protein | 0.09 | 49.28 | 0.005 | - |
| 245.m00039_x_at | XM_645025 | hypothetical protein | 0.47 | 22.34 | 0.014 | - |
| 301.m00039_s_at | XM_644356 | heat shock protein 70, putative | 0.30 | 21.14 | 0.003 | - |
| 81.m00150_s_at | XM_648418 | heat shock protein 101, putative | 1.54 | 20.48 | 0.001 | - |
| 450.m00030_at | XM_643384 | hypothetical protein | 0.20 | 19.96 | 0.027 | Colitis |
| 356.m00029_s_at | XM_643911 | hypothetical protein | 0.18 | 16.2 | 0.008 | - |
| 7.m00453_s_at | XM_651737 | hypothetical protein | 0.30 | 14.84 | 0.001 | - |
| 344.m00044_at | XM_644000 | hypothetical protein | 0.24 | 13.53 | 0.033 | Cysts |
| 295.m00030_at | XM_644416 | conserved hypothetical protein | 0.16 | 13.28 | 0.048 | - |
| 134.m00124_at | XM_646949 | heat shock protein, Hsp20 family, putative | 1.44 | 12.29 | 0.016 | - |
| 33.m00209_x_at | XM_650174 | hypothetical protein | 0.10 | 12.1 | 0.004 | - |
| 141.m00082_at | XM_646820 | protein kinase, putative | 0.54 | 12.09 | 0.027 | - |
| 20.m00272_x_at | XM_650868 | conserved hypothetical protein | 0.17 | 9.86 | 0.043 | - |
| 796.m00013_s_at | XM_642828 | conserved hypothetical protein | 0.23 | 9.69 | 0.015 | - |
Table 5.
Genes down-regulated by DPTA-NONOate in Entamoeba histolytica HM-1:IMSS. The probe ID, accession number, description, baseline expression level, fold-change, p-value, and regulation under other conditions are shown. The 30 most highly regulated genes are listed. HS (heat shock) (adapted from Hackney et al); Colitis (ameba from a mouse model of amebic colitis) (adapted from Gilchrist et al); Trophs (E. histolytica trophozoites) (adapted from Ehrenkaufer et al).
| Probe ID | Accession number | Description | Baseline expression level | Fold-change | p-value | Regulated under other conditions? |
|---|---|---|---|---|---|---|
| 4.m00678_s_at | XM_652013 | hypothetical protein | 41.94 | −49.02 | 0.002 | - |
| 172.m00078_at | XM_646199 | hypothetical protein | 0.86 | −14.43 | 0.001 | HS |
| 99.m00179_at | XM_647858 | inositol polyphosphate kinase, putative | 4.53 | −11.48 | 0.001 | - |
| 341.m00039_s_at | XM_644035 | hypothetical protein | 2.41 | −10.59 | 0.015 | - |
| 229.m00063_at | XM_645215 | hypothetical protein | 5.04 | −9.35 | 0.019 | - |
| 221.m00089_s_at | XM_645315 | hypothetical protein | 3.74 | −9.01 | 0.002 | - |
| 13.m00349_at | XM_651311 | protein kinase, putative | 2.17 | −8.85 | 0.004 | - |
| 65.m00147_at | XM_648922 | hypothetical protein | 2.93 | −8.47 | 0.003 | Trophs + HS |
| 255.m00049_at | XM_644881 | conserved hypothetical protein | 1.25 | −8.2 | 0.022 | - |
| 93.m00151_at | XM_648019 | WD repeat protein | 1.99 | −8.2 | 0.004 | - |
| 22.m00263_at | XM_650739 | hypothetical protein | 1.42 | −7.94 | 0.019 | - |
| 74.m00199_at | XM_648611 | hypothetical protein | 2.53 | −7.87 | 0.001 | - |
| 232.m00071_at | XM_645176 | hypothetical protein | 1.15 | −7.46 | 0.004 | - |
| 4.m00607_at | XM_652082 | protein kinase, putative | 3.19 | −7.46 | 0.002 | HS |
| 408.m00044_at | XM_643584 | hypothetical protein | 0.95 | −7.46 | 0.043 | - |
| 86.m00158_s_at | XM_648244 | DEAD DEAH box helicase, putative | 0.46 | −7.35 | 0.044 | HS |
| 32.m00207_at | XM_650196 | zinc finger protein, putative | 12.62 | −6.94 | 0.009 | - |
| 32.m00202_s_at | XM_650191 | hypothetical protein | 0.72 | −6.9 | 0.008 | - |
| 4.m00640_at | XM_652059 | conserved hypothetical protein | 3.07 | −6.9 | 0.000 | - |
| 113.m00152_at | XM_647500 | BspA-like leucine rich repeat protein, putative | 1.09 | −6.85 | 0.015 | Trophs |
| 289.m00071_s_at | XM_644481 | putative GTPase | 7.01 | −6.29 | 0.000 | - |
| 82.m00139_s_at | XM_648379 | hypothetical protein | 0.59 | −6.21 | 0.005 | - |
| 408.m00045_s_at | XM_643583 | hypothetical protein | 2.43 | −6.17 | 0.007 | HS |
| 2.m00528_at | XM_652349 | protein phosphatise, putative | 0.35 | −5.99 | 0.028 | HS |
| 201.m00110_at | XM_645662 | hypothetical protein | 1.35 | −5.92 | 0.006 | HS |
| 12.m00322_at | XM_651338 | hypothetical protein | 3.67 | −5.81 | 0.030 | HS |
| 48.m00216_s_at | XM_649527 | hypothetical protein | 3.05 | −5.75 | 0.019 | Colitis |
| 98.m00148_s_at | XM_647890 | hypothetical protein | 1.02 | −5.52 | 0.001 | - |
| 66.m00150_at | XM_648887 | hypothetical protein | 0.39 | −5.49 | 0.000 | HS |
| 816.m00009_at | XM_642817 | hypothetical protein | 4.41 | −5.38 | 0.049 | HS |
Figure 1.
Semi-quantitative RT-PCR analysis of selected genes, for validation of array results. Up - Genes induced by exposure to the corresponding stress; Down Genes repressed by exposure to the corresponding stress.
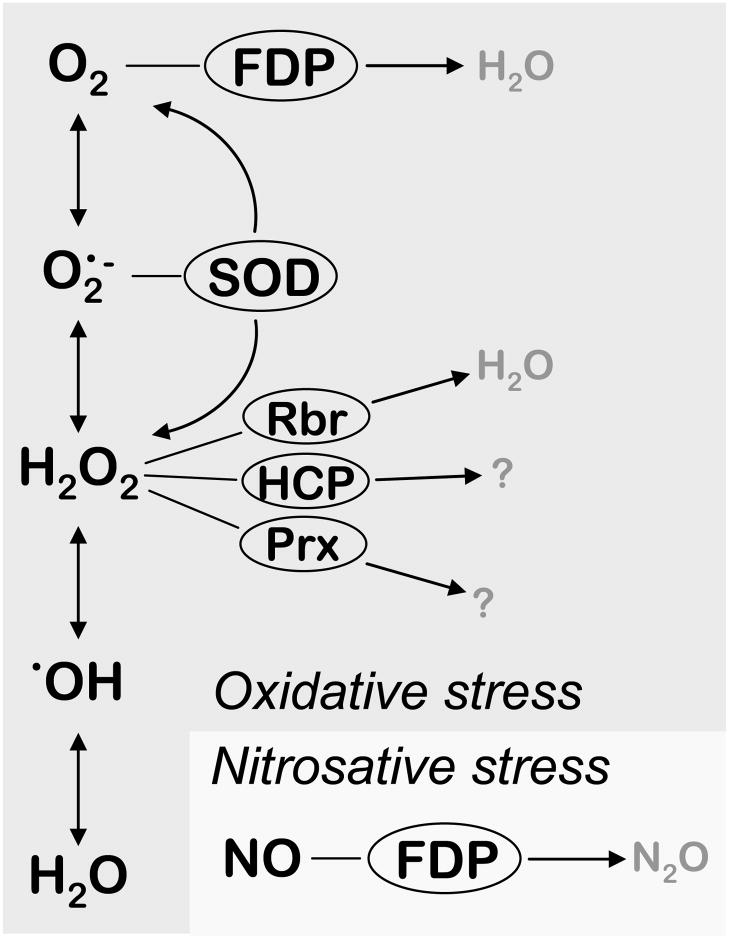
Genes encoding known detoxification systems in E. histolytica trophozoites are not significantly modulated by oxidative or nitrosative stresses
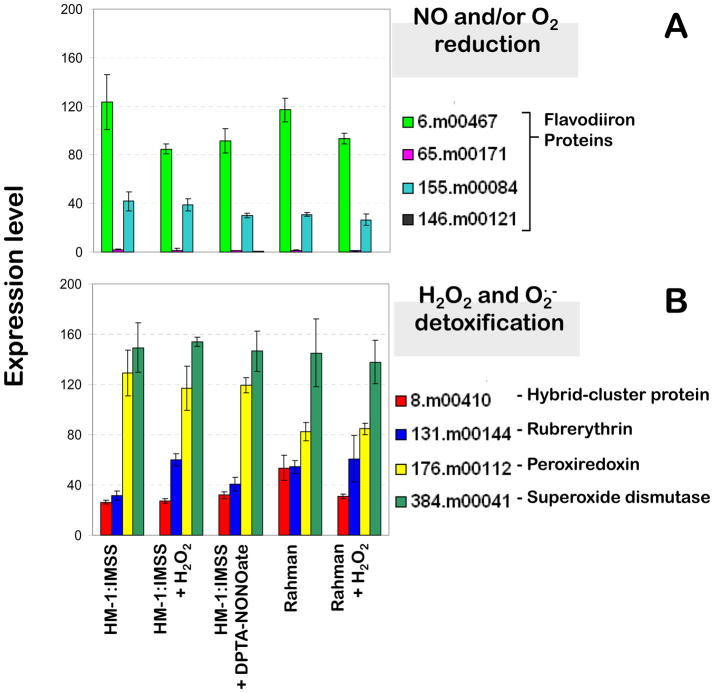
In E. histolytica HM-1:IMSS strain growing under standard axenic culture conditions, the basal transcription levels of genes encoding the enzymatic ROS and RNS detoxification systems are generally high (Figure 2). Putative detoxification systems for reactive oxygen and nitrogen species, which were identified in the genome of E. histolytica are depicted in Scheme 1. Flavodiiron proteins (FDPs) have nitric oxide (NO) and/or molecular oxygen (O2) reductase activities, although the molecular basis for substrate selectivity remains elusive (Vicente et al., 2007). Out of the four copies of genes encoding FDPs, two (6.m00467 and 155.m00084) have much higher transcription levels than the other two homologues (65.m00171 and 146.m00121) (Figure 2A). The gene encoding superoxide dismutase (384.m00041, SOD), the sole known superoxide detoxifying enzyme in E. histolytica’s genome, displays high transcription levels in all conditions tested with no significant changes under either stress condition (Figure 2B). The same is observed for gene products involved in hydrogen peroxide detoxification, since the three distinct scavenging systems identified in its genome also display high and almost invariable expression levels: peroxiredoxin (176.m00112, Prx), rubrerythrin (131.m00144, Rbr), and hybrid-cluster protein (8.m00410, HCP) (Figure 2B). Although these genes do not display significant transcriptional changes upon H2O2 or NO exposure, basal expression levels of some of these genes do vary across Entamoeba: the FDP-encoding 6.m00467 gene, which is the most highly expressed FDP in E. histolytica HM-1:IMSS, has a lower expression in the non-virulent species, E. dispar, and the gene coding for peroxiredoxin, has reduced expression in both E. histolytica Rahman and E. dispar (Davis et al., 2006; MacFarlane and Singh, 2006). Moreover, a proteomic analysis revealed that E. histolytica superoxide dismutase and peroxiredoxin have significantly higher expression levels in the virulent E. histolytica HM-1:IMSS strain as compared to the non-virulent Rahman strain (Davis et al., 2006), indicating a possible contribution by these proteins to amebic virulence.
Figure 2.
Expression levels of genes encoding identified detoxification pathways for reactive oxygen and nitrogen species (top and middle panels) and iron-sulfur center assembly systems (bottom panel) for unchallenged cultures of E. histolytica strains HM-1:IMSS (pathogenic) and Rahman (non-pathogenic), and for cultures challenged with hydrogen peroxide and the nitric oxide releaser DPTA-NONOate.
Scheme 1.
Putative detoxification pathways for reactive oxygen and nitrogen species identified in the genome of E. histolytica HM-1:IMSS. Reactive oxygen species depicted as the sequential one-electron reduced intermediates of oxygen reduction. FDP – flavodiiron proteins, oxygen and/or nitric oxide reductases; SOD – superoxide dismutase; Rbr – rubrerythrin, hydrogen peroxide reductase; HCP – hybrid-cluster protein; Prx – peroxiredoxin.
Although the majority of genes encoding the ROS and RNS stress detoxifying proteins were not significantly changed in parasites exposed to oxidative and nitrosative stresses, some transcriptional changes were noted. Three genes encoding iron-sulfur flavoproteins (35.m00253, 312.m00037 and 41.m00244) that are proposed to be related to the oxidative stress response, but whose function remains undetermined (Cruz and Ferry, 2006), were up-regulated by H2O2. Two homologous genes (187.m00073 and 646.m00021) encoding iron-sulfur flavoproteins were induced by nitrosative stress. Additionally, one of the four flavodiiron proteins (146.m00121) was up-regulated by NO (4.8-fold). Besides FDPs, no other known NO-detoxifying enzyme has thus far been identified in E. histolytica’s genome.
Aside from these limited changes, most of the genes in the ROS and RNS detoxification pathways were not transcriptionally regulated under a number of different conditions that model the host-pathogen interaction (parasites colonizing the mouse intestine) (Gilchrist et al., 2006) or trophozoite to cyst stage conversion (Ehrenkaufer et al., 2007). Thus, contrary to what has been observed in many prokaryotes and a few eukaryotes (Rodionov et al., 2005; Paget and Buttner, 2003), the transcriptional response at the detoxification level appears constitutive as E. histolytica trophozoites have a number of scavenging enzymes that are robustly expressed even under basal conditions.
Genes up-regulated in E. histolytica HM-1:IMSS in response to oxidative stress
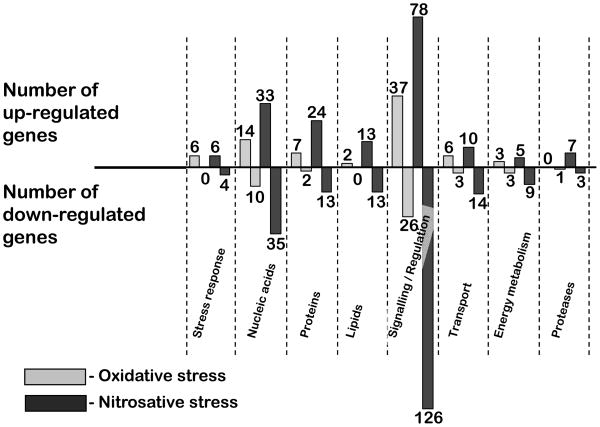
Out of the 185 genes up-regulated by oxidative stress, 107 (58%) are annotated as encoding hypothetical proteins. The remaining genes code for proteins with roles in signaling/regulatory processes, metabolic/repair processes, energy metabolism, stress response, and transport (Figure 3). The categories comprising the largest numbers of up-regulated genes include the repair systems and signaling/regulatory pathways, which are detailed below.
Figure 3.
Transcriptional profiles of E. histolytica HM-1:IMSS exposed to oxidative and nitrosative stress. Genes were grouped according to putative functions inferred from the respective annotations.
Response to DNA damage
DNA is a known cellular target of oxidative stress (reviewed in (D’Autreaux and Toledano, 2007)) and 8% of the genes up-regulated by H2O2 in E. histolytica encode putative nucleic acids metabolism/repair proteins (Figure 3, Table 2). Two genes encoding deoxyuridine triphosphate nucleotidehydrolase (dUTPase) (194.m00123 and 375.m00057), considered to be essential for DNA integrity (Nguyen et al., 2005), are among the genes most highly up-regulated by H2O2 (51 and 22-fold, respectively). A putative homologue of polynucleotide kinase-3-phosphatase (1.m00709, 4-fold) is involved in the repair of nicks and gaps in damaged DNA, including oxidatively generated DNA strand breaks (Blondal et al., 2005; Betti et al., 2001). A similar function in the repair of oxidative damage to DNA has been attributed to the large family of MutS repair proteins (Dzierzbicki et al., 2004; Chang et al., 2002; Khil and Camerini-Otero, 2002). A gene coding for a homologue of MutS DNA mismatch repair proteins in E. histolytica (115.m00144) was up-regulated by oxidative stress (2-fold). We also observed induction by hydrogen peroxide of a gene encoding a homologue of DEAD DEAH box helicase (27.m00240, 3-fold). The homologous gene from the pathogenic fungus Candida albicans is regulated by Cap1p, a transcription factor involved in oxidative stress tolerance (Wang et al., 2006).
Response to protein and lipid damage
Hydrogen peroxide oxidatively damages proteins, mainly by reacting with thiol groups from cysteine side chains, and also with redox cofactors such as metal centers. Moreover, the hydroxyl radical generated in the reaction between H2O2 and free Fe2+ further reacts with amino acid residues, namely methionine (Halliwell and Gutteridge, 2007). In E. histolytica HM-1:IMSS challenged with H2O2, 4% of the up-regulated genes encode homologues of proteins involved in the repair or degradation of misfolded proteins (Figure 3 and Table 2). Genes encoding homologues of chaperone-like heat-shock proteins were up-regulated by oxidative stress, for example HSP40/DnaJ (21.m00247) which stimulates the ATPase activity of HSP70 chaperones (Qiu et al., 2006) and HSP101 (64.m00187). An ubiquitin-conjugating enzyme (142.m00162), which may mark misfolded proteins for degradation, was also up-regulated. A homologue of peptidyl-prolyl cis-trans isomerase (75.m00189), a protein with a role in the repair of oxidatively damaged proteins from plants (Shapiguzov et al., 2006) and mammalian cells (Hong et al., 2002; Santos et al., 2000) was also upregulated.
Oxidative damage exerted on lipids by H2O2 and other ROS may result in loss of membrane integrity (Halliwell and Gutteridge, 2007; Colles and Chisolm, 2000). Accordingly, some of the genes up-regulated by H2O2 have a role in lipid metabolism: phosphatidylcholine transport-like protein (99.m00180), and a phospholipid-transporting P-type ATPase (75.m00173), homologous to aminophospholipid translocases. Increased expression of the gene coding for glucosamine-6-phosphate N-acetyltransferase (34.m00243) suggests a requirement for cell wall repair and/or the assembly of cell wall proteins (Hurtado-Guerrero et al., 2007). It has been reported that a mutant strain of C. albicans with a deletion in this gene displays decreased virulence (Mio et al., 2000).
Signaling and regulatory pathways
The largest group of genes up-regulated by H2O2 (20%) is that encoding proteins that may be related to signaling pathways including protein kinases, phosphatases and acetyltransferases. E. histolytica possesses an intricate phosphorylation network involving a large number of kinases, which have roles in diverse cell processes (Anamika et al., 2008), including effects on parasite virulence (Beck et al., 2005; Batista Ede and de Souza, 2004). Other genes that may be involved in signal transduction mechanisms include those that encode GTPases and related proteins, such as the G protein regulator phosducin (407.m00055) (2.4-fold). The two genes coding for Rab GTPases, RabI1 (20.m00304, 3.6-fold) and RabM1 (103.m00161, 2.0-fold) (Saito-Nakano et al., 2005), are homologues of Rab1B and Rab15, respectively involved in transport and endocytosis. A small GTPase CDC42 (encoded by 219.m00107, 11.3-fold) has a human homologue that regulates adherence and membrane permeability (Broman et al., 2007) and more recently has been proposed to have a role in regulating protein ubiquitination (Shen et al., 2008). In addition, ArfA3 (147.m00113) (Clark et al., 2007), an ADP ribosylation factor (ARF) GTPase, was induced by H2O2 (2.6-fold). ARF GTPases participate in the regulation of organelle structure and vesicular trafficking, besides participating in diverse cellular functions, such as cytokinesis, endocytosis, phagocytosis and cell adhesion (D’Souza-Schorey and Chavrier, 2006). We also observed induction of a gene encoding a putative copine (333.m00053) (2.1-fold), whose homologue from Dictyostelium discoideum plays a role in cytokinesis and contractile vacuole function (Damer et al., 2007). An AIG1 plant-like antibacterial protein (451.m00039) was induced (2.1-fold) and was also significantly up-regulated upon E. histolytica invasion of the mouse intestine (Gilchrist et al., 2006). Two genes encoding BspA-like leucine rich proteins (1.m00705 and 310.m00070) were induced by H2O2 (6.6 and 2.4 fold, respectively). Homologues of these proteins from Tannerella forsythia have been reported to modulate the host response by interfering with interleukin-8 expression and ultimately contributing to invasion of the epithelial barrier (Onishi et al., 2008; Inagaki et al., 2006). In summary, E. histolytica responds to oxidative stress by inducing genes coding for a multitude of signaling/regulatory systems, with roles in diverse cellular functions.
Other pathways
Since E. histolytica redox homeostasis is sustained by free (homo)cysteine, in the absence of glutathione or any related metabolic enzymes, it is noteworthy that sulfur amino-acid metabolism enzymes are regulated by oxidative stress. The gene encoding the MGL1 isotype of methionine-γ-lyase (MGL1, 395.m00028), which in eukaryotes has thus far only been found in plants (Rebeille et al., 2006), E. histolytica (Tokoro et al., 2003) and T. vaginalis (Nozaki et al., 2005) is up-regulated (3.6-fold) by H2O2. This enzyme catalyzes elimination reactions with a wide range of substrates, such as methionine, (homo)cysteine, and substituted (homo)serine homologues (Nozaki et al., 2005). Moreover, the products of methionine degradation by MGL not only supply the energy metabolism, but may also be implicated in amebic pathogenicity since thiols and hydrogen sulfide can become toxic for the host cells by permeating the membrane barrier and interfering with host signaling systems (Nozaki et al., 2005).
Genes down-regulated in E. histolytica HM-1:IMSS in response to oxidative stress
A total of 102 genes were down-regulated by H2O2 exposure; 52 (51%) of which encode hypothetical proteins (Table 3; Supplementary Table 2). As displayed in Figure 3, the majority of the E. histolytica HM-1:IMSS genes repressed by oxidative stress appear to be involved in the same general processes whose components were up-regulated, namely repair systems (mostly for nucleic acids), signaling pathways, and regulatory mechanisms. The numerous genes possibly involved in signaling and regulatory processes encode a number of putative GTPases, one of which (64.m00149) encodes a protein with RhoGEF and ArfGAP domains and has a homologue proposed to be involved in D. discoideum development (Mondal et al., 2007).
Genes up-regulated in E. histolytica HM-1:IMSS in response to nitrosative stress
From the 443 genes up-regulated by nitrosative stress, 248 (56%) are annotated as encoding unknown hypothetical proteins. Similar to what was observed with oxidative stress, the largest groups of genes induced by nitrosative stress are those related with signaling / regulatory processes and repair systems for nucleic acids, proteins and lipids.
Response to DNA damage
Upon exposure to NO, 33 up-regulated genes (7%) encode proteins involved in the metabolism and/or repair of nucleic acids (Figure 3). This reflects the direct damage exerted by reactive nitrogen species on nucleic acids, either NO directly or NO-derived species, such as peroxynitrite (Halliwell and Gutteridge, 2007). In Salmonella enterica, NO impairs DNA replication and arrests growth (Schapiro et al., 2003). Two genes encoding dUTPase (194.m00123 and 375.m00057) are among the genes most highly up-regulated by NO (192 and 82-fold, respectively), and are essential enzymes for DNA integrity (Nguyen et al., 2005). A homologue of MutS DNA mismatch repair proteins (115.m00144), was also up-regulated by nitrosative stress (3.3-fold). We observed induction of genes encoding a panoply of DNA repair enzymes, such as DNA excision repair protein (117.m00190) (4.0-fold), Rad3 DNA repair helicase (197.m00081) (2.1-fold), and Rad50 (102.m00081) (2.0-fold).
Response to protein and lipid damage
NO and its derived species can be harmful to proteins involved in all cellular processes, both by reacting with specific amino acids and/or with redox active cofactors, mostly metal centers. The reaction of RNS with amino acids results in S-nitrosylation or nitration of the corresponding side chains. NO and other RNS can also bind transiently or permanently to protein metal cofactors and either inhibit or definitely inactivate its function. The resulting modifications can be damaging to the overall folding and structural arrangements of proteins, which was confirmed by the significant number of E. histolytica genes with these functions that were up-regulated by NO exposure (5% of all upregulated genes by NO) (Figure 3). The vast majority of these genes encode either heat-shock proteins, HSP20 (134.m00124) (12.3-fold), three homologues of HSP40 (12.m00313, 28.m00338 and 28.m00311) (5.6, 3.8 and 3.3-fold, respectively), two of HSP70 (301.m00039 and 584.m00019) (21.1 and 2.3-fold), and three of HSP101 (81.m00150, 64.m00178 and 64.m00187) (20.5, 8.8 and 6.1-fold, respectively), or ubiquitin-conjugating proteins, which are all involved in degradation / repair of misfolded proteins and often responsive to many types of cellular stress.
Other major targets of NO and RNS reactivity are lipids. While this interaction may damage lipids, NO and RNS may also act as antioxidants, reacting with lipid radicals generated by oxidative stress, thus blocking and terminating harmful lipid radical chain reactions (Halliwell and Gutteridge, 2007). The low polarity of NO allows it to freely diffuse through membranes. Damage to lipids can not only affect metabolic processes, but most importantly may result in cell membrane permeability. The effect of nitrosative stress on lipids in E. histolytica is directly observable by the number of up-regulated genes encoding proteins related to lipid metabolism (3% of all induced genes): a PCTP-like protein (99.m00180) (2.8-fold), a myotubularin lipid phosphatase (35.m00216) (2.6-fold), and a phospholipid-transporting P-type ATPase (75.m00173) (2.6-fold). In addition to affecting the membranous environment, NO reacts directly with membrane proteins, such as ion channels and transport proteins, thus disturbing ion homeostasis. Two genes encoding putative importins (310.m00066 and 1.m00747) were induced by nitrosative stress (4.0 and 2.1-fold, respectively). The nuclear transport pathway mediated by importin in human cells has recently been shown to be impaired by nitrosative stress (Qu et al., 2007). A putative sodium/proton antiporter (152.m00122) was up-regulated by NO (3.5-fold). Nitric oxide has been shown to inhibit Na+/H+ exchange activity, mediated by the cyclic GMP signal transduction pathway (Gill et al., 2002). Genes encoding homologues of amino acid transporters (2.m00499, 46.m00238 and 82.m00146) were also induced by nitrosative stress.
Signaling and regulatory processes induced by nitrosative stress
The largest groups of regulated genes (18%) are those involved in signaling and regulation of cellular processes (Figure 3). Part of this group overlaps with genes found to be up-regulated by oxidative stress. From the 78 genes in this group, at least 21 encode putative protein kinases, 7 code for protein phosphatases, and 5 encode acetyltransferases. A significant number of GTPases (4 Rab-type, 3 Rho family, 1 Rap Ran GTPase activating protein, and 2 Rab GTPase activating proteins) and zinc finger proteins were also up-regulated by nitrosative stress. Reversible inhibition of DNA-binding zinc containing proteins by nitrosative stress has been thought to be part of NO-related DNA replication inhibition in pathogenic bacteria (Schapiro et al., 2003).
Other pathways
A putative FAD-binding NADH oxidoreductase (328.m00064) was induced (2.4-fold) by NO. Homologues of this enzyme are primary electron carriers in electron transfer chains with NO-detoxifying activity (Saraiva et al., 2004). A nitroreductase (13.m00321) was induced upon NO exposure (3.9-fold). Nitroreductases are flavoproteins that catalyze the reduction of nitro groups in a wide range of substrates and are associated with resistance to the anti-parasitic antibiotic metronidazole (Mendz and Megraud, 2002). The gene that codes for methionine-γ-lyase (MGL1, 395.m00028) was also up-regulated by nitrosative stress (4.4-fold). The products of methionine degradation by MGL may permeate the host cells membrane barrier and interfere with signaling systems (Nozaki et al., 2005). Interestingly, three genes encoding putative cysteine proteases were induced by NO (24.m00271, 180.m00101, and 97.m00133) (2.0, 2.1, and 2.1-fold, respectively). Cysteine proteases are an important virulence factor for E. histolytica, and are involved in cytotoxicity and colonic invasion (Stanley, 2003).
Genes downregulated by nitrosative stress
A surprisingly large number of genes had decreased expression upon exposure to DPTA-NONOate: 592 genes of which 366 (62%) are annotated as unknown hypothetical proteins (Table 4; Supplementary Table 3). The profile of down-regulated genes and their putative functions is shown in Figure 3. There is a marked repression of genes involved in the metabolism of nucleic acids (6%), repair and degradation of misfolded proteins (2%) and lipids (2%), and in transport (2%). The largest category of genes is that involved in cell signaling and regulatory processes (21%).
A significant down-regulation of genes encoding proteins involved in RNA metabolism was observed. Genes encoding RNA polymerase subunits were repressed by nitrosative stress (64.m00185, 73.m00160, 59.m00197, 51.m00170, and 406.m00050) (4.0, 2.1, 3.2, 2.5, and 3.2-fold, respectively), suggesting that the overall transcription may be slowed-down. Exposure to nitric oxide led to repression of genes coding for putative proteins related to ubiquitination (three cullin homologues, one ubiquitin and one ubiquitin-conjugating enzyme) and ribosome-related proteins. We also observed down-regulation of genes encoding proteins involved in (glyco)lipid metabolism and glycosylation, such as phosphatidylinositol-glycan biosynthesis class C protein (80.m00142) (3.4-fold), N-acetylglucosaminyl transferase (32.m00239) (3.3-fold) and N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase (52.m00150) (2.5-fold). These genes are likely to participate in the biosynthesis of glycosylphosphatidylinositol (GPI) anchors, which have been proposed to be involved in the regulation of cell growth, endocytosis and the adhesion to target cells by E. histolytica (Vats et al., 2005). The bulk of genes repressed by NO that encode putative transport proteins comprise those coding for importin subunits.
From the extended list of NO repressed genes encoding putative signaling/regulatory proteins, 21 code for protein kinases and 23 for GTPases (9 Rab-type, 6 Ras-type and 8 Rho-type). There is also down-regulation of 10 genes coding for zinc finger proteins, 6 for WD repeat proteins and 10 for leucine rich proteins. Significantly, exposure to NO resulted in repression of gene 43.m00187 (2.4-fold). This gene encodes a key regulatory protein, phosphatidylinositol (PI)-3,4,5-trisphosphate 3-phosphatase (PTEN), which controls diverse cellular processes as the antagonist of PI 3-kinase (constituting the PTEN/PIK signaling pathway). E. histolytica PI 3-kinase inhibition has been shown to impair proliferation, encystation and autophagy (Picazarri et al., 2008).
Common transcriptional responses of E. histolytica HM-1:IMSS to oxidative and nitrosative stress
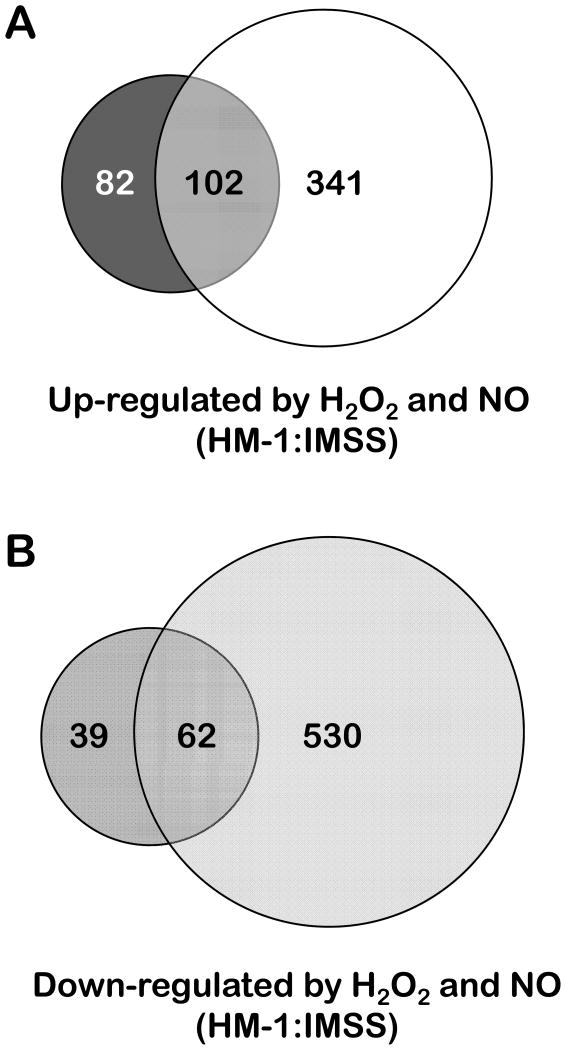
We observed a substantial overlap in genes transcriptionally regulated byoxidative and nitrosative stress with 102 genes up-regulated under both stress conditions including those genes that are the most induced under each of those conditions (Figure 4) (Supplementary Table 5). Of these 102 genes, 63 encode unknown hypothetical proteins. The remaining ones are distributed according to the function profiles observed for the individual stresses (Figure 3). The observation of commonly induced repair systems-encoding genes by both stress types reflects the fact that nucleic acids and proteins are among the cell components that suffer similar damaging reactions upon exposure to both ROS and RNS (Halliwell and Gutteridge, 2007). The two genes showing the highest induction folds by both stress types are deoxyuridine 5-triphosphate nucleotidohydrolase (dUTPase, 194.m00123 and 375.m00057). As mentioned above, this enzyme is involved in nucleotide metabolism and contributes to DNA integrity and has been previously assessed as a potential target for anti-parasitic drugs (Nguyen et al., 2005). Two genes (1.m00709 and 115.m00144) are homologues of a polynucleotide kinase-3-phosphatase and a MutS DNA mismatch repair protein, respectively, and are both involved in the repair of oxidatively damaged DNA (Dzierzbicki et al., 2004; Betti et al., 2001). Both stresses induced genes encoding systems involved in the degradation and repair of misfolded proteins such as heat-shock proteins HSP101 and DnaJ/HSP40 (64.m00187 and 21.m00247) and an ubiquitin-conjugating enzyme (142.m00162).
Figure 4.
Venn diagrams depicting the overlap between the transcriptional changes observed upon exposing E. histolytica HM-1:IMSS to oxidative and nitrosative stress.
We observed induction under oxidative and nitrosative stresses of genes involved in lipid metabolism, transport and glycosylation. Two genes encoding proteins involved in lipid metabolism were phospholipid-transporting P-type ATPase (75.m00173) and phosphatidylcholine transfer protein (99.m00180). An aminophospholipid translocase (75.m00173), upon nitrosative inhibition in apoptotic cells, leads to an accumulation of extra-cellular phosphatidylserine that marks cells for macrophage engulfment (Tyurina et al., 2007). E. histolytica has been reported to recognize externalized phosphatidylserine on the surface of host cells and target these cells for phagocytosis (Boettner et al., 2005; Huston et al., 2003). Genes encoding putative transport proteins (mainly ion transporters) were induced both by oxidative and nitrosative stress, consistent with the observation that nitrosative and oxidative stress disturb ion homeostasis (Beausejour et al., 2007; Orsenigo et al., 2007). A dTDP-D-glucose 4,6-dehydratase (116.m00108) was up-regulated by both types of stress; this enzyme is related to glycosylation in pathogenic bacteria (Allard et al., 2001). Also induced by both stresses was a glucosamine-6-phosphate N-acetyltransferase (34.m00243) whose homologous gene in C. albicans has a role in virulence (Mio et al., 2000).
It is worth noting that all four protein families that respond to oxidative stress in all eukaryotic kingdoms (Vandenbroucke et al., 2008) (heat-shock proteins, ubiquitin-conjugating enzymes, kinases and small GTPases) have homologues which were transcriptionally regulated in E. histolytica by both oxidative and nitrosative stress: HSP101 (64.m00187), an ubiquitin-conjugating enzyme (142.m00162), three protein kinases (199.m00096, 46.m00221 and 275.m00123) and one Rab family GTPase (20.m00304).
In previous sections we emphasized the induction of a gene encoding methionine-γ-lyase (MGL) (395.m00028) by both stress types, since its methionine degradation products may contribute to the permeation and disruption of the host epithelial barrier by E. histolytica. MGL is an attractive drug target which is being actively pursued (Sato et al., 2008; Ali and Nozaki, 2007).
The majority of the genes most significantly repressed by each individual stress were down-regulated under both stresses (Supplementary Table 5). Sixty-two genes were down-regulated by both stresses, 35 (56%) of which encode unknown hypothetical proteins. The remaining genes encode putative proteins mostly involved in signaling and regulatory mechanisms, and also include a few genes encoding nucleic acid metabolism proteins. No genes encoding repair systems for misfolded proteins or lipids were commonly down-regulated under both stresses, although both stresses did regulate genes in these pathways. In summary, the overlap between genes responsive to oxidative and nitrosative stress by E. histolytica is significant and shows common strategies to overcome the cytotoxicity of reactive oxygen and nitrogen species.
We also analyzed the overlap between the transcriptional responses of E. histolytica to oxidative or nitrosative stress and other conditions. As expected, some genes regulated by H2O2 were also regulated by cyst conversion indicating that these genes respond to multiple stress conditions (Ehrenkaufer et al., 2007; Hackney et al., 2007; Weber et al., 2006). A significant fraction of the genes repressed by H2O2 (47%) were also repressed in response to heat-shock (Hackney et al., 2007; Weber et al., 2006). Of the 443 genes up-regulated by DPTA-NONOate, 50 were also induced by heat-shock, some of which encode putative repair systems for damaged nucleic acids and proteins, 25 were also up-regulated in cysts, but only a few were induced upon colonization of the mouse intestine (Gilchrist et al., 2006). Almost half of the 592 genes down-regulated by nitrosative stress were also down-regulated by heat-shock, a much smaller fraction (10%) was down-regulated in cysts, and 12 genes were down-regulated during an in vivo mouse colitis model (Table 5 and Supplementary Table 3). The limited overlap between genes regulated by oxidative and nitrosative stress and the changes seen during colonization of the mouse colon and hepatic invasion deserve further comment (Santi-Rocca et al., 2008; Gilchrist et al., 2006). Whether this represents technical issues (differences in time points of colonic animal model, colonic model represents more colonization than invasion, or differences in the arrays used for the liver invasion model compared to these studies) or biological differences (animal models are more complex and difficult to compare directly to studies utilizing in vitro methods) is not currently clear. However, since the genes and pathways transcriptionally modulated by oxidative or nitrosative stress overlap significantly with changes seen in multiple other systems using the same stress conditions, the approach with the in vitro model of stress exposure is identifying conserved mechanisms of stress response between Entamoeba and other systems.
Differential response to oxidative stress may contribute to the decreased virulence phenotype of the E. histolytica Rahman strain
In order to determine whether virulent and non-virulent amebic strains had differing responses to oxidative stress, we characterized the transcriptional changes of the non-pathogenic E. histolytica Rahman strain to hydrogen peroxide. It has previously been demonstrated that the non-virulent E. histolytica Rahman is more susceptible to hydrogen peroxide than the pathogenic E. histolytica HM-1:IMSS and that higher levels of peroxiredoxin in the virulent strain contribute to E. histolytica’s virulence (Davis et al., 2006) Three arrays using RNA from E. histolytica Rahman exposed to 1 mM H2O2 for one hour were performed and displayed good correlation values (0.97–0.99) (Table 1). Expression was compared to array data from E. histolytica Rahman under standard axenic culture conditions (Ehrenkaufer et al., 2007). Using the same fold-change criteria described in the Materials and Methods (≥2-fold change and FDR<0.5), a total of 153 genes were up-regulated by H2O2 in E. histolytica Rahman and 65 genes were down-regulated (Tables 6 and 7; Supplementary Table 4).
Table 6.
Genes up-regulated by hydrogen peroxide in Entamoeba histolytica Rahman. The probe ID, accession number, description, baseline expression level, fold-change and p-value are shown. The 30 most highly regulated genes are listed. If the gene is regulated and its fold-change in HM-1:IMSS are listed.
| Probe ID | Accession number | Description | Baseline expression level | Fold-change | p-value | Regulated in HM1:1MSS | Fold-change in HM1:IMSS |
|---|---|---|---|---|---|---|---|
| 301.m00039_s_at | XM_644356 | heat shock protein 70, putative | 0.30 | 7.37 | 0.005 | - | |
| 205.m00100_s_at | XM_645582 | hypothetical protein | 8.19 | 6.9 | 0.013 | - | |
| 134.m00124_at | XM_646949 | heat shock protein, Hsp20 family, putative | 1.44 | 6.62 | 0.006 | - | |
| 606.m00014_s_at | XM_642953 | hypothetical protein | 0.28 | 6.59 | 0.043 | - | |
| 64.m00187_s_at | XM_648976 | heat shock protein 101, putative | 36.55 | 6.53 | 0.015 | + | 2.42 |
| 8.m00393_at | XM_651687 | late competence protein, putative | 0.25 | 6.48 | 0.010 | + | 8.85 |
| 264.m00070_x_at | XM_644777 | hypothetical protein | 0.05 | 6.31 | 0.030 | - | |
| 181.m00068_s_at | XM_646040 | hsp101-related protein | 18.45 | 6.1 | 0.009 | - | |
| 562.m00023_at | XM_643023 | protein kinase, putative | 1.18 | 5.74 | 0.022 | - | |
| 256.m00083_x_at | XM_644864 | hypothetical protein | 1.22 | 5.72 | 0.050 | + | 82.85 |
| 188.m00103_at | XM_645894 | hypothetical protein | 0.48 | 5.68 | 0.038 | - | |
| 451.m00037_s_at | XM_643383 | hypothetical protein | 0.07 | 5.59 | 0.016 | - | |
| 81.m00150_s_at | XM_648418 | heat shock protein 101, putative | 1.54 | 5.51 | 0.001 | - | |
| 110.m00118_x_at | XM_647568 | Rho family GTPase | 0.11 | 5.47 | 0.030 | - | |
| 64.m00178_s_at | XM_648975 | heat shock protein 101, putative | 3.41 | 5.24 | 0.011 | - | |
| 654.m00032_x_at | XM_642891 | hypothetical protein | 1.39 | 5.01 | 0.047 | + | 114.90 |
| 42.m00175_at | XM_649778 | hypothetical protein | 0.96 | 4.92 | 0.018 | - | |
| 363.m00049_x_at | XM_643869 | hypothetical protein | 0.17 | 4.9 | 0.030 | + | 89.05 |
| 344.m00045_at | XM_644001 | hypothetical protein | 0.60 | 4.82 | 0.008 | + | 6.21 |
| 442.m00023_at | XM_643432 | hypothetical protein | 0.69 | 4.75 | 0.015 | + | 4.49 |
| 50.m00195_s_at | XM_649449 | hypothetical protein | 0.05 | 4.73 | 0.018 | - | |
| 30.m00249_at | XM_650277 | 1-O-acylceramide synthase, putative | 0.40 | 4.63 | 0.026 | - | |
| 39.m00253_at | XM_649881 | CXXC-rich protein | 8.57 | 4.56 | 0.009 | - | |
| 168.m00119_s_at | pseudogene, N- acetylmuraminidase | 0.35 | 4.55 | 0.029 | - | ||
| 167.m00116_x_at | XM_646306 | hypothetical protein | 0.08 | 4.48 | 0.014 | - | |
| 458.m00058_at | XM_643333 | hypothetical protein | 0.14 | 4.45 | 0.017 | - | |
| 6.m00424_at | XM_651810 | hypothetical protein | 0.22 | 4.26 | 0.012 | + | 3.00 |
| 70.m00178_s_at | XM_648752 | hypothetical protein | 0.24 | 3.96 | 0.027 | - | |
| 245.m00039_x_at | XM_645025 | hypothetical protein | 0.47 | 3.84 | 0.040 | + | 14.39 |
| 7.m00453_s_at | XM_651737 | hypothetical protein | 0.30 | 3.7 | 0.021 | + | 3.46 |
Table 7.
Genes down-regulated by hydrogen peroxide in Entamoeba histolytica Rahman. The probe ID, accession number, description, baseline expression level, fold-change and p-value are shown. The 30 most highly regulated genes are listed. If the gene is regulated and its fold-change in HM-1:IMSS are listed.
| Probe ID | Accession number | Description | Baseline expression level | Fold-change | p-value | Regulated in HM1:1MSS | Fold-change in HM1:IMSS |
|---|---|---|---|---|---|---|---|
| 50.m00168_at | XM_649471 | hypothetical protein | 107.85 | −30.49 | 0.012 | - | |
| 233.m00105_at | XM_645153 | hypothetical protein | 6.57 | −4.13 | 0.005 | + | −4.41 |
| 214.m00072_at | XM_645429 | hypothetical protein | 1.02 | −3.69 | 0.015 | - | |
| 249.m00072_at | XM_644965 | hypothetical protein | 0.24 | −3.5 | 0.042 | - | |
| 459.m00030_at | XM_643329 | hypothetical protein | 74.08 | −3.4 | 0.034 | - | |
| 25.m00245_at | XM_650545 | conserved hypothetical protein | 0.35 | −3.19 | 0.006 | - | |
| 67.m00091_x_at | XM_648866 | protein kinase, putative | 6.08 | −3.1 | 0.046 | - | |
| 113.m00152_at | XM_647500 | hypothetical protein | 1.09 | −2.92 | 0.002 | - | |
| 224.m00085_at | XM_645268 | cullin, putative | 0.44 | −2.82 | 0.033 | - | |
| 41.m00219_s_at | XM_649804 | ABC transporter, putative | 47.84 | −2.74 | 0.006 | - | |
| 380.m00029_at | XM_643755 | hypothetical protein | 0.09 | −2.72 | 0.019 | - | |
| 54.m00183_at | XM_649345 | hypothetical protein | 28.38 | −2.65 | 0.025 | + | −2.30 |
| 129.m00151_at | XM_647089 | hypothetical protein | 0.08 | −2.56 | 0.015 | - | |
| 264.m00067_at | XM_644788 | hypothetical protein | 2.21 | −2.56 | 0.034 | - | |
| 234.m00047_at | XM_645144 | hypothetical protein | 1.04 | −2.51 | 0.007 | - | |
| 232.m00071_at | XM_645176 | hypothetical protein | 1.15 | −2.49 | 0.022 | - | |
| 41.m00243_s_at | XM_649788 | hypothetical protein | 5.24 | −2.46 | 0.000 | - | |
| 310.m00064_at | XM_644295 | RNA polymerase I largest subunit, putative | 26.67 | −2.43 | 0.009 | - | |
| 103.m00165_x_at | XM_647745 | hypothetical protein | 0.14 | −2.4 | 0.034 | - | |
| 291.m00043_at | XM_644448 | leucine rich repeat protein | 0.78 | −2.38 | 0.041 | - | |
| 26.m00293_at | XM_650512 | poly(A) polymerase, putative | 1.35 | −2.37 | 0.029 | - | |
| 247.m00075_at | XM_644998 | LIM domain protein | 1.74 | −2.33 | 0.007 | - | |
| 2.m00522_at | XM_652343 | U6 snRNA-associated Sm-like protein, putative | 0.63 | −2.32 | 0.008 | - | |
| 9.m00420_at | XM_651594 | carbonic anhydrase, putative | 2.67 | −2.32 | 0.031 | - | |
| 129.m00135_at | XM_647073 | PH domain protein | 4.92 | −2.31 | 0.002 | - | |
| 34.m00252_at | XM_650085 | conserved hypothetical protein | 0.29 | −2.3 | 0.030 | - | |
| 25.m00242_at | XM_650542 | hypothetical protein | 7.15 | −2.29 | 0.011 | - | |
| 277.m00057_at | XM_644616 | actobindin homolog, putative | 126.03 | −2.28 | 0.007 | - | |
| 108.m00108_at | XM_647607 | hypothetical protein | 0.50 | −2.26 | 0.019 | - | |
| 59.m00197_x_at | XM_649134 | DNA-directed RNA polymerase subunitN, putative | 7.02 | −2.25 | 0.023 | - |
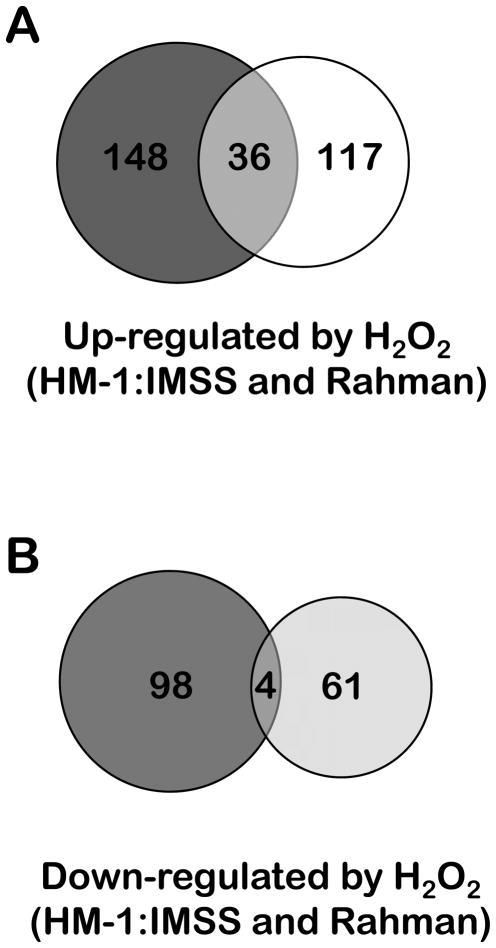
Overall, E. histolytica Rahman had a decreased repertoire of transcriptional changes in response to oxidative stress - both in terms of the numbers of genes regulated but also in the magnitude of their regulation (Figure 5 and Tables 6 and 7; Supplementary Table 4). Of the genes up-regulated in E. histolytica Rahman, only 36 (24%) were also up-regulated in the E. histolytica HM-1:IMSS strain under the same conditions (Figure 5). Furthermore, only 20% of the genes induced by oxidative stress in HM-1:IMSS strain also up-regulated in Rahman. Importantly, even for the genes that overlapped in their expression patterns between E. histolytica HM-1:IMSS and Rahman, the extent of up-regulation was much higher in the pathogenic strain E. histolytica HM-1:IMSS (Supplementary Table 6). Given the degree of genetic identity between E. histolytica HM-1:IMSS and E. histolytica Rahman (Shah et al., 2005), the limited similarity of the response to the same stress condition was unexpected. Indeed, previous comparisons of the two strains under standard culture conditions have identified a limited number of genes with lower expression in Rahman compared to the HM-1:IMSS strain (Davis et al., 2006; MacFarlane and Singh, 2006). The more robust transcriptional response of the virulent strain may contribute to its decreased sensitivity to oxidative stress. Furthermore, the differential response may signify that other genes exclusively up-regulated in the pathogenic E. histolytica HM-1:IMSS strain possibly contribute to this strain’s virulence potential. These include highly induced genes such as those encoding deoxyuridine 5-triphosphate nucleotidohydrolase (194.m00123 and 375.m00057), aminophospholipid translocase (75.m00173), dTDP-D-glucose 4,6-dehydratase (116.m00108) and methionine-γ-lyase (395.m00028). These genes are highly up-regulated by both oxidative and nitrosative stress in the HM-1:IMSS strain but are not regulated at all in the Rahman strain. Moreover, some of these genes have roles in the pathogenicity of other organisms and have been tested as potential novel drug targets (Sato et al., 2008; Ali and Nozaki, 2007; Nguyen et al., 2005).
Figure 5.
Venn diagrams depicting the overlap between the transcriptional changes observed upon exposing a pathogenic and non-pathogenic E. histolytica strain to oxidative stress.
Of the genes induced in E. histolytica Rahman by oxidative stress, 93 (61%) encode unknown hypothetical proteins. Some genes with known functions in response to oxidative stress were regulated in the Rahman strain. One such is a UDP-glucose 4-epimerase (226.m00073) whose homologue from C. albicans contributes to morphology and cell-wall integrity and gene silencing results in fungi that are more susceptible to H2O2-derived oxidative stress (Singh et al., 2007). The remaining genes displayed a different profile from those of the E. histolytica HM-1:IMSS strain challenged with oxidative stress (Figure 3). The number of genes encoding nucleic acids metabolism / repair proteins that were induced by oxidative stress in E. histolytica Rahman was much lower than the number of such genes up-regulated by oxidative stress in the pathogenic HM-1:IMSS strain (Figure 3). Notably, no induction of ubiquitin-conjugating enzymes was observed, contrary to what was observed in E. histolytica HM-1:IMSS.
Overall it appears that the Rahman strain may lack the transcriptional regulatory mechanisms for coping with oxidative damage. During tissue invasion trophozoites are exposed to an oxygen rich environment and the Rahman strain’s limited ability to cope to oxidative stress may contribute to its avirulent phenotype.
CONCLUSIONS
Upon invasion of the host intestinal epithelium, E. histolytica trophozoites are confronted with varying oxygen tensions and cytotoxic reactive oxygen and nitrogen species. The transcriptional changes in E. histolytica HM-1:IMSS upon exposure to oxidative and nitrosative stress were extensive both in the numbers of regulated genes as well as the fold-changes of these genes. A significant fraction of the genes modulated by both stresses code for unknown proteins, which may constitute response mechanisms yet to be unraveled. Among the genes regulated by H2O2 exposure, we identified genes encoding members of four protein families proposed to compose the core of oxidative stress response in eukaryotes: heat-shock proteins, ubiquitin-conjugating enzymes (misfolded protein degradation and repair), protein kinases, and small GTPases (signaling and regulation). Strikingly, genes coding for members of these four families were also induced by nitrosative stress and a significant fraction of these genes responded to both stress types. The common responses thus reflect the overlapping regulatory mechanisms to both stresses by E. histolytica.
Following the premise that an important component of E. histolytica’s pathogenic potential is related to its resistance to ROS and RNS cytotoxicity, we identified a number of genes responsive to either or both stresses, which may contribute to this organism’s virulence. Furthermore, we demonstrated that the non-pathogenic E. histolytica Rahman strain had a marked difference in response to oxidative stress. The differential transcriptional regulation observed for both strains upon exposure to the same oxidative stress conditions suggests that Rahman may experience a higher degree of oxidative damage and these changes could contribute to a decreased virulence phenotype of the Rahman strain.
Overall, our work demonstrates that (i) response to oxidative and nitrosative stress is modulated by a large and complex network of genes in E. histolytica, (ii) a number of known virulence genes are regulated by these stresses, (iii) the decreased virulence phenotype of the non-pathogenic E. histolytica Rahman may be in part due to its limited response to oxidative stress, and (iv) some genes responding to these stress pathways may represent important novel drug targets.
EXPERIMENTAL PROCEDURES
Entamoeba histolytica strains and culture methods
E. histolytica HM-1:IMSS (pathogenic, ATCC 30459) and E. histolytica Rahman (non-pathogenic, ATCC 30886) (Dvorak et al., 2003; Ankri et al., 1999) were obtained from ATCC, strain identity confirmed by PCR and RFLP of known genomic loci (Clark and Diamond, 1993) and grown axenically in TYI-S-33 medium at 36.5°C, as previously described (Diamond et al., 1978).
Sensitivity of E. histolytica trophozoites to oxidative and nitrosative stress
To determine the sensitivity of E. histolytica trophozoites to NO and oxidative stress, parasites from the HM-1:IMSS strain were seeded into 48-well plates (2 × 104 cells per well), each well filled with growth medium and individually sealed with parafilm. After 16–18 h, cells in mid-log phase (50–70% confluent) were exposed to dipropylenetriamine (DPTA)-NONOate (100 μM to 1 mM, nitric oxide releaser with a half-time 180 min−1 at 37°C (Nittler et al., 2005)), or hydrogen peroxide (H2O2, 100 μM to 5 mM) for 1 to 8h. At regular intervals the percentage of rounded up parasites and the number of cells that stained with Trypan blue were determined. For microarray experiments, E. histolytica trophozoites were grown in capped 16-ml Falcon glass tubes to mid-log phase, the medium changed and culture tubes capped. In the case of the nitrosative stress assays, the tube cap included a rubber seal that could be punctured without significantly compromising the anaerobic conditions within the culture. Two hours after replacing the medium, DPTA and H2O2 were added, cultures incubated for 60 min, observed to assess the cell morphology, chilled for 5 min and parasites harvested (spun at 1,000 g, 4°C, 5 min). The supernatant was removed, the cells re-suspended in trypan blue and the percentage of dead cells assessed.
Isolation of RNA and microarray hybridization
RNA was isolated with Trizol reagent (Invitrogen) according to the manufacturer’s protocol, purified with a Qiagen® RNeasy kit, and microarray hybridization performed at the Stanford University Protein and Nucleic Acids facility (http://cmgm.stanford.edu/pan/) using previously published protocols (Ehrenkaufer et al., 2007). A custom generated Affymetrix platform microarray described in (Gilchrist et al., 2006), with probe sets that represents 9,435 open reading frames was used for all studies. A fraction of probe sets are predicted to cross-hybridize with multiple genes and are annotated as follows: probe sets labeled (_at) represent a single gene; probe sets labeled (_x_at) have at least one probe that may cross-hybridize with another gene(s); probe sets labeled (_s_at) are those in which all the probes for a given gene cross-hybridize with another gene(s). Probes for intergenic noncoding regions were excluded from all analyses. The arrays were scanned after hybridization and the probe intensities were calculated using Affymetrix GCOS software (http://www.affymetrix.com/products/software/specific/gcos.affx).
Microarray data normalization and analysis
Analysis was performed as in (Ehrenkaufer et al., 2007). A minimum of three arrays were used for each condition. Standard correlation coefficients were calculated in Genespring. Normalized expression values for each probe set were obtained from raw probe intensities in R 2.2.0 downloaded from the BioConductor project (http://www.bioconductor.org), using robust multi-array averaging with correction for oligosequence (gcRMA) (Wu et al., 2004). To identify differentially expressed genes, we used local pooled error testing along with Benjamini-Hochberg multiple test correction (Benjamini and Hochberg, 1995). In addition, fold-change was calculated in Genespring GX (http://www.chem.agilent.com/scripts/pds.asp?lpage=27881). The threshold for a probe set to be considered differentially expressed was set at a 2-fold change with a false discovery rate (FDR) of < 0.05. Differentially expressed genes that were not annotated as hypothetical proteins were grouped according to their putative functions.
Semi-quantitative reverse transcriptase polymerase chain reaction (RT-PCR)
Total RNA from E. histolytica HM-1:IMSS and Rahman trophozoites was isolated using Trizol reagent (Invitrogen) and further purified with a Qiagen RNeasy kit. cDNA was synthesized from total RNA (2.5μg) with the Universal riboclone cDNA system (Promega), following the manufacturer’s instructions. The cDNA samples were quantified in a Nanodrop spectrophotometer (NanoDrop Technologies, LLC) and PCR reactions performed with 100ng of cDNA (15 cycles at 55°C plus 15 cycles at 50°C for every gene, except for gene 879.m00008, which was amplified with 15 cycles at 50°C plus 15 cycles at 57°C). The PCR products were fractionated on 2% agarose gels and analyzed with a Kodak Digital Science electrophoresis documentation and analysis system 120. The primers used are listed in Supplementary Table 1.
Supplementary Material
Supplementary Table 1. Primers used in RT-PCR analysis. The gene ID, primer direction, and primer sequences are listed.
Supplementary Table 2. Microarray data for all genes for all arrays used in the analysis. The probe ID, gene annotation, locus ID, basal expression level, fold-change and p-values are shown for E. histolytica HM-1:IMSS exposed to H2O2 or DPTA-NONOate and for E. histolytica Rahman exposed to H2O2.
Supplementary Table 3. All genes transcriptionally up-regulated by H2O2 in E. histolytica HM-1:IMSS. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 4. All genes transcriptionally down-regulated by H2O2 in E. histolytica HM-1:IMSS. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 5. All genes transcriptionally down-regulated by DPTA-NONOate in E. histolytica HM-1:IMSS. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 6. All genes transcriptionally up-regulated by DPTA-NONOate in E. histolytica HM-1:IMSS. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 7. All genes transcriptionally up-regulated by H2O2 in E. histolytica Rahman. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 8. All genes transcriptionally down-regulated by H2O2 in E. histolytica Rahman. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Table 1B.
Averaged correlations between DNA microarrays for each experimental condition of E. histolytica HM-1:IMSS.
| Array condition (# of arrays) | HM-1:IMSS (3) | HM-1+H2O2 (3) | HM-1+DPTA (3) |
|---|---|---|---|
| HM-1:IMSS (3) | 0.98 | 0.96 | |
| HM-1 + H2O2 (3) | 0.98 | ||
| HM-1 + DPTA (3) |
Acknowledgments
We gratefully acknowledge all members of the Singh and Teixeira laboratories for helpful discussions. This work is supported by FCT grant SFRH/BPD/26895/2006 for JBV, NIH grants AI-068899 and AI-069382 for GME, FCT project grant POCI/SAU-IMI/56088/2004 to LMS, FCT project grant PTDC/BIA-PRO/67267/2006 to MT, and a grant from the NIAID (AI-053724) to US. We wish to thank Lígia S. Nobre (ITQB, Portugal) for the support with the semi-quantitative RT-PCRs. All array data is available at the GEO database website at NCBI (Accession number GSE11811).
References
- Akbar MA, Chatterjee NS, Sen P, Debnath A, Pal A, Bera T, Das P. Genes induced by a high-oxygen environment in Entamoeba histolytica. Mol Biochem Parasitol. 2004;133:187–196. doi: 10.1016/j.molbiopara.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Ali V, Nozaki T. Current therapeutics, their problems, and sulfur-containing-amino-acid metabolism as a novel target against infections by “amitochondriate” protozoan parasites. Clin Microbiol Rev. 2007;20:164–187. doi: 10.1128/CMR.00019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard ST, Giraud MF, Whitfield C, Graninger M, Messner P, Naismith JH. The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway. J Mol Biol. 2001;307:283–295. doi: 10.1006/jmbi.2000.4470. [DOI] [PubMed] [Google Scholar]
- Anamika K, Bhattacharya A, Srinivasan N. Analysis of the protein kinome of Entamoeba histolytica. Proteins. 2008;71:995–1006. doi: 10.1002/prot.21790. [DOI] [PubMed] [Google Scholar]
- Andersson JO, Hirt RP, Foster PG, Roger AJ. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC Evol Biol. 2006;6:27. doi: 10.1186/1471-2148-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO, Sjogren AM, Davis LA, Embley TM, Roger AJ. Phylogenetic analyses of diplomonad genes reveal frequent lateral gene transfers affecting eukaryotes. Curr Biol. 2003;13:94–104. doi: 10.1016/s0960-9822(03)00003-4. [DOI] [PubMed] [Google Scholar]
- Ankri S, Padilla-Vaca F, Stolarsky T, Koole L, Katz U, Mirelman D. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol Microbiol. 1999;33:327–337. doi: 10.1046/j.1365-2958.1999.01476.x. [DOI] [PubMed] [Google Scholar]
- de Batista EJ, de Souza W. Involvement of protein kinases on the process of erythrophagocytis by Entamoeba histolytica. Cell Biol Int. 2004;28:243–248. doi: 10.1016/j.cellbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Beausejour A, Houde V, Bibeau K, Gaudet R, St-Louis J, Brochu M. Renal and cardiac oxidative/nitrosative stress in salt-loaded pregnant rat. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1657–1665. doi: 10.1152/ajpregu.00090.2007. [DOI] [PubMed] [Google Scholar]
- Beck DL, Boettner DR, Dragulev B, Ready K, Nozaki T, Petri WA., Jr Identification and gene expression analysis of a large family of transmembrane kinases related to the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell. 2005;4:722–732. doi: 10.1128/EC.4.4.722-732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Betti M, Petrucco S, Bolchi A, Dieci G, Ottonello S. A plant 3′-phosphoesterase involved in the repair of DNA strand breaks generated by oxidative damage. J Biol Chem. 2001;276:18038–18045. doi: 10.1074/jbc.M010648200. [DOI] [PubMed] [Google Scholar]
- Blondal T, Hjorleifsdottir S, Aevarsson A, Fridjonsson OH, Skirnisdottir S, Wheat JO, et al. Characterization of a 5′-polynucleotide kinase/3′-phosphatase from bacteriophage RM378. J Biol Chem. 2005;280:5188–5194. doi: 10.1074/jbc.M409211200. [DOI] [PubMed] [Google Scholar]
- Boettner DR, Huston CD, Sullivan JA, Petri WA., Jr Entamoeba histolytica and Entamoeba dispar utilize externalized phosphatidylserine for recognition and phagocytosis of erythrocytes. Infect Immun. 2005;73:3422–3430. doi: 10.1128/IAI.73.6.3422-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan C, Rollinghoff M, Diefenbach A. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr Opin Immunol. 2000;12:64–76. doi: 10.1016/s0952-7915(99)00052-7. [DOI] [PubMed] [Google Scholar]
- Broman MT, Mehta D, Malik AB. Cdc42 regulates the restoration of endothelial adherens junctions and permeability. Trends Cardiovasc Med. 2007;17:151–156. doi: 10.1016/j.tcm.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, Marra G, Chauhan DP, Ha HT, Chang DK, Ricciardiello L, et al. Oxidative stress inactivates the human DNA mismatch repair system. Am J Physiol Cell Physiol. 2002;283:C148–154. doi: 10.1152/ajpcell.00422.2001. [DOI] [PubMed] [Google Scholar]
- Clark CG, Diamond LS. Entamoeba histolytica: a method for isolate identification. Exp Parasitol. 1993;77:450–455. doi: 10.1006/expr.1993.1105. [DOI] [PubMed] [Google Scholar]
- Clark CG, Alsmark UC, Tazreiter M, Saito-Nakano Y, Ali V, Marion S, et al. Structure and content of the Entamoeba histolytica genome. Adv Parasitol. 2007;65:51–190. doi: 10.1016/S0065-308X(07)65002-7. [DOI] [PubMed] [Google Scholar]
- Colles SM, Chisolm GM. Lysophosphatidylcholine-induced cellular injury in cultured fibroblasts involves oxidative events. J Lipid Res. 2000;41:1188–1198. [PubMed] [Google Scholar]
- Cruz F, Ferry JG. Interaction of iron-sulfur flavoprotein with oxygen and hydrogen peroxide. Biochim Biophys Acta. 2006;1760:858–864. doi: 10.1016/j.bbagen.2006.02.016. [DOI] [PubMed] [Google Scholar]
- D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Damer CK, Bayeva M, Kim PS, Ho LK, Eberhardt ES, Socec CI, et al. Copine A is required for cytokinesis, contractile vacuole function, and development in Dictyostelium. Eukaryot Cell. 2007;6:430–442. doi: 10.1128/EC.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PH, Schulze J, Stanley SL., Jr Transcriptomic comparison of two Entamoeba histolytica strains with defined virulence phenotypes identifies new virulence factor candidates and key differences in the expression patterns of cysteine proteases, lectin light chains, and calmodulin. Mol Biochem Parasitol. 2007;151:118–128. doi: 10.1016/j.molbiopara.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Davis PH, Zhang X, Guo J, Townsend RR, Stanley SL., Jr Comparative proteomic analysis of two Entamoeba histolytica strains with different virulence phenotypes identifies peroxiredoxin as an important component of amoebic virulence. Mol Microbiol. 2006;61:1523–1532. doi: 10.1111/j.1365-2958.2006.05344.x. [DOI] [PubMed] [Google Scholar]
- Di Matteo A, Scandurra FM, Testa F, Forte E, Sarti P, Brunori M, Giuffre A. The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J Biol Chem. 2008;283:4061–4068. doi: 10.1074/jbc.M705605200. [DOI] [PubMed] [Google Scholar]
- Diamond LS, Cunnick CC. A serum-free, partly defined medium, PDM-805, for axenic cultivation of Entamoeba histolytica Schaudinn, 1903 and other Entamoeba. J Protozool. 1991;38:211–216. doi: 10.1111/j.1550-7408.1991.tb04430.x. [DOI] [PubMed] [Google Scholar]
- Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- Dvorak JA, Kobayashi S, Nozaki T, Takeuchi T, Matsubara C. Induction of permeability changes and death of vertebrate cells is modulated by the virulence of Entamoeba spp. isolates. Parasitol Int. 2003;52:169–173. doi: 10.1016/s1383-5769(02)00090-9. [DOI] [PubMed] [Google Scholar]
- Dzierzbicki P, Koprowski P, Fikus MU, Malc E, Ciesla Z. Repair of oxidative damage in mitochondrial DNA of Saccharomyces cerevisiae: involvement of the MSH1-dependent pathway. DNA Repair (Amst) 2004;3:403–411. doi: 10.1016/j.dnarep.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, et al. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol Biochem Parasitol. 2006;147:163–176. doi: 10.1016/j.molbiopara.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Gill RK, Saksena S, Syed IA, Tyagi S, Alrefai WA, Malakooti J, et al. Regulation of NHE3 by nitric oxide in Caco-2 cells. Am J Physiol Gastrointest Liver Physiol. 2002;283:G747–756. doi: 10.1152/ajpgi.00294.2001. [DOI] [PubMed] [Google Scholar]
- Gillin FD, Diamond LS. Attachment and short-term maintenance of motility and viability of Entamoeba histolytica in a defined medium. J Protozool. 1980;27:220–225. doi: 10.1111/j.1550-7408.1980.tb04685.x. [DOI] [PubMed] [Google Scholar]
- Hackney JA, Ehrenkaufer GM, Singh U. Identification of putative transcriptional regulatory networks in Entamoeba histolytica using Bayesian inference. Nucleic Acids Res. 2007;35:2141–2152. doi: 10.1093/nar/gkm028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford: Oxford University Press; 2007. [Google Scholar]
- Hong F, Lee J, Song JW, Lee SJ, Ahn H, Cho JJ, et al. Cyclosporin A blocks muscle differentiation by inducing oxidative stress and inhibiting the peptidyl-prolyl-cis-trans isomerase activity of cyclophilin A: cyclophilin A protects myoblasts from cyclosporin A-induced cytotoxicity. Faseb J. 2002;16:1633–1635. doi: 10.1096/fj.02-0060fje. [DOI] [PubMed] [Google Scholar]
- Hurtado-Guerrero R, Raimi O, Shepherd S, van Aalten DM. Glucose-6-phosphate as a probe for the glucosamine-6-phosphate N-acetyltransferase Michaelis complex. FEBS Lett. 2007;581:5597–5600. doi: 10.1016/j.febslet.2007.10.065. [DOI] [PubMed] [Google Scholar]
- Huston CD, Boettner DR, Miller-Sims V, Petri WA., Jr Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect Immun. 2003;71:964–972. doi: 10.1128/IAI.71.2.964-972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia”. Infect Immun. 2006;74:5023–5028. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakov E, Siman-Tov R, Weber C, Guillen N, Ankri S. Trichostatin A regulates peroxiredoxin expression and virulence of the parasite Entamoeba histolytica. Mol Biochem Parasitol. 2008;158:82–94. doi: 10.1016/j.molbiopara.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Justino MC, Vicente JB, Teixeira M, Saraiva LM. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J Biol Chem. 2005;280:2636–2643. doi: 10.1074/jbc.M411070200. [DOI] [PubMed] [Google Scholar]
- Khil PP, Camerini-Otero RD. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol Microbiol. 2002;44:89–105. doi: 10.1046/j.1365-2958.2002.02878.x. [DOI] [PubMed] [Google Scholar]
- Korner H, Sofia HJ, Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- Kurtz DM. Flavo-diiron enzymes: nitric oxide or dioxygen reductases? Dalton Transactions. 2007:4115–4121. [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- MacFarlane RC, Singh U. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect Immun. 2006;74:340–351. doi: 10.1128/IAI.74.1.340-351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Mayol JM, Adame-Navarrete Y, Alarma-Estrany P, Molina-Roldan E, Huete-Toral F, Fernandez-Represa JA. Luminal oxidants selectively modulate electrogenic ion transport in rat colon. World J Gastroenterol. 2006;12:5523–5527. doi: 10.3748/wjg.v12.i34.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendz GL, Megraud F. Is the molecular basis of metronidazole resistance in microaerophilic organisms understood? Trends Microbiol. 2002;10:370–375. doi: 10.1016/s0966-842x(02)02405-8. [DOI] [PubMed] [Google Scholar]
- Mio T, Kokado M, Arisawa M, Yamada-Okabe H. Reduced virulence of Candida albicans mutants lacking the GNA1 gene encoding glucosamine-6-phosphate acetyltransferase. Microbiology. 2000;146(Pt 7):1753–1758. doi: 10.1099/00221287-146-7-1753. [DOI] [PubMed] [Google Scholar]
- Mondal S, Neelamegan D, Rivero F, Noegel AA. GxcDD, a putative RacGEF, is involved in Dictyostelium development. BMC Cell Biol. 2007;8:23. doi: 10.1186/1471-2121-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Nguyen C, Kasinathan G, Leal-Cortijo I, Musso-Buendia A, Kaiser M, Brun R, et al. Deoxyuridine triphosphate nucleotidohydrolase as a potential antiparasitic drug target. J Med Chem. 2005;48:5942–5954. doi: 10.1021/jm050111e. [DOI] [PubMed] [Google Scholar]
- Nittler MP, Hocking-Murray D, Foo CK, Sil A. Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol Biol Cell. 2005;16:4792–4813. doi: 10.1091/mbc.E05-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki T, Ali V, Tokoro M. Sulfur-containing amino acid metabolism in parasitic protozoa. Adv Parasitol. 2005;60:1–99. doi: 10.1016/S0065-308X(05)60001-2. [DOI] [PubMed] [Google Scholar]
- Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D, Hajishengallis G, Sharma A. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect Immun. 2008;76:198–205. doi: 10.1128/IAI.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsenigo MN, Faelli A, Porta C, Sironi C, Laforenza U, Paulmichl M, Tosco M. Oxidative stress reduces transintestinal transports and (Na+, K+) -ATPase activity in rat jejunum. Arch Biochem Biophys. 2007;466:300–307. doi: 10.1016/j.abb.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Paget MS, Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- Picazarri K, Nakada-Tsukui K, Nozaki T. Autophagy during proliferation and encystation in the protozoan parasite Entamoeba invadens. Infect Immun. 2008;76:278–288. doi: 10.1128/IAI.00636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J, Liu GH, Huang B, Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res. 2007;35:2522–2532. doi: 10.1093/nar/gkl1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeille F, Jabrin S, Bligny R, Loizeau K, Gambonnet B, Van Wilder V, et al. Methionine catabolism in Arabidopsis cells is initiated by a gamma-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci U S A. 2006;103:15687–15692. doi: 10.1073/pnas.0606195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito-Nakano Y, Loftus BJ, Hall N, Nozaki T. The diversity of Rab GTPases in Entamoeba histolytica. Exp Parasitol. 2005;110:244–252. doi: 10.1016/j.exppara.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Santi-Rocca J, Weber C, Guigon G, Sismeiro O, Coppee JY, Guillen N. The lysine- and glutamic acid-rich protein KERP1 plays a role in Entamoeba histolytica liver abscess pathogenesis. Cell Microbiol. 2008;10:202–217. doi: 10.1111/j.1462-5822.2007.01030.x. [DOI] [PubMed] [Google Scholar]
- Santos AN, Korber S, Kullertz G, Fischer G, Fischer B. Oxygen stress increases prolyl cis/trans isomerase activity and expression of cyclophilin 18 in rabbit blastocysts. Biol Reprod. 2000;62:1–7. doi: 10.1095/biolreprod62.1.1. [DOI] [PubMed] [Google Scholar]
- Saraiva LM, Vicente JB, Teixeira M. The role of the flavodiiron proteins in microbial nitric oxide detoxification. Adv Microb Physiol. 2004;49:77–129. doi: 10.1016/S0065-2911(04)49002-X. [DOI] [PubMed] [Google Scholar]
- Sarti P, Fiori PL, Forte E, Rappelli P, Teixeira M, Mastronicola D, et al. Trichomonas vaginalis degrades nitric oxide and expresses a flavorubredoxin-like protein: a new pathogenic mechanism? Cellular and Molecular Life Sciences. 2004;61:618–623. doi: 10.1007/s00018-003-3413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato D, Yamagata W, Harada S, Nozaki T. Kinetic characterization of methionine gamma-lyases from the enteric protozoan parasite Entamoeba histolytica against physiological substrates and trifluoromethionine, a promising lead compound against amoebiasis. Febs J. 2008;275:548–560. doi: 10.1111/j.1742-4658.2007.06221.x. [DOI] [PubMed] [Google Scholar]
- Schapiro JM, Libby SJ, Fang FC. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc Natl Acad Sci U S A. 2003;100:8496–8501. doi: 10.1073/pnas.1033133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Chatterjee NS, Akbar MA, Nandi N, Das P. The 29-kilodalton thiol-dependent peroxidase of Entamoeba histolytica is a factor involved in pathogenesis and survival of the parasite during oxidative stress. Eukaryot Cell. 2007;6:664–673. doi: 10.1128/EC.00308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PH, MacFarlane RC, Bhattacharya D, Matese JC, Demeter J, Stroup SE, Singh U. Comparative genomic hybridizations of Entamoeba strains reveal unique genetic fingerprints that correlate with virulence. Eukaryot Cell. 2005;4:504–515. doi: 10.1128/EC.4.3.504-515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A, Edvardsson A, Vener AV. Profound redox sensitivity of peptidyl-prolyl isomerase activity in Arabidopsis thylakoid lumen. FEBS Lett. 2006;580:3671–3676. doi: 10.1016/j.febslet.2006.05.054. [DOI] [PubMed] [Google Scholar]
- Shen Y, Hirsch DS, Sasiela CA, Wu WJ. Cdc42 regulates E-cadherin ubiquitination and degradation through an epidermal growth factor receptor to Src-mediated pathway. J Biol Chem. 2008;283:5127–5137. doi: 10.1074/jbc.M703300200. [DOI] [PubMed] [Google Scholar]
- Singh V, Satheesh SV, Raghavendra ML, Sadhale PP. The key enzyme in galactose metabolism, UDP-galactose-4-epimerase, affects cell-wall integrity and morphology in Candida albicans even in the absence of galactose. Fungal Genet Biol. 2007;44:563–574. doi: 10.1016/j.fgb.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Spiro S. Nitric oxide-sensing mechanisms in Escherichia coli. Biochem Soc Trans. 2006;34:200–202. doi: 10.1042/BST0340200. [DOI] [PubMed] [Google Scholar]
- Stanley SL., Jr Amoebiasis. Lancet. 2003;361:1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- Thum T, Bauersachs J. Microarray-based gene expression profiling to elucidate cellular responses to nitric oxide--a review from an analytical and biomedical point of view. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:3–11. doi: 10.1016/j.jchromb.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Tokoro M, Asai T, Kobayashi S, Takeuchi T, Nozaki T. Identification and characterization of two isoenzymes of methionine gamma-lyase from Entamoeba histolytica: a key enzyme of sulfur-amino acid degradation in an anaerobic parasitic protist that lacks forward and reverse trans-sulfuration pathways. J Biol Chem. 2003;278:42717–42727. doi: 10.1074/jbc.M212414200. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Basova LV, Konduru NV, Tyurin VA, Potapovich AI, Cai P, et al. Nitrosative stress inhibits the aminophospholipid translocase resulting in phosphatidylserine externalization and macrophage engulfment: implications for the resolution of inflammation. J Biol Chem. 2007;282:8498–8509. doi: 10.1074/jbc.M606950200. [DOI] [PubMed] [Google Scholar]
- Vandenbroucke K, Robbens S, Vandepoele K, Inze D, Van de Peer Y, Van Breusegem F. Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol. 2008;25:507–516. doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- Vats D, Vishwakarma RA, Bhattacharya S, Bhattacharya A. Reduction of cell surface glycosylphosphatidylinositol conjugates in Entamoeba histolytica by antisense blocking of E. histolytica GlcNAc-phosphatidylinositol deacetylase expression: effect on cell proliferation, endocytosis, and adhesion to target cells. Infect Immun. 2005;73:8381–8392. doi: 10.1128/IAI.73.12.8381-8392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente JB, Carrondo MA, Teixeira M, Frazao C. Flavodiiron Proteins: Nitric Oxide and/or Oxygen Reductases. In: Messerschmidt A, editor. Handbook of Metalloproteins. John Wiley & Sons, Ltd; 2007. [Google Scholar]
- Wang Y, Cao YY, Jia XM, Cao YB, Gao PH, Fu XP, et al. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic Biol Med. 2006;40:1201–1209. doi: 10.1016/j.freeradbiomed.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Weber C, Guigon G, Bouchier C, Frangeul L, Moreira S, Sismeiro O, et al. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell. 2006;5:871–875. doi: 10.1128/EC.5.5.871-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry R, Gentleman R, Murillo F, Spencer F. Technical Report. John Hopkins University, Department of Biostatistics Working Papers; 2004. A model based background adjustment for oligonucleotide expression arrays. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Primers used in RT-PCR analysis. The gene ID, primer direction, and primer sequences are listed.
Supplementary Table 2. Microarray data for all genes for all arrays used in the analysis. The probe ID, gene annotation, locus ID, basal expression level, fold-change and p-values are shown for E. histolytica HM-1:IMSS exposed to H2O2 or DPTA-NONOate and for E. histolytica Rahman exposed to H2O2.
Supplementary Table 3. All genes transcriptionally up-regulated by H2O2 in E. histolytica HM-1:IMSS. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 4. All genes transcriptionally down-regulated by H2O2 in E. histolytica HM-1:IMSS. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 5. All genes transcriptionally down-regulated by DPTA-NONOate in E. histolytica HM-1:IMSS. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 6. All genes transcriptionally up-regulated by DPTA-NONOate in E. histolytica HM-1:IMSS. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 7. All genes transcriptionally up-regulated by H2O2 in E. histolytica Rahman. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.
Supplementary Table 8. All genes transcriptionally down-regulated by H2O2 in E. histolytica Rahman. The probe ID, fold-change, baseline expression value, gene annotation, TIGR gene number, locus ID, and expression data for each array analyzed are shown.