Abstract
Biotin synthase catalyzes the conversion of dethiobiotin (DTB) to biotin through the oxidative addition of sulfur between two saturated carbon atoms, generating a thiophane ring fused to the existing ureido ring. Biotin synthase is a member of the Radical SAM superfamily, composed of enzymes that reductively cleave S-adenosyl-L-methionine (SAM or AdoMet) to generate a 5′-deoxyadenosyl radical that can abstract unactivated hydrogen atoms from a variety of organic substrates. In biotin synthase, abstraction of a hydrogen atom from the C9 methyl group of DTB would result in formation of a dethiobiotinyl methylene carbon radical, which is then quenched by a sulfur atom to form a new carbon-sulfur bond in the intermediate 9-mercaptodethiobiotin (MDTB). We have proposed that this sulfur atom is the μ-sulfide of a [2Fe-2S]2+ cluster found near DTB in the enzyme active site. In the present work, we show that formation of MDTB is accompanied by stoichiometric generation of a paramagnetic FeS cluster. The electron paramagnetic resonance (EPR) spectrum is modeled as a 2:1 mixture of components attributable to different forms of a [2Fe-2S]+ cluster, possibly distinguished by slightly different coordination environments. Mutation of Arg260, one of the ligands to the [2Fe-2S] cluster, causes a distinctive change in the EPR spectrum. Furthermore, magnetic coupling of the unpaired electron with 14N from Arg260, detectable by electron spin envelope modulation (ESEEM) spectroscopy, is observed in WT enzyme but not in the Arg260Met mutant enzyme. Both results indicate that the paramagnetic FeS cluster formed during catalytic turnover is a [2Fe-2S]+ cluster, consistent with a mechanism in which the [2Fe-2S]2+ cluster simultaneously provides and oxidizes sulfide during carbon-sulfur bond formation.
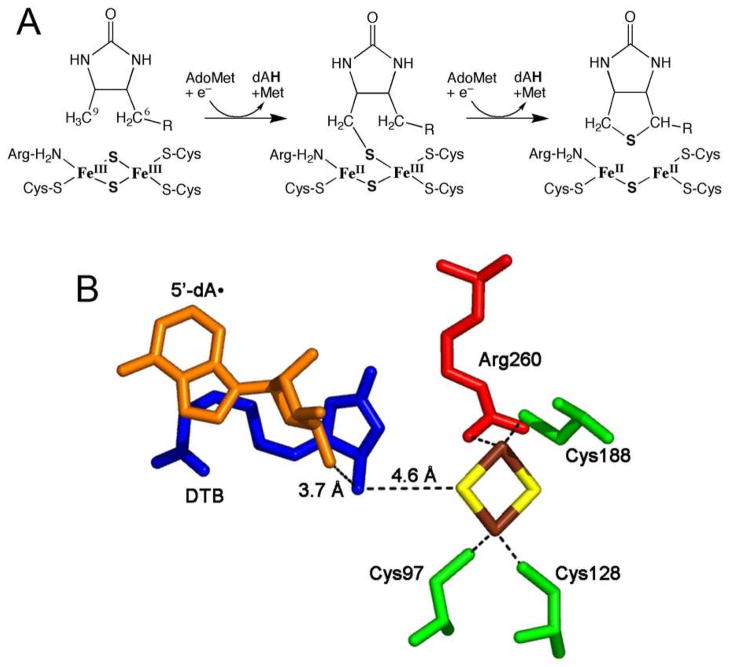
Biotin synthase (BS)1 is an S-adenosyl-L-methionine (AdoMet) radical enzyme that catalyzes the oxidative addition of a sulfur atom between the C6 methylene and C9 methyl groups of dethiobiotin (DTB), generating the thiophane ring of biotin (1–3). In our working mechanism (Figure 1A), catalysis is initiated by one-electron reduction of AdoMet sulfonium, generating methionine and a 5′-deoxy-adenosyl radical (dA•). This reactive radical is a strong oxidant that abstracts a hydrogen atom from the C9 methyl of DTB (4), generating a putative DTB-centered carbon radical that is then quenched by a sulfur atom derived from within the enzyme (5–7). The product of this initial reaction sequence, 9-mercaptodethiobiotin (MDTB) (8), remains tightly bound to the enzyme while 5′-deoxyadenosine (dAH) and methionine dissociate and a second equivalent of AdoMet binds (Figure 1A). A similar reaction sequence directed at the C6 methylene of MDTB closes the thiophane ring. While the overall chemical stoichiometry is now clearly established (9–11), the electron stoichiometry depends on the chemical identity of the sulfur atom incorporated into biotin.
Figure 1.
(A) The proposed biotin synthase reaction sequence. The reductive cleavage of AdoMet to dAH and methionine is coupled to abstraction of a hydrogen atom from C9 of DTB and formation of a new C-S bond in MDTB. We propose that the sulfur donor is the [2Fe-2S]2+ cluster, and that MDTB formation is coupled to transfer of an electron from the μ-sulfide into the cluster. Dissociation of dAH and methionine and binding of a second equiv of AdoMet is followed by a similar reaction sequence that closes the thiophane ring and results in complete reduction to an unstable diferrous cluster. (B) The active site environment of the [2Fe-2S]2+ cluster based on the 3.4 Å resolution crystal structure of E. coli BS in complex with AdoMet and DTB (12). Cys97, Cys128, Cys188 (green) and Arg260 (red) coordinate the tetragonal [2Fe-2S]2+ cluster (Fe3+, brown, S2−, yellow). C9 of DTB (blue) is ~4.6 Å (center-to-center) from the nearest μ-sulfide of this cluster. The C5′ position of the transient dA• radical (orange) is generated ~3.7 Å away from C9 of DTB. Image generated with PyMol (52) using pdb file 1R30.
The structure of the BS homodimer shows that each monomer contains an (αβ)8 barrel that encapsulates the presumed active site, containing AdoMet, DTB, and two FeS clusters (12). In a structural motif shared with other AdoMet radical enzymes, a [4Fe-4S]2+ cluster is coordinated by Cys53, Cys57, and Cys60 within an extended loop between β strand 1 and α helix 1. The fourth Fe atom of this cluster is coordinated by the amine and carboxylate of AdoMet (12, 13), positioning the sulfonium of AdoMet ~4 Å from the cluster. The redox-active [4Fe-4S]2+/+ cluster lies near the surface of the protein, and the presumed role for this cluster is to pass an electron from an external protein donor (flavodoxin in E. coli) into the AdoMet sulfonium, facilitating reductive cleavage of the C5′-S bond and generation of a 5′-deoxyadenosyl radical. In addition to this cluster, a [2Fe-2S]2+ cluster is bound deep within the (αβ)8 barrel, with one Fe coordinated by Cys97 and Cys121, and the other Fe coordinated by Cys188 and Arg260 (12). The [2Fe-2S]2+ cluster is bound ~4.6 Å from DTB (Figure 1B), and we have proposed the μ-sulfide from this cluster is the sulfur incorporated into DTB. Consistent with this proposal, elimination of this FeS cluster by site-directed mutagenesis of Cys97, Cys121, or Cys188 to Ala renders the enzyme inactive (14). Further, BS in which the [2Fe-2S]2+ cluster has been reconstituted with 34S2− or Se2− incorporates ca. 60–70% of the heavy-atom isotope into biotin, with the remainder of biotin incorporating 32S presumably derived from buffer components (6, 7). In addition, formation of biotin is either accompanied by (15), or possibly preceded by (16), destruction of the [2Fe-2S]2+ cluster, which might be expected if sulfide is extracted from the cluster during catalysis.
If sulfide derived from the [2Fe-2S]2+ cluster quenches a dethiobiotinyl C9 methylene radical to generate the new C-S bond in MDTB, then the resulting molecule would contain an extra electron derived from sulfide that must be shed prior to or concurrent with bond formation. This electron could be transferred to either FeS cluster, resulting in the formation of a reduced paramagnetic FeS cluster that could be detected by electron paramagnetic resonance (EPR) spectroscopy. The [4Fe-4S]2+ cluster readily undergoes chemical reduction by dithionite at Em ≈ −500 mV (vs. SHE) (17, 18), generating a [4Fe-4S]+ cluster that exhibits two signals detectable by EPR spectroscopy: a strong high-field S = 1/2 signal with g = [2.04, 1.95, 1.91] (19–21) and a weak low-field S = 3/2 signal at g = 5.6 (20). In the WT enzyme, the [2Fe-2S]2+ cluster is not stable to chemical reduction. Treatment of WT BS containing one [2Fe-2S]2+ cluster per monomer with dithionite leads to irreversible reduction and loss of this cluster with an apparent Ered = −430 mV (18). The product of this reduction process is a substoichiometric amount of the [4Fe-4S]+ cluster (18, 20, 22) formed by dissociation and reassociation of the iron and sulfide ions (21). However, mutation of the nearby residues Asn153 or Asp155 to Ala appears to stabilize the reduced cluster. Chemical reduction of the [2Fe-2S]2+ cluster with dithionite in these mutant enzymes results in formation of a semi-stable [2Fe-2S]+ cluster exhibiting a high-field axial S = 1/2 signal with g = [2.00, 1.91] (23). This EPR signal has not been observed at any point during reduction of the WT enzyme.
A significantly different EPR signal has been observed during catalysis when BS is incubated with DTB, AdoMet, and a flavodoxin/FNR/NADPH reducing system. Ugulava et al. initially reported that room temperature incubation of BS with substrates and reducing system in the presence of excess iron and sulfide in the buffer resulted in slow development of a broad rhombic signal centered at gav = 1.94 (15). Jameson et al. more thoroughly examined changes in the [2Fe-2S] cluster during turnover using EPR and Mössbauer spectroscopy (16), and reported formation and decay of a reduced paramagnetic FeS cluster exhibiting two broad overlapping rhombic signals (g = [2.010, 1.955, 1.880] and g = [2.000, 1.940, 1.845]). Mössbauer spectra obtained in parallel contained a minor component (δ ≈ 0.25 mm/s, ΔEQ ≈ 5.5 mm/s, ~10 – 20 % of total 57Fe species, estimated from Fig. 1 in Jameson, et al. (16)) attributed to a [2Fe-2S]+ cluster. However, formation and decay of this [2Fe-2S]+ cluster occurred on a time-scale that was significantly faster than biotin formation, and the authors concluded that reduction and degradation of the [2Fe-2S]2+ cluster likely occurs before the catalytic reactions that generate biotin (16).
We have since demonstrated that biotin formation proceeds in a stepwise manner, with conversion of DTB to MDTB within the first 5–10 min of turnover at 37 °C (k1 = 0.07 min−1), while conversion of MDTB to biotin lags initially and then proceeds at an apparent rate of kobs = 0.05 min−1 (8, 24). However, a careful stoichiometric analysis suggests that only one monomer within the dimeric enzyme is undergoing turnover to form biotin, while the other monomer binds substrates but does not undergo a reaction; i.e. that the enzyme is half-site active (24). Half-site activity leads to a lower-than-expected initial yield of products as well as a heterogeneous mixture of unreacted, partially reacted, and fully reacted FeS cluster-depleted enzyme monomers. In addition, the apparent rate of biotin formation is also subject to an unusual form of product inhibition (25), in which dAH and methionine generated during the first half-reaction, conversion of DTB to MDTB, can cooperatively compete with AdoMet during the second half-reaction, preventing conversion of MDTB to biotin. This product inhibition would be most severe at high enzyme concentration ([E] ≥ Ki ≈ 20 μM (25)) and would cause a significant disparity between the rate of formation of MDTB vs. biotin. A stepwise mechanism for conversion of DTB to biotin, together with half-site activity and significant product inhibition of the second step, could explain the kinetic discrepancy between biotin formation and transformation of the [2Fe-2S]2+ cluster reported by Huynh and coworkers (16).
If the [2Fe-2S]2+ cluster provides sulfide for conversion of DTB to MDTB, then we propose a mechanism in which the sulfide is simultaneously oxidized by inner-sphere electron transfer into the cluster, generating a transient paramagnetic FeS cluster in which MDTB may remain as a thiolate ligand (Figure 1A) (1, 8, 11). In this scenario, generation of a reduced FeS cluster should kinetically correlate with MDTB formation, not biotin formation. In the present work, we conduct assays with limiting AdoMet to enhance the production of MDTB over biotin. By comparing spin concentrations determined from EPR to MDTB concentrations determined by HPLC analysis, we demonstrate a 1:1 stoichiometric correlation between production of MDTB and generation of a paramagnetic FeS cluster. We also confirm that the observed EPR signal is due to the [2Fe-2S]+ cluster, taking advantage of the unique Arg260 ligand to this cluster. EPR spectra of the active Arg260Met mutant enzyme show significant changes in g tensors consistent with a change in the coordination environment of the [2Fe-2S]+ cluster. ESEEM spectra of the WT enzyme shows hyperfine coupling of 14N from the Arg260 ligand, and this coupling is lost in the Arg260Met mutant enzyme. The dual role of the [2Fe-2S]2+ cluster as both a substrate and redox cofactor is discussed within the broader context of the emerging subfamily of AdoMet radical C-S bond forming enzymes.
Materials and Methods
All reagents were obtained from commercial sources and used without further purification. AdoMet was purchased as the p-toluenesulfonate salt and contains ~15 % 5′-methylthioaden-osine and ~2 % S-adenosyl-L-homocysteine. Expression and purification of wild-type (18) and Arg260Met (17) BS, as well as flavodoxin and His6-FNR (26), was performed as previously described. The concentration of aerobically purified BS was estimated based on the absorbance spectrum of the [2Fe-2S]2+ cluster using ε452 = 8400 M−1 cm−1 and confirmed by Bradford assay using BSA as a standard (18).
Preparation of EPR Samples Collected During BS Turnover
Samples were prepared in a nitrogen-filled anaerobic glove box operated at <1 ppm O2. BS (25 – 125 μM final dimer concentration) was added to vials containing 50 mM Tris HCl, 10 mM KCl, and 5 mM DTT, pH 8.0. The [4Fe-4S]2+ cluster was reconstituted by addition of 4 equiv of Na2S and FeCl3. Flavodoxin (20 μM), FNR (2 μM), NADPH (1 mM) and AdoMet (0 – 5 equiv) were added and the sample incubated for 10 min at room temperature. DTB (1 – 5 equiv) was added to initiate the reaction. Vials were incubated in the anaerobic chamber for 5 – 120 min, either at ambient temperature or in a 37 °C sand bath. At various intervals, samples were collected for HPLC analysis by removing 90 μL of sample and quenching with 10 μL of 4.5 M acetic acid/sodium acetate buffer, pH 4.5. At the same time, 300 μL of sample was transferred to an EPR tube, and the tube was sealed, removed from the anaerobic chamber, and frozen in liquid nitrogen. EPR samples were shipped overnight on dry ice and then stored under liquid N2 until spectra could be collected.
Analysis of Biotin and MDTB by HPLC
Acid-quenched samples were removed from the glove box and chilled on ice for 10 min, and precipitated protein was removed by centrifugation for 10 min at 18,000 ×g. The supernatant was transferred to an HPLC autosampler vial and injected on a Waters Atlantis dC18 reverse-phase column (3.0 × 150 mm, 5 μm) equilibrated with 2% acetonitrile/H2O (0.1 % H3PO4) at a flow rate of 0.7 mL/min. Sample components were eluted with a linear gradient from 2 – 25 % acetonitrile over 25 min in the same buffer and detected by UV absorbance at 210 nm: biotin, tR = 18 min; DTB, tR = 21 min; MDTB, tR = 23.6 min.
The concentration of biotin and DTB in unknown samples was determined by comparison to commercial samples (≥ 98% pure, Sigma), which were lyophilized to remove bound water and accurately weighed prior to preparation of stock solutions in stoichiometric NaOH solution. Extinction coefficients were experimentally determined from UV spectra of multiple sample preparations: DTB, ε210 = 870 M−1 cm−1 and biotin, ε210 = 1860 M−1 cm−1. Since MDTB was not commercially available, the extinction coefficient of MDTB at 210 nm was estimated using the experimentally determined extinction coefficients for the amino acids alanine and cysteine to estimate the molar absorptivity of the thiol functional group. Solutions of each amino acid were gravimetrically prepared in deionized water, UV spectra recorded, and ε210 calculated for each amino acid (note that this is a shoulder region, not a peak, in the UV spectrum). The increased absorbance of cysteine was used to determine Δε210 due to the thiol functional group. The extinction coefficient for MDTB was then calculated as εMDTB = εDTB + (εCys − εAla), yielding ε210 = 1100 M−1 cm−1. This method of estimation is likely to be reasonably accurate, as a similar estimation for biotin, using methionine instead of cysteine, yields εbiotin = εDTB + (εMet − εAla)= 2100 M−1 cm−1, which compares well with the experimental ε210 = 1860 M−1 cm−1. Further, an examination of reference spectra of alkyl thiols and dialkyl thioethers in the NIST chemical database (http://webbook.nist.gov/chemistry) suggests that alkyl thiols have ε210 = 250 – 300 M−1 cm−1, while dialkyl thioethers have ε210 = 800 – 1000 M−1 cm−1. A sum of the extinction coefficients for DTB and alkyl thiols gives a similar estimate for the extinction coefficient MDTB of ε210 = 1120 – 1170 M−1 cm−1. Using these measured and estimated reference values, the concentration of MDTB in enzyme assays was then determined by HPLC analysis, using biotin as a standard, but then dividing the peak area of the biotin standard by the ratio of extinction coefficients: εbiotin/εMDTB (210 nm) = 1860/1100 = 1.69.
Electron Paramagnetic Resonance Spectroscopy
EPR spectra were measured at the CalEPR Center in the Department of Chemistry at the University of California, Davis. Continuous-wave (CW) spectra were collected using a Bruker ECS106 EPR spectrometer operating at X-band frequency (around 9.5 GHz). Typical sweep widths were 320 – 400 mT, although wider sweeps were also performed to rule out high-spin signals. Using a sample with a strong g ≈ 2.04 signal, spectra were collected with sample temperatures from 5 – 80 K and microwave power from 200 nW to 200 mW to examine the relaxation properties of signal components, yielding optimized conditions for observation of the FeS cluster signals at 20 K and 10 mW. Spectral simulations were performed using EasySpin (http://easyspin.org) (11). Pulsed spectra were measured using a Bruker EleXsys E580 spectrometer. Three-pulse ESEEM (π/2-τ-π/2-T-π/2-τ-echo) spectra were acquired with 8 ns π/2 pulses for several τ values. The spin concentration of FeS clusters in BS samples was determined by comparison to samples of 50–200 μM Cu2+-EDTA. For determination of the BS spin concentration, the flavodoxin FMN semiquinone signal (g = 2.005) was saturated at high microwave power, and the residual signal was manually subtracted prior to spin integration.
Results
MDTB is Optimally Produced at Substoichiometric AdoMet Concentrations
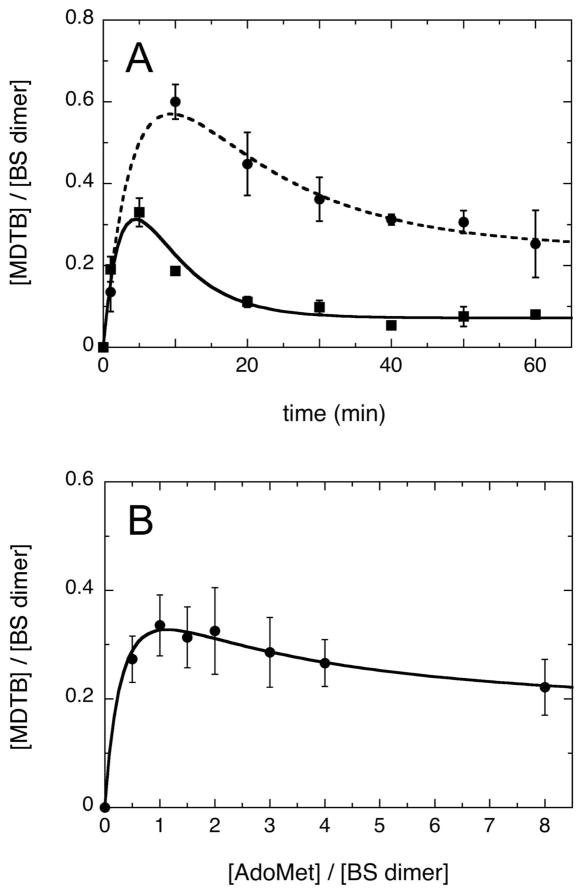
We have previously demonstrated that BS catalyzes a sequential reaction in which DTB is first converted to the intermediate MDTB, which remains tightly bound to the enzyme and is subsequently converted to biotin (8). The reaction cycle requires two sequential hydrogen atom abstractions, first from C9 of DTB and then from C6 of MDTB, by 5′-dA• generated via reductive cleavage of AdoMet. Since we wished to measure changes in the EPR spectrum that might correlate with formation of intermediates, we attempted to develop conditions that would generate a high concentration of the MDTB intermediate and we suspected would also yield a more intense EPR signal. Under typical assay conditions, a large excess of AdoMet (16 equiv per BS dimer) results in low yield of MDTB, with ~0.2 – 0.3 equiv per dimer formed in the first 5 min (Figure 2A, squares), falling off to ~0.1 – 0.2 equiv after 20 min as this intermediate is converted to biotin. A decrease in the excess of AdoMet (8 equiv per dimer) results in an increased yield and prolonged lifetime of the MDTB intermediate (Figure 2A, circles). Initially, we had reasoned that limiting AdoMet to ~1 equiv would result in conversion of DTB to MDTB, but there would be insufficient AdoMet to convert MDTB to biotin. However, when we varied the concentration of AdoMet over the range 0.5 – 8 equiv per dimer in a 60 min assay, we observed that maximally ~0.35 equiv MDTB per dimer is formed when BS is incubated with 1 equiv AdoMet (Figure 2B), but higher concentrations of AdoMet resulted in only a modest suppression of MDTB formation.
Figure 2.
The catalytic intermediate MDTB is optimally formed at short reaction times and at limiting AdoMet concentrations. (A) Time dependence of MDTB formation. BS (12.5 μM dimer) was assayed with DTB (200 μM) and AdoMet (circles, 100 μM; squares, 200 μM) at 37 °C. (B) AdoMet concentration-dependence of MDTB formation. BS (50 μM dimer) was assayed with DTB (200 μM) and AdoMet (25 – 400 μM) for 60 min at 37 °C. MDTB was determined by HPLC with UV detection at 210 nm, and the concentration determined using a biotin standard and the conversion ratio εbiotin/εMDTB (210 nm) = 1.69. Error bars represent the standard deviation of 4 samples.
The inability to generate a stoichiometric amount of MDTB, even in the presence of limiting AdoMet, can be explained if AdoMet from unreacted active sites can rapidly dissociate and preferentially bind to active sites that contain MDTB; in other words, if the Km for AdoMet is lower for the second half-reaction with MDTB than for the first half-reaction with DTB (Figure 1A). Consistent with this interpretation, we have also found that MDTB formation in the first half-reaction is significantly slower in the presence of limiting AdoMet (data not shown), possibly due to incomplete saturation of AdoMet binding during the first half-reaction. Thus we faced a trade-off in which the concentration of the intermediate could be increased somewhat by limiting the concentration of AdoMet, but the turnover rate is slowed under these conditions, possibly resulting in a decrease in the yield of intermediates and products.
BS Turnover is Accompanied by the Growth and Decay of a Multicomponent EPR Signal
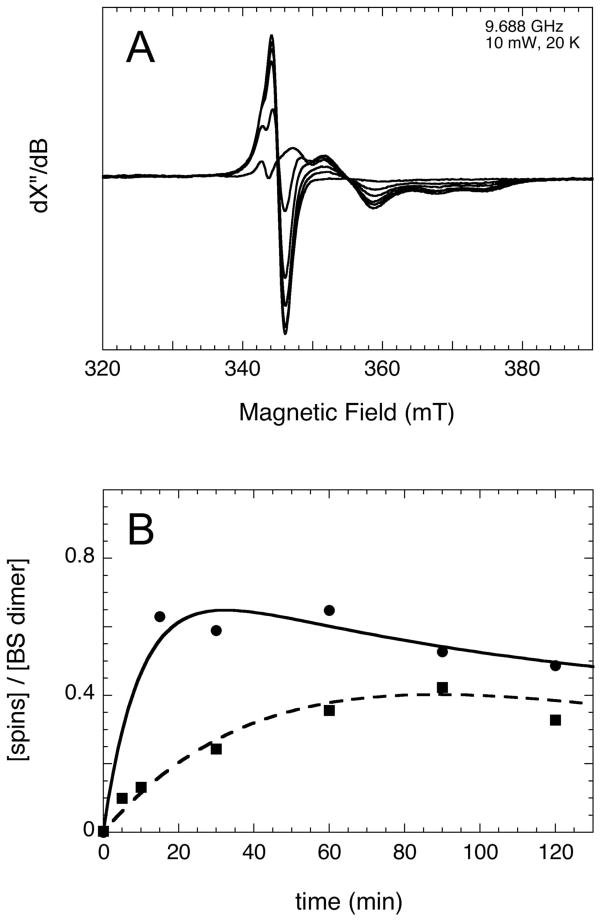
Having established that the BS reaction intermediate MDTB is maximally formed at ~0.3 – 0.6 equiv per BS dimer after incubation with 1 – 2 equiv AdoMet, we examined whether changes in the redox state of the BS FeS clusters might accompany MDTB formation, as predicted by our proposed mechanism. BS (100 μM dimer) was preincubated for 5 min with DTT, FeCl3, and Na2S to reconstitute the [4Fe-4S]2+ cluster, and for 10 min with flavodoxin, FNR, and NADPH to establish reducing conditions conducive to enzyme activity. DTB (400 μM) and AdoMet (100 or 400 μM) were then added to initiate BS turnover as in prior studies (8). To ease handling of the large sample volumes required, the reaction was carried out in an open vial inside an anaerobic glovebox at ambient temperature (~25 °C). At various time intervals (5 – 120 min), samples were collected in parallel for EPR and HPLC analysis: a 300 μl sample was transferred to a quartz EPR tube, tightly capped, and frozen in liquid N2 until spectra could be collected several days later, while a 90 μl sample was quenched with acid, the protein precipitate removed by centrifugation, and the supernatant analyzed for MDTB and biotin content by HPLC as described in the Materials and Methods. Within 5 – 10 min, the assay mixture developed a complex EPR spectrum in the range g = 1.84 to 2.02 (gav ≈ 1.94), most likely due to overlapping signals from several spin systems (Figure 3A), including a narrow isotropic signal at g = 2.005 from the flavin semiquinone radical in flavodoxin. A control sample with all reaction components except BS shows only this flavin semiquinone radical. While the flavin component gradually decays in intensity throughout the assay, the other components of the spectrum grow and decay in parallel, with strong intensity early in the time course and gradually weakening intensity at later times. The broad EPR spectrum with an approximately rhombic lineshape, together with a relatively low gav value of ~1.94, suggests that the spectrum is predominantly due to one or more [2Fe-2S]+ or [4Fe-4S]+ clusters.
Figure 3.
Formation of a reduced FeS cluster during the BS assay. (A) BS (100 μM dimer) was incubated with DTB (400 μM) and AdoMet (100 μM) at ambient temperature (~25 °C) in an anaerobic glovebox, with other components of the assay as described in the Materials and Methods. At varying intervals (5 – 120 min), samples were transferred to an EPR tube and frozen in liquid N2. Spectra were recorded at a 9.688 GHz, 10 mW, and 20 K. (B) EPR spectra from above (squares), as well as spectra of BS assayed with 400 μM AdoMet (circles, 9.4733 GHz, 10 mW, 20 K), were double integrated after manually subtracting the flavin semiquinone contribution, and the resulting spectra compared to Cu2+-EDTA standards to determine spin concentration.
To quantify the spin concentration in each sample, we compared double integrals of the spectra to those obtained from standards of Cu2+-EDTA, with all spectra acquired under identical nonsaturating conditions. Prior to integration, the flavodoxin semiquinone contribution to each spectrum (~2 – 5 μM spin, mostly saturated at 10 mW microwave power) was manually subtracted using the control spectrum. The resulting spin concentrations (Figure 3B) correspond to the slow formation and partial decay of overlapping signals attributable to one or more reduced FeS clusters. At higher concentrations of AdoMet (4 equiv per BS dimer), the rate of formation of the EPR signal (kobs ≈ 0.1 min−1 at 25 °C) appears to roughly correlate with the previously reported rate of MDTB formation (kobs ≈ 0.07 min−1 at 37 °C (8)). However, at lower AdoMet concentrations (1 equiv per BS dimer), the apparent rate of development of the EPR signal is significantly slowed (kobs ≈ 0.02 min−1) possibly due to incomplete occupancy of the enzyme active sites with limiting AdoMet.
Formation of the Reduced FeS Cluster Parallels Production of MDTB
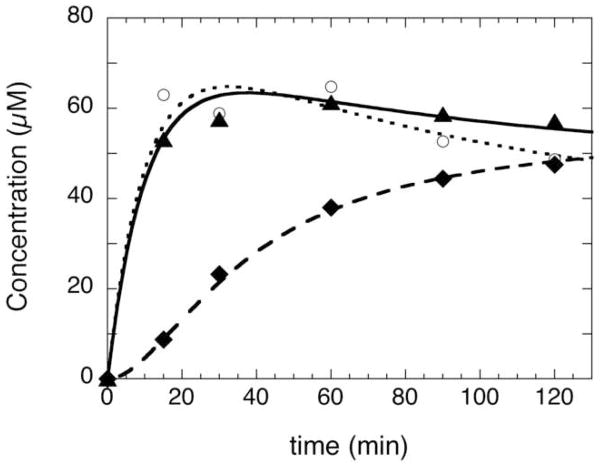
In parallel with the collection of the EPR samples described above, additional samples were also collected and quenched with acid to denature the protein and release any bound DTB, MDTB, and biotin. The production of MDTB was analyzed by HPLC with UV detection (8) and is compared to the EPR signal intensity (open circles) in Figure 4. We observe that MDTB (filled triangles) forms rapidly (k1 ≈ 0.1 min−1), as also observed in Figure 2 and previously reported, but decays at a slower rate (k2 ≈ 0.02 min−1), possibly due to the lower concentration of AdoMet and incubation temperature (~25 °C). When compared to the concentration of reduced FeS clusters (open circles), the formation of MDTB parallels FeS cluster reduction, both in magnitude (maximally ~60 μM or ~0.6 equiv per dimer) and in the approximate rate of formation and decay. The only notable discrepancy is the apparent steady-state concentration of these putative intermediates, ~50 μM MDTB vs. ~40 μM reduced FeS cluster (although this range is within the estimated error of the experiment). In contrast, biotin formation (filled diamonds) initially lags behind both MDTB formation and formation of the reduced FeS cluster, consistent with prior studies that demonstrated that MDTB is an intermediate that is transformed into biotin (8, 10, 27). Thus we would suggest that MDTB and the reduced FeS cluster are both catalytic intermediates that are formed at equivalent concentrations in the same reaction step.
Figure 4.
Formation and decay of the reduced FeS cluster parallels formation and decay of the reaction intermediate MDTB. BS (100 μM dimer) was incubated with DTB (400 μM) and AdoMet (400 μM) at ambient temperature (~25 °C) in an anaerobic glovebox, with other components of the assay as described in the Materials and Methods. Samples were collected for EPR analysis and the FeS cluster spin concentration (circles, dotted curve) determined as described in Figure 3B. Parallel samples were also collected for HPLC analysis, and the concentration of MDTB (triangles, solid curve) and biotin (diamonds, dashed curve) was determined. The curves represent a fit to a two-step sequential reaction sequence (8), in which MDTB and the reduced FeS cluster are formed at k1 ≈ 0.1 min−1 and then decay at a rate of k2 ≈ 0.02 min−1 to steady-state concentration of 40–50 μM. Differences between MDTB and FeS cluster fits are due to small differences in the maximum and steady-state concentrations. Biotin exhibits a short lag followed by a burst (kburst ≈ 0.02 min−1) and is then formed with kSS ≈ 0.002 min−1.
It should be noted that this comparison has some associated uncertainty: the concentration of MDTB was determined using biotin as a standard and using an estimate of the extinction coefficient for MDTB based on similar thiol and thioether containing compounds (see Materials and Methods for a more detailed description). In addition, the spin quantitation procedure has an associated uncertainty of ca. 10–20%.
EPR Spectra are Simulated by Two Overlapping Rhombic Signals
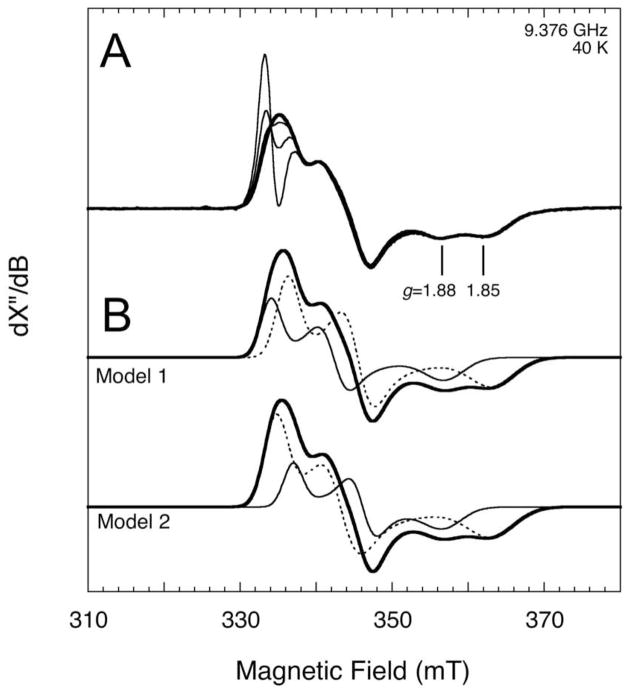
The EPR spectrum detected during BS turnover contains multiple components suggesting an overlap of signals arising from two or more spin systems. To further characterize the spectrum, a sample of BS (100 μM dimer) was prepared as described above, except that the concentration of flavodoxin used in the assay mixture was reduced to 2 μM to minimize the overlapping flavin semiquinone signal. The EPR spectrum was recorded as a function of microwave power, and the spectra were scaled so as to superimpose the high-field region from 340–370 mT. As shown in Figures 5A and 6B, the low intensity and narrow flavin contribution is easily saturated relative to the other components. At a power of 100 mW, it is sufficiently saturated such that its contribution to the total spectrum is negligible, and the resulting spectrum (thick solid line in Figure 5A) represents the signal from only the iron-sulfur cluster components. The spectrum has two high-field troughs at g = 1.88 and 1.85 that suggest an overlap of two FeS cluster signals, and can be simulated as the sum of two rhombic S = ½ FeS cluster signals. However, the composition and g tensors of the components can be modeled with two equally accurate sets of parameters (Figure 5B): Model 1: g = [1.9947, 1.9410, 1.8458], 64% and g = [2.0079, 1.9590, 1.8787], 36%, or Model 2: g = [2.0037, 1.9533, 1.8469], 74% and g = [1.9906, 1.9375, 1.8796], 26%. In model 1, the g tensors of the two components differ in their mean, but not much in their anisotropy, whereas in model 2, the g tensors differ in their anisotropy but not their mean. Common to both models is that the component with g ≈ 1.85 is the major component (64% and 74%), and the component with g ≈ 1.88 is the minor component (36% and 26%). The first set of parameters is similar to those reported by Huynh and coworkers (16), with similar g values and only slight differences in the respective ratios of the individual components. Of particular interest, both studies report that two overlapping signals are formed in parallel following incubation under assay conditions, and we further have observed that the ratio of these two signals does not change as the signals grow and decay over time (Figure 3A) or as the sample temperature is altered (Figure 6A). It might be conceivable that, instead of two components, the spectrum consists of only one S = 1/2 component with the two high-field troughs resulting from hyperfine splitting due to a proton, possibly from the Arg260 ligand. However, we were not able to accurately fit the spectrum with this assumption; specifically, hyperfine splitting by a proton would have generated a spectrum in which the trough at g ≈ 1.88 is deeper than the trough at g ≈ 1.85.
Figure 5.
EPR spectra of BS frozen during turnover. BS (100 μM dimer) was incubated with DTB (400 μM), AdoMet (400 μM), and flavodoxin (2 μM) for 90 min at 37 °C and then transferred to an EPR tube and frozen in liquid nitrogen. (A) Scaled experimental spectra recorded at 9.376 GHz and 40 K and varying microwave power (thin lines: 0.1 mW, 1 mW, 10 mW; thick line: 100 mW) show the progressive saturation of the flavin signal at 334 mT and two components of the iron-sulfur signal. (B) Two possible two-component models that accurately fit the 100 mW spectrum (thick solid). Model 1: g = [1.9947, 1.9410, 1.8458], 64% (dashed) and g = [2.0079, 1.9590, 1.8787], 36% (thin solid). Model 2: g = [2.0037, 1.9533, 1.8469], 74% (dashed) and g = [1.9906, 1.9375, 1.8796], 26% (thin solid).
Figure 6.
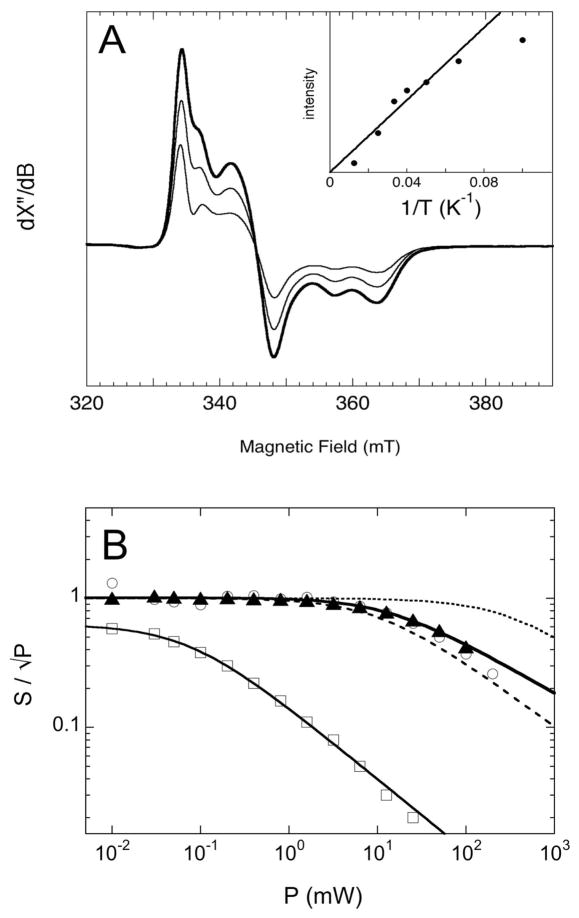
Temperature and microwave power dependence of EPR spectra of BS (100 μM dimer) incubated with DTB (400 μM) and AdoMet (400 μM) for 60 min at 25 °C, with other assay conditions as described in the Materials and Methods. (A) Spectra were recorded at varying sample temperatures (10 – 80 K), with the microwave power held constant at 1 mW, and the total integrated intensity of the absorption spectrum is plotted vs. inverse temperature, showing approximate Curie law dependence (inset). Spectra are shown for 15 (thick curve), 25, and 40 K. (B) Spectra recorded at varying microwave power (0.01 – 150 mW), with the sample temperature held constant at 20 K, and the intensity of the signal at 348 mT divided by the square root of the microwave power (▲) is plotted vs. power. Similar data were collected for the Arg260Met mutant (○) and flavodoxin FMN semiquinone (□). Data are fit to power saturation curves using P1/2 = 19 mW for BS and P1/2 = 0.07 mW for flavodoxin (solid curves). Simulated power saturation curves at 20 K for the H. halobium ferredoxin [2Fe-2S]+ cluster (P1/2 = 10 mW, dashed curve) and the B. stearothermophilus ferredoxin [4Fe-4S]+ cluster (P1/2 = 330 mW, dotted curve) are shown for reference (29).
The observed EPR spectra are not similar to spectrum of the [2Fe-2S]+ cluster observed by Lotierzo, et al. in the chemically reduced Asn153Ala mutant enzyme (23). The [2Fe-2S]+ cluster produced by chemical reduction exhibits an axial spectrum with g⊥ ≈ 2.00 and g|| ≈ 1.91, while the reduced cluster observed during enzyme turnover exhibits a rhombic spectrum with gx,y,z ≈ [2.00, 1.95, 1.85] for the major component as described above. Minimally, the dissimilarity of the spectra observed following chemical reduction vs. enzyme turnover suggests that the paramagnetic cluster generated during turnover does not correspond to simple reduction of the [2Fe-2S] cluster by electron transfer. The increased dispersion of g tensors during catalytic turnover could result from decreased structural symmetry, as might be expected if MDTB coordinates to and distorts the [2Fe-2S]+ cluster.
The Relaxation Properties of the EPR Signals are Consistent With a [2Fe-2S]+ Cluster
The lineshape and g values associated with the observed EPR signal are consistent with either [2Fe-2S]+ or [4Fe-4S]+ clusters. Since the initial active BS sample contains both [2Fe-2S]2+ and [4Fe-4S]2+ clusters (18, 28), electron transfer during formation of MDTB could lead to reduction of one or both clusters. To distinguish both the number and type of reduced FeS clusters, we probed the spin relaxation properties of the cluster by examining the signal intensity as a function of both sample temperature and microwave power. Samples of WT BS and the active Arg260Met mutant (100 μM dimer) were prepared by incubating with DTB (400 μM) and AdoMet (400 μM) for 90 min, in a similar manner as described above. The sample temperature was varied (10 – 80 K) at a constant moderate microwave power (1 mW), and the most intense spectra were observed at 15 – 20 K (Figure 6A). The decrease in signal intensity was as predicted by the Curie law (Figure 6A inset) except at very low temperature (10 K), where partial signal saturation was observed due to the moderately high microwave power. The microwave power was then varied (0.01 – 200 mW) at a constant sample temperature (20 K), and EPR spectra show that the signal observed in both the WT and Arg260Met BS is only partially saturated at moderate-to-high power (P1/2 = 19 mW, Figure 6B). For comparison, the [2Fe-2S]+ cluster of Halobacterium halobium ferredoxin exhibits a maximum signal at 20 – 40 K and is saturated at a similar moderate power (P1/2 = 10 mW, Figure 6B, dashed curve) (29). In contrast, [4Fe-4S]+ clusters typically are very fast relaxing and show saturation only at very high microwave powers (> 50 mW) and very low temperatures (< 10 K). For example, the [4Fe-4S]+ cluster in B. stearothermophilus ferredoxin has a half-saturation power of P1/2 = 330 mW (Figure 6B, dotted curve) (29). In contrast, the isotropic signal at g = 2.005 arising from the flavin semiquinone is easily saturated at very low power (P1/2 = 0.07 mW).
Johnson and coworkers have noted that the dithionite-reduced [4Fe-4S]+ cluster in BS has intermediate relaxation properties and is also partially saturated above 10 mW at 20 K (20). However, the [4Fe-4S]+ cluster spectrum exhibits g values of 2.044, 1.944, and 1.914 (20) that are significantly higher than those reported in this study. Therefore, on the basis of spin relaxation properties and g values, the signal we observe during BS turnover can be most likely attributed to a somewhat fast-relaxing [2Fe-2S]+ cluster. As noted previously, Jameson et al. have characterized a similar EPR-detectable signal as a minor component of the total cluster content of BS incubated with substrates, and have attributed the signal to a [2Fe-2S]+ cluster on the basis of Mössbauer parameters (16). It should also be noted that the major features of the complex EPR signal respond to changes in sample temperature and microwave power in a similar manner, suggesting that these have identical spin relaxation properties and arise from the same type of FeS cluster within the enzyme.
The Arg260Met Mutation Alters the g Tensors in the EPR Spectrum
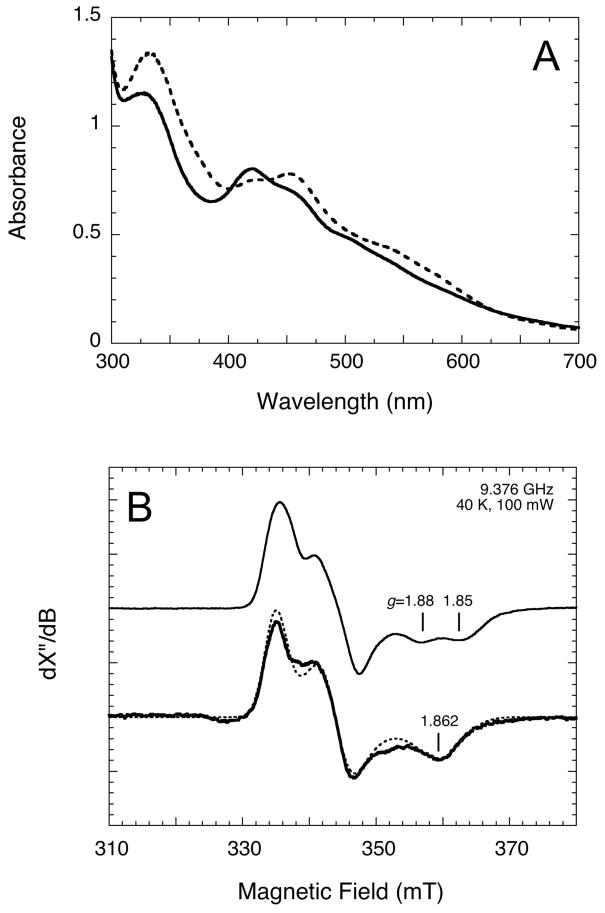
Arginine is an unusual metal ligand (30), particularly for an FeS cluster, yet in BS it is a highly conserved residue that is coordinated to via the guanidino group to the [2Fe-2S] cluster (12). In a previous study, we demonstrated that the enzyme retains nearly full catalytic activity when Arg260 is replaced by several other amino acid residues, including alanine, histidine, and methionine (but not cysteine) (31). Replacement of arginine by alanine may leave a pocket in the active site that allows coordination of water to the [2Fe-2S]2+ cluster in place of arginine, while histidine and methionine side-chains are predicted to be sufficiently long that they can coordinate directly to the [2Fe-2S]2+ cluster. Consistent with this hypothesis, there is a significant change in the UV/visible spectrum of the [2Fe-2S]2+ cluster in the Arg260Met mutant as compared to WT (Figure 7A), suggesting changes in the relative energies of ligand-to-metal charge transfer bands.
Figure 7.
The Arg260Met mutant enzyme exhibits altered spectroscopic properties for the reduced FeS cluster. (A) UV/visible spectra of WT (dashed curve) and Arg260Met BS (solid curve, 50 μM dimer) aerobically purified containing one [2Fe-2S]2+ cluster per monomer. Shifts in the position and intensity of the major ligand-to-metal charge transfer bands in the UV/visible spectrum suggest that the methionine sulfur atom replaces the arginine nitrogen atom as a ligand to the [2Fe-2S]2+ cluster. (B) EPR spectra of the [2Fe-2S]+ clusters in WT and Arg260Met mutant enzymes (100 μM dimer), experimental (solid) and simulated (dashed). Aliquots were incubated with DTB (400 μM) and AdoMet (400 μM) for 90 min, with other assay conditions as described in the Materials and Methods. Samples were frozen in liquid N2 and EPR spectra were collected at 9.376 GHz, 100 mW, 40 K. Simulation parameters for the EPR spectrum of the Arg260Met mutant enzyme: g = [2.000, 1.947, 1.862].
Since Arg260 is coordinated to the [2Fe-2S]2+ cluster, changes in the EPR spectrum of the Arg260Met mutant are expected if the spectrum is due to the [2Fe-2S]+ cluster. The Arg260Met mutant enzyme was incubated with DTB and AdoMet under assay conditions for 90 min, as described above for the WT enzyme, and a sample frozen in liquid N2. Similar to WT enzyme, the EPR spectrum of the Arg260Met enzyme was recorded under conditions (40 K, 100 mW) that saturate the flavin semiquinone radical and minimize the contribution of this signal to the EPR spectra. In contrast to the WT enzyme, the EPR spectrum of the Arg260Met mutant enzyme shows only a single S = 1/2 component with a rhombic g tensor. The high-field component at g ≈ 1.862 falls directly between the two high-field components of the WT enzyme spectrum. The other g tensor values are not significantly different from the values obtained from simulations of the WT enzyme. The observation of a significant change in the high-field g tensor upon mutation of a coordinating ligand supports the assignment of the EPR signal to a reduced state of the [2Fe-2S] cluster.
Coupling of the Unpaired Electron to Nitrogen is Observed in WT BS, but not in the Arg260Met Mutant
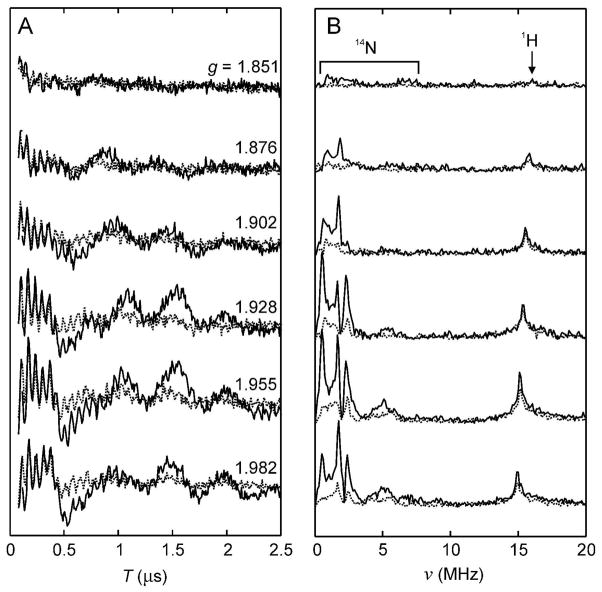
Typical FeS clusters in non-isotopically labeled proteins exhibit small hyperfine coupling to nearby hydrogen atoms (1H) bonded to the FeS cluster ligands (eg. cysteine Hβ) and to nitrogen atoms (14N) in nearby functional groups. Weak 14N coupling has been attributed to through-space coupling to nearby amide nitrogen atoms, while strong 14N coupling implies a close proximity that is only achieved by covalent coordination of a nitrogen-containing amino acid ligand to the FeS cluster. ESEEM is a pulsed EPR technique that allows detection of hyperfine coupling of unpaired electrons to nearby nuclear spins (32). Since WT BS contains a single arginine ligand to the [2Fe-2S]2+ cluster that is eliminated in the Arg260Met mutant, ESEEM spectra should provide additional confirmation that the EPR spin is localized on the [2Fe-2S] cluster. Three-pulse X-band ESEEM spectra were recorded at several magnetic field positions across the EPR spectrum of WT enzyme (Figure 8, solid curves), and the spectra show peaks due to coupling to 1H nuclei at 15 and 30 MHz and peaks due to coupling to 14N nuclei at 2 – 6 MHz. In contrast, ESEEM spectra of the Arg260Met mutant enzyme measured under the same conditions (Figure 8, dotted curves), have only very weak intensity in the 1 – 6 MHz region, most likely due to through-space coupling to the Cys97, Cys128, or Cys188 amide nitrogen atoms (5 – 6 Å Fe-N distance) or the nearby Arg95 side chain (5 – 7 Å Fe-N distance).
Figure 8.
Comparison of ESEEM spectra of WT and Arg260Met mutant enzymes. (A) Three-pulse ESEEM decay envelopes with τ = 160 ns measured at 10 K, 9.71 GHz and magnetic field strengths corresponding to g values of 1.851, 1.876, 1.902, 1.928, 1.955, and 1.982. WT (solid) and Arg260Met (dotted). (B) Peaks corresponding to electron-nuclear coupling to 1H appear at around 15 MHz and are present in both WT (solid) and Arg260Met (dotted), but peaks corresponding to electron-nuclear coupling to 14N appear at 2 to 6 MHz only for WT BS with the Arg260 ligand to the [2Fe-2S]+ cluster, and are largely lost in spectra of the Arg260Met mutant, which contains only sulfur ligands to the [2Fe-2S]+ cluster.
The ESEEM spectra in Figure 8 show three sharp peaks at 0.5, 1.6 – 1.8 and 2.3 – 2.4 MHz, due to 14N in the cancellation regime (33). We can estimate the 14N quadrupole parameters from these frequencies and get |K| = |e2Qq/4h| ≈ 0.65–0.70 MHz and η ≈ 0.36–0.54. Histidine can be excluded as a source of these signals, as the closest one (His152) is >10 Å away from the [2Fe-2S] cluster. The value of |K| is inconsistent with a primary aliphatic amine (|K| ≈ 1 MHz) (34), and it is lower than the values observed for backbone amide nitrogens in peptides (|K| ≈ 0.75–0.85 MHz) (35, 36). The values fall in the range of those assigned to Arg side chains in nitrogenase (|K| ≈ 0.54 MHz, η ≈ 0.59) (37) and ethanolamine ammonia-lyase (|K| ≈ 0.76 MHz, η ≈ 0.55) (38). Therefore, the 14N peaks in Figure 8 are consistent with an assignment to Arg260. In addition, the fact that the weak ESEEM peaks in the R260M mutant are at similar frequencies than in the WT supports arginine assignments in both variants (Arg260 in the WT, and Arg95 in the mutant). However, further study is required to confirm this and to identify the particular 14N in the Arg260 side chain that is responsible for the ESEEM modulations.
Discussion
The chemical properties and biochemical function of the iron-sulfur clusters in BS have been the subject of numerous investigations (reviewed in (1)). When aerobically purified, recombinant BS contains a [2Fe-2S]2+ cluster (19) that is also observed as the primary cofactor component in vivo using whole-cell Mössbauer spectroscopy (39, 40). In addition, a [4Fe-4S]2+/+ cluster can be chemically reconstituted by incubation with Fe2+/3+, S2−, and a chemical reductant (18, 41, 42). On the basis of spectroelectrochemical titrations (18) and Mössbauer spectroscopy of differentially-reconstituted 57Fe protein (28), the [2Fe-2S]2+ and [4Fe-4S]2+/+ clusters were found to occupy different sites within the protein, a feature later confirmed by the x-ray crystal structure of the reconstituted protein with substrates bound (12). Most importantly, only protein that contains both the [2Fe-2S]2+ cluster and the [4Fe-4S]2+ cluster is significantly active in vitro (15).
The chemical properties the [2Fe-2S]2+ cluster and the role of this cluster in biotin formation have remained somewhat controversial. The x-ray crystal structure shows that this cluster is bound to Cys97, Cys121, Cys188, and Arg260, and resides ~4.6 Å away from DTB. The proximity of the [2Fe-2S]2+ cluster to DTB has led us to suggest that this cluster plays a role in sulfur insertion (12, 15). Several studies have examined the fate of the [2Fe-2S]2+ cluster under in vitro assay conditions. Using UV/visible spectra of reconstituted BS, we have shown that at least a portion of the [2Fe-2S]2+ cluster is reduced or lost at a rate that is similar to the rate of biotin formation (15). Marquet and coworkers subsequently examined the transformation of the FeS clusters in partially purified 57Fe-labeled BS using Mössbauer spectroscopy (43). Prior to the addition of substrates, the Mössbauer spectrum of the initial protein is best modeled by an approximate 1:1 (±0.1) ratio of [2Fe-2S]2+ and [4Fe-4S]2+ clusters. While incubation with DTB alone did not affect the ratio of FeS clusters, incubation with both DTB and AdoMet decreased the [2Fe-2S]2+ to [4Fe-4S]2+ cluster ratio to 0.6:1, while simultaneously increasing the concentration of high-spin Fe3+ species, most likely in the buffer (43). Huynh and coworkers undertook a more detailed analysis of changes in the [2Fe-2S]2+ cluster during enzyme turnover by combining EPR and Mössbauer spectroscopy of BS that was 57Fe-labeled in only the [2Fe-2S]2+ cluster (16). They also report a significant decrease in the concentration of the [2Fe-2S]2+ cluster, and report the transient formation of a [2Fe-2S]+ cluster followed by an increase in the concentration of high-spin Fe2+. Although in principle these reports are consistent with the involvement of the [2Fe-2S]2+ cluster in providing sulfur for biotin formation, Huynh and coworkers noted a significant discrepancy between the rate of [2Fe-2S]2+ cluster degradation (0.13 min−1) and the rate of biotin formation (0.012 min−1). They suggested that an alternative mechanism in which the cluster degrades to provide a persulfide sulfur for biotin formation could not be ruled out.
In the present work, we show that reduction of the [2Fe-2S]2+ cluster to a [2Fe-2S]+ cluster is kinetically correlated with the production of MDTB, with an approximate 1:1 stoichiometric ratio (Figure 4). However, given the experimental uncertainties involved, and the relatively slow progress of both MDTB and biotin formation, we cannot necessarily distinguish whether reduction of the [2Fe-2S]2+ cluster occurs in the same chemical step as MDTB formation or occurs immediately prior to or after C-S bond formation. However, since a reduced [2Fe-2S]+ cluster is never observed in the absence of DTB, despite the presence of the complete flavodoxin reducing system, we conclude that it is DTB, or more specifically, the quenching of the DTB carbon radical that triggers reduction of this cluster (8).
The apparent complexity of the resulting EPR spectrum is due to the overlap of two rhombic EPR signals, both of which are fairly typical of reduced FeS clusters. The same ~2:1 ratio of signals has now been observed in two different laboratories with different preparations of enzyme. The ratio of these two signals is also not sensitive to incubation time, sample temperature, or microwave power, suggesting the two signals arise simultaneously in the assay and have nearly identical spin relaxation properties. The origin of the two overlapping signals is not presently understood. One possible explanation for the occurrence of two components is differential valence localization on the two iron atoms within the [2Fe-2S]+ cluster. The electronic state of the reduced cluster can be described as a high-spin Fe2+ site (S = 2) antiferromagnetically coupled to a high-spin Fe3+ site (S = 5/2). If there is a preference for Fe2+ to reside at one site over the other, as for example has been observed in Rieske-type [2Fe-2S]+ proteins with Fe2+ coordinated by two His residues and Fe3+ coordinated by two Cys residues, then the unpaired electron spin is not delocalized over the entire cluster (44). In our system, there could be only a small energy difference between the two electronic states (2:1 ratio corresponds to ~0.1 kJ at 20 K), such that both possible spin valence-localized states are occupied in different FeS clusters within the frozen sample. A more detailed analysis of hyperfine coupling constants that arise from coupling of the electron spin to 14N/15N of Arg260 using ESEEM and ENDOR spectroscopy would provide a more complete electronic description of the reduced cluster.
An alternative explanation for the observation of two distinct EPR signals could be structural heterogeneity within the active site of the intermediate. For example, the MDTB thiolate could equilibrate between coordination to the two opposing Fe atoms within the intermediate cluster. However, assuming that the general scheme depicted in Figure 1A is correct, then the quenching of a DTB carbon radical by the μ-sulfide of the [2Fe-2S]2+ cluster would initially generate a reduced cluster in which the MDTB thiolate is a symmetric bridging ligand. This cluster would be best described as a [Fe2(μ-S)(μ-S-MDTB)(S-Cys)3(NH2-Arg)]− cluster. A somewhat similar synthetic cluster, [Fe2(μ-S)(μ-S-Et)(NO)4]2− was recently characterized by various spectroscopic methods and by high resolution x-ray crystallography, and the core was described as a slightly nonplanar rhombus (dihedral angle = 171.7°) with equivalent bond distances between the Fe atoms and the bridging EtS− ligand (45). This cluster had physical properties that were only slightly altered from the values for the corresponding [Fe2(μ-S)2(NO)4]2− cluster, and in most respects behaved in a manner that would be indistinguishable from a typical FeS cluster. Based on this chemical precedent, we would predict that structure of the BS intermediate FeS cluster likely includes a symmetric bridging MDTB thiolate ligand (Figure 1A). From a cursory examination of the initial active site structure, containing AdoMet, DTB, and the [2Fe-2S]2+ cluster (Figure 1B), it appears that re-positioning of MDTB as a symmetric bridging thiolate ligand would require significant reorganization of the active site, and that the current structural data may not be sufficient to describe this intermediate or the subsequent ring closing reaction.
Another possible origin of structural heterogeneity, and consequently a possible reason for the two spectral components, could be the positioning of the arginine ligand relative to the [2Fe-2S]+ cluster. The guanidine group has two terminal nitrogen atoms that could coordinate the iron. From the x-ray crystal structure, Fe-Nω distances of 2.35 and 3.15 Å were found, but it is possible that this geometry rearranges upon reaction of DTB with the bridging sulfur and concomitant changes in the geometry of the FeS cluster core. Lastly, the two spectral components could possibly arise from two different protonation states of the guanidine group of Arg260. Clearly, a more precise structural and electronic description of the intermediate state will be required to distinguish between these various hypotheses.
Several AdoMet radical enzymes have now been identified that catalyze the formation of new carbon-sulfur bonds using FeS clusters as the probable source of sulfur atoms. E. coli lipoyl synthase (LipA) catalyzes the addition of two sulfur atoms to octanoyl-E2 protein or octanoyl-H protein to generate lipoyl-protein, with both sulfur atoms most likely derived from a [4Fe-4S] cluster within a single enzyme monomer (46–48). S. solfataricus LipA will accept a octanoyl-tripeptide substrate and generates a C6 monothiolated intermediate prior to addition of the second sulfur atom (49). T. maritima MiaB catalyzes the methylthiolation at the C2 position of (N6-isopentenyl)-adenosine-37 in certain tRNAs, with the sulfur atom possibly derived from a [4Fe-4S] cluster (50). RimO catalyzes the methylthiolation of an aspartate residue in the ribosomal protein S12, and the presence of two [4Fe-4S] clusters in the enzyme again suggests the sulfur atom is derived from one of these clusters (51). Presumably these sulfur insertion reactions will share some features of the biotin synthase reaction mechanism. In particular, formation of a carbon-sulfur bond by attack of a carbon radical on sulfide must be accompanied by one electron oxidation of the sulfur atom, and presumably this would be accompanied by transfer of this electron into the FeS cluster. In most cases, these other enzymes have been assayed in the presence of excess dithionite, resulting in an equilibrium concentration of chemically reduced FeS clusters that obscures observation of paramagnetic enzyme intermediates and may also inhibit enzyme activity. If assay conditions could be developed with enzyme initially containing only diamagnetic [4Fe-4S]2+ clusters, and in which the natural reducing system supports AdoMet radical generation, then we would predict that paramagnetic [4Fe-4S]+ clusters would be generated during catalysis, either as an intermediate in LipA, or as an enzyme-bound product in MiaB or RimO.
Acknowledgments
We would like to thank Dr. Christine Farrar for supplying data incorporated into Figure 2, and Corey Fugate for supplying data incorporated into Figure 6.
Footnotes
This research has been supported by grants from the NSF (MCB 09-23829 to J.T.J.) and the NIH (GM73789 to R.D.B.).
Abbreviations: AdoMet, S-adenosyl-L-methionine; BS, biotin synthase; dAH, 5′-deoxyadenosine; dA•, 5′-deoxyadenosyl radical; DTB, dethiobiotin; DTT, dithiothreitol; EPR, electron paramagnetic resonance; ESEEM, electron spin echo envelope modulation; Fld, flavodoxin; FNR, ferredoxin(flavodoxin): NADP+ oxidoreductase; MDTB, 9-mercaptodethio-biotin; Tris, tris(hydroxymethyl)aminomethane;
References
- 1.Jarrett JT. The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase. Arch Biochem Biophys. 2005;433:312–321. doi: 10.1016/j.abb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Marquet A. Enzymology of carbon-sulfur bond formation. Curr Opin Chem Biol. 2001;5:541–549. doi: 10.1016/s1367-5931(00)00249-0. [DOI] [PubMed] [Google Scholar]
- 3.Marquet A, Florentin D, Ploux O, Tse Sum Bui B. In vivo formation of C-S bonds in biotin. An example of radical chemistry under reducing conditions. J Phys Org Chem. 1998;11:529–535. [Google Scholar]
- 4.Escalletes F, Florentin D, Tse Sum Bui B, Lesage D, Marquet A. Biotin Synthase Mechanism: Evidence for Hydrogen Transfer from the Substrate into Deoxyadenosine. J Am Chem Soc. 1999;121:3571–3578. [Google Scholar]
- 5.Gibson KJ, Pelletier DA, Turner IM., Sr Transfer of sulfur to biotin from biotin synthase (BioB protein) Biochem Biophys Res Commun. 1999;254:632–635. doi: 10.1006/bbrc.1998.9991. [DOI] [PubMed] [Google Scholar]
- 6.Tse Sum Bui B, Florentin D, Fournier F, Ploux O, Mejean A, Marquet A. Biotin synthase mechanism: on the origin of sulphur. FEBS Lett. 1998;440:226–230. doi: 10.1016/s0014-5793(98)01464-1. [DOI] [PubMed] [Google Scholar]
- 7.Tse Sum Bui B, Mattioli TA, Florentin D, Bolbach G, Marquet A. Escherichia coli biotin synthase produces selenobiotin. Further evidence of the involvement of the [2Fe-2S]2+ cluster in the sulfur insertion step. Biochemistry. 2006;45:3824–3834. doi: 10.1021/bi052388m. [DOI] [PubMed] [Google Scholar]
- 8.Taylor AM, Farrar CE, Jarrett JT. 9-Mercaptodethiobiotin is formed as a competent catalytic intermediate by Escherichia coli biotin synthase. Biochemistry. 2008;47:9309–9317. doi: 10.1021/bi801035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guianvarc’h D, Florentin D, Tse Sum Bui B, Nunzi F, Marquet A. Biotin synthase, a new member of the family of enzymes which uses S-adenosylmethionine as a source of deoxyadenosyl radical. Biochem Biophys Res Commun. 1997;236:402–406. doi: 10.1006/bbrc.1997.6952. [DOI] [PubMed] [Google Scholar]
- 10.Shaw NM, Birch OM, Tinschert A, Venetz V, Dietrich R, Savoy LA. Biotin synthase from Escherichia coli: isolation of an enzyme-generated intermediate and stoichiometry of S-adenosylmethionine use. Biochem J. 1998;330:1079–1085. doi: 10.1042/bj3301079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrar CE, Jarrett JT. Encyclopedia of the Life Sciences. John Wiley & Sons, Ltd; Chichester: 2007. Radical S-Adenosylmethionine (SAM) Superfamily. [Google Scholar]
- 12.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosper MM, Jameson GN, Davydov R, Eidsness MK, Hoffman BM, Huynh BH, Johnson MK. The [4Fe-4S]2+ cluster in reconstituted biotin synthase binds S-adenosyl-L-methionine. J Am Chem Soc. 2002;124:14006–14007. doi: 10.1021/ja0283044. [DOI] [PubMed] [Google Scholar]
- 14.Hewitson KS, Ollagnier-de Choudens S, Sanakis Y, Shaw NM, Baldwin JE, Munck E, Roach P, Fontecave M. The iron-sulfur center of biotin synthase: site-directed mutants. J Biol Inorg Chem. 2002;7:83–93. doi: 10.1007/s007750100268. [DOI] [PubMed] [Google Scholar]
- 15.Ugulava NB, Sacanell CJ, Jarrett JT. Spectroscopic changes during a single turnover of biotin synthase: destruction of a [2Fe-2S] cluster accompanies sulfur insertion. Biochemistry. 2001;40:8352–8358. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jameson GN, Cosper MM, Hernandez HL, Johnson MK, Huynh BH. Role of the [2Fe-2S] cluster in recombinant Escherichia coli biotin synthase. Biochemistry. 2004;43:2022–2031. doi: 10.1021/bi035666v. [DOI] [PubMed] [Google Scholar]
- 17.Jarrett JT, Wan JT. Thermal inactivation of reduced ferredoxin (flavodoxin):NADP+ oxidoreductase from Escherichia coli. FEBS Lett. 2002;529:237–242. doi: 10.1016/s0014-5793(02)03349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ugulava NB, Gibney BR, Jarrett JT. Biotin synthase contains two distinct iron-sulfur cluster binding sites: chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry. 2001;40:8343–8351. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanyal I, Cohen G, Flint DH. Biotin synthase: purification, characterization as a [2Fe-2S] cluster protein, and in vitro activity of the Escherichia coli bioB gene product. Biochemistry. 1994;33:3625–3631. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- 20.Duin EC, Lafferty ME, Crouse BR, Allen RM, Sanyal I, Flint DH, Johnson MK. [2Fe-2S] to [4Fe-4S] Cluster Conversion in Escherichia coli Biotin Synthase. Biochemistry. 1997;36:11811–11820. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- 21.Ugulava NB, Gibney BR, Jarrett JT. Iron-sulfur cluster interconversions in biotin synthase: dissociation and reassociation of iron during conversion of [2Fe-2S] to [4Fe-4S] clusters. Biochemistry. 2000;39:5206–5214. doi: 10.1021/bi9926227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosper MM, Jameson GN, Hernandez HL, Krebs C, Huynh BH, Johnson MK. Characterization of the cofactor composition of Escherichia coli biotin synthase. Biochemistry. 2004;43:2007–2021. doi: 10.1021/bi0356653. [DOI] [PubMed] [Google Scholar]
- 23.Lotierzo M, Bui BT, Leech HK, Warren MJ, Marquet A, Rigby SE. Iron-sulfur cluster dynamics in biotin synthase: a new [2Fe-2S](1+) cluster. Biochem Biophys Res Commun. 2009;381:487–490. doi: 10.1016/j.bbrc.2009.02.089. [DOI] [PubMed] [Google Scholar]
- 24.Farrar CE, Siu KKW, Howell PL, Jarrett JT. Biotin Synthase Exhibits Burst Kinetics and Multiple Turnovers in the Absence of Inhibition by Products and Product-Related Biomolecules. Biochemistry. 2010;49:9985–9996. doi: 10.1021/bi101023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farrar CE, Jarrett JT. Protein residues that control the reaction trajectory in S-adenosylmethionine radical enzymes: mutagenesis of asparagine 153 and aspartate 155 in Escherichia coli biotin synthase. Biochemistry. 2009;48:2448–2458. doi: 10.1021/bi8022569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan JT, Jarrett JT. Electron acceptor specificity of ferredoxin (flavodoxin):NADP+ oxidoreductase from Escherichia coli. Arch Biochem Biophys. 2002;406:116–126. doi: 10.1016/s0003-9861(02)00421-6. [DOI] [PubMed] [Google Scholar]
- 27.Tse Sum Bui B, Lotierzo M, Escalettes F, Florentin D, Marquet A. Further investigation on the turnover of Escherichia coli biotin synthase with dethiobiotin and 9-mercaptodethiobiotin as substrates. Biochemistry. 2004;43:16432–16441. doi: 10.1021/bi048040t. [DOI] [PubMed] [Google Scholar]
- 28.Ugulava NB, Surerus KK, Jarrett JT. Evidence from Mössbauer spectroscopy for distinct [2Fe-2S]2+ and [4Fe-4S]2+ cluster binding sites in biotin synthase from Escherichia coli. J Am Chem Soc. 2002;124:9050–9051. doi: 10.1021/ja027004j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rupp H, Rao KK, Hall DO, Cammack R. Electron spin relaxation of iron-sulphur proteins studied by microwave power saturation. Biochim Biophys Acta. 1978;537:255–260. doi: 10.1016/0005-2795(78)90509-3. [DOI] [PubMed] [Google Scholar]
- 30.Di Costanzo L, Flores LV, Jr, Christianson DW. Stereochemistry of guanidine-metal interactions: implications for L-arginine-metal interactions in protein structure and function. Proteins. 2006;65:637–642. doi: 10.1002/prot.21127. [DOI] [PubMed] [Google Scholar]
- 31.Broach RB, Jarrett JT. Role of the [2Fe-2S]2+ cluster in biotin synthase: mutagenesis of the atypical metal ligand arginine 260. Biochemistry. 2006;45:14166–14174. doi: 10.1021/bi061576p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chasteen ND, Snetsinger PA. ESEEM and ENDOR Spectroscopy. In: Que J, Lawrence, editors. Physical Methods in Bioinorganic Chemistry: Spectroscopy and Magnetism. University Science Books; Sausalito, CA: 2000. pp. 187–231. [Google Scholar]
- 33.Flanagan HL, Singel DJ. Analysis of 14N ESEEM patterns of randomly oriented solids. J Chem Phys. 1987;87:5606–5616. [Google Scholar]
- 34.Dikanov SA, Tsvetkov YD. Electron Spin Echo Envelope Modulation (ESEEM) Spectroscopy. CRC Press, Inc; Boca Raton, Florida: 1992. [Google Scholar]
- 35.McCracken J, Vassiliev IR, Yang EC, Range K, Barry BA. ESEEM studies of peptide nitrogen hyperfine coupling in tyrosyl radicals and model peptides. J Phys Chem B. 2007;111:6586–6592. doi: 10.1021/jp071402x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yap LL, Samoilova RI, Gennis RB, Dikanov SA. Characterization of the exchangeable protons in the immediate vicinity of the semiquinone radical at the QH site of the cytochrome bo3 from Escherichia coli. J Biol Chem. 2006;281:16879–16887. doi: 10.1074/jbc.M602544200. [DOI] [PubMed] [Google Scholar]
- 37.Lee HI, Thrasher KS, Dean DR, Newton WE, Hoffman BM. 14N electron spin-echo envelope modulation of the S = 3/2 spin system of the Azotobacter vinelandii nitrogenase iron-molybdenum cofactor. Biochemistry. 1998;37:13370–13378. doi: 10.1021/bi980956a. [DOI] [PubMed] [Google Scholar]
- 38.Sun L, Groover OA, Canfield JM, Warncke K. Critical role of arginine 160 of the EutB protein subunit for active site structure and radical catalysis in coenzyme B12-dependent ethanolamine ammonia-lyase. Biochemistry. 2008;47:5523–5535. doi: 10.1021/bi702366e. [DOI] [PubMed] [Google Scholar]
- 39.Benda R, Tse Sum Bui B, Schunemann V, Florentin D, Marquet A, Trautwein AX. Iron-sulfur clusters of biotin synthase in vivo: a Mössbauer study. Biochemistry. 2002;41:15000–15006. doi: 10.1021/bi026590q. [DOI] [PubMed] [Google Scholar]
- 40.Cosper MM, Jameson GNL, Eidsness MK, Huynh BH, Johnson MK. Recombinant Escherichia coli biotin synthase is a [2Fe-2S]2+ protein in whole cells. FEBS Lett. 2002;529:332–336. doi: 10.1016/s0014-5793(02)03390-2. [DOI] [PubMed] [Google Scholar]
- 41.Ollagnier-de Choudens S, Sanakis Y, Hewiston KS, Roach P, Baldwin JE, Munck E, Fontecave M. Iron-Sulfur Center of Biotin Synthase and Lipoate Synthase. Biochemistry. 2000;39:4165–4173. doi: 10.1021/bi992090u. [DOI] [PubMed] [Google Scholar]
- 42.Tse Sum Bui B, Florentin D, Marquet A, Benda R, Trautwein AX. Mössbauer studies of Escherichia coli biotin synthase: evidence for reversible interconversion between [2Fe-2S]2+ and [4Fe-4S]2+ clusters. FEBS Lett. 1999;459:411–414. doi: 10.1016/s0014-5793(99)01300-9. [DOI] [PubMed] [Google Scholar]
- 43.Tse Sum Bui B, Benda R, Schunemann V, Florentin D, Trautwein AX, Marquet A. Fate of the (2Fe-2S)2+ cluster of Escherichia coli biotin synthase during reaction: a Mössbauer characterization. Biochemistry. 2003;42:8791–8798. doi: 10.1021/bi034426c. [DOI] [PubMed] [Google Scholar]
- 44.Gurbiel RJ, Batie CJ, Sivaraja M, True AE, Fee JA, Hoffman BM, Ballou DP. Electron-nuclear double resonance spectroscopy of 15N-enriched phthalate dioxygenase from Pseudomonas cepacia proves that two histidines are coordinated to the [2Fe-2S] Rieske-type clusters. Biochemistry. 1989;28:4861–4871. doi: 10.1021/bi00437a051. [DOI] [PubMed] [Google Scholar]
- 45.Lu TT, Huang HW, Liaw WF. Anionic mixed thiolate-sulfide-bridged Roussin’s red esters [(NO)2Fe(mu-SR)(mu-S)Fe(NO)2]- (R = Et, Me, Ph): a key intermediate for transformation of dinitrosyl iron complexes (DNICs) to [2Fe-2S] clusters. Inorg Chem. 2009;48:9027–9035. doi: 10.1021/ic9012679. [DOI] [PubMed] [Google Scholar]
- 46.Cicchillo RM, Booker SJ. Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J Am Chem Soc. 2005;127:2860–2861. doi: 10.1021/ja042428u. [DOI] [PubMed] [Google Scholar]
- 47.Cicchillo RM, Iwig DF, Jones AD, Nesbitt NM, Baleanu-Gogonea C, Souder MG, Tu L, Booker SJ. Lipoyl synthase requires two equivalents of S-adenosyl-L-methionine to synthesize one equivalent of lipoic acid. Biochemistry. 2004;43:6378–6386. doi: 10.1021/bi049528x. [DOI] [PubMed] [Google Scholar]
- 48.Cicchillo RM, Lee KH, Baleanu-Gogonea C, Nesbitt NM, Krebs C, Booker SJ. Escherichia coli lipoyl synthase binds two distinct [4Fe-4S] clusters per polypeptide. Biochemistry. 2004;43:11770–11781. doi: 10.1021/bi0488505. [DOI] [PubMed] [Google Scholar]
- 49.Douglas P, Kriek M, Bryant P, Roach PL. Lipoyl synthase inserts sulfur atoms into an octanoyl substrate in a stepwise manner. Angew Chem Int Ed Engl. 2006;45:5197–5199. doi: 10.1002/anie.200601910. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez HL, Pierrel F, Elleingand E, Garcia-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. MiaB, a bifunctional radical-S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe-4S] clusters. Biochemistry. 2007;46:5140–5147. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
- 51.Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, Roberts RJ, Krebs C, Booker SJ. Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry. 2009;48:10162–10174. doi: 10.1021/bi900939w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano WL. The PyMOL Molecular Graphics System. 2002. [Google Scholar]