Abstract
Biotin synthase catalyzes formation of the thiophane ring through stepwise substitution of a sulfur atom for hydrogen atoms at the C9 and C6 positions of dethiobiotin. Biotin synthase is a Radical SAM enzyme that reductively cleaves S-adenosylmethionine, generating 5′-deoxyadenosyl radicals that initially abstract a hydrogen atom from the C9 position of dethiobiotin. We have proposed that the resulting dethiobiotinyl radical is quenched by the μ-sulfide of the nearby [2Fe-2S]2+ cluster, resulting in coupled formation of 9-mercaptodethiobiotin and a reduced [2Fe-2S]+ cluster. This reduced FeS cluster is observed by electron paramagnetic resonance spectroscopy as a mixture of two orthorhombic spin systems. In the present work, we use isotopically labeled 9-mercaptodethiobiotin and enzyme to probe the ligand environment of the [2Fe-2S]+ cluster in this reaction intermediate. HYSCORE spectra exhibit strong cross-peaks demonstrating strong isotropic coupling of the nuclear spin with the paramagnetic center. The hyperfine coupling constants are consistent with a structural model for the reaction intermediate in which 9-mercaptodethiobiotin is covalently coordinated to the remnant [2Fe-2S]+ cluster.
The formation of new C–S bonds at unactivated carbon centers represents a particularly challenging reaction for an enzyme catalyst.1–5 The unactivated carbon center must become oxidized and have one or more hydrogen atoms removed, a process that might typically involve molecular oxygen and a metallocofactor. However, sulfur tends to be preferentially oxidized instead of carbon, and for this reason metal-oxygen chemistry is not generally used to activate the carbon center during the formation of C–S bonds.6
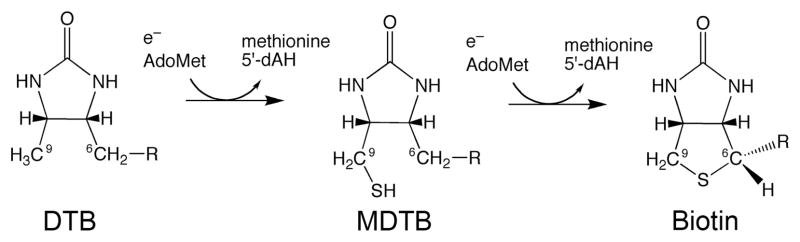
Recently, a number of enzymes in the Radical SAM superfamily have been identified that catalyze formation of C–S bonds.1,3,7 Radical SAM enzymes catalyze the one-electron reduction of the sulfonium of S-adenosylmethionine (SAM or AdoMet), generating methionine and a transient 5′-deoxyadenosyl radical (5′-dA•).8 This radical is a strong oxidant that can remove a hydrogen atom from a variety of organic substrates, generating high-energy substrate radicals that can undergo further reactions to generate a variety of products.8 Two Radical SAM enzymes catalyze addition of sulfur at unactivated aliphatic carbon atoms: lipoyl synthase catalyzes the addition of two thiol groups at the C6 methylene and C8 methyl positions of octanoyl-E2 protein or H protein,10,11 and biotin synthase (BS) catalyzes formation of a thioether between the C6 methylene and C9 methyl positions of dethiobiotin (DTB).12–14
With BS, we have demonstrated that the addition of a single sulfur atom proceeds in a stepwise manner.15 During the first half turnover, reductive cleavage of AdoMet produces a transient 5′-dA• radical that abstracts a hydrogen atom from the C9 position of DTB.16 The resulting 9-dethiobiotinyl radical adds a sulfur atom to generate 9-mercaptodethiobiotin (MDTB) as a stable chemical intermediate that is not released from the enzyme.15 Exchange of methionine and 5′-deoxyadenosine (5′-dAH) for a second equiv of AdoMet, followed by generation of a second 5′-dA• radical, facilitates abstraction of a hydrogen atom from the C6 position and thiophane ring closure. Isotopic labeling studies suggest that during the first turnover, the sulfur atom incorporated into the thiophane ring likely derives from one of the sulfides of a [2Fe-2S]2+ cluster tightly bound within the core of the as-purified enzyme.17
Formation of a carbon-sulfur bond between a dethiobiotinyl carbon radical and sulfide requires concurrent one-electron oxidation. If the sulfide is donated by a [2Fe-2S]2+ cluster, the excess electron is likely shed into the cluster, resulting in reduction to a paramagnetic FeIIFeIII spin system. Using electron paramagnetic resonance (EPR) spectroscopy, we have observed formation and decay of a paramagnetic species exhibiting an overlap of two orthorhombic spectra.18,19 The total spin concentration approximately correlates with the concentration of MDTB at various time intervals, consistent with the simultaneous formation of these two reaction intermediates.19
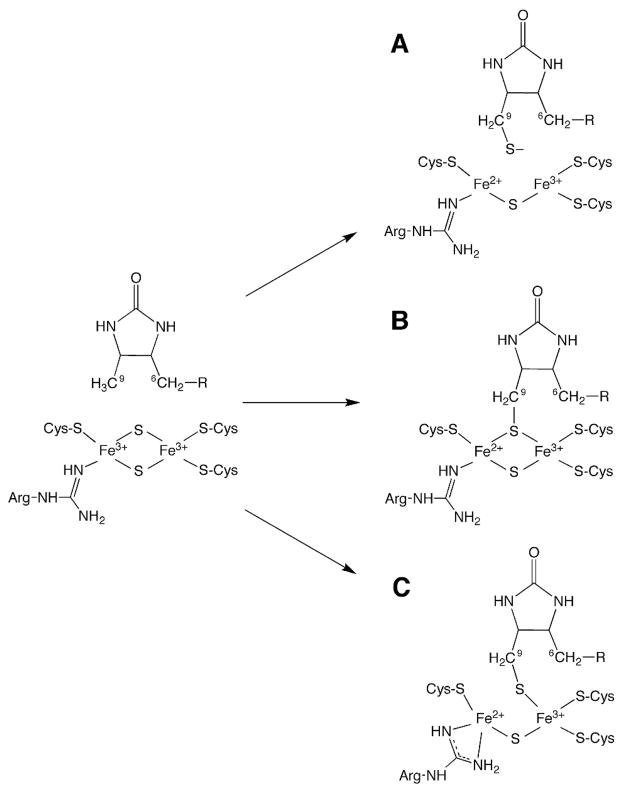
Although these experimental results are consistent with an intermediate state in which the enzyme contains both tightly bound MDTB and some form of a [2Fe-2S]+ cluster, there is considerable uncertainty regarding the precise structure and electronic state of this transient intermediate. In the structure of E. coli BS with DTB and AdoMet bound,20 the C9 position of DTB is located ~4.6 Å (center-to-center) from the nearest μ-sulfide of the [2Fe-2S]2+ cluster, while a typical carbon–sulfur bond length is ~1.7 Å. We have previously proposed that the enzyme intermediate incorporates MDTB as a bridging thiolate ligand to the [2Fe-S(RS)]+ cluster (Figure 1B),15 which would require a significant conformational change, as the C9 carbon atom would need to move by ~2.9 Å to form the C–S bond. Alternatively, the sulfide could be released from the cluster and migrate to the C9 position of DTB, generating MDTB that is proximal to but not covalently attached to the remnant [2Fe-1S]3+ cluster (Figure 1A). Other scenarios can be envisioned in which MDTB remains as a ligand to only one Fe within the cluster (Figure 1C).
Figure 1.
Three alternative structures for the BS intermediate state. (A) Formation of MDTB as a distal free thiolate that is ion-paired with the remnant [2Fe-1S]3+ cluster. (B) Formation of MDTB as a symmetric bridging ligand in the [2Fe-S(RS)]+ cluster. (C) Formation of MDTB as a ligand to one Fe within the remnant [2Fe-1S]3+ cluster.
To further probe the structure of the BS reaction intermediate, we produced two isotopically labeled samples that incorporate nuclear spins that would be in the vicinity of the reduced FeS cluster on the enzyme. Starting from (3-methyl-13C)-L-alanine and pimelic acid, we used BioW from B. sphaericus and BioF, BioA, and BioD from E. coli, along with the appropriate substrates and cofactors, to produce (9-methyl-13C)-DTB (>99% isotopic incorporation, see Supporting Information for a detailed protocol). NMR and LCMS analyses confirmed the structure and the level of isotopic incorporation. Starting with (guanidino-15N2)-L-arginine fed to a ΔargH E. coli expression strain, we expressed and purified BS in which all arginine residues are labeled in the terminal guanidino positions (>98% isotopic incorporation). The isotopic incorporation was confirmed by MALDI MS analysis of peptides produced from labeled BS by digestion with trypsin
Samples of BS were trapped in an intermediate state containing MDTB and a paramagnetic FeS cluster in a manner similar to previous reports.15,19 Briefly, the catalytic [4Fe-4S]2+ cluster in BS was reconstituted by incubation with Fe3+, S2−, and dithiothreitol. The enzyme was then pre-incubated with DTB and the biological reducing system consisting of flavodoxin, ferredoxin(flavodoxin):NADP+ oxidoreductase, and NADPH. Finally, AdoMet (1 equiv per BS monomer) was added to initiate the reaction. After 30 – 60 min, samples were transferred to EPR tubes and frozen in liquid N2 for spectroscopic analysis. In parallel, samples were also quenched in acid for HPLC analysis of MDTB and biotin formation.
Hyperfine sublevel correlation spectroscopy (HYSCORE) is a two-dimensional pulse EPR technique that correlates nuclear spin-flip transition frequencies in one electron-spin manifold to those in another connected to the first by an EPR transition.21 Correlations are typically observed between nuclear frequencies of the same nucleus, which, in the case of spin systems with multiple hyperfine-coupled nuclei, can greatly aid in the assignment of spectral features.
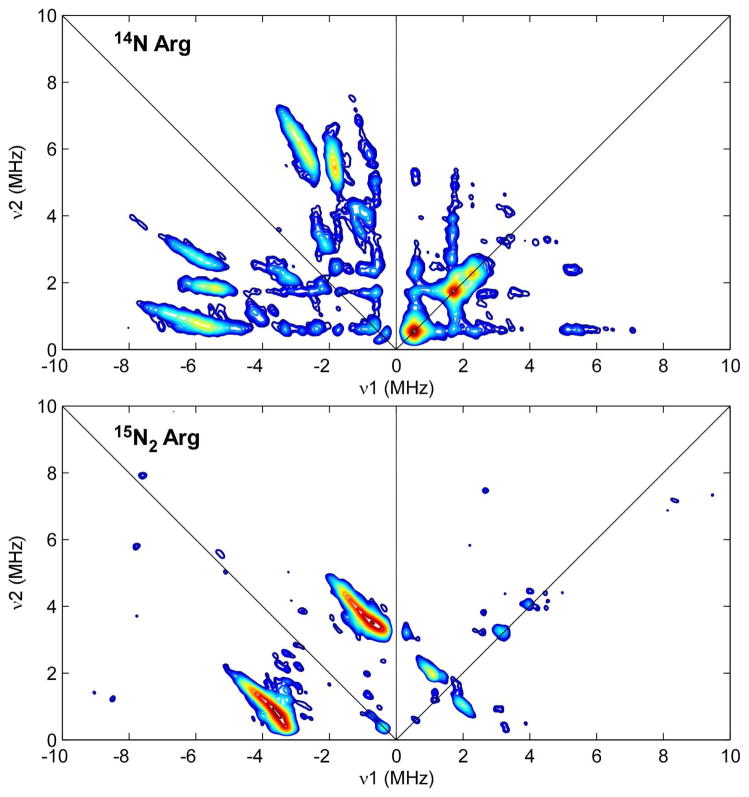
In the HYSCORE spectrum of the paramagnetic intermediate generated upon mixing natural abundance BS with AdoMet and DTB in the presence of the flavodoxin reducing system, several intense correlation ridges are observed with frequencies similar to those measured using three-pulse electron spin echo envelope modulation (ESEEM) spectroscopy (Figure 2, top).19 These features in the ESEEM spectra were previously assigned as arising from nitrogen atoms in the guanidino group of Arg260, a conserved ligand to the [2Fe-2S] cluster,20 based on the absence of these features in spectra collected under identical conditions for the active Arg260Met mutant enzyme.19 This assignment is confirmed by the HYSCORE spectrum of the BS intermediate obtained using wild-type enzyme isolated from a ΔargH E. coli expression strain grown in media supplemented with (guanidino-15N2)-L-arginine (Figure 2, bottom). Two sets of correlation ridges are present: one set centered at (0.82, 3.56 MHz) in the +− quadrant and one set centered at (1.08, 1.97 MHz) in the ++ quadrant that are respectively diagnostic of the presence of both strongly and weakly hyperfine-coupled 15N nuclei. The more strongly coupled class of 15N nuclei likely corresponds to the iron-ligated nitrogen atom(s) of the guanidino group of Arg260 (Figure 1). It is possible that both guanidino nitrogen atoms lie in a similar coordination environment relative to the FeS cluster, as depicted in Figure 1C, giving rise to a single set of cross peaks in the +− quadrant. The weakly coupled 15N nuclei could be due to the nearby Arg95 residue, which is hydrogen bonded to Arg260 in the crystal structure (dFe–N = 5–7 Å).20 This assignment would perhaps explain the prior observation of residual 14N peaks in the three-pulse ESEEM spectrum of the Arg260Met mutant enzyme.19 However, based on the crystal structure of the oxidized form of wild-type BS, the guanidino nitrogens of Arg95 are too distant to account for the observed hyperfine anisotropy (see Supporting Information) without invoking a substantial reorganization of the active site upon formation of the paramagnetic intermediate. Alternatively, the weakly coupled 15N nucleus could correspond to the uncoordinated nitrogen of the guanidino group as depicted in Figure 1B. This assignment is supported by the small Aiso = 0.57 MHz determined for these features (see Figure S10 in Supporting Information).
Figure 2.
HYSCORE spectra of the BS paramagnetic intermediate produced in BS purified from E. coli grown with natural abundance arginine (top) and (guanidino-15N2)-L-arginine (bottom). Spectrometer settings: (top) excitation frequency = 9.363 GHz; τ = 136 ns; (bottom) excitation frequency = 9.419 GHz; τ = 132 ns. All other settings were identical: temperature = 10 K; B0 = 347.5 mT; tπ/2 = tπ = 16 ns; t1 = t2 = 100 ns; Δt = 20 ns.
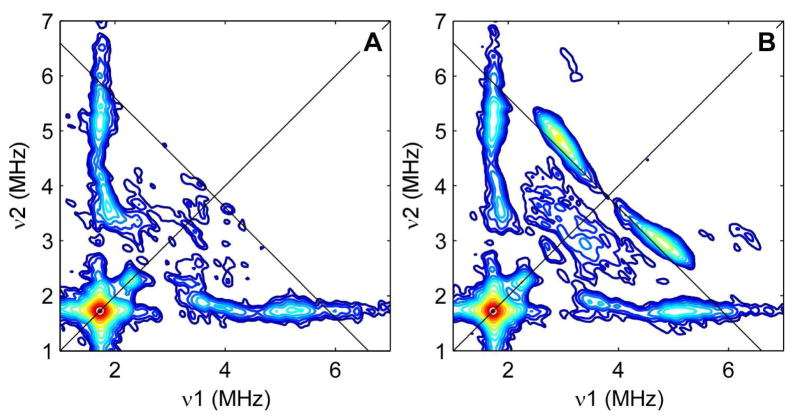
The HYSCORE spectrum of the paramagnetic intermediate generated using natural abundance BS and (9-methyl-13C)-DTB (Figure 3B) possesses correlation ridges centered at (2.9, 4.7 MHz) and (4.7, 2.9 MHz) that extend for nearly one MHz and have no counterpart in the corresponding natural abundance spectrum (Figure 3A). The position and field-dependence of these features are well-simulated using an axial 13C hyperfine interaction (HFI) of [1.2, 1.2, 5.7] MHz and electron g-values = [1.995, 1.941, 1.849], determined previously for the major spectral component (64 – 74%) of the EPR spectrum (see Figure S9).19 Although the EPR spectra (Figure S12) contain 2 components that can be simulated using up to 4 unique sets of g tensors,19 the HFI is not strongly dependent on the particular set of g values used in the simulation (see Table S1). The nature of the structural or electronic difference between major and minor components in the EPR spectrum is presently not understood. Field-dependent HYSCORE spectra show no evidence of two distinct sets of 13C correlation ridges that would suggest the presence of two species with different hyperfine couplings for C9 of MDTB. This finding could indicate that (i) the 13C hyperfine interaction is identical for both forms of the [2Fe-2S]+ cluster observed as major and minor components in the EPR spectra, or (ii) for one of the forms, there are no observable 13C couplings (i.e., MDTB is not bound or the coupling is too inhomogeneous to observe).
Figure 3.
HYSCORE spectra of the BS paramagnetic intermediate prepared with (A) natural abundance DTB and (B) (9-methyl-13C)-DTB. Spectrometer settings: excitation frequency = 9.75 GHz; temperature = 10 K; B0 = 355 mT; tπ/2 = tπ = 16 ns; τ = 132 ns; t1 = t2 = 100 ns; Δt = 20 ns.
Multinuclear coherences lead to the weak features centered at (3.0, 6.3) and (6.3, 3.0) in the (9-methyl-13C)-DTB/BS spectrum. The 6.3 MHz frequency is a combination of one of the 13C frequencies (4.7 MHz) with the 14N-associated frequency (1.7 MHz). This behavior results when nuclear transition frequencies from different nuclei but within the same electron spin manifold (e.g., mS = +1/2) are correlated to a frequency in the opposite electron spin manifold (e.g., mS = −1/2).22 That these multinuclear coherences appear requires that both nuclei are hyperfine-coupled to the same electron spin; thus, C9 is interacting directly with the [2Fe-2S]+ cluster that is hyperfine-coupled to an arginine residue, presumably Arg260, as depicted in Figures 1B or 1C.
The strength of the 13C hyperfine interaction is significant as disclosed by the relatively large isotropic contribution to the hyperfine tensor (Aiso = 2.7 MHz). For comparison, in the Radical SAM superfamily member pyruvate formate lyase-activating enzyme (PFL-AE), the methyl group of AdoMet is spatially close to but not bound to the [4Fe-4S]+ cluster (dFe—C = 3.5 Å),23, and ENDOR spectra exhibit an effective Aiso < 0.3 MHz.23 Alternatively, carbon atoms that are covalently bonded to heteroatoms coordinated to the Fe atoms of FeS clusters can have Aiso values ranging from 3 to 17 MHz (see Table 1). The anisotropic contribution to the 13C hyperfine interaction is also relatively large (T = 1.5 MHz), corroborating the close association of C9 with the [2Fe-2S] cluster. The effective 13C isotropic hyperfine interaction reported here for (9-methyl-13C)-DTB/BS is most similar to that found for the β-C of the cysteine ligands to the FeIIFeIII form of ferredoxin.25 This would be consistent with a structural model of the BS reaction intermediate in which MDTB is formed as a ligand to the remnants of the [2Fe-2S] cluster.
Table 1.
13C Hyperfine Couplings in Fe Cluster Spin Systems.
| System | Mox | 13C HFI (MHz) | ref. |
|---|---|---|---|
| MMOH + DMSOa | FeIIFeIII | ~1.3 | 26 |
| PFL-AE + SAMb | [4Fe-4S]+ | [−0.6, +0.4, −0.5] | 24 |
| IspG + MEcPPc | [4Fe-4S]+ | C2 = [14.5, 12.0, 26.5] C3 = [1.8, 2.0, 5.1] |
27 |
| Hox-COd | FeIFeII | C1 = [15.6, 16.6, 19.2] C2 = [8.5, 9.8, 3.9] C3 = [3.2, 3.7, 4.4] |
28 |
| Fde | [2Fe-2S]+ | C1 = 0.76–1.2 C2 = 1.9–2.1 |
25 |
| BS + 13C9-DTB | [2Fe-2S]+ | [1.2, 1.2, 5.7] | this study |
13C-DMSO + methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath)
(methyl-13C)-S-adenosylmethionine + pyruvate formate-lyase activating enzyme from E. coli.
Reaction intermediate of (2,3-13C)-2-C-methylerythritol-cyclo-2,4-diphosphate with IspG from E. coli.
13CO-inhibited form of [FeFe] hydrogenase from Desulfovibrio desulfuricans
13C-enriched ferredoxin from Anabaena
The spectroscopic data are most consistent with a mechanism in which 5′-dA• abstracts a hydrogen atom from the C9 position of dethiobiotin, and the dethiobiotinyl radical moves ~2.9 Å to attack the μ-sulfide of the [2Fe-2S]2+ cluster. Since DTB is held in place primarily through hydrogen bonds between the DTB imidazolidinone, Asn151, Asn153, and Asn222,20,29 this attack could be accomplished through a simple hinge motion around these hydrogen bonds. The resulting MDTB intermediate then remains as a ligand to the remnant FeS cluster, likely resulting in a significant conformational rearrangement of the active site. Additional structural information regarding the precise positioning of MDTB relative to the FeS cluster and AdoMet is needed to provide a complete mechanistic description of biotin thiophane ring formation.
Supplementary Material
Scheme 1.
Stepwise Reaction Catalyzed by Biotin Synthase.
Acknowledgments
This research has been supported by grants from the NSF (MCB 09-23829 to J.T.J.) and the NIH (GM73789 to R.D.B.). EPR data were collected at the CalEPR facility at UC Davis funded by the NIH (S10-RR021075) and the University of California.
ABBREVIATIONS
- AdoMet
S-adenosylmethionine
- BS
biotin synthase
- DTB
dethiobiotin
- dA•
5′-deoxyadenosyl radical
- dAH
5′-deoxyadenosine
- ENDOR
electron-nuclear double resonance
- EPR
electron paramagnetic resonance
- ESEEM
electron-spin-echo envelope modulation
- HYSCORE
hyperfine sublevel correlation spectroscopy
- MALDI MS
matrix-assisted laser-desorption-ionization mass spectrometry
- MDTB
9-mercaptodethiobiotin, SAM, S-adenosyl-methionine
Footnotes
Supporting Information. Experimental procedures for production, purification, and characterization of (9-methyl-13C)-DTB and (guanidino-15N2)-arginine biotin synthase, procedures for generation of the paramagnetic enzyme intermediate, CW EPR spectra of all samples, and additional magnetic field-dependent HYSCORE spectra are included in the supporting information. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Booker SJ, Cicchillo RM, Grove TL. Curr Opin Chem Biol. 2007;11:543. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marquet A, Florentin D, Ploux O, Tse Sum Bui B. J Phys Org Chem. 1998;11:529. [Google Scholar]
- 3.Fugate CJ, Jarrett JT. Biochim Biophys Acta. 2012 in press. [Google Scholar]
- 4.Fontecave M, Ollagnier-de-Choudens S, Mulliez E. Chem Rev. 2003;103:2149. doi: 10.1021/cr020427j. [DOI] [PubMed] [Google Scholar]
- 5.Marquet A. Curr Opin Chem Biol. 2001;5:541. doi: 10.1016/s1367-5931(00)00249-0. [DOI] [PubMed] [Google Scholar]
- 6.The non-heme iron enzyme, isopenicillin-N-synthase, is a rare example of a metal-oxygen system that can form a C-S bond. Roach PL, Clifton IJ, Hensgens CM, Shibata N, Schofield CJ, Hajdu J, Baldwin JE. Nature. 1997;387:827. doi: 10.1038/42990.
- 7.Atta M, Arragain S, Fontecave M, Mulliez E, Hunt JF, Luff JD, Forouhar F. Biochim Biophy Acta. 2012 doi: 10.1016/j.bbapap.2011.11.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey PA, Booker SJ. Adv Prot Chem. 2001;58:1. doi: 10.1016/s0065-3233(01)58001-8. [DOI] [PubMed] [Google Scholar]
- 9.Frey PA, Hegeman AD, Ruzicka FJ. Crit Rev Biochem Mol Biol. 2008;43:63. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 10.Cicchillo RM, Booker SJ. J Am Chem Soc. 2005;127:2860. doi: 10.1021/ja042428u. [DOI] [PubMed] [Google Scholar]
- 11.Douglas P, Kriek M, Bryant P, Roach PL. Angew Chem Int Ed Engl. 2006;45:5197. doi: 10.1002/anie.200601910. [DOI] [PubMed] [Google Scholar]
- 12.Mejean A, Bui BT, Florentin D, Ploux O, Izumi Y, Marquet A. Biochem Biophys Res Commun. 1995;217:1231. doi: 10.1006/bbrc.1995.2900. [DOI] [PubMed] [Google Scholar]
- 13.Sanyal I, Cohen G, Flint DH. Biochemistry. 1994;33:3625. doi: 10.1021/bi00178a020. [DOI] [PubMed] [Google Scholar]
- 14.Ugulava NB, Sacanell CJ, Jarrett JT. Biochemistry. 2001;40:8352. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor AM, Farrar CE, Jarrett JT. Biochemistry. 2008;47:9309. doi: 10.1021/bi801035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escalletes F, Florentin D, Tse Sum Bui B, Lesage D, Marquet A. J Am Chem Soc. 1999;121:3571. [Google Scholar]
- 17.Farrar CE, Siu KK, Howell PL, Jarrett JT. Biochemistry. 2010;49:9985. doi: 10.1021/bi101023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jameson GN, Cosper MM, Hernandez HL, Johnson MK, Huynh BH. Biochemistry. 2004;43:2022. doi: 10.1021/bi035666v. [DOI] [PubMed] [Google Scholar]
- 19.Taylor AM, Stoll S, Britt RD, Jarrett JT. Biochemistry. 2011;50:7953. doi: 10.1021/bi201042r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Science. 2004;303:76. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofer P, Grupp A, Nebenfuhr H, Mehring M. Chem Phys Lett. 1986;132:279. [Google Scholar]
- 22.Stoll S, Calle C, Mitrikas G, Schweiger A. J Magn Reson. 2005;177:93. doi: 10.1016/j.jmr.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 23.Vey JL, Yang J, Li M, Broderick WE, Broderick JB, Drennan CL. Proc Nat Acad Sci USA. 2008;105:16137. doi: 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM. J Am Chem Soc. 2002;124:3143. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 25.Houseman ALP, Oh BH, Kennedy MC, Fan C, Werst MM, Beinert H, Markley JL, Hoffman BM. Biochemistry. 1992;31:2073. doi: 10.1021/bi00122a026. [DOI] [PubMed] [Google Scholar]
- 26.DeRose VJ, Liu KE, Lippard SJ, Hoffman BM. J Am Chem Soc. 1996;118:121. [Google Scholar]
- 27.Wang W, Wang K, Li J, Nellutla S, Smirnova TI, Oldfield E. J Am Chem Soc. 2011;133:8400. doi: 10.1021/ja200763a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silakov A, Wenk B, Reijerse E, Albracht SPJ, Lubitz W. J Biol Inorg Chem. 2009;14:301. doi: 10.1007/s00775-008-0449-5. [DOI] [PubMed] [Google Scholar]
- 29.Farrar CE, Jarrett JT. Biochemistry. 2009;48:2448. doi: 10.1021/bi8022569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.