Abstract
The promiscuous IncPα plasmids RK2 and R995 encode a broad-host-range partition system, whose essential components include the incC and korB genes and a DNA site (OB) to which the korB product binds. IncC2, the smaller of the two incC products, is sufficient for stabilization of R995ΔincC. It is a member of the type Ia ParA family of partition ATPases. To better understand the role of ATP in partition, we constructed three alanine-substitution mutants of IncC2. Each mutation changed a different residue of the Walker-like ATP-binding and hydrolysis motif, including a lysine (K10) conserved solely among members of the ParA and MinD families. All three IncC2 mutants were defective in plasmid partition, but they differed from one another in other respects. The IncC2 T16A mutant, predicted to be defective in Mg2+ coordination, was severely impaired in all activities tested. IncC2 K10A, predicted to be defective in ATP hydrolysis, mediated enhanced incompatibility with R995 derivatives. IncC2 K15A, predicted to be defective in ATP binding, exhibited two distinct incompatibility properties depending on the genotype of the target plasmid. When in trans to plasmids carrying a complementable incC deletion, IncC2 K15A caused dramatic plasmid loss, even at low levels of expression. In trans to wild-type R995 or to R995ΔincC carrying a functional P1 partition system, IncC2 K15A-mediated incompatibility was significantly less than that caused by wild-type IncC2. All three Walker-like A box mutants were also defective for the host toxicity that normally results from co-overexpression of incC and korB. The phenotypes of the mutants support a model in which nucleotide hydrolysis is required for separation of paired plasmid complexes and possible interaction with a host factor.
Keywords: KorB, partition, segregation, RK2, R995, ATPase
1. Introduction
Active partition is the process of DNA segregation in bacteria. The prototypical partition locus consists of an autoregulated operon of two genes and at least one cis-acting DNA element functionally analogous to the eukaryotic centromere (Gerdes et al., 2004; Hayes and Barilla, 2006; Leonard et al., 2005b; Schumacher, 2008). The genes specify a weak ATPase and a DNA-binding protein specific for the cis-acting element. The locus constitutes a functional cassette sufficient to stabilize heterologous, unstable plasmids (Austin and Abeles, 1983a; Gerdes and Molin, 1986; Kalnin et al., 2000; Kwong et al., 2001; Ogura and Hiraga, 1983).
Partition systems have been identified in numerous Eubacteria and Archaea. The majority can be divided into two distinct and evolutionarily unrelated families that are most easily distinguished by the ATPases they encode (Gerdes et al., 2000). Members of the type I group encode Walker-type ATPases and type II members encode ATPases of the actin/Hsp70 family. Type II systems use actin-like filaments to drive plasmid partition (Garner et al., 2007; Moller-Jensen et al., 2003; Moller-Jensen et al., 2002; Salje and Lowe, 2008). In contrast, the mechanism of type I partition remains unclear. In an early step, molecules of the DNA-binding protein attach to the cis-acting site to generate a large nucleoprotein structure, termed the partition complex. Subsequent events have yet to be clearly defined. According to a recent model (Soberon et al., 2011), the partition complex recruits the ATPase, in the presence of which the complex undergoes a change in conformation to generate the segregation apparatus. DNA molecules are paired via their partition complexes at this juncture, although plasmid pairing may also occur prior to ATPase recruitment (Edgar et al., 2001; Ringgaard et al., 2007). Separation may occur via polymerization of the partition ATPase or may be driven by oscillation of the protein, possibly within spiral-shaped structures in the cell (Barilla et al., 2005; Bouet et al., 2007a; Ebersbach and Gerdes, 2004; Hatano et al., 2007; Lim et al., 2005; Ringgaard et al., 2009).
Plasmids of incompatibility group P (IncP) are capable of replication and stable maintenance in a variety of gram-negative bacteria (Kolatka et al., 2010; Thomas and Helinski, 1989). They consist of two distinct groups: IncPα plasmids, such as RK2 and R995 with virtually identical genetic backbones, and IncPβ plasmids, which include R751 (Pansegrau et al., 1994; Thorsted et al., 1998). Stable inheritance of the IncPα prototype, RK2, depends heavily on a broad-host-range active partition system with strong similarities to, and notable differences from, other well-characterized plasmid and chromosomal partition systems (Bignell et al., 1999; Motallebi-Veshareh et al., 1990; Rosche et al., 2000; Siddique and Figurski, 2002). The system comprises incC, which encodes a weak ATPase (Batt et al., 2009) (A. Siddique and D. H. Figurski, unpublished results); korB, which encodes a DNA-binding protein (Balzer et al., 1992; Khare et al., 2004) that interacts with IncC (Rosche et al., 2000); and OB, the 13-bp DNA target for KorB (Balzer et al., 1992; Khare et al., 2004; Rosche et al., 2000; Williams et al., 1998). The plasmids RK2 and R995 encode identical incC genes (Siddique and Figurski, 2002) that express two polypeptide products: full-length IncC1 (38.1 kDa) and the shorter IncC2 (27.5 kDa), which is initiated from an internal translation start site in incC (Kornacki et al., 1984; Thomas and Smith, 1986) (Fig. 1). Both incC1 and incC2 are members of the large family of genes that are predicted to encode partition ATPases related to the type I ParA protein of plasmid P1 (Gerdes et al., 2000; Hayes, 2000; Motallebi-Veshareh et al., 1990). KorB is a bifunctional protein that acts both as a transcriptional repressor of several IncP operons and as an integral component of the plasmid partition apparatus (Bechhofer et al., 1986; Jagura-Burdzy et al., 1999; Pansegrau et al., 1994; Rosche et al., 2000; Williams et al., 1998). Whereas most other partition systems harbor a single partition site, the IncPα plasmid RK2 carries 12 OB sites (OB 1-12) located throughout its genome (Pansegrau et al., 1994). Six are positioned at or near promoters and are required for KorB-mediated transcriptional regulation (Jagura-Burdzy et al., 1999; Pansegrau et al., 1994), while the other six are in locations with no obvious function. Unlike other plasmid partition systems, in which three components are sufficient to stabilize a heterologous unstable replicon, the three analogous components known to be required for IncP plasmid partition (incC, korB and a single OB site) are not, indicating that at least one other gene or OB site is necessary for the system to function properly (Rosche et al., 2000; Verheust and Helinski, 2007).
Fig. 1.
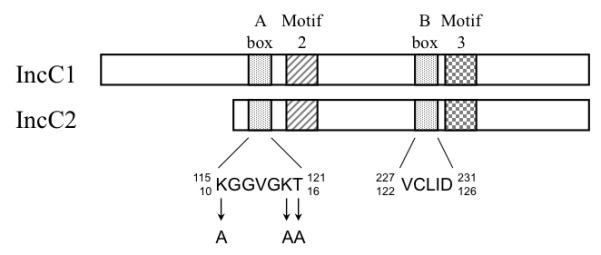
ATP-binding motifs in the IncC1 and IncC2 proteins. The Walker-like A and B boxes and motifs 2 and 3 are represented on a schematic of the IncC1 and IncC2 proteins. The Walker-like A and B boxes are described in the text. Motifs 2 and 3, also known as the A’ and B’ boxes, respectively, are conserved motifs that are also predicted to be part of the nucleotide-binding pocket (Koonin, 1993; Leonard et al., 2005a; Motallebi-Veshareh et al., 1990). The amino acid sequences of the core segments of the Walker-like A and B boxes are shown below the schematic. Upper numbers that flank the sequences refer to IncC1 amino acid residues; lower numbers refer to IncC2 residues. The residues targeted for mutation within IncC2 are lysine 10, lysine 15 and threonine 16. All were mutated to alanines. The schematic is drawn approximately to scale.
The defining feature of type I partition ATPases is a short glycine/lysine-rich sequence termed the Walker-like A box (KGGXXK[T/S]) (Koonin, 1993; Motallebi-Veshareh et al., 1990), which is similar to, but distinct from, the classical Walker A box motif found in many prokaryotic and eukaryotic proteins (Walker et al., 1982). The Walker-like A box and another conserved sequence, the Walker-like B box, form critical portions of the ATP-binding and catalysis pocket. Residues of the Walker-like A box form a flexible loop-like structure (known as the P-loop) that contacts the phosphate groups of the bound nucleotide (Hayashi et al., 2001; Leonard et al., 2005a; Pratto et al., 2008). The last two residues in the Walker-like A motif are highly conserved among Walker-type proteins. The lysine seems to be important for the binding of ATP (Hishida et al., 1999; Mitchell and Oliver, 1993; Panagiotidis et al., 1993; Seefeldt et al., 1992) and the threonine/serine residue is involved in Mg2+ coordination, which is essential for catalysis (Hayashi et al., 2001; Hishida et al., 1999; Jang et al., 2000; Story and Steitz, 1992).
In the partition ATPases of plasmids P1 (ParA) and F (SopA), residue changes within the A and B boxes abolish in vivo partitioning, alter autoregulatory properties and plasmid localization patterns, and impair ATPase activity (Davis et al., 1996; Fung et al., 2001; Hatano and Niki, 2010; Li et al., 2004; Libante et al., 2001). Biochemical studies have revealed that the P1 ParA protein requires ATP for partition complex formation but prefers ADP for operator-binding and transcriptional regulation, indicating that ATP hydrolysis regulates a switch from a form of the protein active for partition to one required for transcriptional regulation (Bouet and Funnell, 1999). A number of P1 ParA homologs form dynamic ATP-dependent filaments (Barilla et al., 2005; Bouet et al., 2007a; Ebersbach and Gerdes, 2004; Ebersbach et al., 2006; Lim et al., 2005). ATP is also required for the non-specific binding of F SopA and P1 ParA to DNA (Bouet et al., 2007a; Castaing et al., 2008; Vecchiarelli et al., 2010). Thus, adenine nucleotides play multiple roles in the biology of the ParA protein. To gain further insight into the role of nucleotide in partition, we have undertaken a genetic analysis of the Walker-like A box of IncC2. The mutants generated during this study were found to have different phenotypes. These phenotypes indicate distinct roles for the two lysine residues in the Walker-like A box of IncC.
2. Material and Methods
2.1. Bacterial strains
The Escherichia coli strains used in this study are DF4063 (thr-1 leuB6 lacY1 thi-1 tonA21 supE44 rfbD1 ΔtrpE5 gyrA) (Ayres et al., 1993), EKA13 (hsdR lacY leuB6 ΔtrpE5 recA1 gyrA) a spontaneous nalidixic acid mutant of JA221 (a gift from Charles Yanofsky), and TOP10 (F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlac(lacZΔM15) ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG) (Invitrogen, Carlsbad, CA).
2.2. Plasmids
A list of plasmids used in this study is presented in Table 1. Construction of unpublished plasmids is described below. For pR9410, the parABS locus of plasmid P1 was excised from pALA1557 (a gift from Stuart Austin) and inserted into the BamHI site of a cloned ΩAp HindIII fragment (Fellay et al., 1987). The resulting P1 parABS-ΩAp cassette was excised with HindIII and inserted into the unique HindIII site within the aphA (kanamycin resistance) gene of pR9401 (Siddique and Figurski, 2002). In vitro insertion of the EZ::TN <KAN-2> transposon (Epicentre, Madison, WI) into the tetA (tetracycline resistance) gene of the pR9401 derivative generated a kanamycin-resistant, tetracycline-sensitive version of the plasmid, designated pR9410. pRK22429 was constructed by PCR amplification of incC2 from pRK2178 (Bechhofer et al., 1986), cloning of the PCR product into pCR2.1 (Invitrogen), and ligation of the XbaI- and HindIII-digested incC2-T7 fragment from pCR2.1 to pJAK16 (J. A. Kornacki, unpublished) digested with the same enzymes. For pRK22455, the 5′-half of incC2 was amplified by PCR from pRK2178 (Bechhofer et al., 1986) and the resulting PCR product, which incorporates an AAA→1GCA change at codon 10, was cloned into pCR2.1. A NotI digest released a 5′-fragment of incC2 K10A that was inserted into a NotI deletion-derivative of pRK22429. The resulting plasmid was digested with XbaI and HindIII, and the incC2 K10A-T7 fragment was ligated to XbaI- and HindIII-digested pJAK16 to generate pRK22455. pRK22456 and pRK22457 were constructed by the same method as for pRK22455. The cloned incC2-T7 genes incorporate an AAG→GCG change at codon 15 (for pRK22456) and an AAG→TCG change at codon 16 (for pRK22457).
Table 1.
Plasmids
| Plasmid | Markers | Relevant Genotype | Description | Reference or source |
|---|---|---|---|---|
| pDB6 | Kmr | - | P15A replicon; vector control for pRK2300 | Bechhofer & Figurskia |
| pJAK16 | Cmr | lacIq tacp | IncQ replicon; expression vector | Kornackib |
| pR9401 | Kmr Tcr | Δ incC | R995 with 465-bp in-frame deletion within incC | (Siddique and Figurski, 2002) |
| pR9410 | Kmr Apr Tcs | ΔincC P1parABS+ | pR9401 with P1 parABS locus and an Apr marker inserted into the aphA (Kmr) gene, and an EZ::TN <KAN-2> transposon inserted in the tetA-tetR locus; parAB reads in the same direction as aphA |
This study |
| pRK2300 | Kmr |
korA+ ΔincC korB+ korF+ OB1− |
P15A replicon; carries RK2 korA and korB genes; has 465-bp in-frame deletion within incC |
(Bechhofer et al., 1986) |
| pRK22324 | Apr |
korA+ ΔincC korB+ OB1+ |
P15A replicon; carries region of RK2 from korAp to korB with 465-bp in-frame deletion within incC (Fig. 2) |
(Rosche et al., 2000) |
| pRK22330 | Apr |
korA+ ΔincC korB+ korF+ korG+ kfrA+ upf54.8+ OB1+ OB2+ OB3+ |
P15A replicon; carries region of RK2 from korAp to upf54.8 with 465-bp in-frame deletion within incC (Fig. 2) |
(Rosche et al., 2000) |
| pRK22429 | Cmr |
lacIq ϕ[tacp-incC2- T7•Tag] |
pJAK16 with incC2-T7 coding region expressed from tacp |
This study |
| pRK22455 | Cmr |
lacIq ϕ[tacp-incC2 K10A- T7•Tag] |
pJAK16 with incC2 K10A-T7 coding region expressed from tacp |
This study |
| pRK22456 | Cmr |
lacIq ϕ[tacp-incC2 K15A- T7•Tag] |
pJAK16 with incC2 K15A-T7 coding region expressed from tacp |
This study |
| pRK22457 | Cmr |
lacIq ϕ[tacp-incC2 T16A- T7•Tag] |
pJAK16 with incC2 T16A-T7 coding region expressed from tacp |
This study |
| R995 | Kmr Tcr | incC + | Natural IncPα plasmid (Fig. 2) | (Villarroel et al., 1983) |
D. H. Bechhofer and D. H. Figurski, unpublished results
J. A. Kornacki, unpublished results
2.3. Media
All strains were grown on Luria-Bertani (LB) medium at 37°C (Sambrook et al., 1989). The following antibiotics were used at the indicated concentrations: ampicillin, 50 μg/ml; penicillin, 150 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 50 μg/ml; tetracycline, 30 μg/ml (on plates) and 15 μg/ml (in broth); and nalidixic acid, 20 μg/ml. Expression from the tac promoter was induced with the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to solid or liquid medium.
2.4. Molecular procedures
DNA manipulations using restriction endonucleases, T4 DNA ligase, calf intestinal alkaline phosphatase, and E. coli DNA polymerase I Klenow fragment were done according to manufacturers’ recommendations (New England BioLabs, Beverly, MA and Fermentas Inc., Hanover, MD). Amplification of DNA by PCR was done with Taq DNA polymerase (Qiagen, Valencia, CA) or Vent DNA polymerase (New England BioLabs). All cloned PCR products were confirmed by nucleotide sequencing. Preparation of plasmid DNA was done either by a protocol involving alkaline lysis, phenol-chloroform extraction and ethanol precipitation (Ausubel et al., 1989) or by one involving alkaline lysis followed by column purification (Qiagen).
2.5. Plasmid stability assay
E. coli DF4063 cells carrying the appropriate plasmid(s) were grown in broth at 37°C with selection for the resident plasmid(s) until the cultures reached saturation. The cultures were diluted 105-fold into medium without selection for the test plasmid, grown to stationary phase, then diluted 105-fold again into fresh non-selective medium. This cycle was repeated until loss of the test plasmid was observed or until the strains had grown approximately 70 – 90 generations. At the time of each dilution, serial dilutions of the cultures were plated on medium without selection for the test plasmid. Plasmid retention was measured by picking 100 colonies from the non-selective plates onto plates with selection for the test plasmid. Each experiment was independently performed at least twice.
3. Results
3.1. Construction of Walker-like A box mutants of IncC2
The 27.5-kDa polypeptide product of the RK2 incC gene, IncC2, is sufficient to confer full partition function on R995ΔincC, a R995 derivative lacking a functional and identical incC gene (Fig. 2) (Siddique and Figurski, 2002). Consequently, our analysis of IncP plasmid partition has focused on IncC2, which is smaller and simpler to use than full length IncC1.
Fig. 2.
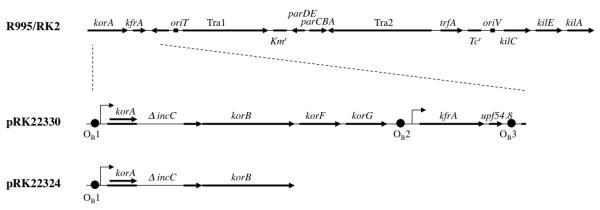
Linear schematic of key plasmids used to study the incompatibility properties of the incC2 mutants. The IncPα backbone for plasmids RK2 and R995 is shown at the top with landmark genetic determinants for reference. The primary difference between the IncPα plasmids, R995 and RK2, is transposon Tn1 in the kilC operon, which is present only in RK2 and is excluded from this schematic. The bold horizontal arrows in the R995/RK2 schematic refer to genetic loci or operons; in the pRK22330 and pRK22324 schematics, the arrows indicate genes. oriV and oriT refer to the plasmid origin of replication and the origin of conjugal transfer, respectively. Kmr denotes the gene for kanamycin resistance; Tcr denotes the tetracycline resistance locus. The angled arrows indicate promoters. The black circles, referred to as OB1, OB2 and OB3, indicate binding sites for KorB. The korA gene lies within the incC coding sequence, but in a different reading frame. ΔincC refers to a 465-bp in-frame deletion within incC (see text for details). The incC deletion present on R995ΔincC is the same as the one depicted here in the schematics of pRK22330 and pRK22324.
To facilitate detection of IncC2, we generated a T7 epitope-tagged version of the protein. The incC2-T7 fusion is indistinguishable from wild-type incC2 in its ability to complement R995ΔincC for stable maintenance and in its incompatibility and host toxicity properties, as described below.
To investigate the in vivo roles of ATP-binding and hydrolysis in partition, we constructed mutants of the Walker-like A box of IncC2. Three highly conserved residues in the region were targeted for substitution with alanine: lysine 10 (K10), lysine 15 (K15), and threonine 16 (T16) (Fig. 1). By analogy with Walker-like A boxes of related proteins, K10 is predicted to be involved in nucleotide hydrolysis; K15, in nucleotide binding; and T16, in Mg2+ coordination (see Discussion for details).
The Walker-like A box mutations were generated in the context of incC2-T7 by PCR; and, like wild-type incC2-T7, the mutant genes were inserted downstream of the tac promoter in an expression vector (pJAK16). Western blot analysis showed that the IncC2 K10A-T7 and IncC2 K15A-T7 proteins are expressed at levels similar to that of wild-type IncC2-T7. IncC2 T16A-T7 is expressed at a level significantly lower (approximately 5-fold) than that of wild type (data not shown).
3.2. Complementation and incompatibility with R995ΔincC
We first tested the T7-tagged version of incC2 for the ability to complement R995ΔincC in trans in E. coli. Uninduced expression of incC2-T7 or expression induced with a low level of IPTG (0.001 mM) was able to restore R995ΔincC stability to wild-type levels, indicating that IncC2-T7 and IncC2 have equivalent stabilization properties [Fig. 3B, data not shown; compare to Fig. 3A in (Siddique and Figurski, 2002)].
Fig. 3.
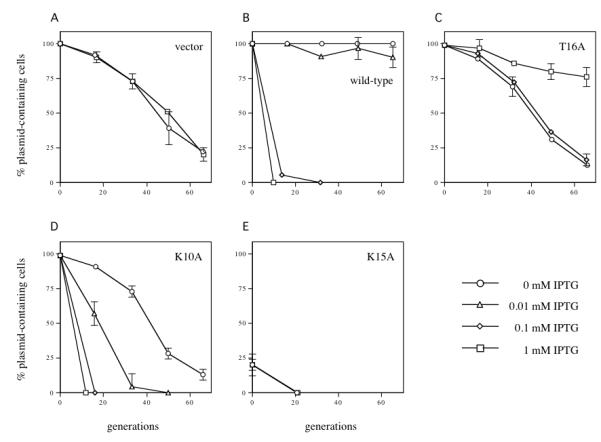
The effect of incC2 mutant gene expression on R995ΔincC stability. DF4063 strains containing the plasmids listed below were grown to saturation in media with selection for all resident plasmids. At T=0, the cultures were diluted 105-fold into media with selection only for the incC2-expressing plasmid and grown to stationary phase, at which point the cultures were diluted again. The procedure was repeated until the strains had grown approximately 70 generations. Colonies were screened for the presence of the target plasmid (R995ΔincC) on medium containing kanamycin. All strains contain pR9401 (R995ΔincC). In addition they contain pJAK16 (vector), pRK22429 (tacp-incC2-T7), pRK22457 (tacp-incC2 T16A-T7), pRK22455 (tacp-incC2 K10A-T7) or pRK22456 (tacp-incC2 K15A-T7). Circles: no IPTG; triangles: 0.01 mM IPTG; diamonds: 0.1 mM IPTG; squares: 1 mM IPTG. Error bars refer to the standard error of the mean (SEM). For the sake of clarity, error bars that are less than ±2% are not shown. For each point, the sample size is three (n=3), except for a few points along the curves where the cultures were sampled twice.
Elevated expression of incC causes the increased loss of IncP plasmids (“incompatibility”) (Rosche et al., 2000; Siddique and Figurski, 2002). Therefore, we expected that increased expression of incC2-T7 would also destabilize R995ΔincC. Indeed, induction of incC2-T7 with elevated IPTG levels (0.1 mM or 1 mM) caused rapid loss of R995ΔincC (Fig. 3B). The rates of loss are indistinguishable from those caused by the equivalent expression of wild-type incC2, indicating that the two proteins have similar incompatibility properties (compare to Fig. 3A in (Siddique and Figurski, 2002)). In contrast, the pJAK16 vector had no discernable effect on R995ΔincC stability at any IPTG induction level (Fig. 3A). Having shown the equivalence of wild-type IncC2 and its epitope-tagged version with regard to the phenotypes of stable maintenance and incompatibility, we have simplified the following text by using “IncC2” or “wild-type IncC2” in place of “IncC2-T7”.
We tested the three incC2 mutants for complementation of R995ΔincC stability and for incompatibility with R995ΔincC. Induction of the incC2 T16A allele (plasmid pRK22457) with concentrations of IPTG up to 0.1 mM had no detectable effect on R995ΔincC stability (Fig. 3C). However, partial complementation of R995ΔincC stability was observed with maximal induction of IPTG (1 mM). The inability of incC2 T16A to complement at the lower IPTG levels is not due to inadequate intracellular protein concentration. The amount of IncC2 T16A protein after induction with 0.1 mM IPTG is at least 10-fold greater than that of uninduced wild-type IncC2 (data not shown).
The incC2 K10A allele is unable to complement R995ΔincC stability at any IPTG induction level. Low level expression from the incC2 K10A plasmid, pRK22455, had no effect on R995ΔincC stability, while higher expression levels (≥ 0.01 mM IPTG) destabilized the plasmid (Fig. 3D). Thus, IncC2 K10A is unable to partition but can still generate incompatibility against R995ΔincC.
The incC2 K15A mutant provided a surprising phenotype. E. coli transformants containing both R995ΔincC and the incC2 K15A plasmid, pRK22456, grew poorly on double selection. In stability assays, R995ΔincC was lost so rapidly that only 20% of the colonies tested from the T=0 plates contained the plasmid (Fig. 3E). The plasmid was undetectable by the subsequent time point (T=20 generations). The rapid loss of R995ΔincC in the presence of the incC2 K15A allele occurred even in the absence of any induction, implying that a very low level of protein is sufficient to trigger loss of R995ΔincC. Thus, the phenotype, although consistent with the concept of incompatibility, is distinctly different from that caused by the overexpression of wild-type incC2. We refer to this phenotype as “super-incompatibility.”
The results show that all three Walker-like A box mutants of IncC2 are incapable of partition. Two of the mutants (incC2 K10A and incC2 K15A) display incompatibility phenotypes, indicating that they possess some degree of function in vivo.
3.3. Incompatibility with R995
To examine the dominance or recessiveness of the mutant alleles with respect to incompatibility, we tested the ability of the mutants to destabilize wild-type R995 (incC+). As expected, wild-type incC2 expression in trans to R995 caused loss of the plasmid (Fig. 4). The rate of loss correlated with the degree of induction. IPTG concentrations below 0.01 mM had no discernable effect on R995 maintenance, while higher levels caused increasing plasmid destabilization (Fig. 4, data not shown). With 0.1 mM (Fig. 4B) and 1 mM IPTG (not shown), R995 was lost from the host population within approximately 15 generations, similar to what was observed earlier with R995ΔincC as the target plasmid (Fig. 3B). The pJAK16 vector control had no effect on R995 stability at any IPTG induction level.
Fig. 4.
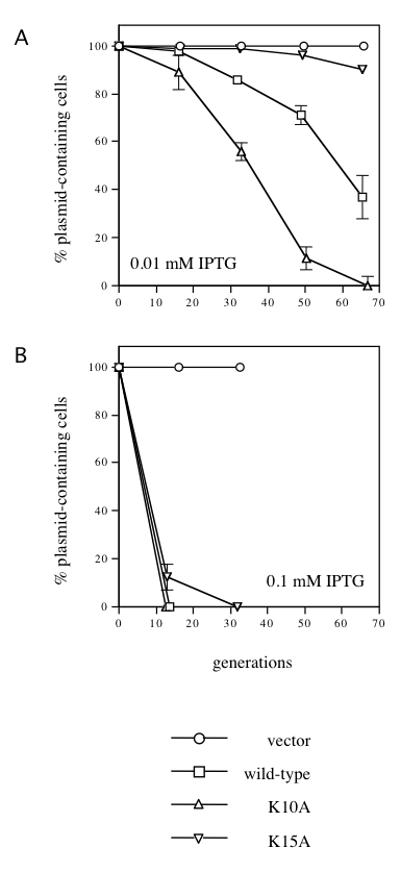
IncC2 K10A- and IncC2 K15A-mediated incompatibility against R995. Stability assays were performed as described in Fig. 3. All strains harbor R995. In addition, they contain pJAK16 (vector) (circles), pRK22429 (tacp-incC2-T7) (squares), pRK22455 (tacp-incC2 K10A-T7) (upward-pointing triangles) or pRK22456 (tacp-incC2 K15A-T7) (downward-pointing triangles). (A) 0.01 mM IPTG, (B) 0.1 mM IPTG. SEM bars are indicated as in Fig. 3.
Induction of the incC2 T16A allele had no discernable effect on R995 maintenance, even at the highest IPTG induction level (1 mM) (data not shown). Thus, incC2 T16A is incapable of causing incompatibility with R995.
Surprisingly, induction of incC2 K10A in trans to R995 caused greater plasmid destabilization than that caused by wild-type incC2 expressed at the same level, indicating possible genetic dominance of the mutant allele with respect to incompatibility. It is most obvious at the 0.01 mM IPTG induction level (Fig. 4A). At higher IPTG concentrations (≥ 0.1 mM), R995 destabilization by both wild-type incC2 and incC2 K10A was so severe that the loss curves are indistinguishable (Fig. 4B).
In striking contrast to earlier results with R995ΔincC, uninduced expression of the incC2 K15A allele had no destabilizing effect on R995 (data not shown). This implies that the incC2 K15A allele is recessive, since the native wild-type incC allele on R995 was able to abolish the super-incompatibility phenotype displayed by the mutant. Increased expression of incC2 K15A resulted in some loss of R995, but the degree of destabilization (visible at intermediate IPTG induction levels) was considerably less than that caused by wild-type incC2 expressed at the same levels (Fig. 4A).
3.4. Requirements for IncC2 K10A-mediated incompatibility and IncC2 K15A-mediated super-incompatibility are different
Since the incompatibility phenotypes conferred by the incC2 K10A and incC2 K15A alleles differed from those of wild-type incC2 and from each other, we wished to know whether the three incC alleles have the same or different genetic determinants for sensitivity to incompatibility. The minimal genetic requirements for sensitivity to incompatibility by wild-type incC are the korB gene and a single OB site (Rosche et al., 2000). These minimal determinants are present on plasmid pRK22324, a P15A derivative carrying a region of RK2 that contains OB1, korA, korB, and the ΔincC allele that is present in R995ΔincC (Fig. 2). As expected, expression of wild-type incC2 in trans destabilized pRK22324, whereas the pJAK16 vector had no effect (Fig. 5). Therefore, we used pRK22324 to test the incompatibility requirements of the incC2 mutants.
Fig. 5.
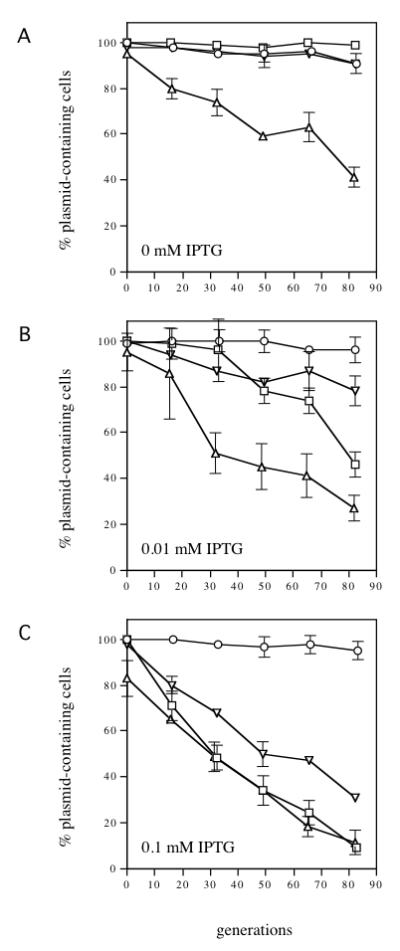
IncC2 K10A- and IncC2 K15A-mediated incompatibility against pRK22324. Stability assays were performed as described in Fig. 3, except that ampicillin was used to screen for the presence of the target plasmid (pRK22324). All strains harbor pRK22324 (ΔincC korB+ OB1+; Fig. 2). In addition, they contain pJAK16 (vector) (circles), pRK22429 (tacp-incC2-T7) (squares), pRK22455 (tacp-incC2 K10A-T7) (upward-pointing triangles) or pRK22456 (tacp-incC2 K15A-T7) (downward-pointing triangles). The symbols are the same as those used in Fig. 4. (A) no IPTG, (B) 0.01 mM IPTG, (C) 0.1 mM IPTG. SEM bars are indicated as in Fig. 3.
Induction of incC2 K10A caused greater destabilization of pRK22324 than did equivalent induction of wild-type incC2 (Fig. 5). Even uninduced expression of incC2 K10A destabilized pRK22324, whereas uninduced expression of wild-type incC2 had no effect on the stability of the plasmid (Fig. 5A). Thus, incC2 K10A displays enhanced incompatibility with pRK22324, as it does with R995. This indicates that the determinants necessary for the incompatibility phenotype of incC2 K10A are present on pRK22324.
In contrast, the incC2 K15A allele did not display its super-incompatibility phenotype with pRK22324. Instead, the incompatibility was weaker than that caused by equivalent expression of wild-type incC2 (Fig. 5). Thus, the components on pRK22324 are not sufficient for sensitivity to IncC2 K15A-mediated super-incompatibility.
3.5. Requirements for super-incompatibility
The super-incompatibility phenotype of incC2 K15A was observed with R995ΔincC but not with pRK22324. Yet the cloned region on pRK22324 contains the same incC allele present on R995ΔincC and possesses the necessary components for incompatibility with wild-type incC. One significant difference between the plasmids is the potential to partition. The presence of wild-type incC2 in trans allows for efficient stabilization of R995ΔincC, but it has no stabilizing effect on pRK22324 at any IPTG level (T. M. Rosche and D. H. Figurski, unpublished). It is possible, therefore, that the super-incompatibility phenotype conferred by incC2 K15A requires not only the components necessary for incompatibility, but also determinants that provide the target plasmid with the capacity to partition. If so, a ΔincC plasmid that can be stabilized by wild-type incC in trans (unlike pRK22324) should be sensitive to super-incompatibility by incC2 K15A.
One such plasmid is pRK22330, a derivative similar to pRK22324, but harboring a region of RK2 stretching from the korA promoter to upf54.8 (Fig. 2). This plasmid, like pRK22324, contains the ΔincC allele of R995ΔincC, and korB. However, unlike pRK22324, it has three OB sites (OB1-3) and additional genes. Plasmid pRK22330 is unstable, as expected; but, in contrast to pRK22324, it can be stabilized by wild-type incC2 in trans. After overnight growth in the absence of selection, only 4% of cells lacked pRK22330 with incC2 in trans, whereas 52% of cells with the isogenic vector lacked pRK22330. Stabilization by incC2 in trans occurred in the absence of IPTG induction, as it does for R995ΔincC. In contrast, pRK22330 was strongly destabilized by the incC2 K15A allele in trans (87% of cells lacked pRK22330, even without induction of the tac promoter). Thus, pRK22330, unlike pRK22324, is sensitive to IncC2 K15A-mediated super-incompatibility. The difference in sensitivities of pRK22324 and pRK22330 to IncC2 K15A may indicate that the protein causes the destabilization of plasmids by interacting in an abnormal manner with an IncP plasmid partition apparatus that has all the components necessary for partition except IncC.
3.6. Suppression of super-incompatibility by P1parABS
The previous result predicts that an inactive IncP partition system would not be sensitive to IncC2 K15A-mediated super-incompatibility. One possible way to inhibit the partition system on R995 is to insert a heterologous functional partition system to create a dicentric plasmid. Studies have revealed that the presence of a second partition system on a plasmid does not destabilize it as one might expect if the two partition systems were to compete (Austin and Nordstrom, 1990; Austin, 1984; Bignell et al., 1999; Bouet et al., 2005; Ebersbach and Gerdes, 2001). The results have been interpreted to indicate that the formation of one partition complex is inhibitory to the function of a second complex on the same plasmid, possibly as a result of topological constraints induced by the first complex (Austin, 1984).
To test whether the presence of an independent partition system on R995Δ incC would prevent IncC2 K15A-mediated super-incompatibility, we constructed an R995ΔincC derivative carrying the parABS partition locus of the P1 plasmid. The P1 partition system fully stabilizes R995ΔincC, indicating that it is functional in this context (Fig. 6). Stabilization is not the result of complementation of the ΔincC allele, since parABS has no effect in trans (data not shown). We then tested the R995ΔincC/P1parABS+ plasmid for sensitivity to super-incompatibility mediated by the incC2 K15A allele. Remarkably, uninduced expression of incC2 K15A had no effect on R995ΔincC/P1parABS+ maintenance, indicating that the presence of the P1 partition system is able to overcome IncC2 K15A-mediated super-incompatibility (Fig. 6A). At higher levels of IPTG, the incC2 K15A allele did cause some incompatibility, but the degree of R995ΔincC/P1parABS+ destabilization was less than that caused by equivalent expression of wild-type incC2 (Fig. 6B, data not shown). We conclude that the presence of a functional partition system on R995ΔincC can suppress the super-incompatibility phenotype associated with the incC2 K15A mutant.
Fig. 6.
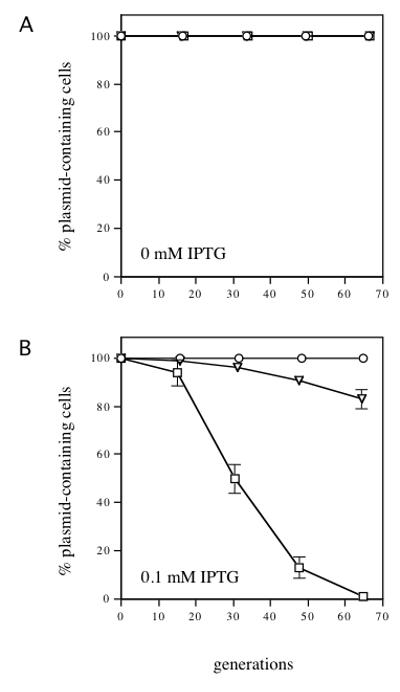
IncC2 K15A-mediated incompatibility against R995ΔincC/P1parABS+. Stability assays were performed as described in Fig. 3. Strains contain pR9410 (R995ΔincC/P1parABS+) and pJAK16 (vector) (circles), pR9410 and pRK22429 (tacp-incC2-T7) (squares), and pR9410 and pRK22456 (tacp-incC2 K15A-T7) (downward-pointing triangles). The symbols are the same as those used in Fig. 4. (A) no IPTG, (B) 0.1 mM IPTG. SEM bars are indicated as in Fig. 3.
3.7. Walker-like A box mutants of IncC2 are defective for host toxicity
We previously demonstrated that elevated expression of incC and korB together, but not individually, is toxic to E. coli cells (Rosche et al., 2000). We suggested that IncC-KorB complexes interact with and titrate or block an essential host component required for partition, thereby compromising cell viability. If ATPase activity is required for this activity, Walker-like A box mutants of IncC2 would be expected to exhibit reduced toxicity when overexpressed with KorB. We therefore examined the mutants for toxicity in E. coli strains harboring pRK2300, which expresses elevated levels of KorB (Rosche et al., 2000). Induction of incC2-T7 in trans to pRK2300 caused toxicity similar to that caused by wild-type incC2 (EOP = ~10−4) (Table 2). In contrast, all three mutants were defective for toxicity (Table 2). Induction of incC2 K15A or incC2 T16A had no effect on the numbers of colony-forming units or their size. Expression of incC2 K10A caused weak toxicity characterized by smaller colonies on IPTG-containing selective plates. Thus, the Walker-like A box mutants of IncC2 cause little or no toxicity in E. coli.
Table 2.
Walker-like A box mutants of IncC2 are defective for toxicity
| Plasmid 1a | Relevant properties | Plasmid 2a | Relevant properties | Relative EOP on double selectionb |
|---|---|---|---|---|
| pDB6 | vector | pJAK16 | vector | 1.1 |
| pRK2300 | korA+ korB+ korF+ | pJAK16 | vector | 0.8 |
| pDB6 | vector | pRK22429 | tacp-incC2-T7 | 1.1 |
| pRK2300 | korA+ korB+ korF+ | pRK22429 | tacp-incC2-T7 | 1.3×10−4 |
| pDB6 | vector | pRK22455 | tacp-incC2 K10A-T7 | 0.8 |
| pRK2300 | korA+ korB+ korF+ | pRK22455 | tacp-incC2 K10A-T7 | 0.5c |
| pDB6 | vector | pRK22456 | tacp-incC2 K15A-T7 | 1.1 |
| pRK2300 | korA+ korB+ korF+ | pRK22456 | tacp-incC2 K15A-T7 | 1.1 |
| pDB6 | vector | pRK22457 | tacp-incC2 T16A-T7 | 1.0 |
| pRK2300 | korA+ korB+ korF+ | pRK22457 | tacp-incC2 T16A-T7 | 1.3 |
EKA13 is the host strain
Relative EOP (efficiency of plating) = CFU[+IPTG]/CFU[−IPTG]
colonies on IPTG-containing plates are significantly smaller than those on plates lacking IPTG
4. Discussion
The phenomenon of plasmid incompatibility has been documented for more than four decades, yet the molecular basis of partition-related incompatibility remains unclear. What is likely is that the molecular mechanism is different depending on the plasmid involved and the circumstances under which it is destabilized (Bouet et al., 2007b; Bouet et al., 2005; Dabrazhynetskaya et al., 2005; Lemonnier et al., 2000). A widely accepted explanation for the phenomenon is that partition-related incompatibility results from competition between plasmids for pairing (Austin and Abeles, 1983b; Nordstrom and Austin, 1989). Our previous analysis of IncC-mediated destabilization of IncP plasmids, in which excess IncC appeared to induce indiscriminate pairing between plasmids via IncC-KorB complexes at multiple OB sites on RK2, is consistent with this interpretation (Rosche et al., 2000). Intermolecular pairing of ParB (ω2)-bound parS sites by the ParA homolog (α2) of plasmid pSM19035 has been demonstrated by atomic force microscopy (Pratto et al., 2009). Increasing the ratio of α2 to ω2 resulted in the appearance of higher-order structures containing 3 or more paired parS sites. These structures could be similar to the RK2 aggregates postulated to form in the presence of excess IncC.
The lysine residue equivalent to K15 in IncC2 is highly conserved among Walker family ATPases (including Walker-like ATPases). Mutation of this residue typically impairs ATP-binding (Hishida et al., 1999; Mitchell and Oliver, 1993; Panagiotidis et al., 1993; Seefeldt et al., 1992). Among partition ATPases, alteration of this residue has a detrimental effect on partition-related functions, although the defects vary depending on the type of substitution made (Fung et al., 2001; Hatano et al., 2007; Libante et al., 2001; Vecchiarelli et al., 2010). Not surprisingly, IncC2 K15A is defective for plasmid partition. It is also defective for host toxicity. Its unusual incompatibility properties depend on the target plasmid.
Against R995, incC2 K15A exhibited reduced incompatibility relative to wild-type incC2. Since IncC-mediated incompatibility appears to be associated with plasmid pairing (see above), the phenotype may indicate that the mutant protein is deficient in the ability to pair plasmids. This would imply that there is a requirement for ATP binding in plasmid pairing or in an earlier step, such as nucleoprotein complex-formation. In vitro work with the P1 ParA protein showed that the interaction of ParA with a ParB-parS complex is strictly dependent on the presence of ATP (Bouet and Funnell, 1999). Our results agree with these data and with recent evidence demonstrating ATP-dependent coupling of centromere-carrying plasmids by the type Ib partition system of pSM19035 (Pratto et al., 2008).
In trans to R995ΔincC, the incC2 K15A allele triggered remarkably rapid loss of the plasmid from cells (i.e., super-incompatibility). This loss occurred even in the absence of induction, indicating that the destabilization phenotype is distinctly different from the incompatibility typically generated by overexpression of incC2. The phenomenon is reminiscent of the strong partition defects observed with certain Walker-like A box and motif 2 mutants of P1 ParA and with the P1 parAM314I mutant (Fung et al., 2001; Youngren and Austin, 1997). The super-incompatibility phenotype was abolished by the presence of the wild-type incC allele. It also appeared to require that the target plasmid have the potential to form a functional IncP partition complex (see section 3.5.). If sensitivity to super-incompatibility requires a potentially functional IncP partition system, this phenotype may provide a powerful assay to identify the missing components of the IncP plasmid partition apparatus.
What is the mechanism of IncC2 K15A-mediated super-incompatibility? One possibility is that IncC2 K15A triggers silencing of essential plasmid genes or sites. Partition-related silencing, well-documented upon elevated expression of P1 ParB, F SopB or Bacillus subtilis Spo0J (Breier and Grossman, 2007; Lynch and Wang, 1995; Rodionov et al., 1999), is caused by the binding of the ParB/SopB/Spo0J protein to the target partition site followed by polymerization of the protein along the DNA in both directions from the site of initial binding (Lynch and Wang, 1995; Rodionov et al., 1999). Interestingly, certain mutants of P1 ParA are able to generate strong ParB-dependent transcriptional blocks on genes adjacent to a parS site probably by a mechanism similar to silencing (Sawitzke et al., 2002). In the case of IncC2 K15A, the need for an IncP plasmid system with the potential for partition may reflect the requirement for a “correct” KorB-OB complex to be present before the mutant protein can trigger polymerization and silencing. Further studies are required to test this hypothesis.
Unlike the residues corresponding to K15 and T16, the K10 equivalent residue is generally absent from the classical Walker A box, yet it is highly conserved in the Walker-like A box. Alteration of the corresponding residue in the Walker-like protein MinD resulted in loss of in vivo function (Hayashi et al., 2001). Structural analysis of the B. subtilis ParA homolog (Soj) dimer revealed that the nucleotide-binding pocket forms part of the interface between interacting subunits and that the K10 equivalent residue of one monomer interacts with the α and γ phosphates of the ATP molecule bound to the second monomer (Leonard et al., 2005a). Similarly, in the dimeric nitrogenase Fe-protein, the equivalent lysine contacts the terminal oxygen of the β-phosphate of the nucleotide bound to the adjacent subunit and is proposed to stabilize the leaving group during hydrolysis (Schindelin et al., 1997). These data indicate a possible role for the lysine residue corresponding to K10 in ATP hydrolysis and an association between nucleotide modification and protein multimerization.
In our study, IncC2 K10A mediated enhanced incompatibility against all plasmids tested, including R995ΔincC/P1parABS+ (data not shown). The simplest interpretation of this result is that IncC2 K10A can pair plasmids but is unable to separate them. We hypothesize that the K10 residue is required for the separation of plasmid pairs or the release of separated plasmids from the segregation machinery. Since ATP is required for partition complex formation, plasmid pairing and the formation of ParA protein filaments (Barilla et al., 2005; Bouet et al., 2007a; Bouet and Funnell, 1999; Ebersbach et al., 2006; Lim et al., 2005; Pratto et al., 2008), but nucleotide hydrolysis is necessary for the directed separation of plasmids (Barilla et al., 2007; Pratto et al., 2009), the phenotype of the incC2 K10A mutant may be indicative of a role for the K10 residue in ATP hydrolysis.
Might the host play a role in plasmid segregation? While many current models discount an active role for a host factor in plasmid partition, RK2 might make a case for it. We previously showed that IncC2 and KorB are toxic to the host when co-expressed, although neither one alone is toxic (Rosche et al., 2000). In this work, the involvement of the host is strengthened by the finding that partition-defective mutants of IncC2 are also toxicity-defective. In addition, localization studies have revealed that RK2 is highly mobile in the absence of a partitioning system, but the plasmids become fixed to sites at the quarter positions of the cell in the presence of partitioning proteins (Anand and Khan, 2010; Derman et al., 2008). One interpretation is that a host factor anchors RK2 to a specific site(s) in the cell. In this context, the relatively weak host toxicity of the IncC2 K10A mutant might indicate an inability to interact with the host. If IncC2 K10A is unable to hydrolyze ATP, the toxicity phenotype of incC2 K10A would imply that ADP, not ATP, is required by IncC for productive interaction with the host. The nature of the putative host factor is a mystery. However, the association of many ParA-type proteins with the nucleoid might indicate that the host factor is a nucleoid-associated protein or a site on the chromosome itself.
Highlights.
We constructed three mutants of the partition protein IncC2 from IncP plasmid R995.
The mutants change single amino acids in the ATPase motif of IncC2.
We examined the mutants for partition, incompatibility, and host toxicity.
Each IncC2 mutant has a different array of phenotypes.
The results indicate that IncC2 ATPase activity is not required for incompatibility.
Acknowledgements
The research was supported by NIH grant R01-GM29085 to D.H.F. and by Columbia University. D.H.F. deeply appreciates the help of Saul Silverstein, Aaron Mitchell and Vincent Racaniello.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand SP, Khan SA. Plasmid segregation: birds of a feather try not to flock together. J. Bacteriol. 2010;192:1171–1174. doi: 10.1128/JB.01551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S, Abeles A. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J. Mol. Biol. 1983a;169:353–372. doi: 10.1016/s0022-2836(83)80055-2. [DOI] [PubMed] [Google Scholar]
- Austin S, Abeles A. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J. Mol. Biol. 1983b;169:373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- Austin S, Nordstrom K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- Austin SJ. Bacterial plasmids that carry two functional centromere analogs are stable and are partitioned faithfully. J. Bacteriol. 1984;158:742–745. doi: 10.1128/jb.158.2.742-745.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. J. Wiley & Sons; New York City, NY: 1989. [Google Scholar]
- Ayres EK, Thomson VJ, Merino G, Balderes D, Figurski DH. Precise deletions in large bacterial genomes by vector-mediated excision (VEX). The trfA gene of promiscuous plasmid RK2 is essential for replication in several gram-negative hosts. J. Mol. Biol. 1993;230:174–185. doi: 10.1006/jmbi.1993.1134. [DOI] [PubMed] [Google Scholar]
- Balzer D, Ziegelin G, Pansegrau W, Kruft V, Lanka E. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 1992;20:1851–1858. doi: 10.1093/nar/20.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilla D, Carmelo E, Hayes F. The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc. Natl. Acad. Sci. USA. 2007;104:1811–1816. doi: 10.1073/pnas.0607216104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barilla D, Rosenberg MF, Nobbmann U, Hayes F. Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO. 2005;24:1453–1464. doi: 10.1038/sj.emboj.7600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batt SM, Bingle LEH, Dafforn TR, Thomas CM. Bacterial genome partitioning: N-terminal domain of IncC protein encoded by broad-host-range plasmid RK2 modulates oligomerisation and DNA binding. J. Mol. Biol. 2009;385:1361–1374. doi: 10.1016/j.jmb.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer DH, Kornacki JA, Firshein W, Figurski DH. Gene control in broad host range plasmid RK2: expression, polypeptide product, and multiple regulatory functions of korB. Proc. Natl. Acad. Sci. USA. 1986;83:394–398. doi: 10.1073/pnas.83.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell CR, Haines AS, Khare D, Thomas CM. Effect of growth rate and incC mutation on symmetric plasmid distribution by the IncP-1 partitioning apparatus. Mol. Microbiol. 1999;34:205–216. doi: 10.1046/j.1365-2958.1999.01565.x. [DOI] [PubMed] [Google Scholar]
- Bouet J-Y, Ah-Seng Y, Benmeradi N, Lane D. Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol. Microbiol. 2007a;63:468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- Bouet J-Y, Funnell BE. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO. 1999;18:1415–1424. doi: 10.1093/emboj/18.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet J-Y, Nordstrom K, Lane D. Plasmid partition and incompatibility - the focus shifts. Mol. Microbiol. 2007b;65:1405–1414. doi: 10.1111/j.1365-2958.2007.05882.x. [DOI] [PubMed] [Google Scholar]
- Bouet J-Y, Rech J, Egloff S, Biek DP, Lane D. Probing plasmid partition with centromere-based incompatibility. Mol. Microbiol. 2005;55:511–525. doi: 10.1111/j.1365-2958.2004.04396.x. [DOI] [PubMed] [Google Scholar]
- Breier AM, Grossman AD. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 2007;64:703–718. doi: 10.1111/j.1365-2958.2007.05690.x. [DOI] [PubMed] [Google Scholar]
- Castaing J-P, Bouet J-Y, Lane D. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol. Microbiol. 2008;70:1000–1011. doi: 10.1111/j.1365-2958.2008.06465.x. [DOI] [PubMed] [Google Scholar]
- Dabrazhynetskaya A, Sergueev K, Austin S. Species and incompatibility determination within the P1par family of plasmid partition elements. J. Bacteriol. 2005;187:5977–5983. doi: 10.1128/JB.187.17.5977-5983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Radnedge L, Martin KA, Hayes F, Youngren B, Austin SJ. The P1 ParA protein and its ATPase activity play a direct role in the segregation of plasmid copies to daughter cells. Mol. Microbiol. 1996;21:1029–1036. doi: 10.1046/j.1365-2958.1996.721423.x. [DOI] [PubMed] [Google Scholar]
- Derman AI, Lim-Fong G, Pogliano J. Intracellular mobility of plasmid DNA is limited by the ParA family of partitioning systems. Mol. Microbiol. 2008;67:935–946. doi: 10.1111/j.1365-2958.2007.06066.x. [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc. Natl. Acad. Sci. USA. 2001;98:15078–15083. doi: 10.1073/pnas.261569598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K. Bacterial mitosis: partitioning protein ParA oscillates in spiral-shaped structures and positions plasmids at mid-cell. Mol. Microbiol. 2004;52:385–398. doi: 10.1111/j.1365-2958.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Ringgaard S, Moller-Jensen J, Wang Q, Sherratt DJ, Gerdes K. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol. Microbiol. 2006;61:1428–1442. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- Edgar R, Chattoraj DK, Yarmolinsky M. Pairing of P1 plasmid partition sites by ParB. Mol. Microbiol. 2001;42:1363–1370. doi: 10.1046/j.1365-2958.2001.02717.x. [DOI] [PubMed] [Google Scholar]
- Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- Fung E, Bouet J-Y, Funnell BE. Probing the ATP-binding site of P1 ParA: partition and repression have different requirements for ATP binding and hydrolysis. EMBO. 2001;20:4901–4911. doi: 10.1093/emboj/20.17.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Molin S. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J. Mol. Biol. 1986;190:269–279. doi: 10.1016/0022-2836(86)90001-x. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Moller-Jensen J, Ebersbach G, Kruse T, Nordstrom K. Bacterial mitotic machineries. Cell. 2004;116:359–366. doi: 10.1016/s0092-8674(04)00116-3. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Moller-Jensen J, Jensen RB. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- Hatano T, Niki H. Partitioning of P1 plasmids by gradual distribution of the ATPase ParA. Mol. Microbiol. 2010;78:1182–1198. doi: 10.1111/j.1365-2958.2010.07398.x. [DOI] [PubMed] [Google Scholar]
- Hatano T, Yamaichi Y, Niki H. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol. Microbiol. 2007;64:1198–1213. doi: 10.1111/j.1365-2958.2007.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I, Oyama T, Morikawa K. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO. 2001;20:1819–1828. doi: 10.1093/emboj/20.8.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily distinct subgroup of the ParA superfamily. Mol. Microbiol. 2000;37:528–541. doi: 10.1046/j.1365-2958.2000.02030.x. [DOI] [PubMed] [Google Scholar]
- Hayes F, Barilla D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- Hishida T, Iwasaki H, Yagi T, Shinagawa H. Role of Walker motif A of RuvB protein in promoting branch migration of Holliday junctions. Walker motif A mutations affect ATP binding, ATP hydrolyzing, and DNA binding activities of RuvB. J. Biol. Chem. 1999;274:25335–25342. doi: 10.1074/jbc.274.36.25335. [DOI] [PubMed] [Google Scholar]
- Jagura-Burdzy G, Macartney DP, Zatyka M, Cunliffe L, Cooke D, Huggins C, Westblade L, Khanim F, Thomas CM. Repression at a distance by the global regulator KorB of promiscuous IncP plasmids. Mol. Microbiol. 1999;32:519–532. doi: 10.1046/j.1365-2958.1999.01365.x. [DOI] [PubMed] [Google Scholar]
- Jang SB, Seefeldt LC, Peters JW. Insights into nucleotide signal transduction in nitrogenase: structure of an iron protein with MgADP bound. Biochemistry. 2000;39:14745–14752. doi: 10.1021/bi001705g. [DOI] [PubMed] [Google Scholar]
- Kalnin K, Stegalkina S, Yarmolinsky M. pTAR-encoded proteins in plasmid partitioning. J. Bacteriol. 2000;182:1889–1894. doi: 10.1128/jb.182.7.1889-1894.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare D, Ziegelin G, Lanka E, Heinemann U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 2004;11:656–663. doi: 10.1038/nsmb773. [DOI] [PubMed] [Google Scholar]
- Kolatka K, Kubik S, Rajewska M, Konieczny I. Replication and partitioning of the broad-host-range plasmid RK2. Plasmid. 2010;64:119–134. doi: 10.1016/j.plasmid.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Koonin EV. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- Kornacki JA, West AH, Firshein W. Proteins encoded by the trans-acting replication and maintenance regions of broad host range plasmid RK2. Plasmid. 1984;11:48–57. doi: 10.1016/0147-619x(84)90006-4. [DOI] [PubMed] [Google Scholar]
- Kwong SM, Yeo CC, Poh CL. Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site, parS. Mol. Microbiol. 2001;40:621–633. doi: 10.1046/j.1365-2958.2001.02405.x. [DOI] [PubMed] [Google Scholar]
- Lemonnier M, Bouet J-Y, Libante V, Lane D. Disruption of the F plasmid partition complex in vivo by partition protein SopA. Mol. Microbiol. 2000;38:493–503. doi: 10.1046/j.1365-2958.2000.02101.x. [DOI] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer - a conserved biological switch. EMBO. 2005a;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard TA, Moller-Jensen J, Lowe J. Towards understanding the molecular basis of bacterial DNA segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005b;360:523–535. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dabrazhynetskaya A, Youngren B, Austin S. The role of Par proteins in the active segregation of the P1 plasmid. Mol. Microbiol. 2004;53:93–102. doi: 10.1111/j.1365-2958.2004.04111.x. [DOI] [PubMed] [Google Scholar]
- Libante V, Thion L, Lane D. Role of the ATP-binding site of SopA protein in partition of the F plasmid. J. Mol. Biol. 2001;314:387–399. doi: 10.1006/jmbi.2001.5158. [DOI] [PubMed] [Google Scholar]
- Lim GE, Derman AI, Pogliano J. Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl. Acad. Sci. USA. 2005;102:17658–17663. doi: 10.1073/pnas.0507222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Wang JC. SopB protein-mediated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc. Natl. Acad. Sci. USA. 1995;92:1896–1900. doi: 10.1073/pnas.92.6.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol. Microbiol. 1993;10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol. Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J, Jensen RB, Lowe J, Gerdes K. Prokaryotic DNA segregation by an actin-like filament. EMBO. 2002;21:3119–3127. doi: 10.1093/emboj/cdf320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motallebi-Veshareh M, Rouch DA, Thomas CM. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol. Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- Nordstrom K, Austin SJ. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Ogura T, Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Panagiotidis CH, Reyes M, Sievertsen A, Boos W, Shuman HA. Characterization of the structural requirements for assembly and nucleotide binding of an ATP-binding cassette transporter. J. Biol. Chem. 1993;268:23685–23696. [PubMed] [Google Scholar]
- Pansegrau W, Lanka E, Barth PT, Figurski DH, Guiney DG, Haas D, Helinski DR, Schwab H, Stanisich VA, Thomas CM. Complete nucleotide sequence of Birmingham IncPα plasmids. Compilation and comparative analysis. J. Mol. Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- Pratto F, Cicek A, Weihofen WA, Lurz R, Saenger W, Alonso JC. Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res. 2008;36:3676–3689. doi: 10.1093/nar/gkn170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto F, Suzuki Y, Takeyasu K, Alonso JC. Single-molecule analysis of protein-DNA complexes formed during partition of newly replicated plasmid molecules in Streptococcus pyogenes. J. Biol. Chem. 2009;284:30298–30306. doi: 10.1074/jbc.M109.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, Lowe J, Gerdes K. Centromere pairing by a plasmid-encoded type 1 ParB protein. J. Biol. Chem. 2007;282:28216–28225. doi: 10.1074/jbc.M703733200. [DOI] [PubMed] [Google Scholar]
- Ringgaard S, Zon J. v., Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc. Natl. Acad. Sci. USA. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov O, Lobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283:546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- Rosche TM, Siddique A, Larsen MH, Figurski DH. Incompatibility protein IncC and global regulator KorB interact in active partition of promiscuous plasmid RK2. J. Bacteriol. 2000;182:6014–6026. doi: 10.1128/jb.182.21.6014-6026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salje J, Lowe J. Bacterial actin: architecture of the ParMRC plasmid DNA partitioning complex. EMBO. 2008;27:2230–2238. doi: 10.1038/emboj.2008.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sawitzke JA, Li Y, Sergueev K, Youngren B, Brendler T, Jones K, Austin S. Transcriptional interference by a complex formed at the centromere-like partition site of plasmid P1. J. Bacteriol. 2002;184:2447–2454. doi: 10.1128/JB.184.9.2447-2454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Structure of ADP.AIF4−-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- Schumacher MA. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 2008;412:1–18. doi: 10.1042/BJ20080359. [DOI] [PubMed] [Google Scholar]
- Seefeldt LC, Morgan TV, Dean DR, Mortenson LE. Mapping the site(s) of MgATP and MgADP interaction with the nitrogenase of Azotobacter vinelandii. Lysine 15 of the iron protein plays a major role in MgATP interaction. J. Biol. Chem. 1992;267:6680–6688. [PubMed] [Google Scholar]
- Siddique A, Figurski DH. The active partition gene incC of IncP plasmids is required for stable maintenance in a broad range of hosts. J. Bacteriol. 2002;184:1788–1793. doi: 10.1128/JB.184.6.1788-1793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soberon NE, Lioy VS, Pratto F, Volante A, Alonso JC. Molecular anatomy of the Streptococcus pyogenes pSM19035 partition and segrosome complexes. Nucleic Acids Res. 2011;39:2624–2637. doi: 10.1093/nar/gkq1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- Thomas CM, Helinski DR. Vegetative replication and stable inheritance of IncP plasmids. In: Thomas CM, editor. Promiscuous Plasmids of Gram-negative Bacteria. Academic Press; San Diego, California: 1989. [Google Scholar]
- Thomas CM, Smith CA. The trfB region of broad host range plasmid RK2: the nucleotide sequence reveals incC and key regulatory gene trfB/korA/korD as overlapping genes. Nucleic Acids Res. 1986;14:4453–4469. doi: 10.1093/nar/14.11.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsted PB, Macartney DP, Akhtar P, Haines AS, Ali N, Davidson P, Stafford T, Pocklington MJ, Pansegrau W, Wilkins BM, Lanka E, Thomas CM. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J. Mol. Biol. 1998;282:969–990. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- Vecchiarelli AG, Han Y-W, Tan X, Mizuuchi M, Ghirlando R, Biertumpfel C, Funnell BE, Mizuuchi K. ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol. Microbiol. 2010;78:78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheust C, Helinski DR. The incC korB region of RK2 repositions a mini-RK2 replicon in Escherichia coli. Plasmid. 2007;58:195–204. doi: 10.1016/j.plasmid.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Villarroel R, Hedges RW, Maenhaut R, Leemans J, Engler G, Montagu MV, Schell J. Heteroduplex analysis of P-plasmid evolution: the role of insertion and deletion of transposable elements. Mol. Gen. Genet. 1983;189:390–399. doi: 10.1007/BF00325900. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the α- and β–subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DR, Macartney DP, Thomas CM. The partitioning activity of the RK2 central control region requires only incC, korB and KorB-binding site OB3 but other KorB-binding sites form destabilizing complexes in the absence of OB3. Microbiology. 1998;144:3369–3378. doi: 10.1099/00221287-144-12-3369. [DOI] [PubMed] [Google Scholar]
- Youngren B, Austin S. Altered ParA partition proteins of plasmid P1 act via the partition site to block plasmid propagation. Mol. Microbiol. 1997;25:1023–1030. doi: 10.1046/j.1365-2958.1997.4761842.x. [DOI] [PubMed] [Google Scholar]


