Abstract
Background
The reduction-sensitive cationic polymer is a promising nonviral carrier for gene delivery. Until now, disulfide bonds have been the only golden standard for its design. The aim of this research was to develop a novel reduction-responsive cationic polymer as a gene carrier.
Methods
Polycationic carriers were synthesized by addition of branched oligoethylenimine 800 Da (OEI800) via an active ester containing diselenide bonds. Disulfide bonds cross-linked with OEI800-SSx and monoselenide bonds linked with OEI800-Sex were synthesized and compared. Their molecular weights and degradation properties were determined using gel permeation chromatography. Changes in particle size, morphology, and DNA binding were investigated by dynamic light scattering, transmission electron microscopy, and electrophoresis assay in a reduction environment. Cytotoxicity and transfection in vitro were evaluated in a murine melanoma cell line (B16F10) and a human cervical epithelial carcinoma cell line (HeLa), while intracellular degradation and dissociation with DNA were studied by confocal laser scanning microscopy with FITC-labeled OEI800 derivatives and Cy5-labeled DNA.
Results
Diselenide-conjugated OEI800 (OEI800-SeSex) polymer carriers of high molecular weight were successfully synthesized. After compacting with DNA, the OEI800-SeSex polymers formed nanoparticles with an average size of 140 nm at an adequate C/P ratio. OEI800-SeSex showed reduction-responsive degradation properties similar to those of the OEI800-SSx via gel permeation chromatography, dynamic light scattering, and transmission electron microscopy. OEI800-SeSex showed much lower cytotoxicity than PEI25k, and significantly higher transfection efficiency than OEI800 in both B16F10 and HeLa cells. Transfection of luciferase in the OEI800-SeSex group was comparable with that of standard PEI25k and traditional reduction-sensitive polymer OEI800-SSx groups. Furthermore, intracellular degradation of OEI800-SeSex and dissociation with DNA were also confirmed by confocal laser scanning microscopy.
Conclusion
The OEI800-SeSex obtained was able to bind plasmid DNA efficiently to yield nanosized particles and had reduction sensitivity which is as efficient as that for OEI800-SSx. In vitro experiments confirmed its low cytotoxicity and high transfection ability. Diselenide bonds can be used as effective and novel reduction-sensitive linkages for gene delivery.
Keywords: diselenide, oligoethylenimine, reduction-sensitive, gene carriers
Introduction
Cationic polymer-mediated nonviral gene delivery has emerged as a viable alternative to viral vectors because of its ease of manufacturing, no limitation on DNA size, lesser inflammatory response, and lack of immunogenicity.1,2 However, cationic polymers are problematic in terms of the delivery process. Between the time when the complexes are assembled and when they are taken up by endosomes, the nucleic acids need to be well compacted within the cationic polymers to form complexes, thus requiring strong binding interactions to prevent gene degradation as a result of mechanical stress and enzymatic activity.3 On the other hand, after entry into the cell or endosomal escape, the complexes should dissociate to enable efficient gene release, which involves movement of DNA into the nucleus for transcription or targeting siRNA to mRNA for RNA interference.1,4,5 Moreover, high transfection efficiency is often associated with high toxicity of the vector. A typical example is polyethylenimine (PEI), which is one of the most effective nonviral gene delivery carriers due to its DNA-condensing capability, proton sponge effect for osmotic swelling, and physical rupture of the endosome.6–10 PEI with its high molecular weight is necessary for effective gene transfection, but its presence also results in high cytotoxicity. In contrast, low molecular weight PEI has low toxicity but cannot condense DNA effectively and has poor transfection ability.11 Therefore, one approach is to design biodegradable polycations which allow efficient gene transfer and stimulus-responsive degradation into low molecular weight products with minimal toxicity.
Intracellular and extracellular environments differ in their pH, enzymatic activity, and redox potential. For example, some intracellular compartments, such as late endosomes and lysosomes, are more acidic than normal physiological pH.12,13 High matrix metalloproteinase expression is observed in tumor cells,14 and glutathione levels are higher intracellularly than extracellularly,15–17 being approximately 1000-fold higher than the concentration in plasma.18 Furthermore, other intracellular reductive reagents, such as nicotinamide adenosine dinucleotide phosphate (NADPH), can change oxidative glutathione to reduced glutathione.19 These factors can be used as triggers for disassociation of polycation and gene payload. Various chemical linkers responsive to different triggers have been designed, including acetals,20–22 hydrazone bonds,23–26 esters,6 disulfides,27–33 and matrix metalloproteinase substrate peptide-containing linkers.14 Among them, disulfide bonds have been widely used as the only golden standard for reductive design in gene and drug delivery.27,29,34–37 After it was found that low molecular weight oligoethylenimine (OEI) cross-linked by dithiobis (succinimidylpropionate) or dimethyl · 3,3′-dithiobispropionimidate · 2HCl achieved highly efficient transfection in vitro and had lower cytotoxicity,34 much research attention has been focused on modification of different disulfide bonds containing linkers27–29,35 and effect of the molecular weight of cross-linked OEI.35
Inspired by the success of disulfide bonds, the selenium (Se) element listed in the same family as sulfur (S) in the periodic table of elements has attracted attention. Diselenide bonds have also been considered as potential candidates for stimuli-sensitive design. As reported recently, diselenide bonds possess reductive biodegradable ability similar to that of disulfide bonds.19,40 Micelles formed by polymers containing diselenide bonds were quite stable under physiological conditions, but were sensitive to reductive stimuli.36 In addition, disulfide bonds were too fragile for the reductive environment, but can be cleaved by a reductive reagent in low concentration.35 This property is not beneficial for transfection in vivo, because the disulfide may be destroyed in the circulation, so the loaded genes cannot be delivered to their desired location.
In this study, diselenide bonds were introduced into a nonviral gene delivery system for the first time. The diselenide bonds were designed to be cleavable due to reduction, resulting in release of DNA inside the cell. Specifically, branched OEI 800 Da (OEI800) was conjugated with diselenide bonds and with disulfide bonds, and their potential for reduction sensitivity was tested and compared.
Materials and methods
Materials
Selenium powder and sodium borohydride (NaBH4) were obtained from Kelong Chemical Company (Chengdu, China). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum were obtained from Life Technologies Corporation (Gibco, Grand Island, NY), and 3-chloropropanoic acid, 3, 3′-disulfanediyldipropanoic acid (DSDPA), reduced glutathione, antibiotics (penicillin and streptomycin), branched polyethylenimine 25 kDa (PEI25k), oligoethylenimine (OEI800), and fluorescein isothiocyanate (FITC) were obtained from Sigma-Aldrich (Shanghai, China). N-hydroxysuccinimide (NHS) and 1-ethyl-3-[3-(dimethylamino)-propyl] carbodiimide (EDC) were purchased from AstaTech Pharmaceutical (Chengdu, China). 1,4-dithiothreitol (DTT) and 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) were commercially available from Amresco (Solon, OH). Cell lysate and the luciferase reporter gene assay kit were purchased from Promega (Madison, WI). A bicinchoninic acid protein assay kit was purchased from Pierce (Rockford, IL). The DNA labeling kit, Label IT®Cy5, was from Mirus Bio Corporation (Madison, WI), plasmid pGL3 (encoding Photinus pyralis luciferase under control of the SV40 enhancer/promoter) and enhanced green fluorescent protein encoding plasmid (pEGFP) was purified using the EndoFree plasmid kit from Qiagen (Hilden, Germany). All buffers were prepared in MilliQ ultrapure water and filtered (0.22 μm) prior to use. All other chemicals were purchased from Sigma-Aldrich and used as received.
Synthesis of DSeDPA and SeDPA
3, 3′-Diselanediyldipropanoic acid (DSeDPA) was synthesized in the same manner as in previous studies, with some modifications.37,38 Selenium powder (2.37 g, 30 mmol) in 10 mL of water was added to a three-necked flask under a nitrogen atmosphere. NaBH4 (2.27 g, 60 mmol) dissolved in 25 mL of cold H2O was syringed dropwise into the selenium suspension. The reaction mixture was stirred at 0°C and reacted until colorless for complete dissolution of the selenium powder. Another quantity of selenium powder (2.37 g, 30 mmol) was added, and the mixture was heated to 105°C for 20 minutes until reddish brown. 3-Chloropropanoic acid (6.5 g, 60 mmol) was dissolved in 15 mL of water, and its pH was adjusted to 8.0 with sodium carbonate (Na2CO3). This was added to the reddish brown solution, with stirring overnight at room temperature under nitrogen. After another 4 hours of stirring and exposure to the atmosphere, the reaction mixture was filtered. The yellow supernatant was adjusted to pH 3–4 using 1 mol/L HCl solution and extracted twice with ethyl acetate. The combined organic layers were washed with water, dried with anhydrous magnesium sulfate, filtered, and recrystallized from ethyl acetate to give a product of 5.29 g (a 58% yield). 1H-NMR (400 MHz, d6-DMSO): δ 2.71 (t, 2H), 3.05 (t, 2H). 13C-NMR (d6-DMSO): δ 23.93, 35.41, 173.05. 77Se-NMR (400 MHz, d6-DMSO): δ 318.98. MS m/z: 302/306 (78Se/80Se) (shown in Supplementary Figure 1).
Synthesis of 3, 3′-selenodipropanoic acid (SeDPA) was similar to the process used for DSeDPA, but with a different ratio of selenium powder to NaBH4. Selenium powder (2.37 g, 30 mmol) was mixed with NaBH4 (2.27 g, 60 mmol) to obtain a colorless solution without adding another quantity of selenium powder, and then reacted with 3-chloropropanoic acid, with the same procedure followed after treatment. A final product of 3.85 g was obtained (a 57% yield). 1H-NMR (400 MHz, d6-DMSO): δ2.62 (t, 2H), 2.69 (t, 2H). 13C-NMR (d6-DMSO): δ17.70, 35.40, 173.26 (as shown in Supplementary Figure 2).
Synthesis of disulfide, diselenide and monoselenide bonds for conjugation with OEI
DSeDPA (0.365 g, 1.2 mmol), DSDPA (0.252 g, 1.2 mmol), or SeDPA (0.270 g, 1.2 mmol) and NHS (0.331 g, 2.88 mmol) dissolved in 5 mL of anhydrous tetrahydrofuran were added to a three-necked flask under nitrogen with magnetic stirring. EDC (0.445 g, 2.88 mmol) dissolved in 5 mL of anhydrous tetrahydrofuran was added dropwise into the mixture at 0°C. The reaction mixture was stirred overnight at room temperature. After filtration, evaporation, and redissolution in 0.5 mL of anhydrous dimethyl sulfoxide, the active ester solution (DSeDPA-NHS, DSDPA-NHS or SeDPA-NHS) obtained was ready for further use.
Prior to cross-linking, 0.8 g (1 mmol) of oligoethylenimine (OEI800) was dissolved in water with pH adjusted to 7.4 by HCl solution, and lyophilized. It was redissolved in 2 mL of dimethyl sulfoxide, and mixed with various active ester solutions (DSeDPA-NHS, DSDPA-NHS, or SeDPA-NHS) under nitrogen. The solution was stirred continuously for 2 days at 35°C, and dialyzed against water using cellulose dialysis bags (molecular weight cutoff 7 kDa). The final products of OEI800-SeSex, OEI800-SSx, and OEI800-Sex were obtained after lyophilization.
NMR characterization
NMR spectra for 1H, 13C, and 77Se were recorded on a Bruker Avance II NMR spectrometer at 400 MHz using DMSO-d6 as solvents, with 0.5% tetramethylsilane as the internal standard.
Molecular weight characterization
The molecular weight of the products was determined by gel permeation chromatography (Waters, Milford, MA). A Waters 2690D high-pressure liquid chromatography system was equipped with Ultrahydrogel 1000 and 120 columns, as well as a refractive index detector. NaCl solution 0.1 M with pH adjusted to 2.8 by HCOOH was used as the eluent at a flow rate of 1.0 mL per minute. The external and column temperatures were kept at 35°C. Pullulan of a different molecular weight (1 mg/mL) was used as the standard for determination of the calibration curve.
Measurement of degradation
To measure degradation, 5 mg of OEI800-SeSex or OEI800-SSx polymer dissolved in 0.9 mL of water was treated with 0.1 mL of glutathione (1.0 mol/L), DTT (1.0 mol/L), or NaBH4 (1.0 mol/L) at 37°C for 2 hours, respectively. OEI800-Sex, PEI25k, and OEI800 were used as controls.27 Degradation of OEI800 and other analogs cross-linked with disulfide and diselenide in reductive conditions was evaluated by gel permeation chromatography.
Complex formation, particle size, zeta potential, and morphology
A cationic polymer solution in HEPES buffered glucose (20 mM HEPES and 5% [w/v] glucose, pH 7.4) was gently mixed with plasmid DNA HEPES buffered glucose (100 μg/mL) for preparation of complexes with different C/P ratios (ie, weight ratios of cationic polymer to plasmid DNA). The nanoparticle suspension was incubated at room temperature for 20 minutes before use.
Particle sizes and zeta potentials of the complexes were characterized using a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, UK). The incubated complexes (C/P 1.2, 3, or 6) were diluted to 1 mL with MilliQ water to a final plasmid DNA concentration of 3 μg/mL. For measurement in a reductive environment, DTT or NaBH4 25 mM was added to the complexes and incubated at 37°C for 30 minutes prior to dilution. The morphology of the complexes was characterized by transmission electron microscopy. A drop of complex (C/P 6) suspension incubated with or without a reductive reagent was deposited on amorphous carbon-coated copper grids, dried, dyed using 2% (w/w) phosphomolybdic acid, and detected using a JEOL JEM-100CX electron microscope (Tokyo, Japan).
DNA binding determined by electrophoresis assay
Suspensions of various complexes (OEI800-SeSex, OEI800-SSx, OEI800-Sex, PEI25k, or OEI800) with different C/P ratios (0.02–1.4) were loaded onto 1% agarose gel. Electrophoresis was performed in Tris-acetate-EDTA buffer (pH 8) running at 85 V for 40 minutes. The gel was stained with ethidium bromide and analyzed on an ultraviolet illuminator (BioRad, Hercules, CA) to visualize the location of the DNA bands. Compaction of the complexes under reduction conditions was evaluated after incubation with DTT at a final concentration of 25 mM at 37°C for 30 minutes.
Cell culture and cytotoxicity assay
Murine melanoma cells (B16F10) and human cervical epithelial carcinoma cells (HeLa) obtained from the Shanghai Institute for Biological Science (Shanghai, China) were used for the cytotoxicity study. The B16F10 and HeLa cells were cultured in DMEM containing penicillin 100 U/mL, streptomycin 100 μg/mL, and fetal bovine serum 10% at 37°C in the presence of 5% CO2.
Relative cell viability when incubated with the cationic polymer solution was determined by MTT assay. The B16F10 and HeLa cells were seeded into a 96-well tissue culture plate at a density of 8 × 103 cells per well in 100 μL of DMEM. A designated amount of cationic polymer solution (OEI800-SeSex, OEI800-SSx OEI800-Sex, PEI25k, and OEI800 at a final concentration of 5–50 μg/mL) were added once cell confluence had reached 70%–80%. After 24 hours of incubation, the cytotoxicity measurement was performed according to the usual laboratory protocol, and the ultraviolet value of the dissolved formazan crystals was recorded at 570 nm using a BioRad 550 microplate reader.
Gene transfection of pEGFP and pGL3
pEGFP and pGL3 was used to determine the transfection efficiency of the OEI800 derivatives. B16F10 and HeLa cells were seeded into 96-well plates at a density of 8 × 103 cells per well in 100 μL of DMEM, and the cells were incubated overnight for attachment. Directly before transfection, the medium in each well was replaced with 100 μL of fresh serum-free DMEM containing various amount of complexes (200 ng of plasmid DNA per well, C/P ratios 4–14). After 4 hours of incubation, the medium was replaced with 100 μL of fresh serum-containing medium. Qualitative evaluation of pEGFP transfection was observed under a fluorescence microscope (Leica, Germany) after further incubation for 48 hours. Subsequently, the cells were washed with ice-cold phosphate-buffered solution, harvested, and placed in 1 mL of phosphate-buffered solution containing 10% fetal bovine serum. The percentage of transfected cells was determined by flow cytometry (BD FACSAria™ II, Franklin Lakes, NJ), and was obtained by determining the statistics of cells fluorescing above the control level, where nontransfected cells were used as the control. Quantitative measurement using the luciferase assay was carried out after 24 hours of incubation according to the manufacturer’s protocol. Relative light units were measured using a BioRad 550 microplate reader while total protein was measured using a bicinchoninic acid protein assay kit. Luciferase activity was expressed as relative fluorescence intensity per mg protein.
Intracellular fate of nanoparticles by confocal laser scanning microscopy
B16F10 cells were seeded at a density of 2 × 104 cells per well in a 35 × 12 mm glass-bottomed chamber (NEST, Wuxi, China) the day before use. Cy5-labeled pEGFP DNA was prepared according to the DNA labeling kit (Label IT®Cy5), while FITC-labeled cationic polymer was obtained via conjugation of FITC to the cationic polymer.39 Complexes containing 40% Cy5-labeled pEGFP and FITC-labeled cationic polymer with a C/P ratio of 6 were added to serum-free fresh medium at 300 ng of plasmid per chamber. At the desired incubation time points (0.5, 1, 2, 4 and 8 hours), the cells were washed twice with phosphate-buffered solution and observed with a confocal laser scanning microscope (Leica TCS SP5, Germany). The Cy5 fluorophore was excited at 543 nm and emission was detected using a 565–615 nm band pass filter, while FITC was excited at 495 nm and emission was detected using a 515–545 nm band pass filter. Data were analyzed using LAS AF Lite 2.3 software (Leica, Germany).40
Data and statistical analysis
Group data were reported as the mean ± standard deviation. Differences between groups were analyzed by two-tailed heteroscedastic t-tests using Microsoft Excel 2010, and statistical significance was accepted at P < 0.05.
Results
Polymer synthesis and structural analysis
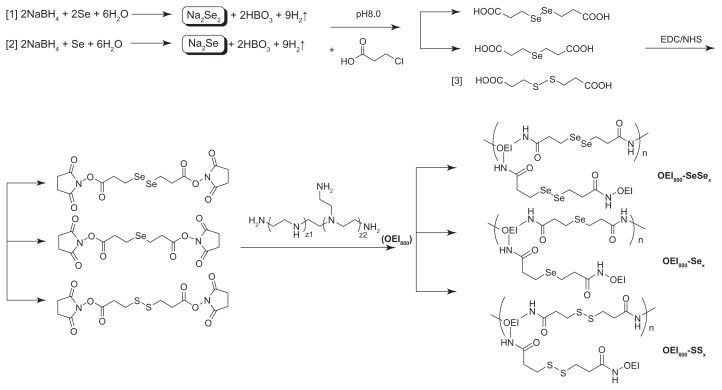
The synthetic route of various cross-linked OEI800 (reduction-responsive and stable derivatives) is shown in Figure 1. Diselenide bonds cross-linked with OEI800 (OEI800-SeSex) was started by preparation of diselenide bonds containing the linker DSeDPA. The carboxyl group at the terminal end of DSeDPA was activated by NHS, using EDC as the dehydrolyzing agent in tetrahydrofuran. The active ester reacted with the primary amine group of the OEI800, resulting in diselenide bonds containing OEI800-SeSex. For controls, the corresponding stable analog OEI800-Sex, and the standard reduction-responsive polymer OEI800-SSx containing disulfide bonds, were prepared (Figure 1). All the critical steps were confirmed by 1H NMR. The chemical environment of the hydrogen close to selenium was different between DSeDP and SeDPA. The peak at 3.05 ppm reflects the signal of CH2 in DSeDPA, while the peak of 2.62 ppm is assigned to the protons of CH2 in SeDPA.41 Accordingly, the peak of CH2 close to selenium in DSeDPA detected by 13C-NMR is 23.93 ppm, while that in SeDPA is 17.70 ppm. (Supplementary Figures 1 and 2).
Figure 1.
Synthetic schema for OEI800-SeSex (1), OEI800-Sex (2), and OEI800-SSx (3).
Abbreviations: OEI, oligoethylenimine; NHS, N-hydroxysuccinimide; EDC, 1-ethyl-3-[3-(dimethylamino)-propyl] carbodiimide.
Molecular weight and degradation of diselenide, monoselenide, and disulfide bonds conjugated with OEI
The molecular weights of the reduction-sensitive OEI800-SeSex and OEI800-SSx polymers and the stable OEI800-Sex polymer were detected by gel permeation chromatography in the absence or presence of the reduction substance (glutathione, DTT, or NaBH4, Table 1). Standard PEI25k and OEI800 showed average molecular weights of 16 kDa and 0.76 kDa, while the molecular weights of the cross-linked OEI800-SSx and OEI800-SeSex polymers were about 12 kDa and 18 kDa, respectively. OEI800-Sex was also cross-linked as a high molecular weight polymer of about 15 kDa. After incubation with the reductive reagents, the peak shift of the different polymers in gel permeation chromatography became diversified (Supplementary Figure 3). The peaks of OEI800-SSx and OEI800-SeSex obviously shifted to a longer time point, indicating a decrease in molecular weight due to degradation, while the molecular weight of OEI800-Sex remained constant. It was interesting that the molecular weight of OEI800-SSx decreased to 1.6 kDa after treatment with glutathione, but to 0.99 kDa after DTT treatment, which is very close to the molecular weight of OEI800 (0.76 kDa). The molecular weight of OEI800-SeSex reduced to 8.1, 3.9, and 1.4 kDa after being incubated with glutathione, DTT and NaBH4, respectively.
Table 1.
Molecular weights of various polymers detected by gel permeation chromatography in the absence or presence of reduction reagents
| Polymers | Mw/103 Da | Mn/103 Da | PDI |
|---|---|---|---|
| OEI800-SSx | 12 | 3.9 | 3.1 |
| OEI800-SSx + GSH | 1.6 | 0.78 | 2.1 |
| OEI800-SSx + DTT | 0.99 | 0.75 | 1.3 |
| OEI800-SeSex | 18 | 5.3 | 3.4 |
| OEI800-SeSex + GSH | 8.1 | 2.5 | 3.2 |
| OEI800-SeSex + DTT | 3.9 | 2.6 | 1.5 |
| OEI800-SeSex + NaBH4 | 1.4 | 0.79 | 1.8 |
| OEI800-Sex | 15 | 5.7 | 2.8 |
| OEI800-Sex + NaBH4 | 15 | 5.8 | 2.7 |
| PEI25k | 16 | 4.5 | 3.6 |
| OEI800 | 0.76 | 0.52 | 1.5 |
Abbreviations: DTT, 1,4-dithiothreitol; Mw, weight-average molecular weight; Mn, number-average molecular weight; PDI, polydispersity index; OEI, oligoethylenimine; PEI, polyethylenimine; GSH, glutathione.
Characterization of particle size, zeta potential, and morphology
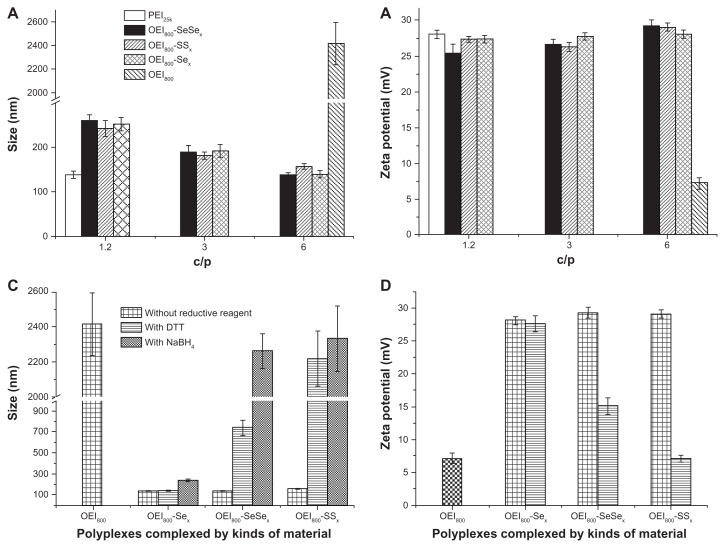
Dynamic light scattering measurements were used to examine the hydrodynamic particle size of the complexes (Figure 2A). The polymers of PEI25k, OEI800-SeSex, OEI800-SSx, and OEI800-Sex were able to condense plasmid DNA in the form of complexes (C/P 1.2 for PEI25k, and C/P 1.2–6 for OEI800 derivatives), with an approximate diameter of 140–250 nm and a narrow distribution. The size of the complexes with OEI800 derivatives depended on the C/P ratio. For instance, the sizes of the OEI800-SeSex complexes decreased from 250 nm, to 190 nm, to about 140 nm, as the C/P increased from 1.2, to 3, to 6. In contrast, the complexes formed by OEI800 and plasmid DNA tended to aggregate or disperse at a diameter over 2000 nm, even at a high C/P ratio of 6. Accordingly, the well compacted nanoparticles composed of high molecular weight polycations (PEI25k and OEI800 derivatives) and DNA were all positively charged, with about +27 mV zeta potential, while that of low molecular weight OEI800 complexes was only about +7 mV (Figure 2B). After being treated with different reductive reagents, the size of the OEI800-SeSex, OEI800-SSx, and OEI-Sex complexes was detected at an appropriate C/P of 6 (Figure 2C). The average diameter of OEI800-SeSex/DNA complexes with NaBH4 and OEI800-SSx/DNA complexes in DTT and NaBH4 was obviously increased to over 2200 nm, and the size of OEI800-SeSex/DNA (C/P 6) treated with DTT increased to about 740 nm. As controls, the stable analog OEI800-Sex/DNA (C/P 6) complexes showed no obvious changes after being treated with reductive reagents. Changes in the zeta potential were also observed after treatment with DTT (Figure 2D). The mean surface charge of well compacted nanoparticles containing disulfide bonds and diselenide bonds reduced to approximately 15 mV and 7 mV, respectively, while no change was detectable in the OEI800-Sex group (P > 0.05).
Figure 2.
Sizes and zeta potentials of various complexes determined by Zetasizer Nano ZS. (A) Sizes of complexes composed of PEI25k, OEI800, or OEI800 derivatives and genes at different C/P ratios, (B) zeta potentials of complexes composed of PEI25k, OEI800 or OEI800 derivatives and genes at different C/P ratios, (C) changes in size with or without the reductive reagent (C/P 6), and (D) zeta potential changes with or without the reductive reagent (C/P 6, n = 3).
Abbreviations: DTT, 1,4-dithiothreitol; OEI, oligoethylenimine; PEI, polyethylenimine.
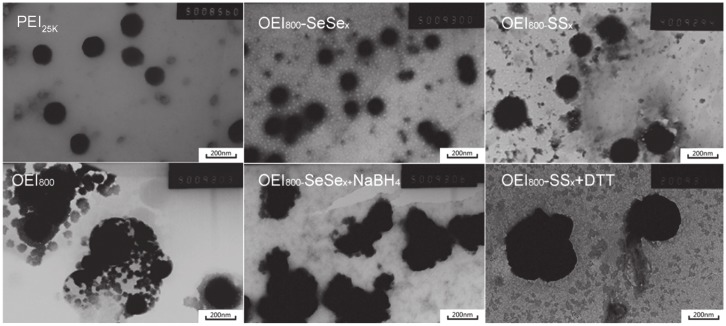
In addition, the morphology of the complexes investigated by transmission electron microscopy was in agreement with the size measurement by dynamic light scattering (Figure 3). PEI25k, OEI800-SeSex, and OEI800-SSx could compact plasmid DNA to form nanoparticles at an appropriate C/P ratio. All diameters of the PEI25k/DNA (C/P 1.2), OEI800-SeSex/DNA (C/P 6), and OEI800-SSx/DNA (C/P 6) complexes were less than 200 nm, whereas samples of DNA condensed by OEI800 was partly aggregated and dispersed. After treatment with NaBH4, the diameter of OEI800-SeSex/DNA increased to more than 400 nm, which was similar to what phenomena happened to OEI800-SSx/DNA upon treatment with DTT. It was obvious that the OEI800-SeSex/DNA and OEI800-SSx/DNA complexes became looser after incubation with the reductive substance, indicating that both OEI800-SeSex and OEI800-SSx had reduction-sensitive ability and would degrade to low molecular weight OEI in the reductive environment.
Figure 3.
Transmission electron microscopy results for PEI25k, OEI800, and OEI800-derived complexes dropped onto amorphous carbon-coated copper grids, dried, and dyed using phosphomolybdic acid.
Note: Bar = 200 nm.
Abbreviations: DTT, 1,4-dithiothreitol; OEI, oligoethylenimine; PEI, polyethylenimine.
DNA binding ability in presence and absence of reduction reagents
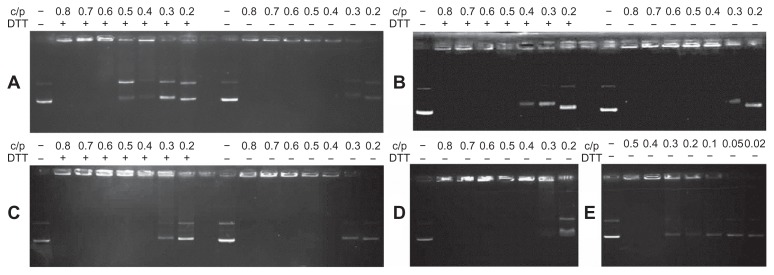
The gel retardation assay verified that all the cationic polymers could retard DNA at an appropriate C/P ratio, and as the polymer concentration increased, their capacity to integrate with plasmid DNA was enhanced (Figure 4). Polycations containing high molecular weight polymers, including OEI800-SSx (Figure 4A), OEI800-SeSex (Figure 4B), OEI800-Sex (Figure 4C), and PEI25k (Figure 4E), could completely retard DNA migration at a C/P ratio of 0.4, while OEI800 formed stable complexes with DNA only at C/P ratios higher than 0.6. This indicated that cross-linked OEI800 can condense plasmid DNA at C/P ratios lower than that of the original monomer. After incubation with DTT, the reduction-responsive polymers of OEI800-SeSex and OEI800-SSx released part of their DNA at C/P ratios of 0.4 and 0.5, respectively, returning to a condensation ability similar to that of OEI800. On the other hand, the corresponding stable analog (OEI800-Sex) showed no detectable changes after treatment with DTT.
Figure 4.
Gel retardation assay of various cationic polymers complexed with plasmid DNA. (A) OEI800-SSx, (B) OEI800-SeSex, (C) OEI800-Sex, (D) OEI800, and (E) PEI25k.
Notes: Polymers and plasmid DNA were mixed at various mass ratios and incubated at 37°C for 30 minutes with or without DTT. The mixed suspension was subsequently applied to the agarose gel and electrophoresed at 85 V for 40 minutes.
Abbreviations: DTT, 1,4-dithiothreitol; OEI, oligoethylenimine; PEI, polyethylenimine.
Cytotoxicity of DNA-free copolymers
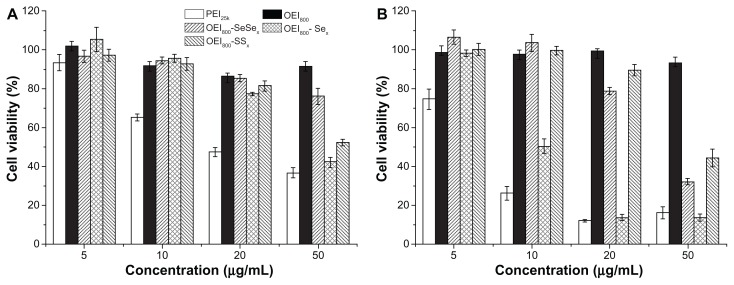
The cytotoxicity of the OEI800 derivatives in B16F10 and HeLa cells was tested by MTT assay, with OEI800 and PEI25k as controls (Figure 5). PEI25k was the most toxic polymer, while cells treated with OEI800 retained more than 90% of their metabolic activity. Both reduction-sensitive bonds of cross-linked copolymers (OEI800-SeSex and OEI800-SSx) showed lower cytotoxicity than PEI25k and stable bonds of cross-linked polymers (OEI800-Sex) in B16F10 cells (Figure 5A), with the same trend observed in HeLa (Figure 5B). For instance, at the same concentration of 50 μg/mL, only 40%–50% viability was found after treatment with PEI25k and OEI800-Sex in B16F10 cells, while that for OEI800-SeSex and OEI800-SSx remained at 76% and 52%, respectively.
Figure 5.
Cytotoxicity assays of the OEI800 derivatives, OEI800, and PEI25k in (A) B16F10 and (B) HeLa cells.
Note: The polymers were incubated with the cells for 24 hours (n = 4).
Abbreviations: OEI, oligoethylenimine; PEI, polyethylenimine.
EGFP and luciferase reporter gene expression in carcinoma cells
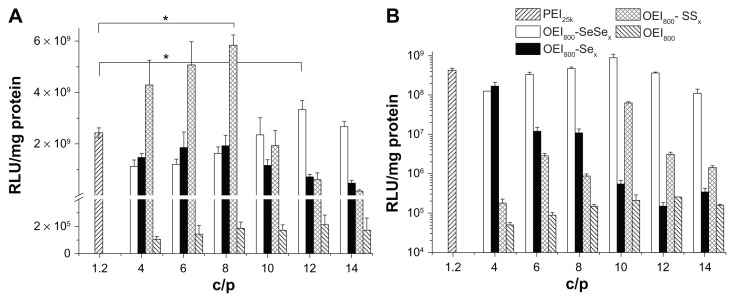
The transfection activity of OEI800-derived nanoparticles, including OEI800-SeSex, OEI800-SSx, and OEI800-Sex, was evaluated in B16F10 (Figure 6A) and HeLa (Figure 6B) cells using pEGFP (Figure 7) and pGL3 (Figure 6). All of the OEI800-derived nanoparticles had significantly higher transfection ability than that of OEI800 alone. Incubation of cells with OEI800-SeSex and OEI800-SSx nanoparticles resulted in slightly higher luciferase expression than that for PEI25k in B16F10. For example, pGL3 transfection of the reduction-sensitive nanoparticles (OEI800-SeSex and OEI800-SSx) was 3.3 × 109 and 5.8 × 109 relative light units/mg protein at the optimum C/P ratio in B16F10 cells, respectively, which was four orders of magnitude higher than that for the OEI800 nanoparticles (only about 1.0 × 105 relative light units/mg protein). In addition, transfection of OEI800-SeSex and OEI800-SSx nanoparticles was up to about 1.5-fold and 2.0-fold higher compared with that of PEI25k (C/P 1.2, 2.4 × 109 relative light units/mg protein, P < 0.05). Interestingly, transfection of OEI800-SeSex was higher than that of OEI800-SSx at C/P ≥ 10, and was lower than OEI800-SSx at C/P ≤ 8 in B16F10 cells. For example, luciferase expression in the OEI800-SeSex group was up to approximately 5-fold higher compared with that in the OEI800-SSx group at C/P 12 (Figure 6A). On the other hand, transfection of OEI800-SeSex was higher than that of OEI800-SSx in HeLa cells at all C/P ratios in the experiment. In particular, it was three orders of magnitude higher than that of OEI800-SSx at C/P 4 (Figure 6B).
Figure 6.
Luciferase transfection of the polycation-pGL3 complexes in (A) B16F10 cells and (B) HeLa cells.
Notes: All the complexes were prepared in HEPES buffered glucose, and 200 ng of pGL3 per well was used for the 96-well tissue culture plate. The data show the average for experiments performed at least three times (n = 3, *P < 0.05).
Abbreviations: OEI, oligoethylenimine; PEI, polyethylenimine; RLU, relative light units.
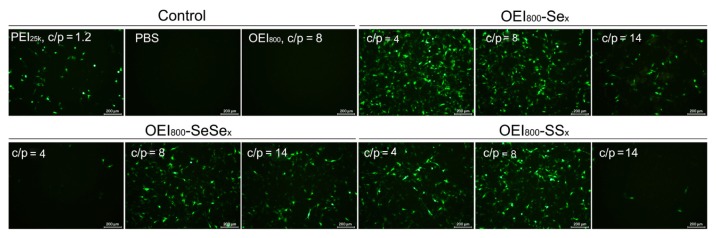
Figure 7.
In vitro transfection efficiency of the complexes of pEGFP with OEI800-SeSex, OEI800-Sex, OEI800-SSx, PEI25k, and OEI800 in B16F10 cell lines at 48 hours.
Note: Bar = 200 μm.
Abbreviations: OEI, oligoethylenimine; PEI, polyethylenimine; pEGFP, enhanced green fluorescent protein encoding plasmid.
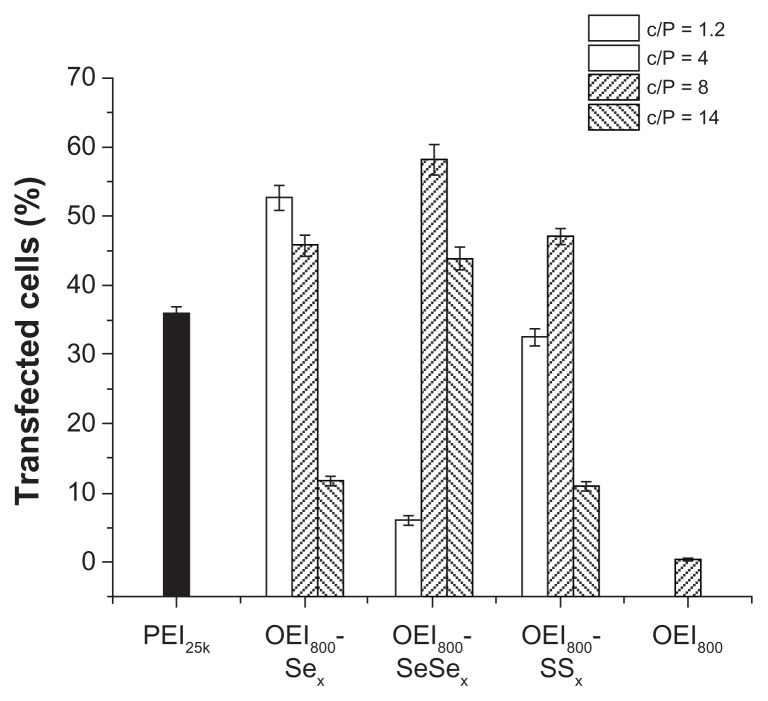
The results of qualitative studies on transfection of plasmid DNA encoding EGFP with various OEI800-derived nanoparticles were consistent with those for luciferase expression, which was visualized with a fluorescent microscope in B16F10 cells. After 48 hours of transfection, cells incubated with OEI800 derivatives (OEI800-SeSex, OEI800-SSx, OEI800-Sex) and PEI25k nanoparticles showed more bright green fluorescent spots, especially at a C/P ratio of 8, for OEI800-SeSex or OEI800-SSx, and a C/P ratio of 4 for OEI800-Sex (Figure 7), indicating more expression of green fluorescent protein. In addition, there were almost no fluorescence spots in the OEI800-SSx group at C/P 14, while a large number of fluorescent spots was found in the OEI800-SeSex group. The percentage of transfected cells detected by flow cytometry showed the same trend as the results for fluorescent microscopy (Figure 8 and Supplementary Figure 4).
Figure 8.
Percentages of transfected cells for the complexes of pEGFP with OEI800-SeSex, OEI800-Sex, OEI800-SSx, PEI25k, and OEI800 quantified by flow cytometry analysis in B16F10 cells at 48 hours
Note: n = 3.
Abbreviations: OEI, oligoethylenimine; PEI, polyethylenimine; pEGFP, enhanced green fluorescent protein encoding plasmid.
Intracellular investigation of labeled DNA and cationic polymers
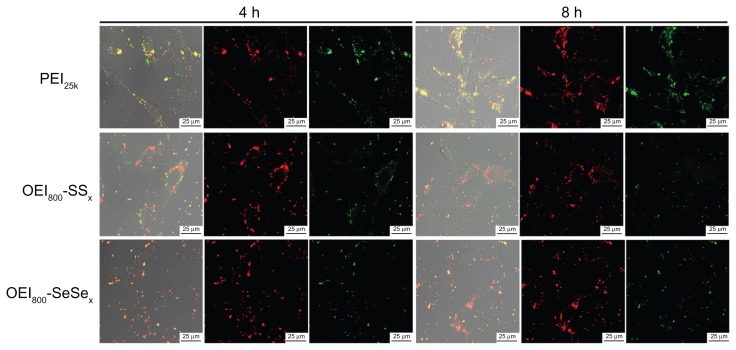
Dual-labeled nanoparticles were prepared using Cy5-labeled DNA and FITC-labeled cationic polymers. Interaction between the polymers, DNA, and cells was monitored by confocal laser scanning microscopy. For ease of graphical representation, images of B16F10 cells incubated with different types of nanoparticles (including PEI25k, OEI800-SeSex and OEI800-SSx) for 4 hours and 8 hours are shown (Figure 9). At 4 hours, approximately 80% of the nanoparticles assembled around the cell membrane, while more fluorescent spots located inside the cells after 8 hours of incubation. This shows that nanoparticles tend to enter the cytoplasm from the surface of cell along with the time. Before 4 hours, good compaction of the polycations and negatively charged DNA is indicated by the orange color, caused by overlay of colors from the green FITC and red Cy5. Importantly, at the 8-hour time point, the intracellular fluorescent dots in the PEI25k group remained orange, but more dots in the OEI800-SSx and OEI800-SeSex groups were red. These changes were not seen on the membrane of cells in the OEI800-derived group.
Figure 9.
Labeled DNA and cationic polymer for intracellular investigation in B16F10 cells. Red dots: Cy5-labeled DNA; green dots: FITC-labeled PEI or OEI800 derivatives.
Note: Bar = 25 μm.
Abbreviations: OEI, oligoethylenimine; PEI, polyethylenimine.
Discussion
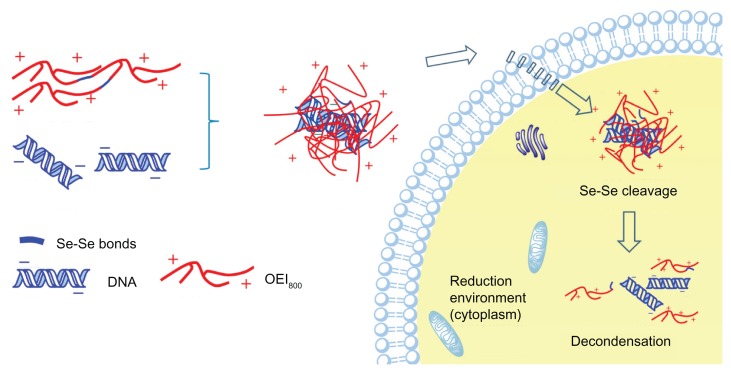
For decades, disulfide bonds have been intensively explored in drug and gene delivery. No other reduction-sensitive chemical linkage has been developed that could work as a substitution for it until now. Recently, diselenide bonds have been reported for the dual redox-sensitive design,36,42 but their utility has not been extended to gene carrier systems. In the current study, diselenide bonds for cross-linked OEI800 were synthesized in order to construct reduction-sensitive gene carriers with high transfection ability and low cytotoxicity. According to the design (Figure 10), cross-linked OEI800-SeSex with a high molecular weight should efficiently compact with DNA to form cationic complexes. After entering an environment of higher redox potential, such as the cytoplasm, the diselenide bonds would be expected to be cleaved, breaking into subunits of OEI800 and leading to dissociation of the complexes. This would help to move DNA into the nucleus for transcription, and keep a balance between high transfection ability and low cytotoxicity on delivery.
Figure 10.
Schematic illustration of decondensation of reduction-sensitive complexes in the intracellular environment.
In order to achieve cross-linking of diselenide bonds with OEI800, DSeDPA linkage was undertaken. The corresponding reduction-stable linker (SeDPA) was prepared by a similar method and was used as a negative control (Figure 1). DSeDPA and SeDPA were synthesized by reacting 3-chloropropanoic acid with Na2Se2 and Na2Se, respectively. The key to obtaining distinct Na2Se2 and Na2Se depended on different ratios of NaBH4 to selenium powder. When the molecular ratio of NaBH4 to selenium was 2, colorless Na2Se was given as the product, and reddish brown Na2Se2 was obtained at a ratio of 1.38 To avoid H2Se2 gas production as a side reaction, the pH of 3-chloropropanoic acid should be adjusted to 8.0. After reacting with 3-chloropropanoic acid overnight, Na2Se2 or Na2Se residues should be oxidized to selenium by exposure to the atmosphere for easy purification via filtration. Unreacted DSeDPA and SeDPA was removed by adjusting the pH of the supernatant for formation of a precipitate at pH 3–4.38 In view of the instability of diselenide bonds in an alkaline environment (pH > 10),43 the alkaline OEI800 solution should be neutralized to pH 7.4 prior to cross-linking. At the cross-linking step, the concentration of OEI800 must be controlled, and is significantly related to the molecular weight of the final product.35 The higher molecular weight of the cross-linked OEI800 would be obtained, because the intermolecular reaction occurs easily at high OEI800 concentrations. In our synthesis reaction, the OEI800 concentration was 25% (w/w).
According to molecular weight measurement by gel permeation chromatography, OEI800-SeSex (about 18 kDa), OEI800-SSx (about 12 kDa), and OEI800-Sex (about 16 kDa) of high molecular weight were synthesized successfully. It was seen that only the OEI800-SeSex and OEI800-SSx degraded after being treated with reductive reagents (Table 1 and Supplementary Figure 3). Interestingly, OEI800-SSx could degrade to OEI800 after treatment with DTT or glutathione. However, under the same conditions, the OEI800-SeSex did not degrade completely, and would degrade to smaller OEI800 pieces only when treated with a much more efficient reductive reagent, ie, NaBH4. After complexing with DNA, dynamic light scattering measurements of the complexes showed concordant results (Figure 2). The size of particles formed by OEI800-SSx increased to over 2000 nm after incubation with DTT, while that of the OEI800-SeSex group increased to approximately 740 nm. The increased size in the OEI800-SSx and OEI800-SeSex groups reached over 2000 nm only after treatment with NaBH4. In addition, the reduction-sensitive ability of the diselenide bonds was demonstrated by characterization of morphology (Figure 3 and Table 1). OEI800-SeSex could complex with plasmid DNA efficiently to form nanosized particles, and the results of transmission electron microscopy indicated the mean diameter of the particles to be less than 200 nm. OEI800-SeSex nanoparticles in NaBH4 showed similar swelling with OEI800-SSx in DTT. After treatment with reduction reagents, changes detected by transmission electron microscopy were not as obvious as the changes determined by dynamic light scattering. This difference was due to the fact that the dynamic light scattering method gives the hydration diameter rather than the actual (dry) diameter detected by transmission electron microscopy.44 The changes in molecular weight, particle size, and morphology all confirmed that the disulfide and diselenide bonds in OEI800-SeSex and OEI800-SSx were cleavable in a reductive environment, and that cleavage of the diselenide bonds needs a more efficient reducible reagent. The likely reason for this is that diselenide bonds are more stable than disulfide bonds, with a lower redox potential.19 This property is beneficial for transfection in vivo because disulfide bonds may be cleaved by glutathione at the concentrations present in blood, while diselenide bonds remain stable.18,35
In the gel retardation assay, the ability of OEI800-SeSex and OEI800-SSx to complex with the DNA decreased after treatment with DTT, which implies a reduction-sensitive ability for diselenide bonds and that the reductive reaction between DTT and OEI800-SeSex was already efficient enough for release of DNA (Figure 4). Accordingly, the MTT assay of OEI800-SeSex and OEI800-SSx showed similarly lower toxicity in comparison with stable OEI800-Sex and PEI25k complexes (Figure 5). Although the diselenide bonds were much more stable than disulfide bonds, in the cytotoxicity assay, there was not only glutathione present as a reductive reagent in the cytoplasm but also high amounts of NADPH. As reported elsewhere, selenoglutathione can oxidize the common biological cofactor NADPH, and reacts strongly with glutathione reductase, which is an NADPH-dependent enzyme.19 Therefore, diselenide bonds can be efficiently cleaved in a cell reduction environment and have the same reduction-sensitive ability as disulfide bonds.
In the in vitro transfection assay, OEI800-SeSex had much higher transfection ability than others based on OEI800, and even similar or higher transfection ability than PEI25k (Figure 6). Compared with OEI800, OEI800-SeSex with its high molecular weight possessed a better ability to condense plasmid DNA, and could form tight nanoparticles and endocytose easily. It was hypothesized that the reduction-sensitive disulfide and diselenide bonds could be cleaved in the intracellular environment, triggered by glutathione or NADPH,19 and the OEI800-SeSex and OEI800-SSx degraded to pieces of OEI800, which led to release of plasmid DNA to get high transfection ability and lower cytotoxicity. These results are in agreement with those of other studies of OEI800 cross-linked by degradable bonds.6,28,29 In addition, the optimum C/P of cross-linked OEI800 for transfection was found to be higher than for PEI25k, which may be because the OEI content was lower in cross-linked OEI800 or because of the increased hydrophobicity.6 At a high C/P ratio (C/P ≥ 10 in quantitative transfection and C/P 14 in qualitative transfection), transfection of OEI800-SeSex was higher than that of OEI800-SSx in B16F10 cells (Figures 6A and 7), which might have been because of the cytotoxicity of OEI800-SSx is higher than that of OEI800-SeSex at high concentrations (Figure 5A). The difference in effect between OEI800-SeSex and OEI800-SSx in B16F10 and HeLa cells indicates the cell-dependent reduction sensitivity of diselenide. Based on these results, the diselenide bond-conjugated OEI is more suitable as a reduction-sensitive gene carrier than disulfide bond-conjugated OEI. Moreover, they had small differences in stability,19 which might affect transfection ability. Further studies will be carried out on the sensitivity of diselenide and disulfide under physiological conditions and the time-dependence of response to a reducing agent. Like disulfide bonds, diselenide bonds can be used widely in smart drugs and for gene delivery.
In order to explore the intracellular fate of the nanoparticles, three kinds of cationic polymer-plasmid DNA complexes were investigated. It was obvious that they could be endocytosed easily, and many red or orange dots were observed in the cytoplasm (Figure 9). More red dots were observed in the groups containing reduction bonds and more orange dots were seen in the groups containing stable bonds. The number of red fluorescent dots increased with the passage of time in the OEI800-SeSex and OEI800-SSx groups, while no significant change was observed in the PEI25k group during the experiment. The PEI25k group contained stable bonds which could condense DNA tightly and showed orange fluorescent dots which were the overlay of the red from Cy5 and the green from FITC. For the OEI800-SeSex and OEI800-SSx groups, which contained reduction-sensitive bonds, cleavage of the disulfide bonds or diselenide bonds would cause parts of the FTIC to be released from the nanoparticles, and more red fluorescent dots were observed with the passage of time. This phenomenon implies that diselenide and disulfide bonds are reduction-sensitive.
Conclusion
This study describes the design and synthesis of diselenide bonds cross-linked to OEI800 for use as reduction-sensitive gene carriers. The results show that diselenide-containing OEI800-SeSex was able to bind plasmid DNA efficiently to yield nanosized particles and had the same reduction sensitivity as OEI800-SSx. In vitro experiments demonstrated that the reducible OEI800-SeSex and OEI800-SSx had much lower cytotoxicity and higher transfection activity compared with the nondegradable PEI25k control. The transfection ability of OEI800-SeSex was higher than that of OEI800-SSx in B16F10 cells at a C/P ratio ≥ 10, while the transfection ability of OEI800-SeSex was higher than that of OEI800-SSx in HeLa cells at all C/P ratios used in this experiment. The reduction sensitivity of diselenide was cell-dependent. The results of our confocal laser scanning microscopy experiments demonstrated directly that both OEI800-SeSex and OEI800-SSx could be cleavable in a cell-reductive environment. Our results imply that OEI conjugated with diselenide bonds is suitable for use as a reduction-sensitive gene carrier.
Supplementary figures
NMR spectrum of DSeDPA. (A) 77Se NMR spectrum; (B) 1H NMR spectrum; (C) 13C NMR spectrum.
Abbreviation: DSeDPA, 3, 3’-Diselanediyldipropanoic acid; NMR, nuclear magnetic resonance.
NMR spectrum of SeDPA. (A) 1H NMR spectrum; (B) 13C NMR spectrum.
Abbreviation: SeDPA, 3, 3’-selenodipropanoic acid; NMR, nuclear magnetic resonance.
GPC traces of OEI800-SSx, OEI800-SeSex, OEI800-Sex and the OEI800 derivatives in the absence or presence of reduction treatment.
Abbreviations: GPC, gel permeation chromatography; OEI, oligoethylenimine; DTT, 1,4-dithiothreitol; GSH, glutathione.
The percentage of transfected cell for the complexes of pEGFP with OEI800-SeSex, OEI800-Sex, OEI800-SSx, PEI25k and OEI800 was quantified by flow cytometry analysis in B16F10 cell lines at 48 hours.
Note: n = 3.
Abbreviations: pEGFP, enhanced green fluorescent protein encoding plasmid; PEI, polyethylenimine; OEI, oligoethylenimine.
Acknowledgments
This study was supported by the National 973 programs, 2011CB606206, National Natural Science Foundation of China (51133004, 81000657, 50830105, 31070849), Ministry of Science and Technology (2010DFA 51550), Research Fund for the Doctoral Program of Higher Education of China (20100181120075), and International Cooperative Foundation of Sichuan Province (2009HH0001)
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2008;109(2):259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 2.Sun YX, Xiao W, Cheng SX, Zhang XZ, Zhuo RX. Synthesis of (Dex-HMDI)-g-PEIs as effective and low cytotoxic nonviral gene vectors. J Control Release. 2008;128(2):171–178. doi: 10.1016/j.jconrel.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Nie Y, Zhang ZR, Duan YR. Combined use of polycationic peptide and biodegradable macromolecular polymer as a novel gene delivery system: a preliminary study. Drug Deliv. 2006;13(6):441–446. doi: 10.1080/10717540600640302. [DOI] [PubMed] [Google Scholar]
- 4.Wang XL, Nguyen T, Gillespie D, Jensen R, Lu ZR. A multifunctional and reversibly polymerizable carrier for efficient siRNA delivery. Biomaterials. 2008;29(1):15–22. doi: 10.1016/j.biomaterials.2007.08.048. [DOI] [PubMed] [Google Scholar]
- 5.Pouton CW, Seymour LW. Key issues in non-viral gene delivery. Adv Drug Deliv Rev. 2001;46(1–3):187–203. doi: 10.1016/s0169-409x(00)00133-2. [DOI] [PubMed] [Google Scholar]
- 6.Russ V, Elfberg H, Thoma C, Kloeckner J, Ogris M, Wagner E. Novel degradable oligoethylenimine acrylate ester-based pseudodendrimers for in vitro and in vivo gene transfer. Gene Ther. 2008;15(1):18–29. doi: 10.1038/sj.gt.3303046. [DOI] [PubMed] [Google Scholar]
- 7.Bonnet ME, Erbacher P, Bolcato-Bellemin AL. Systemic delivery of DNA or siRNA mediated by linear polyethylenimine (L-PEI) does not induce an inflammatory response. Pharm Res. 2008;25(12):2972–2982. doi: 10.1007/s11095-008-9693-1. [DOI] [PubMed] [Google Scholar]
- 8.Coll JL, Chollet P, Brambilla E, Desplanques D, Behr JP, Favrot M. In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Hum Gene Ther. 1999;10(10):1659–1666. doi: 10.1089/10430349950017662. [DOI] [PubMed] [Google Scholar]
- 9.Wightman L, Kircheis R, Rössler V, et al. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J Gene Med. 2001;3(4):362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 10.Intra J, Salem AK. Characterization of the transgene expression generated by branched and linear polyethylenimine-plasmid DNA nanoparticles in vitro and after intraperitoneal injection in vivo. J Control Release. 2008;130(2):129–138. doi: 10.1016/j.jconrel.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen Y, Pan S, Luo X, Zhang X, Zhang W, Feng M. A biodegradable low molecular weight polyethylenimine derivative as low toxicity and efficient gene vector. Bioconjug Chem. 2009;20(2):322–332. doi: 10.1021/bc800428y. [DOI] [PubMed] [Google Scholar]
- 12.Breunig M, Bauer S, Goepferich A. Polymers and nanoparticles: intelligent tools for intracellular targeting? Eur J Pharm Biopharm. 2008;68(1):112–128. doi: 10.1016/j.ejpb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Yang SR, Lee HJ, Kim JD. Histidine-conjugated poly(amino acid) derivatives for the novel endosomolytic delivery carrier of doxorubicin. J Control Release. 2006;114(1):60–68. doi: 10.1016/j.jconrel.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama H, Akita H, Kogure K, et al. Development of a novel systemic gene delivery system for cancer therapy with a tumor-specific cleavable PEG-lipid. Gene Ther. 2007;14(1):68–77. doi: 10.1038/sj.gt.3302843. [DOI] [PubMed] [Google Scholar]
- 15.Yeh CC, Hou MF, Wu SH, et al. A study of glutathione status in the blood and tissues of patients with breast cancer. Cell Biochem Funct. 2006;24(6):555–559. doi: 10.1002/cbf.1275. [DOI] [PubMed] [Google Scholar]
- 16.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30(11):1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 17.Verma A, Simard JM, Worrall JWE, Rotello VM. Tunable reactivation of nanoparticle-inhibited β-galactosidase by glutathione at intracellular concentrations. J Am Chem Soc. 2004;126(43):13987–13991. doi: 10.1021/ja046572r. [DOI] [PubMed] [Google Scholar]
- 18.Jones DP, Carlson JL, Samiec PS, et al. Glutathione measurement in human plasma evaluation of sample collection, storage and derivatization conditions for analysis of dansyl derivatives by HPLC. Clin Chim Acta. 1998;275(2):175–184. doi: 10.1016/s0009-8981(98)00089-8. [DOI] [PubMed] [Google Scholar]
- 19.Beld J, Woycechowsky KJ, Hilvert D. Selenoglutathione: efficient oxidative protein folding by a diselenide. Biochemistry. 2007;46(18):5382–5390. doi: 10.1021/bi700124p. [DOI] [PubMed] [Google Scholar]
- 20.Chen W, Meng F, Cheng R, Zhong Z. pH-sensitive degradable polymersomes for triggered release of anticancer drugs: a comparative study with micelles. J Control Release. 2010;142(1):40–46. doi: 10.1016/j.jconrel.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Liu R, Zhang Y, Zhao X, Agarwal A, Mueller LJ, Feng P. pH-responsive nanogated ensemble based on gold-capped mesoporous silica through an acid-labile acetal linker. J Am Chem Soc. 2010;132(5):1500–1501. doi: 10.1021/ja907838s. [DOI] [PubMed] [Google Scholar]
- 22.Chan Y, Bulmus V, Zareie MH, Byrne FL, Barner L, Kavallaris M. Acid-cleavable polymeric core-shell particles for delivery of hydrophobic drugs. J Control Release. 2006;115(2):197–207. doi: 10.1016/j.jconrel.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 23.Prabaharan M, Grailer JJ, Pilla S, Steeber DA, Gong SQ. Amphiphilic multi-arm-block copolymer conjugated with doxorubicin via pH-sensitive hydrazone bond for tumor-targeted drug delivery. Biomaterials. 2009;30(29):5757–5766. doi: 10.1016/j.biomaterials.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 24.Fella C, Walker GF, Ogris M, Wagner E. Amine-reactive pyridylhydrazone-based PEG reagents for pH-reversible PEI polyplex shielding. Eur J Pharm Sci. 2008;34(4–5):309–320. doi: 10.1016/j.ejps.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Braunová A, Pechar M, Ulbrich K. Biodegradable high-molecular-weight multiblock polymers based on poly(ethylene glycol) for drug delivery. J Control Release. 2008;132(3):e20–e22. [Google Scholar]
- 26.Sawant RM, Hurley JP Salmaso S. et al. “Smart” drug delivery systems: double-targeted pH-responsive pharmaceutical nanocarriers. Bioconjug Chem. 2006;17(4):943–949. doi: 10.1021/bc060080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Jiang X, Xu L, Wang X, Hennink WE, Zhuo R. Novel reduction-responsive cross-linked polyethylenimine derivatives by click chemistry for nonviral gene delivery. Bioconjug Chem. 2010;21(10):1827–1835. doi: 10.1021/bc100191r. [DOI] [PubMed] [Google Scholar]
- 28.Peng Q, Hu C, Cheng J, Zhong Z, Zhuo R. Influence of disulfide density and molecular weight on disulfide cross-linked polyethylenimine as gene vectors. Bioconjug Chem. 2009;20(2):340–346. doi: 10.1021/bc800451j. [DOI] [PubMed] [Google Scholar]
- 29.Peng Q, Zhong Z, Zhuo R. Disulfide cross-linked polyethylenimines (PEI) prepared via thiolation of low molecular weight PEI as highly efficient gene vectors. Bioconjug Chem. 2008;19(2):499–506. doi: 10.1021/bc7003236. [DOI] [PubMed] [Google Scholar]
- 30.Son S, Singha K, Kim WJ. Bioreducible BPEI-SS-PEG-cNGR polymer as a tumor targeted nonviral gene carrier. Biomaterials. 2010;31(24):6344–6354. doi: 10.1016/j.biomaterials.2010.04.047. [DOI] [PubMed] [Google Scholar]
- 31.Sun YX, Zeng X, Meng QF, Zhang XZ, Cheng SX, Zhuo RX. The influence of RGD addition on the gene transfer characteristics of disulfide-containing polyethyleneimine/DNA complexes. Biomaterials. 2008;29(32):4356–4365. doi: 10.1016/j.biomaterials.2008.07.045. [DOI] [PubMed] [Google Scholar]
- 32.Takae S, Miyata K, Oba M, et al. PEG-detachable polyplex micelles based on disulfide-linked block catiomers as bioresponsive nonviral gene vectors. J Am Chem Soc. 2008;130(18):6001–6009. doi: 10.1021/ja800336v. [DOI] [PubMed] [Google Scholar]
- 33.Abdullah-Al-Nahain Lee H, Lee YS, Lee KD, Park SY. Development of disulfide core-crosslinked pluronic nanoparticles as an effective anticancer-drug-delivery system. Macromol Biosci. 2011;11(9):1264–1271. doi: 10.1002/mabi.201100083. [DOI] [PubMed] [Google Scholar]
- 34.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjug Chem. 2001;12(6):989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 35.Yu H, Russ V, Wagner E. Influence of the molecular weight of bioreducible oligoethylenimine conjugates on the polyplex transfection properties. AAPS J. 2009;11(3):445–455. doi: 10.1208/s12248-009-9122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma N, Li Y, Xu H, Wang Z, Zhang X. Dual redox responsive assemblies formed from diselenide block copolymers. J Am Chem Soc. 2010;132(2):442–443. doi: 10.1021/ja908124g. [DOI] [PubMed] [Google Scholar]
- 37.Xue Z, Kwong DWJ, Xue LW, Liu Q, Hou AX, Wong WK. Synthesis of novel diselenide-linked porphyrin dimers under phase-transfer catalysis condition and their interactions with DNA. Chem Biodivers. 2009;6(7):1131–1143. doi: 10.1002/cbdv.200800182. [DOI] [PubMed] [Google Scholar]
- 38.Koch T, Suenson E, Henriksen U, Buchardt O. The oxidative cleavability of protein cross-linking reagents containing organoselenium bridges. Bioconjug Chem. 1990;1(4):296–304. doi: 10.1021/bc00004a012. [DOI] [PubMed] [Google Scholar]
- 39.Saovapakhiran A, D’Emanuele A, Attwood D, Penny J. Surface modification of PAMAM dendrimers modulates the mechanism of cellular internalization. Bioconjug Chem. 2009;20(4):693–701. doi: 10.1021/bc8002343. [DOI] [PubMed] [Google Scholar]
- 40.Nie Y, Gunther M, Gu Z, Wagner E. Pyridylhydrazone-based PEGylation for pH-reversible lipopolyplex shielding. Biomaterials. 2011;32(3):858–869. doi: 10.1016/j.biomaterials.2010.09.032. [DOI] [PubMed] [Google Scholar]
- 41.Kumbhare LB, Jain VK, Phadnis PP, Nethaji M. Palladium(II) and platinum(II) 2-(methoxycarbonyl)ethylselenolates: synthesis, spectroscopy, structures and their conversion into metal selenide. J Organomet Chem. 2007;692(7):1546–1556. [Google Scholar]
- 42.Ma N, Xu H, An L, Li J, Sun Z, Zhang X. Radiation-sensitive diselenide block co-polymer micellar aggregates: toward the combination of radiotherapy and chemotherapy. Langmuir. 2011;27(10):5874–5878. doi: 10.1021/la2009682. [DOI] [PubMed] [Google Scholar]
- 43.Cha W, Meyerhoff ME. Catalytic generation of nitric oxide from S-nitrosothiols using immobilized organoselenium species. Biomaterials. 2007;28(1):19–27. doi: 10.1016/j.biomaterials.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 44.Nie Y, Yuan WM, Gong T, Lu J, Fu Y, Zhang ZR. Investigation on characterization and transfection of a novel multi-polyplex gene delivery system. J Appl Polym Sci. 2007;106(2):1028–1033. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NMR spectrum of DSeDPA. (A) 77Se NMR spectrum; (B) 1H NMR spectrum; (C) 13C NMR spectrum.
Abbreviation: DSeDPA, 3, 3’-Diselanediyldipropanoic acid; NMR, nuclear magnetic resonance.
NMR spectrum of SeDPA. (A) 1H NMR spectrum; (B) 13C NMR spectrum.
Abbreviation: SeDPA, 3, 3’-selenodipropanoic acid; NMR, nuclear magnetic resonance.
GPC traces of OEI800-SSx, OEI800-SeSex, OEI800-Sex and the OEI800 derivatives in the absence or presence of reduction treatment.
Abbreviations: GPC, gel permeation chromatography; OEI, oligoethylenimine; DTT, 1,4-dithiothreitol; GSH, glutathione.
The percentage of transfected cell for the complexes of pEGFP with OEI800-SeSex, OEI800-Sex, OEI800-SSx, PEI25k and OEI800 was quantified by flow cytometry analysis in B16F10 cell lines at 48 hours.
Note: n = 3.
Abbreviations: pEGFP, enhanced green fluorescent protein encoding plasmid; PEI, polyethylenimine; OEI, oligoethylenimine.