Abstract
Background:
Curcumin, the principal curcuminoid of the popular Indian spice turmeric, has a wide spectrum of pharmaceutical properties such as antitumor, antioxidant, antiamyloid, and anti-inflammatory activity. However, poor aqueous solubility and low bioavailability of curcumin is a major challenge in its development as a useful drug. To enhance the aqueous solubility and bioavailability of curcumin, attempts have been made to encapsulate it in liposomes, polymeric nanoparticles (NPs), lipid-based NPs, biodegradable microspheres, cyclodextrin, and hydrogels.
Methods:
In this work, we attempted to entrap curcumin in novel self-assembled dipeptide NPs containing a nonprotein amino acid, α, β-dehydrophenylalanine, and investigated the biological activity of dipeptide-curcumin NPs in cancer models both in vitro and in vivo.
Results:
Of the several dehydrodipeptides tested, methionine-dehydrophenylalanine was the most suitable one for loading and release of curcumin. Loading of curcumin in the dipeptide NPs increased its solubility, improved cellular availability, enhanced its toxicity towards different cancerous cell lines, and enhanced curcumin’s efficacy towards inhibiting tumor growth in Balb/c mice bearing a B6F10 melanoma tumor.
Conclusion:
These novel, highly biocompatible, and easy to construct dipeptide NPs with a capacity to load and release curcumin in a sustained manner significantly improved curcumin’s cellular uptake without altering its anticancer or other therapeutic properties. Curcumin-dipeptide NPs also showed improved in vitro and in vivo chemotherapeutic efficacy compared to curcumin alone. Such dipeptide-NPs may also improve the delivery of other potent hydrophobic drug molecules that show poor cellular uptake, bioavailability, and efficacy.
Keywords: anticancer, curcumin, dipeptide, hydrophobic, nanoparticle, self-assembly
Introduction
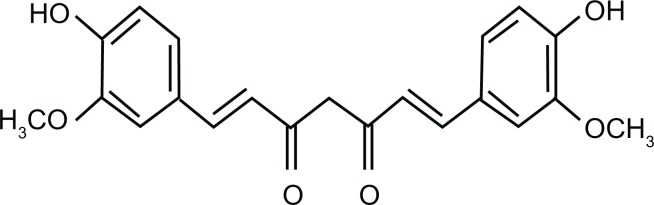
Curcumin (diferuloyl methane) (Figure 1) is a low-molecular-weight natural polyphenol isolated from rhizome of the herb Curcuma longa, which is widely used in traditional Indian and Chinese medicine. Curcumin has a wide range of pharmacological activities, including antitumor, antioxidant, antiamyloid, and anti-inflammatory properties as well as low intrinsic toxicity.1–3 Several studies have unequivocally demonstrated the ability of curcumin to inhibit carcinogenesis in a variety of cell lines such as breast, cervical, colon, gastric, hepatic, leukemia, oral epithelial, ovarian, pancreatic, and prostate cancers.4–7 Despite its association with a number of therapeutic or preventive activities, the potential of curcumin as a useful drug and as an anticancer agent has not been examined due to its hydrophobic nature, which leads to poor aqueous solubility and thus low bioavailability.8 To increase its aqueous solubility and bioavailability, curcumin has been encapsulated in various nanoparticles (NPs), such as liposomes, polymeric NPs, lipid-based NPs, biodegradable microspheres, cyclodextrin, and hydrogels.9–12 While, these efforts have led to enhanced solubility and bioavailability of curcumin, several limitations prohibit their development as successful carriers of the molecule. These include low in vivo stability as well as high drug-leaching properties in liposomes, toxicity of metallic, inorganic, and synthetic polymer-based NPs, and the possibility of an undesirable immune response.13,14
Figure 1.
Molecular structure of curcumin (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione.
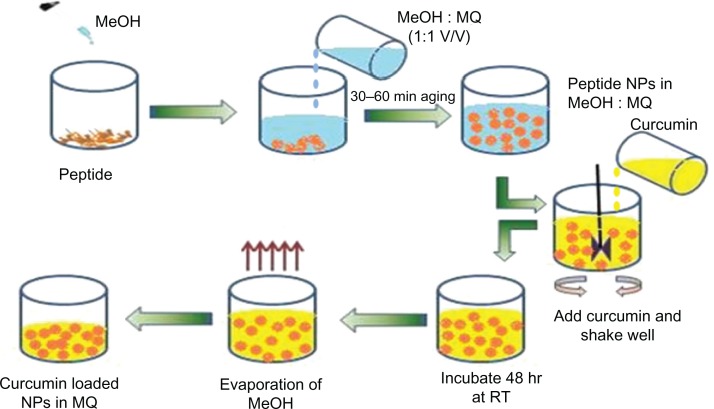
In the present work, we report the design, synthesis, and characterization of novel self-assembled NPs, derived from the dipeptide methionine-dehydrophenylalanine (MΔF), which contains the modified amino acid α,β,-dehydrophenylalanine (ΔPhe), an unsaturated analog of the naturally occurring aromatic amino acid phenylalanine. Incorporation of dehydrophenylalanine in the peptide was expected to provide NPs with enhanced assembly properties due to less spatial freedom as well as increased stability against enzymatic degradation.15–17 MΔF nanoparticles can be assembled in a mixture of aqueous and organic phase and used for the loading and release of the hydrophobic curcumin. Curcumin-loaded MΔF nanoparticles [(Cm)NPs] were well-dispersed in aqueous environments and showed increased cellular availability with enhanced cytotoxicity in various cancerous cell lines as well delayed tumor growth with enhanced survival time in a B6F10 melanoma mouse model.
Materials and methods
Materials
Curcumin (diferuloyl methane), tetrahydrofuran, N-methyl morpholine, isobutyl chloroformate, trifluoroacetic acid, methanol, 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP), phenol, and DL-threo-phenylserine were from Sigma-Aldrich (Munich, Germany). N-[(tert-butoxy)carbonyl]-L-methionine, N-[(tert-butoxy)carbonyl]-L-leucine, N-[(tert-butoxy)carbonyl]-L-and isoleucine, L-phenylalanine were from Novabiochem (Merck, Darmstadt, Germany). Sodium acetate, ethyl acetate, and acetonitrile were from Spectrochem Pvt Ltd (Mumbai, India). Anhydrous sodium sulfate and citric acid were from Merck (Munich, Germany). RPMI-1640, Dulbecco’s modified Eagle’s medium, fetal bovine serum (FBS), penicillin, and streptomycin were from Invitrogen (Carlsbad, CA, USA). 3-(4,5-Dimethylth-iazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) was from Sigma-Aldrich. The apoptosis assay kit was from Invitrogen. HeLa (human cervical cancer), MCF-7 (human breast cancer), Huh-7 (human hepatoma), and L-929 (human fibroblast) cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). HeLa and L-929 cells were cultured in RPMI medium containing 10% FBS and MCF-7 and Huh-7 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. B6F10 cells (mouse melanoma cells) cells were a generous gift from Dr Kundu’s laboratory (NCCS, Pune, India) and were maintained in RPMI medium containing 10% FBS. All culture media were supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were maintained on tissue culture-treated polystyrene plates (Corning, Corning, NY) at 37μC in a humidified atmosphere containing 5% CO2.
Synthesis of dipeptides
Synthesis of methionine-dehydrophenylalanine (MΔF), leucine-dehydrophenylalanine (LΔF), and isoleucine-dehydrophenylalanine (IΔF)
The peptide was synthesized by using a method described previously.18 Details of the synthesis are described in Supplementary Information 1.
Synthesis of methionine-phenylalanine (MF)
The peptide was synthesized by using a method described previously.19 Details of the synthesis are described in Supplementary Information 2.
Preparation of dipeptide NPs
Stock solutions were prepared by dissolving 2 mg of peptide(s) in 50 μL of methanol. In some cases, mild sonication or heating was necessary to dissolve the peptides. Assembly was initiated by addition of 1 mL of aqueous methanol (50% v/v) to the stock. Samples were incubated for 24 hours before experiments.
Characterization of NPs using dynamic light-scattering (DLS)
Particle size and size distribution of NPs before and after drug loading was analyzed using DLS. Light scattering studies were performed in a Photocor complex (Photocor, Moscow, Russia) using a multiple tau digital correlator. All studies were conducted at room temperature at an angle of 90μ using a 632 nm laser. Results are presented as the mean of three independent readings.
Characterization of NPs using transmission electron microscopy (TEM)
TEM of NPs was carried out using uranyl acetate negative staining method. Briefly, after evaporating the methanol, drops of NPs were adsorbed on 300 mesh copper grids with carbon-coated formvar support (TAAB Laboratories Equipment, Berks, UK) and stained with 1% uranyl acetate (Sigma-Aldrich). Excess fluid was removed from the grid surface with a filter paper (Whatman No 1; Whatman, Kent, England) and the grid was air-dried at room temperature before being loaded onto the microscope. Pictures of NPs were acquired in a TEM (Tecnai 12 BioTWIN; FEI, Eindhoven, Netherlands) operating at 120 kV. Photomicrographs were digitally recorded using a Megaview III (SIS, Münster, Germany) digital camera. Image analysis to measure particle dimensions was carried out using the iTem software package (FEI, Hillsboro, OR).
Loading of curcumin in dipeptide NPs and determination of curcumin-loading efficiency and drug-loading content
Curcumin solution in methanol was added to preformed NPs at a ratio of 1:1 w/w, and the mixtures were incubated for 48 hours at room temperature with constant shaking. Methanol was evaporated under a vacuum and free curcumin was separated from that entrapped in NPs by centrifugation at 800 × g for 15 minutes in a MultiScreen 96-well filter plate (Millipore, Billerica, MA). Curcumin content in the filtrate was quantified using UV-vis spectroscopy at 426 nm. The percentage entrapped curcumin in dipeptide NPs was determined using the following equation:
To determine the exact quantity of curcumin in dipeptide NPs, curcumin was loaded in NPs as described above (1:1 w/w of curcumin in 1 mL of dipeptide NPs). Free curcumin was separated from that entrapped in NPs by centrifugation at 800 × g for 15 minutes in a MultiScreen 96-well filter plate (Millipore). Samples were washed three times with PBS and NPs were then dissolved by adding 200 μL of neat methanol. Curcumin content was determined by comparing the sample with curcumin standard curve in methanol using the following equation:
Analysis of photophysical properties of curcumin and curcumin loaded in dipeptide NPs
To determine whether entrapment in dipeptide NPs had an effect on curcumin’s photophysical properties, spectroscopic analysis was condcuted.20 The absorbance and fluorescence spectra of both native curcumin and (Cm)NPs at a concentration of 1 mg/mL were measured in a 50:50 (v/v) methanol: water solution. Absorption spectra were recorded from 350 to 600 nm (Lambda 35; Perkin Elmer, Waltham, MA) and fluorescence emission spectra were recorded from 450 to 700 nm with an excitation wavelength of 420 nm (LS 55; Perkin Elmer).
In vitro curcumin release
Curcumin release studies were carried out using the dialysis bag method.21 A known quantity (1.0 mg/mL) of (Cm)NPs was placed in a dialysis membrane tube with a molecular weight cut-off of 3000 Da (SnakeSkin™ Pleated Dialysis Tubing; Thermo Scientific, Rockford, IL). Because of the low solubility of curcumin in aqueous media, the release study was carried out by immersing the tube in a beaker containing 50 mL of 50:50 (v/v) methanol: water solution to facilitate sink-like conditions and stirred constantly at room temperature for 36 hours. At various time points, 1 mL of solution was withdrawn from the release medium and replaced with fresh PBS. Samples were lyophilized and dissolved in 100 μL of methanol to determine curcumin content using UV-vis spectroscopy (Lambda 35; Perkin Elmer).
Fluorescence and flow cytometric analysis for cellular uptake of NPs
Taking advantage of the intrinsic green fluorescence of curcumin, we studied its cellular uptake by fluorescence microscopy and fluorescence-activated cell sorting (FACS) analysis. HeLa cells were grown in 6-well culture plates at 37μC in a 5% CO2 incubator up to 80% confluency. Cells were then treated with 30 μM native curcumin, (Cm)NPs, and void NPs for 24 hours and analyzed for curcumin uptake. For microscopic analysis, cells were fixed using fixative (Sigma-Aldrich), washed three times with PBS, and analyzed under a fluorescence microscope (Eclipse TE2000-U; Nikon, Tokyo, Japan) to measure intracellular curcumin fluorescence. For FACS analysis, cells were trypsinized, washed with PBS three times, and resuspended in 500 μL sheath fluid (Becton Dickinson, Franklin Lakes, NJ) and examined using FACS-Caliber flow cytometer (Becton Dickinson).
Cytotoxicity of (Cm)NPs towards different cancerous cell lines
HeLa, MCF-7, or HUH-7 cells were plated (1 × 104 cells/well) in triplicate in 96-well sterile microtiter plates (Corning) and allowed to grow for 12 hours for proper cell adherence. Cells were then treated with different concentrations (10–30 μM of free curcumin, (Cm)NPs, and void dipeptide NPs for 24 hours. Cells incubated with 20 μL of PBS were used as a control. After incubation, the media was discarded and replaced with 180 μL of fresh growth medium. Subsequently, 20 μL of MTT reagent, (5 mg/mL) was added to each well. The plate was then incubated at 37μC under 5% CO2 for 4 hours. The medium was removed, and 100 μL of dimethylsulfoxide was added to solubilize the formazan crystals. Optical density was then taken at 572 nm, subtracting for plate absorbance at 690 nm. The percentage of viable cells was calculated based on the following equation:
where “At” is the absorbance of cells treated with void NPs, free curcumin, or (Cm)NPs and “Ac” is the absorbance of untreated cells. Each value is expressed as the mean ± standard deviation.
Cytotoxicity of NPs towards L-929 (human fibroblast) cells
L-929 cells were plated (1 × 104 cells/well) in triplicate in 96-well sterile microtiter plates (Corning) and allowed to grow for 12 hours for proper cell adherence. Cells were then treated with different concentrations (20–50 μM) of void dipeptide NPs for 24 hours. Cells incubated with 20 μL of PBS were used as a control. After incubation, the media was discarded and replaced with 180 μL of fresh growth medium. Subsequently, 20 μL of MTT reagent (5 mg/mL) was added to each well. The plate was then incubated at 37μC under 5% CO2 for 4 hours. The medium was then removed and 100 μL of dimethylsulfoxide was added to solubilize the formazan crystals. Optical density was then taken at 572 nm, subtracting for plate absorbance at 690 nm. The percentage of viable cells was calculated based on the following equation:
where “At” is the absorbance of cells treated with void NPs and “Ac” is the absorbance of the untreated cells. Each value is expressed as the mean ± standard deviation.
Apoptosis assay
HeLa cells were seeded (2.5–3.0 × 105 cells/plate) on 60-mm plates and incubated until 80% confluency was reached. Cells were then treated with 30 μM of free curcumin and the same amount of (Cm)NPs based on the cytotoxicity results, which was used to determine the maximum cell killing efficacy at a concentration of 30 μM. As a control, cells were incubated with complete growth medium. After 24 hours, cells were harvested by quick trypsinization to minimize potentially high annexin V background levels in adherent cells, washed twice with cold PBS, and stained with annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) in binding buffer as directed by the Vybrant apoptosis assay kit (V13241; Invitrogen). Stained cells were placed on ice and protected from light until reading using flow cytometry. Processed single-cell suspensions were analyzed on a FAC-SCaliber flow cytometer with a laser excitation wavelength of 488 nm. The green signal from fluorescein isothiocyanate/annexin V was measured at 525 nm and the red signal from PI was measured at 620 nm. Cells that stained negative for both annexin V and PI were viable. Cells that were annexin V+/PI− were in early apoptosis, whereas cells that were necrotic or in late apoptosis were annexin V+/PI+.
In vivo antitumor efficacy of curcumin-loaded NPs
Male mice (Balb/c), 4–6 weeks old, weighing 18–22 g were housed in a temperature- and light-controlled room at the animal maintenance facility of International Centre of Genetic Engineering and Biotechnology at our institution. Animal experiments were approved by the Institutional Animal Care and Use Committee and were in compliance with all regulatory guidelines. Cultured tumor cells B6F10 (mouse melanoma cells) were detached from flasks using trypsin/EDTA followed by neutralization with RPMI containing 10% FBS and pelleted at 300 × g for 5 minutes. The cells were then washed three times with cold PBS and resuspended in PBS. To initiate tumors, a total of 5.0 × 106 cells (0.1 mL) were injected subcutaneously into the abdomen of 4–6-week-old mice. Palpable solid tumors developed within 10–12 days post-tumor cell inoculation. Once tumor masses became established (as soon as tumor volume reached ∼125 mm3), the animals were randomized to receive either physiological saline, void NPs (at a dose of 20 mg/kg body weight), curcumin-NPs in aqueous solution (at a dose of 25 mg/kg body weight), curcumin alone [at a dose of 25 mg/kg body weight in methanol: water solution (50:50; v/v)] in 100 μL for 28 days via intravenous injection (at 3-day intervals). Tumor sizes were measured using calipers and tumor volume was calculated using the equation:
where L is the length and W is the width of the tumor.
In vivo toxicity studies of dipeptide NPs
Animal were intravenously administered with physiological saline and void NPs in 100 μL (equivalent to the therapeutic dose) for 28 days via intravenous injection (at 2-day intervals) and monitored continuously for any behavioral changes and body mass loss. After these animals were sacrificed, organs were excised, sectioned, and studied for morphological changes using hematoxylin and eosin staining.
Statistical analysis
The results are presented as the mean ± SD calculated over at least three data points.
Results and discussion
Preparation and characterization of NPs
Self-assembly can be used to produce a variety of NPs with characteristics such as high stability, biocompatibility, and high therapeutic loading of drug molecules, which is useful for various biological applications.22–27 Several reports, including our own studies, have shown that dipeptides, either free or with N- or C- terminal modifications, can self-assemble into well-defined NPs.18,19,28–30 However, most of these studies have shown assembly in an aqueous environment and may not be suitable for delivering hydrophobic bioactive molecules. Thus, we attempted to develop dipeptide-based nanostructures, which can be prepared in organic or aqueous–organic environments, for potential loading of hydrophobic drugs. Curcumin was the model hydrophobic drug for this study.
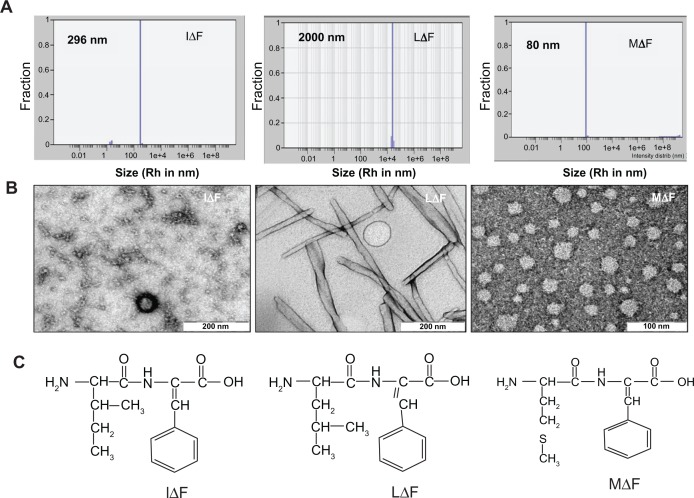
HFIP is a commonly used solvent for preparing small peptide-based self-assembled nanostructures.19,29 Although this solvent is excellent for dissolving peptides, it may not be suitable if the self-assembled structures are to be developed for in vivo studies due to high toxicity.31 Therefore, we sought to replace HFIP with a more suitable solvent. Since curcumin is soluble and retains all of its activities in metha-nol,32,33 which is a pharmaceutically acceptable solvent and has been used to prepare various drug nanoformulations,34,35 we first investigated whether ΔPhe-containing dipeptides would self-assemble in methanol. For this purpose, we selected previously described self-assembling dehydrodipeptides, such as phenylalanine–dehydrophenylalanine (FΔF), glutamic acid-dehydrophenylalanine (EΔF), and lysine-dehydrophenylalanine (KΔF)18,19,29 as well as newly synthesized molecules such as MΔF, IΔF, and LΔF (Table S1) and examined their assembly behavior using DLS. Surprisingly, none of these peptides assembled in neat methanol; however, all assembled when their HFIP solution was diluted with water. While the mechanistic details of self- assembly are not well-understood, it is possible that hydrophobic clustering followed by nucleation at organic-water interfaces formed by addition of amphiphilic peptides molecules to an aqueous/organic mixture of water miscible organic solvents36 may lead to peptide self-assembly. However, while some reports have shown demonstrated self-assembly of relatively longer peptides in methanol and in other organic solvents,37,38 we speculate that simple dipeptides may not provide enough hydrophobicity for self-assembly in methanol. We then used several combinations of methanol and water for peptide assembly, in which curcumin also showed reasonable solubility, and a mixture of methanol: water (50:50, v/v) (Figure 2). Of all the dehydrodipeptides tested, only MΔF, IΔF, and LΔF formed stable, self-assembled, and monodispersed NPs (Table S2 and Figure 3A) in a mixture of methanol and water (50:50; v/v). LΔF formed large, visible aggregates with hydrodynamic radii (Rh) in the micrometer range, whereas IΔF formed NPs with Rh of approximately 290 nm and a polydispersity index (PDI) of 0.16. The mean radius of MΔF nanoparticles was found to be approximately 80 nm, with a narrow size distribution and PDI of 0.15.
Figure 2.
Scheme showing formation of curcumin-loaded dipeptide NPs.
Abbreviations: MeOH, Methanol; MQ, Milli-Q water; NPs, nanoparticles; RT, room temperature.
Figure 3.
Self-assembly of dipeptides into NPs in aqueous solution containing 50% methanol (v/v). (A) Mean particle size of MΔF, LΔF, and IΔF measured using DLS [Rh (MΔF) = 80 nm; Rh (IΔF) = 296 nm; Rh (LΔF) = 2000 nm]. (B) TEM images of dipeptide NPs. (C) Molecular structure of the dipeptides.
Abbreviations: DLS, dynamic light scattering; IΔF, isoleucine-dehydrophenylalanine; LΔF, leucine-dehydrophenylalanine; MΔF, methionine-dehydrophenylalanine; NPs, nanoparticles; Rh, hydrodynamic radius; TEM, transmission electron microscopy.
TEM showed that under the same assembly conditions, all three dipeptides exhibited different assembly and morphology (Figure 3B). IΔF formed micelle-like structures with an average diameter of 20 nm, whereas LΔF, which has an identical molecular mass and differs only in the position of a methyl group on the side chain of the N-terminal amino acid, assembled into fibrillar structures, with diameters in the nanometer range. MΔF formed relatively large vesicular structures with a mean diameter of ∼40 nm, and, like other reported polymeric NPs of similar dimensions,39–42 may be suitable for curcumin-loading and delivery. In most cases, DLS sizes of dipeptide NPs were higher than their TEM sizes. This is reasonable because in light-scattering studies, the average hydrodynamic radii of particles are often affected by a particle’s surface charge and interactions with the water shell. These factors contribute to an overall increase in particle size, resulting in different particle size measurements using TEM and DLS.43,44
Curcumin loading and release from dipeptide NPs
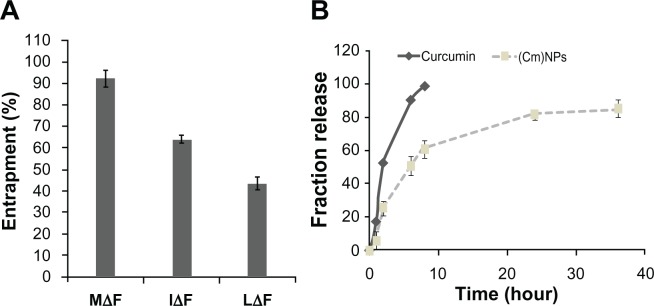
The efficacy of highly hydrophobic curcumin appears to depend directly on its encapsulation in a nanocarrier.9,12 After morphological characterization, dipeptide NPs were investigated for their ability to load curcumin. We found that the three dipeptide NPs could successfully load curcumin, but the extent of drug-loading and loading efficiency varied (Figure 4A). NPs of LΔF showed ∼8% (w/w) loading, with an efficiency of ∼40% compared to ∼28% (w/w) loading with ∼60% efficiency in NPs constructed from IΔF. However, MΔF nanoparticles showed a maximum drug loading of ∼30% (w/w), with an efficiency of (∼92%) (Table 1). Curcumin loading in dipeptide NPs after NP formation may be related to possible hydrophobic interactions between the drug and the dipeptides, as well as aromatic interactions between curcumin and the ΔPhe residue of the dipeptides. Moreover, the percentage of curcumin loading and efficiency of dipeptide NPs also appeared to depend on their overall morphology; vesicular structures formed by MΔF were more suitable for entrapping the hydrophobic drug compared to the tubular or micelle-like structures formed by LΔF and IΔF, respectively, but the exact reasons for the observed difference in drug entrapment are not clear. However, there are several reports of carbon nanotubes and other polymeric tubular and vesicular NPs used as carriers.45–48 Drug entrapment in peptide-based NPs is a relatively unexplored area, and detailed investigations are required to understand the structure–function relationship of peptide-based NPs as carriers of drug-like molecules. Due to high drug-loading efficiency, curcumin-loaded MΔF nanoparticles remained completely dispersed in aqueous media even after the evaporation of methanol, in contrast to native curcumin that precipitated in the water upon removal of the methanol from the mixture. Similar enhancement in aqueous dispersion of curcumin has been demonstrated in several previous studies based on polymeric NP-mediated curcumin delivery.42,49,50
Figure 4.
Entrapment and release of curcumin in different dipeptide NPs. (A) Percentage entrapment of curcumin in dipeptide NPs. Out of the three dipeptide NPs, MΔF showed highest degree of entrapment at ∼92%. (B) Time-dependent release of curcumin from (Cm)NPs.
Notes: Error bars represent mean ± standard deviation (±SD) (n = 3).
Abbreviations: (Cm)NPs, curcumin-loaded nanoparticles; MΔF, methionine-dehydrophenylalanine; NPs, nanoparticles.
Table 1.
Percentage drug-loading and loading efficiency in dipeptide NPs
| S no | Dipeptide | Curcumin loading (% w/w) | Loading efficiency (%) |
|---|---|---|---|
| 1 | MΔF | 35 | 92 |
| 2 | IΔF | 28 | 60 |
| 3 | LΔF | 8 | 40 |
Notes: Drug loading was measured after dissolving curcumin-loaded NPs in methanol and estimating curcumin concentration using UV-vis spectroscopy at 425 nm. Values are the average of three independent readings.
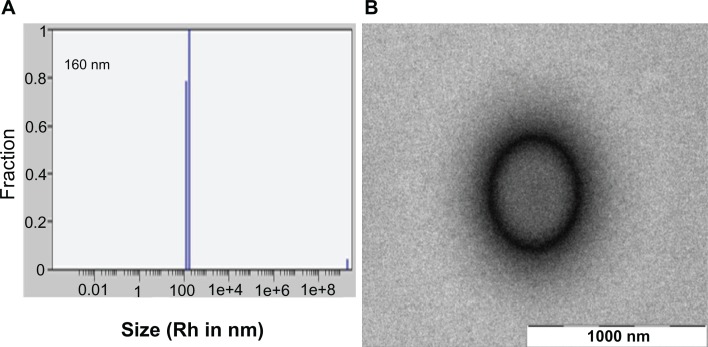
Generally dipeptide NPs are obtained by the process of molecular self-assembly, mainly driven by noncovalent interactions such as van der Waals, electrostatic, and hydrophobic interactions as well as hydrogen bonds.19,29,51 Hydrophobic interactions in particular can play a crucial role in stabilizing peptide-based nanostructures.52,53 Thus, it is possible that curcumin, a hydrophobic molecule, may influence the assembly and overall morphology of MΔF nanoparticles. To address this issue, the stability of (Cm)NPs was determined using DLS and TEM. Light scattering studies showed that addition of curcumin did not affect stability, the particulate nature, or polydispersity index of the dipeptide NPs. In DLS studies, (Cm)NPs exhibited an Rh of approximately 160 nm and PDI of 0.1 (Figure 5A), which is comparable to other reported nanoformulations of curcumin used for delivery purposes.39–42 Since tumors, due to their leaky vasculature and poor lymphatic drainage, usually take up 10–500-nm particles,26,54 the given size of (Cm) NPs is advantageous for their passive targeting to tumor tissues. TEM analysis further revealed that (Cm)NPs were much more regular and stable compared to those without the drug (Figures 5B vs 3B). The hydrophobic nature of curcumin may have further stabilized the nanostructures rather than disrupting them. However, this hypothesis must be confirmed by further studies.
Figure 5.
DLS and TEM analysis of (Cm)NPs. (A) Size distribution histogram and distribution analysis of (Cm)NPs determined using DLS. (B) TEM image of (Cm)NPs.
Note: Scale bar, 1000 nm.
Abbreviations: (Cm)NPs, curcumin-loaded nanoparticles; DLS, dynamic light scattering; TEM, transmission electron microscopy.
A nanoparticle should not only exhibit good drug-loading efficiency but should also release the drug in a controlled manner in order to provide sustained delivery.55 Release of curcumin from (Cm)NPs and from methanolic curcumin solution was followed over a period of 40 hours. A methanol: water solution of curcumin showed fast release from the dialysis bag, with almost 90% of the drug released by 6 hours, and complete release within 8 hours, whereas the release of curcumin from NPs occurred in a controlled manner, with just 50% drug being released by 6 hours. Even after 36 hours, the NPs retained nearly 15% of the drug, suggesting that curcumin molecules were well-encapsulated within the dipeptide NPs (Figure 4B). Such a slow release pattern is desirable as has been demonstrated in other polymeric or liposomal nanoformulations of curcumin.42,49
To address whether the ΔF in MΔF plays any crucial role in the formation, stability,18,19 and subsequent high degree of curcumin entrapment in MΔF NPs, we synthesized MF, which contains L-phenylalanine rather than ΔF and investigated its self-assembly behavior as well as other characteristics. DLS showed that MF formed some nanostructures (Rh of 2 nm) in a 50% methanol: water solution (Figure S1); few small irregular nanostructures were observed in TEM (data not shown). Results of curcumin entrapment experiments showed that MF nanoparticle could only entrap approximately 20% (w/w) of the total drug added, compared to approximately 90% entrapment for MΔF nanoparticles. Clearly, the presence of ΔF in MΔF remarkably not only promoted formation and stability of self-assembled dipeptide NPs, but also enhanced curcumin entrapment efficiency, signifying the importance of the ΔF residue in promoting dipeptide self-assembly18,19 and enhancing the potential of the resulting NPs as drug carriers.
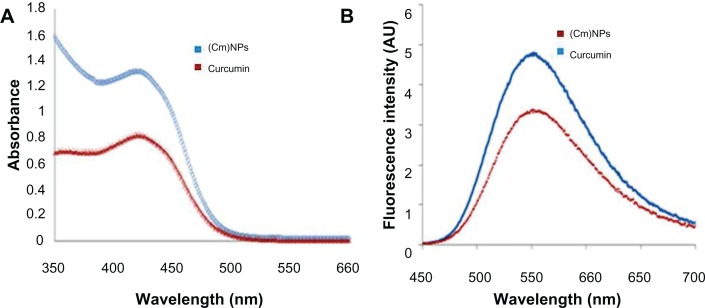
Spectroscopic studies were carried out to investigate the effect of MΔF-nanoparticles on the photophysical properties of curcumin. UV-vis spectroscopy of a methanol: water solution (50:50; v/v) of curcumin showed an absorption maximum at 423 nm. An aqueous methanol solution of (Cm) NPs also showed a similar spectrum (absorption maximum at 421 nm), suggesting that entrapment in MΔF-nanoparticles had no effect on the photophysical properties of curcumin (Figure 6A). The fluorescence spectra of both curcumin and (Cm)NPs (from 450 to 700 nm) in methanol: water (50% v/v) at an excitation wavelength of 420 nm showed sharp emission peaks at 548 nm (Figure 6B). However, the observed change in the fluorescence intensity of (Cm)NPs compared to native curcumin (3.4 × 106 vs 4.8 × 106) suggested robust entrapment of the molecule inside the hydrophobic domains of MΔF NPs.56,57
Figure 6.
(A) Ultraviolet–visible absorbance spectra of curcumin and (Cm)NPs in aqueous methanol (50% v/v). (B) Fluorescence emission spectra of curcumin and (Cm)NPs in an aqueous solution containing 50% methanol excited at 420 nm.
Note: These results show that curcumin retains its photophysical properties when entrapped in MΔF-nanoparticles.
Abbreviations: (Cm)NPs, curcumin-loaded nanoparticles; MΔF, methionine-dehydrophenylalanine.
Cellular uptake of dipeptide NPs
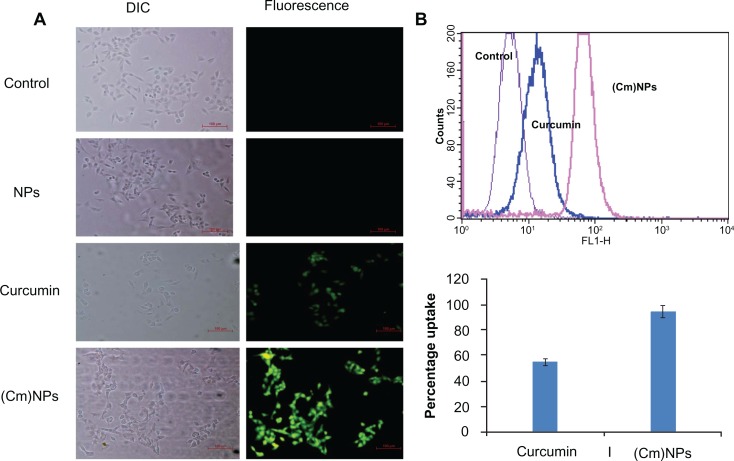
In addition to its many therapeutic uses, such as its antioxidant properties and anti-Alzheimer and antimalarial activities,1,3,7,11,58,59 curcumin has also shown to have promising chemopreventive and chemotherapeutic activities against cancer.4,5,7 Although Phase I clinical trials have shown curcumin to be a safe drug even at high doses, poor bioavailability and suboptimal pharmacokinetics largely restrained its anticancer activity in preclinical and clinical models.60,61 While free curcumin has been shown to penetrate cancerous cells to some extent, like many other potent drugs, its cellular uptake is limited due to its highly hydrophobic nature.9,62 To test the potential of MΔF-nanoparticles as carriers for enhancing cellular availability of curcumin, uptake of (Cm)NPs was investigated. HeLa cells were incubated with curcumin-loaded NPs as well as free curcumin for a period of 24 hours. Since curcumin retained its intrinsic fluorescence activity when entrapped in dipeptide NPs (Figure 6B), its cellular uptake was analyzed using fluorescence microscopy and flow cytometric analysis. Fluorescence microscope images showed that cells treated with (Cm)NPs exhibited high fluorescence intensity compared to curcumin alone, suggesting enhanced curcumin uptake (Figure 7A). Quantitative analysis of cellular uptake, carried out using flow cytometry, showed twofold higher uptake of (Cm)NPs compared to free curcumin (∼95% vs ∼50%; Figure 7B). The observed higher uptake of (Cm)NPs may be due to endocytotic pathways, which have been shown to function in the cellular uptake of other nanoparticulate materials.9,49,59
Figure 7.
Cellular uptake of (Cm)NPs and native curcumin. (A) Fluorescence microscopic image. (B) Flow cytometric analysis showing cellular uptake of (Cm)NPs and native curcumin at a concentration of 30 μM. Loading curcumin in the MΔF-nanoparticles enhanced its cellular uptake by twofold.
Note: Error bars represent the mean ± SD (n = 3).
Abbreviations: (Cm)NPs, curcumin-loaded nanoparticles; MΔF, methionine-dehydrophenylalanine.
Effect of curcumin-loaded NPs on the rate of proliferation/survival of different carcinoma cell lines
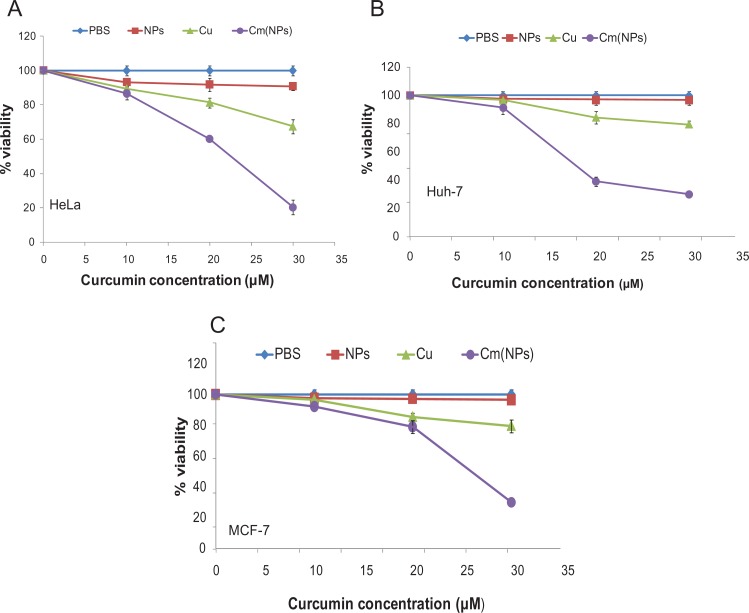
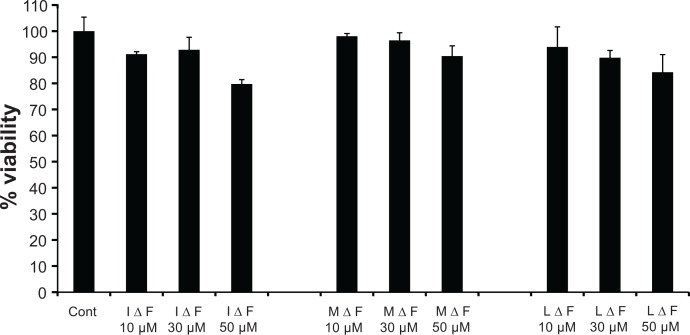
For any NP-based therapy to be effective, it is necessary that the drug molecules entrapped in NPs should retain their intrinsic activity. To test this, the efficacy of (Cm)NPs was determined in various cancer cell lines. Cells were incubated with (Cm)NPs for 24 hours, and their viability was determined using the MTT assay. MTT is a pale yellow substrate which is cleaved by living cells to yield dark blue formazan to reflect active cell proliferation/survival. Free curcumin and void NPs were used as controls. Cancer cells treated with increasing (Cm)NP concentrations (10–30 μM) showed that (Cm)NPs exhibited concentration-dependent toxicity (Figure 8). HeLa, MCF-7, and Huh-7 cells treated with 30 μM of (Cm)NPs showed significant growth inhibition, with percentage viabilities in range of 20%–30%, whereas those treated with curcumin alone showed high percentage viabilities in all cases (HeLa, ∼70%; MCF-7, ∼80%; Huh, ∼80%). At similar concentration (10–30 μM), void peptide NPs were nontoxic to the tested cancer cell lines. The NPs also exhibited no overall cytotoxic effect on human fibroblast (Figure 9) cells treated with void NPs at similar or higher concentration (ranging from 10–50 μM), demonstrating their suitability as a drug carrier. This enhanced cytotoxic effect by (Cm)NPs can be attributed to their differential cellular uptake compared to native curcumin. Generally, internalization of native curcumin is diffusion-mediated, which is restricted after achieving saturation in the cytoplasm, leading to a low drug uptake and a short activity period.63 It is likely that the possible involvement of the endocytotic pathway mediated enhanced uptake of (Cm)NPs, along with their sustained drug-release behavior, resulting in an increased period of the drug’s cellular availability and elevated activity. Although similar enhancement in the cytotoxicity of curcumin-loaded NPs has been reported previously,9–12 the described NPs were mostly derived from synthetic polymers, which themselves can be toxic to cellular systems.64 The MΔF-nanoparticles described here may offer an added advantage of high biocompatibility with no cellular toxicity.
Figure 8.
Toxicity of native curcumin and (Cm)NPs towards different cancer cells. Cells were treated with different concentrations of curcumin, (Cm)NPs, PBS, and void NPs for 24 h and tested for cell viability using the MTT assay (see Materials and methods). (A) HeLa cells; (B) Huh-7 cells; (C) MCF-7 cells.
Notes: Error bars represent (mean ± SD) (n = 3). Curcumin when loaded in dipeptide NPs exhibited enhanced toxicity to cancer cells compared to the free drug.
Abbreviations: (Cm)NPs, curcumin-loaded nanoparticles; NPs, nanoparticles.
Figure 9.
Toxicity of dipeptide NPs towards human fibroblast (L-929) cell line.
Abbreviation: NPs, nanoparticles.
Mode of (Cm)NPs-induced cancer cell death
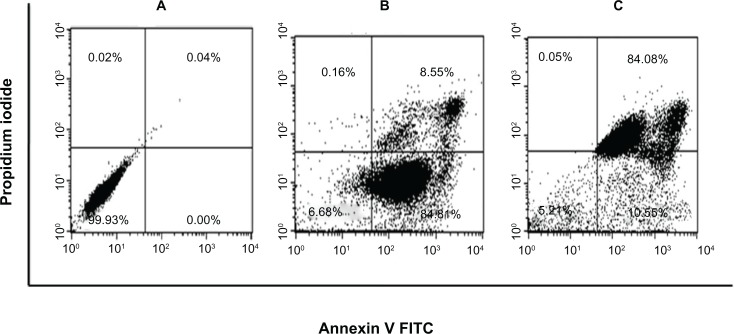
Curcumin is known to exhibit its antitumor activity by inducing apoptotic pathways.65–67 To investigate whether (Cm)NPs also induced cell death in tumor tissues through the same mechanism, we investigated the apoptosis-inducing ability of (Cm)NPs in HeLa cells using annexin V/PI staining.68 Annexin V binds to cells that express phosphatidylserine on the outer layer of the cell membrane, a characteristic feature of cells entering apoptosis.69 This allows discrimination of live cells (unstained by both fluorochromes) from apoptotic cells (stained with annexin V) and necrotic or late apoptotic pathway (stained with both annexin V and PI). Native cur-cumin treated cells demonstrated that ∼84% of cells were in the apoptotic stage and ∼8% of the cells were in late apoptosis, whereas those treated with (Cm)NPs showed that only ∼11% of cells were in apoptosis versus 84% in a late apoptotic stage (Figure 10). These results suggest that like native curcumin, (Cm)NPs induced apoptotic pathways. However, cells treated with native curcumin showed a delayed apoptotic response compared to (Cm)NPs. Moreover, the finding that much higher population of cells were found in the late apoptotic stage in the case of (Cm)NP-treated cells, (Cm)NPs showed enhanced cytotoxicity compared to native curcumin. A similar mode of curcumin-induced death of HepG2 cells was observed previously when curcumin was loaded into chitosan/poly(butyl cyanoacrylate) NPs.55
Figure 10.
Induction of apoptosis in HeLa cells treated with (Cm)NPs and native curcumin. (A) PBS, (B) Free curcumin, (C) (Cm)NPs.
Notes: Results showed that similarly to native curcumin, (Cm)NPs induces apoptosis in a higher population of cells in the late apoptotic stage.
Abbreviations: (Cm)NPs, curcumin-loaded NPs; PBS, phosphate-buffered saline.
Tumor regression studies in B6F10 melanoma model
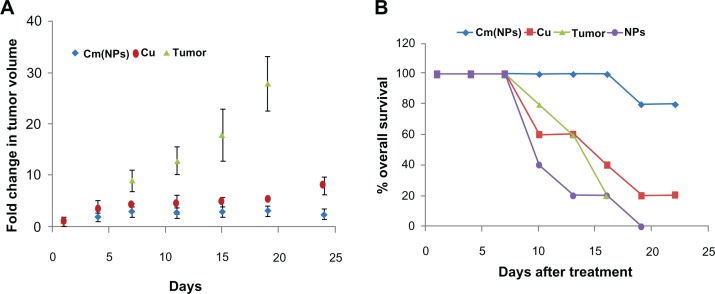
Tumor growth was induced by subcutaneous injection of Balb/c mice with B6F10 melanoma cells70 and animals were treated with (Cm)NPs, native curcumin, void NPs, and saline. The results of tumor volume changes as a function of time are shown in Figure 11A. Though treatment with (Cm)NPs did not result in tumor abolition, B6F10 xenograft growth was retarded compared to the results observed with native curcumin. However, treatment with (Cm)NPs significantly increased the percentage overall survival of melanoma tumor-bearing animals. We could not compare tumor regression results of (Cm)NPs with control or mice treated with void dipeptide NPs because of the death of these animals during the treatment period (Figure 11A).
Figure 11.
Treatment with (Cm)NPs inhibits tumor growth in mice without toxicity. To initiate tumors, a total of 5.0 × 106 cells (0.1 mL) were injected subcutaneously into the abdomen of 4–6 weeks old Balb/c mice. Palpable solid tumors developed within 10–12 days post-tumor cell inoculation. Once tumor masses became established, animals were treated with intravenous injections (at 3-day intervals) with either saline, void NPs, free curcumin, or (Cm)NPs (n = 5 mice/group). (A) Tumor volume was calculated from measurements obtained by calipers on each day of treatment. (B) Percentage survival of animals treated with the above formulations. Animals receiving (Cm)NPs, showed retarded tumor growth and higher percentage of survival.
Notes: Error bars represent the mean ± SD (n = 3).
Abbreviations: (Cm)NPs, curcumin-loaded nanoparticles; NPs, nanoparticles.
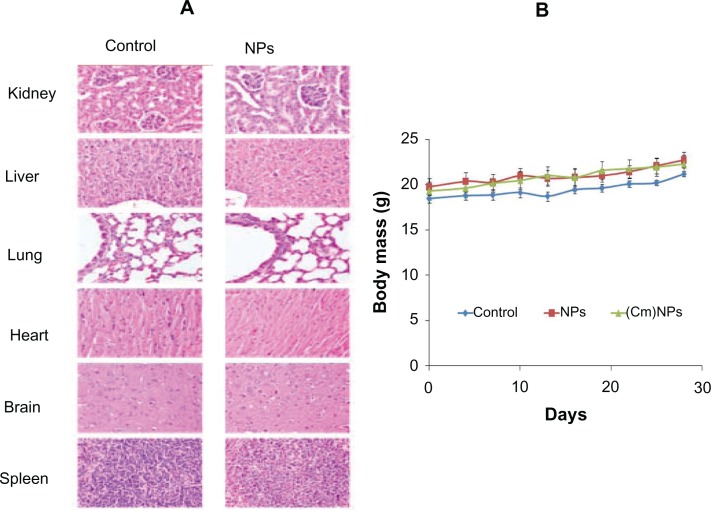
No behavioral abnormalities or weight loss was observed during the course of therapy in any of the mice receiving empty dipeptide NPs (Figure 12). Moreover, since only a dipeptide was used, there is a minimal chance of eliciting an immune response or undesirable side effect using these systems alone. Nanofibers of even longer peptides (10–30 residues) have been shown to elicit no immune response by themselves.71 The main advantages of dipeptide NPs are that they are easy to make, cost-effective, biocompatible, and biodegradable. Treatment with curcumin NPs resulted in retarded tumor growth and increased longevity of melanoma tumor-bearing animals.
Figure 12.
In vivo toxicity study of MΔF-nanoparticles towards Balb/c mice. Animals were intravenously administered (every third day) with 100 μL (equivalent to therapeutic dose) of physiological saline, void NPs, or (Cm)NPs at 100 μL for 28 days. After these animals were sacrificed, their organs were excised, sectioned, and examined for morphological changes. (A). Histological analysis of explanted organs kidney, liver, lung, heart, brain, and spleen using hematoxylin and eosin staining of control group and NP-treated group indicated no signs of cellular or tissue damage. (B). Change in body weight of Balb/c mice treated with the dipeptide NPs. Animals treated with NPs and (Cm)NPs showed no behavioral changes or weight loss during the treatment.
Abbreviations: (Cm)NPs, curcumin-loaded nanoparticles; MΔF, methionine-dehydrophenylalanine; NPs, nanoparticles.
Conclusion
In the present work, we described the synthesis and characterization of novel self-assembled dipeptide NPs prepared in aqueous methanol (50% v/v). The main advantage of these biocompatible NPs is that they can be synthesized in a single-step process under conditions which allow adsorption and loading of a model hydrophobic drug such as curcumin. Additionally, curcumin-loaded NPs showed improved cellular uptake and enhanced anti-proliferative activity towards different cancerous cell lines. Thus, these novel dipeptide NPs have the ability to improve curcumin’s bioavailability, making it amenable to intravenous dosing without alteration of its anticancer properties. Moreover, treatment with curcumin NPs resulted in retarded tumor growth and increased longevity of melanoma tumor-bearing animals. Because it is peptidic in origin, the dipeptide NPs described here offer an added advantage of high biocompatibility. These properties, combined with the ease of synthesis, which dipeptide NPs attractive alternatives for future development as effective nanocarriers of drugs, particularly those with high hydrophobicity.
Acknowledgments
SA thanks Indian Council of Medical Research for fellowship and JJP thanks Unesco-Lo’real for Young Women in Science Program for providing a fellowship. The authors thank the core funding at the International Centre for Genetic Engineering and Biotechnology, New Delhi, Department of Biotechnology and Department of Science and Technology, India, for financial assistance. We also thank Ankur Varshaney, ICGEB, New Delhi, for help with mass spectrometric analysis.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
Supplementary information
Supplementary information 1.
Synthesis of methionine-dehydrophenylalnine (MΔF), leucine-dehydrophenylalanine (LΔF), and isoleucine-dehydrophenylalanine (IΔF)
Briefly, Boc-Met-OH (5 mM; 1.24 g) was dissolved in dry THF, chilled to −20μC and stirred in an ice-salt bath. Equivalent moles of N-methylmorpholine (5 mM; 0.655 mL) and isobutyl chloroformate (5 mM; 0.69 mL) were added to the solution. After 10 min, a precooled aqueous solution of DL-threo-β-phenylserine (5.5 mM; 0.99 g) and sodium hydroxide (5.5 mM; 0.22 g) were added. The reaction mixture was stirred overnight and concentrated in vacuo. The residue was acidified with citric acid and extracted using ethyl acetate. The ethyl acetate layer was washed three times with water and saturated sodium chloride, dried over anhydrous sodium sulfate, and evaporated to obtain Boc-Met-DL-threo-β-phenylserine, which was then mixed with anhydrous sodium acetate (6.5 mM; 0.58 g) in freshly distilled acetic anhydride (50 mL) and stirred for 36 hours. The thick slurry was poured over crushed ice and the precipitate was filtered, washed with 5% NaHCO3 followed by cold water, and dried in vacuo. The resulting compound (Boc-Met-ΔPhe-azalactone) was dissolved in methanol, treated with 1.5 equivalents of 1 N NaOH solution for 3–4 hours, and concentrated in vacuo. Acidification with solid citric acid liberated the peptide, which was extracted in ethyl acetate to yield Boc-Met-ΔPhe-COOH. The Boc group was deprotected using anhydrous tetrahydrofuran (TFA). The peptide was purified on a preparative reverse-phase C18 column (Delta-Pak, Waters Co, Milford, MA; 15 μm, internal diameter, 300 × 19 mm) using an acetonitrile-water linear gradient 5%–95% acetonitrile (0.1% TFA)/water (0.1% TFA) at a flow rate of 4 mL/minute over 55 minute on a high-performance liquid chromatography (HPLC) system [LC-6 AD, liquid chromatograph, Shimadzu, Kyoto, Japan)]. The purified peptide was reinjected into an analytical reverse phase C18 column (Phenomenex, Hyderabad, India, C18, 5 μm, internal diameter, 250 × 4.6 mm) using an acetonitrile-water linear gradient of 5%–95% acetonitrile (0.1% TFA)/water (0.1% TFA) at a flow rate of 1 mL/min over 45 min on a HPLC system (LC-10 AD; Shimadzu) and was found to be 98% pure with a retention time of 23 min. The purified peptide was analyzed using mass spectrometry [AppliedBiosystems QStar (Q-TOF)]. Observed mass: 295.139 Da; expected mass: 294.407 Da.
Table S1.
Yield of synthesis, retention time, and observed and expected masses of MΔF, LΔF, IΔF, and MF
| Dipeptide | Yield | Retention time (min) | Expected mass (Da) | Observed mass (Da) |
|---|---|---|---|---|
| MΔF | 1.5 g, 60% | 23 | 294.407 | 295.139 |
| IΔF | 1.1 g, 42% | 45 | 279 | 276.370 |
| LΔF | 1.2 g, 40% | 24 | 276 | 276.23 |
| MF | 0.6 g, 48% | 21 | 296.3 | 296 |
Table S2.
Morphology and average size of nanoparticles determined using dynamic light scattering and transmission electron microscopy analysis
| S no | Dipeptide | DLS (Rh in nm) | PDI | TEM (mean D in nm) |
|---|---|---|---|---|
| 1 | MΔF | 80 | 0.15 | Vesicle (40) |
| 2 | IΔF | 296 | 0.16 | Vesicle (20) |
| 3 | LΔF | 2000 | 0.25 | Tube (D 30 nm and L 2100 nm) |
Abbreviations: DLS, dynamic light-scattering; PDI, Polydispersity Index; TEM, transmission electron microscopy.
Other dipeptides such as leucine-dehydrophenylalanine (LΔF) and isoleucine-dehydrophenylalanine (IΔF) were synthesized and purified using the same procedure described above, starting from Boc-Leu-OH and Boc-Ile-OH respectively (Supplementary Table 1).
Supplementary information 2.
Synthesis of methionine-phenylalanine (MF)
Briefly, Boc-Met-OH (5 mM; 1.24 g) was dissolved in dry THF, chilled to −20μC, and stirred in an ice-salt bath. Equivalent moles of N-methylmorpholine and isobutyl chloroformate were added to the solution. After 10 min, a precooled aqueous solution of NH2-Phe-OH (5.5 mM; 0.90 g) and sodium hydroxide (5.5 mM; 0.22 g) were added. The reaction mixture was stirred overnight and concentrated in vacuo. The residue was acidified with citric acid and extracted with ethyl acetate. The ethyl acetate layer was washed three times with water and saturated sodium chloride, dried over anhydrous sodium sulfate, and evaporated to obtain Boc-Met-Phe. The Boc group was deprotected using anhydrous tetrahydrofuran. The peptide was purified on a preparative reverse-phase C18 column as described above. The purified peptide was analyzed using mass spectrometry [AppliedBiosystems QStar (Q-TOF)]. Observed mass: 296 Da; expected mass: 296.3 Da.
Mean particle size of methionine–phenylalanine (MF) in methanol:water solution (1:1) measured using dynamic light-scattering (Rh = 2 nm).
References
- 1.Brouet I, Ohshima H. Curcumin, an anti-tumour promoter and anti-inflammatory agent, inhibits induction of nitric oxide synthase in activated macrophages. Biochem Biophys Res Commun. 1995;206(2):533–540. doi: 10.1006/bbrc.1995.1076. [DOI] [PubMed] [Google Scholar]
- 2.Ammon HP, Wahl MA. Pharmacology of Curcuma longa. Planta Med. 1991;57(1):1–7. doi: 10.1055/s-2006-960004. [DOI] [PubMed] [Google Scholar]
- 3.Maheshwari RK, Singh AK, Gaddipati J, Srimal RC. Multiple biological activities of curcumin: a short review. Life Sci. 2006;78(18):2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Rao CV, Rivenson A, Simi B, Reddy BS. Chemoprevention of colon carcinogenesis by dietary curcumin, a naturally occurring plant phenolic compound. Cancer Res. 1995;55(2):259–266. [PubMed] [Google Scholar]
- 5.Moragoda L, Jaszewski R, Majumdar AP. Curcumin induced modulation of cell cycle and apoptosis in gastric and colon cancer cells. Anticancer Res. 2001;21(2A):873–878. [PubMed] [Google Scholar]
- 6.Mehta K, Pantazis P, McQueen T, Aggarwal BB. Antiproliferative effect of curcumin (diferuloylmethane) against human breast tumor cell lines. Anticancer Drugs. 1997;8(5):470–481. doi: 10.1097/00001813-199706000-00010. [DOI] [PubMed] [Google Scholar]
- 7.Kuttan R, Bhanumathy P, Nirmala K, George MC. Potential anticancer activity of turmeric (Curcuma longa) Cancer Lett. 1985;29(2):197–202. doi: 10.1016/0304-3835(85)90159-4. [DOI] [PubMed] [Google Scholar]
- 8.Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007;4(6):807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 9.Kunwar A, Barik A, Pandey R, Priyadarsini KI. Transport of liposomal and albumin loaded curcumin to living cells: an absorption and fluorescence spectroscopic study. Biochim Biophys Acta. 2006;1760(10):1513–1520. doi: 10.1016/j.bbagen.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Sou K, Inenaga S, Takeoka S, Tsuchida E. Loading of curcumin into macrophages using lipid-based nanoparticles. Int J Pharm. 2008;352(1–2):287–293. doi: 10.1016/j.ijpharm.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 11.Kumar V, Lewis SA, Mutalik S, Shenoy DB, Venkatesh, Udupa N. Biodegradable microspheres of curcumin for treatment of inflammation. Indian J Physiol Pharmacol. 2002;46(2):209–217. [PubMed] [Google Scholar]
- 12.Tiyaboonchai W, Tungpradit W, Plianbangchang P. Formulation and characterization of curcuminoids loaded solid lipid nanoparticles. Int J Pharm. 2007;337(1–2):299–306. doi: 10.1016/j.ijpharm.2006.12.043. [DOI] [PubMed] [Google Scholar]
- 13.Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007;2(8):469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 14.Hoet PH, Brüske-Hohlfeld I, Salata OV. NPs – known and unknown health risks. J Nanobiotechnology. 2004;2(1):12. doi: 10.1186/1477-3155-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendel D, Ellman JA, Chang Z, Veenstra DL, Kollman PA, Schultz PG. Probing protein stability with unnatural amino acids. Science. 1992;256(5065):1798–1802. doi: 10.1126/science.1615324. [DOI] [PubMed] [Google Scholar]
- 16.English ML, Stammer CH. The enzyme stability of dehydropeptides. Biochem Biophys Res Commun. 1978;83(4):1464–1467. doi: 10.1016/0006-291x(78)91385-2. [DOI] [PubMed] [Google Scholar]
- 17.Adessi C, Soto C. Converting a peptide into a drug: strategies to improve stability and bioavailability. Curr Med Chem. 2002;9(9):963–978. doi: 10.2174/0929867024606731. [DOI] [PubMed] [Google Scholar]
- 18.Panda JJ, Kaul A, Alam S, Babbar AK, Mishra AK, Chauhan VS. Designed peptides as model self-assembling nanosystems: characterization and potential biomedical applications. Ther Deliv. 2011;2:193–204. doi: 10.4155/tde.10.93. [DOI] [PubMed] [Google Scholar]
- 19.Mishra A, Panda JJ, Basu A, Chauhan VS. Nanovesicles based on self-assembly of conformationally constrained aromatic residue containing amphiphilic dipeptides. Langmuir. 2008;24(9):4571–4576. doi: 10.1021/la7034533. [DOI] [PubMed] [Google Scholar]
- 20.Sahu A, Bora U, Kasoju N, Goswami P. Synthesis of novel biodegradable and self-assembling methoxy poly(ethylene glycol)-palmitate nanocarrier for curcumin delivery to cancer cells. Acta Biomater. 2008;4(6):1752–1761. doi: 10.1016/j.actbio.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Mulik R, Mahadik K, Paradkar A. Development of curcuminoids loaded poly(butyl) cyanoacrylate nanoparticles: Physicochemical characterization and stability study. Eur J Pharm Sci. 2009;37(3–4):395–404. doi: 10.1016/j.ejps.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Koria P, Yagi H, Kitagawa Y, et al. Self-assembling elastin-like peptides growth factor chimeric nanoparticles for the treatment of chronic wounds. Proc Natl Acad Sci U S A. 2011;108(3):1034–1039. doi: 10.1073/pnas.1009881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CH, In HJ, Takahashi S, et al. Assembling nanoparticle catalysts with nanospheres for periodic carbon nanotube structure growth. Nanotechnology. 2011;22(3):035301. doi: 10.1088/0957-4484/22/3/035301. [DOI] [PubMed] [Google Scholar]
- 24.Daniel MC, Tsvetkova IB, Quinkert ZT, et al. Role of surface charge density in nanoparticle-templated assembly of bromovirus protein cages. ACS Nano. 2010;4(7):3853–3860. doi: 10.1021/nn1005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chhabra R, Sharma J, Liu Y, Rinker S, Yan H. DNA self-assembly for nanomedicine. Adv Drug Deliv Rev. 2010;62(6):617–625. doi: 10.1016/j.addr.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Ko YT, Kale A, Hartner WC, Papahadjopoulos-Sternberg B, Torchilin VP. Self-assembling micelle-like nanoparticles based on phospholipid-polyethyleneimine conjugates for systemic gene delivery. J Control Release. 2009;133(2):132–138. doi: 10.1016/j.jconrel.2008.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zharov VP, Kim JW, Curiel DT, Everts M. Self-assembling nanoclusters in living systems: application for integrated photothermal nanodiagnostics and nanotherapy. Nanomedicine. 2005;1(4):326–345. doi: 10.1016/j.nano.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300(5619):625–627. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- 29.Gupta M, Bagaria A, Mishra A, et al. Self-assembly of a dipeptide-containing conformationally restricted dehydrophenylalanine residue to form ordered nanotubes. Adv Mater. 2007;19:858–861. [Google Scholar]
- 30.Mahler A, Reches M, Rechter M, Cohen S, Gazit E. Rigid, self-assembled hydrogel composed of a modified aromatic dipeptide. Adv Mater. 2006;18:1365–1370. [Google Scholar]
- 31.Capone R, Quiroz FG, Prangkio P, et al. Amyloid-beta-induced ion flux in artificial lipid bilayers and neuronal cells: resolving a controversy. Neurotox Res. 2009;16(1):1–13. doi: 10.1007/s12640-009-9033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandaey A, Gupta RK, Shrivastava R. Curcumin -the yellow magic. Asian J Applied Sci. 2011;4:343–354. [Google Scholar]
- 33.Mahindu SN. Food additives: characteristics, detection and estimation. APH Publishing Corporation; New Delhi-India: 2008. [Google Scholar]
- 34.Das S, Suresh PK. Nanosuspension: a new vehicle for the improvement of the delivery of drugs to the ocular surface. Application to amphotericin B. Nanomedicine. 2011;7(2):242–247. doi: 10.1016/j.nano.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Liu J, Yang X, Zhao X, Xu H. Oleanolic acid nanosuspensions: preparation, in-vitro characterization and enhanced hepatoprotective effect. J Pharm Pharmacol. 2005;57(2):259–264. doi: 10.1211/0022357055407. [DOI] [PubMed] [Google Scholar]
- 36.Takamuku T, Wada H, Kawatoko C, Shimomura T, Kanzaki R, Takeuchi M. Amide-induced phase separation of hexafluoroisopropanol-water mixtures depending on the hydrophobicity of amides. Phys Chem Chem Phys. 2012;14(23):8335–8347. doi: 10.1039/c2cp40669a. [DOI] [PubMed] [Google Scholar]
- 37.Krysmann MJ, Castelletto V, McKendrick JE, et al. Self-assembly of peptide nanotubes in an organic solvent. Langmuir. 2008;24(15):8158–8162. doi: 10.1021/la800942n. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Zhang F, Zhang Y, He J, Hu J. Organic solvents mediate self-assembly of GAV-9 peptide on mica surface. Acta Biochim Biophys Sin (Shanghai) 2007;39(4):285–289. doi: 10.1111/j.1745-7270.2007.00278.x. [DOI] [PubMed] [Google Scholar]
- 39.Anand P, Nair HB, Sung B, et al. Design of curcumin-loaded PLGA nanoparticles formulation with enhanced cellular uptake, and increased bioactivity in vitro and superior bioavailability in vivo. Biochem Pharmacol. 2010;79(3):330–338. doi: 10.1016/j.bcp.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Cartiera MS, Ferreira EC, Caputo C, Egan ME, Caplan MJ, Saltzman WM. Partial correction of cystic fibrosis defects with PLGA nanoparticles encapsulating curcumin. Mol Pharm. 2010;7(1):86–93. doi: 10.1021/mp900138a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta V, Aseh A, Rios CN, Aggarwal BB, Mathur AB. Fabrication and characterization of silk fibroin-derived curcumin nanoparticles for cancer therapy. Int J Nanomedicine. 2009;4:115–122. doi: 10.2147/ijn.s5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das RK, Kasoju N, Bora U. Encapsulation of curcumin in alginate-chitosanpluronic composite nanoparticles for delivery to cancer cells. Nanomedicine. 2010;6(1):153–160. doi: 10.1016/j.nano.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 43.Regev O, Gohy J, Lohmeijer BGG, et al. Dynamic light scattering and cryogenic transmission electron microscopy investigations on metallosupramolecular aqueous micelles: evidence of secondary aggregation. Colloid Polym Sci. 2004;282(4):407–411. [Google Scholar]
- 44.Chaleawlert-umpon S, Pimpha N. Morphology study of superparamagnetic iron oxide-chitosan nanoparticles. Journal of Microscopy Society of Thailand. 2009;23(1):62–65. [Google Scholar]
- 45.Prato M, Kostarelos K, Bianco A. Functionalized carbon nanotubes in drug design and discovery. Acc Chem Res. 2008;41:60–68. doi: 10.1021/ar700089b. [DOI] [PubMed] [Google Scholar]
- 46.Liu Z, Chen K, Davis C, et al. Drug delivery with carbon nanotubes for in vivo cancer treatment. Cancer Res. 2008;68(16):6652–6660. doi: 10.1158/0008-5472.CAN-08-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem Int Ed Engl. 2006;45(48):8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 48.Farokhzad OC, Cheng J, Teply BA, et al. Targeted nanoparticleaptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci U S A. 2006;103(16):6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohanty C, Sahoo SK. The in vitro stability and in vivo pharmacokinetics of curcumin prepared as an aqueous nanoparticulate formulation. Biomaterials. 2010;31(25):6597–6611. doi: 10.1016/j.biomaterials.2010.04.062. [DOI] [PubMed] [Google Scholar]
- 50.Bisht S, Feldmann G, Soni S, et al. Polymeric nanoparticle-encapsulated curcumin (“nanocurcumin”): a novel strategy for human cancer therapy. J Nanobiotechnology. 2007;5:3. doi: 10.1186/1477-3155-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295(5564):2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 52.de Groot NS, Parella T, Aviles FX, Vendrell J, Ventura S. Ilephe dipeptide self-assembly: clues to amyloid formation. Biophys J. 2007;92(5):1732–1741. doi: 10.1529/biophysj.106.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scanlon S, Aggeli A. Self-assembling peptide nanotubes. Nano Today. 2008;3:22–30. [Google Scholar]
- 54.Torchilin V. Tumor delivery of macromolecular drugs based on the EPR effect. Adv Drug Deliv Rev. 2011;63(3):131–135. doi: 10.1016/j.addr.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 55.Duan J, Zhang Y, Han S, et al. Synthesis and in vitro/in vivo anti-cancer evaluation of curcumin-loaded chitosan/poly(butyl cyanoacrylate) nanoparticles. Int J Pharm. 2010;400(1–2):211–220. doi: 10.1016/j.ijpharm.2010.08.033. [DOI] [PubMed] [Google Scholar]
- 56.Sahu A, Kasoju N, Bora U. Fluorescence study of the curcumin-casein micelle complexation and its application as a drug nanocarrier to cancer cells. Biomacromolecules. 2008;9(10):2905–2912. doi: 10.1021/bm800683f. [DOI] [PubMed] [Google Scholar]
- 57.Missirlis D, Kawamura R, Tirelli N, Hubbell JA. Doxorubicin encapsulation and diffusional release from stable, polymeric, hydrogel nanoparticles. Eur J Pharm Sci. 2006;29(2):120–129. doi: 10.1016/j.ejps.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Nayak AP, Tiyaboonchai W, Patankar S, Madhusudhan B, Souto EB. Curcuminoids-loaded lipid nanoparticles: novel approach towards malaria treatment. Colloids Surf B Biointerfaces. 2010;81(1):263–273. doi: 10.1016/j.colsurfb.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 59.Reddy RC, Vatsala PG, Keshamouni VG, Padmanaban G, Rangarajan PN. Curcumin for malaria therapy. Biochem Biophys Res Commun. 2005;326(2):472–474. doi: 10.1016/j.bbrc.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 60.Dhillon N, Aggarwal BB, Newman RA, et al. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14(14):4491–4499. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 61.Sharma RA, Euden SA, Platton SL, et al. Phase I clinical trial of oral curcumin: biomarkers of systemic activity and compliance. Clin Cancer Res. 2004;10(20):6847–6854. doi: 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 62.Hsu YC, Weng HC, Lin S, Chien YW. Curcuminoids-cellular uptake by human primary colon cancer cells as quantitated by a sensitive HPLC assay and its relation with the inhibition of proliferation and apoptosis. J Agric Food Chem. 2007;55(20):8213–8222. doi: 10.1021/jf070684v. [DOI] [PubMed] [Google Scholar]
- 63.Ma Z, Haddadi A, Molavi O, Lavasanifar A, Lai R, Samuel J. Micelles of poly(ethylene oxide)-b-poly(epsilon-caprolactone) as vehicles for the solubilization, stabilization, and controlled delivery of curcumin. J Biomed Mater Res A. 2008;86(2):300–310. doi: 10.1002/jbm.a.31584. [DOI] [PubMed] [Google Scholar]
- 64.Wyllie AH, Kerr JF, Currie AR. Cell death: the significance of apoptosis. Int Rev Cytol. 1980;68:251–306. doi: 10.1016/s0074-7696(08)62312-8. [DOI] [PubMed] [Google Scholar]
- 65.Fryer RA, Galustian C, Dalgleish AG. Recent advances and developments in treatment strategies against pancreatic cancer. Curr Clin Pharmacol. 2009;4(2):102–112. doi: 10.2174/157488409788185007. [DOI] [PubMed] [Google Scholar]
- 66.Roy M, Chakraborty S, Siddiqi M, Bhattacharya RK. Induction of apoptosis in tumor cells by natural phenolic compounds. Asian Pac J Cancer Prev. 2002;3(1):61–67. [PubMed] [Google Scholar]
- 67.Karunagaran D, Rashmi R, Kumar TR. Induction of apoptosis by curcumin and its implications for cancer therapy. Curr Cancer Drug Targets. 2005;5(2):117–129. doi: 10.2174/1568009053202081. [DOI] [PubMed] [Google Scholar]
- 68.Acharya S, Dilnawaz F, Sahoo SK. Targeted epidermal growth factor receptor nanoparticle bioconjugates for breast cancer therapy. Biomaterials. 2009;30(29):5737–5750. doi: 10.1016/j.biomaterials.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 69.van Engeland M, Nieland LJ, Ramaekers FC, Schutte B, Reutelingsperger CP. Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 1998;31(1):1–9. doi: 10.1002/(sici)1097-0320(19980101)31:1<1::aid-cyto1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 70.Kluza E, Yeo SY, Schmid S, et al. Anti-tumor activity of liposomal glucocorticoids: The relevance of liposome-mediated drug delivery, intratumoral localization and systemic activity. J Control Release. 2011;151(1):10–17. doi: 10.1016/j.jconrel.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 71.Rudra JS, Sun T, Bird KC, et al. Modulating adaptive immune responses to peptide self-assemblies. ACS Nano. 2012;6(2):1557–1564. doi: 10.1021/nn204530r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Yield of synthesis, retention time, and observed and expected masses of MΔF, LΔF, IΔF, and MF
| Dipeptide | Yield | Retention time (min) | Expected mass (Da) | Observed mass (Da) |
|---|---|---|---|---|
| MΔF | 1.5 g, 60% | 23 | 294.407 | 295.139 |
| IΔF | 1.1 g, 42% | 45 | 279 | 276.370 |
| LΔF | 1.2 g, 40% | 24 | 276 | 276.23 |
| MF | 0.6 g, 48% | 21 | 296.3 | 296 |
Table S2.
Morphology and average size of nanoparticles determined using dynamic light scattering and transmission electron microscopy analysis
| S no | Dipeptide | DLS (Rh in nm) | PDI | TEM (mean D in nm) |
|---|---|---|---|---|
| 1 | MΔF | 80 | 0.15 | Vesicle (40) |
| 2 | IΔF | 296 | 0.16 | Vesicle (20) |
| 3 | LΔF | 2000 | 0.25 | Tube (D 30 nm and L 2100 nm) |
Abbreviations: DLS, dynamic light-scattering; PDI, Polydispersity Index; TEM, transmission electron microscopy.
Mean particle size of methionine–phenylalanine (MF) in methanol:water solution (1:1) measured using dynamic light-scattering (Rh = 2 nm).