Abstract
During recent decades there have been remarkable advances and profound changes in cancer therapy. Many therapeutic strategies learned at the bench, including monoclonal antibodies and small molecule inhibitors, have been used at the bedside, leading to important successes. One of the most important advances in biology has been the discovery that small interfering RNA (siRNA) is able to regulate the expression of genes, by a phenomenon known as RNA interference (RNAi). RNAi is one of the most rapidly growing fields of research in biology and therapeutics. Much research effort has gone into the application of this new discovery in the treatment of various diseases, including cancer. However, even though these molecules may have potential and strong utility, some limitations make their clinical application difficult, including delivery problems, side effects due to off-target actions, disturbance of physiological functions of the cellular machinery involved in gene silencing, and induction of the innate immune response. Many researchers have attempted to overcome these limitations and to improve the safety of potential RNAi-based therapeutics. Nanoparticles, which are nanostructured entities with tunable size, shape, and surface, as well as biological behavior, provide an ideal opportunity to modify current treatment regimens in a substantial way. These nanoparticles could be designed to surmount one or more of the barriers encountered by siRNA. Nanoparticle drug formulations afford the chance to improve drug bioavailability, exploiting superior tissue permeability, payload protection, and the “stealth” features of these entities. The main aims of this review are: to explain the siRNA mechanism with regard to potential applications in siRNA-based cancer therapy; to discuss the possible usefulness of nanoparticle-based delivery of certain molecules for overcoming present therapeutic limitations; to review the ongoing relevant clinical research with its pitfalls and promises; and to evaluate critically future perspectives and challenges in siRNA-based cancer therapy.
Keywords: small interfering RNA, nanoparticles, cancer therapy, delivery strategies, biological barriers, clinical trials
Introduction
During recent years, cancer therapy has undergone important changes and improvements. Learning from translational medicine, new therapeutic strategies, such as small molecule inhibitors and monoclonal antibodies, have been introduced into clinical practice.1–3 These successes have encouraged the research field, and regulation of gene expression is one of the most frequently addressed issues in translational medicine.
Traditionally, RNA in cells has been thought to have two fundamental functions. Until 1980, single-stranded messenger RNA (mRNA) was considered to be no more than a passive transducer of information between DNA and protein synthesis. The catalytic role of RNA was highlighted in the early 1980s, and in 1986 the concept of “the RNA world” was proposed by Walter Gilbert,4 whereby ribosomal and transfer RNA were described to have structural, enzymatic, and information-decoding roles in the process of protein synthesis.
In 1998, Fire et al5 described the phenomenon of RNA interference (RNAi) for the first time. They discovered the ability of double-stranded RNA (dsRNA) to silence gene expression in the nematode worm, Caenorhabditis elegans. Three years later, Elbashir et al6 opened up the field of RNAi applications in the treatment of human disease by demonstrating that synthetic small interfering RNA (siRNA) could knock down a gene in a sequence-specific manner in several mammalian cell lines. The in vivo therapeutic potential of this new technology for transgene expression was rapidly shown in mice by effective targeting of a sequence from hepatitis C virus.7 Since then, there has been an explosion of interest in use of this technology for clinical applications throughout the scientific world, and many papers have been published in the field.8–12 siRNAs show promise for future control of diseases caused by activity of one or several genes. The list of human diseases which could potentially be treated using this approach is long, and includes viral infections,13–17 dominant genetic disorders,18–20 autoimmune disease,21 and cancer.9–11,22–24
In the case of cancer therapy, the appeal of miRNAs is becoming par ticularly strong as the molecular mechanisms underlying tumorigenesis become better defined and various molecular targets are identified. These molecular targets afford a good chance to develop new drugs, in that siRNA molecules can, in theory, be designed to hit almost any target, including those that are difficult or impossible to address at present. Small RNAs are indeed being vigorously pursued as therapies, and development of siRNA therapeutics is now rapidly gaining momentum.
RNAi is one of the fastest advancing fields in biology. In 2004, only six years after the discovery of RNAi, the first siRNA-based agent developed as a treatment for wet age-related macular degeneration entered Phase I clinical trials in humans.25,26 However, recent research has highlighted the need to overcome important issues concerning the safety and feasibility of siRNA-based drugs before their clinical use, and a major advance has been made in this regard with the advent of nanomedicine technology. Because of their tunable features, nanoparticles are being investigated in an attempt to overcome the safety and delivery issues associated with siRNA-based drugs.26,27 In 2010, the first-in-human Phase I clinical trial was started,28 in which siRNAs were systemically administered to patients with solid tumors using a targeted nanoparticle delivery system.
Given the exceptional promise of these combined technologies, this review aims to address fully the main strengths and weaknesses of their applications, highlighting the potential problems and solutions that one day could transform RNAi into a conventional treatment for human malignancy. First, we provide an overview of the mechanism for siRNA, focusing on the applications of siRNA-based cancer therapy. Second, we discuss the possible use of siRNA in the clinic, highlighting several challenges that could limit their applications, with particular emphasis on siRNA delivery and biological barriers. Third, we review the wide range of strategies proposed for siRNA delivery, with attention paid to the advantages and current limitations of the nanoparticle-based approach. Further, we discuss strategies which have shown in vivo success in humans, reporting the recent promising results from clinical trials in cancer and other diseases. Finally, we provide a critical evaluation of future prospects and challenges for siRNA-based therapy in the treatment of cancer.
Principles of siRNA-based cancer therapy
RNAi is a fundamental regulatory pathway for most eukaryotic cells. It consists of complex enzymatic machinery able to control post-transcriptional gene expression through homology-dependent degradation of target mRNA by siRNA, ie, nontranslated dsRNA molecules.26,27 siRNA molecules are 21–23 nucleotides in length with a characteristic and highly specific structure (2–3 nucleotide 3′ overhangs and 5′ phosphate and 3′ hydroxyl groups) to prevent erroneous gene silencing.29 They also contain a sense (passenger) strand and an antisense (guide) strand.30
In mammalian cells, RNAi is triggered by molecules of long dsRNA, which may be of exogenous or endogenous origin, and are cleaved into siRNA by Dicer, an RNase III endonuclease. Once cleaved, siRNAs interact withArgonaute-2, a multifunctional protein, and become incorporated into RNA-induced silencing complexes. Here the duplex RNA is unwound and Argonaute-2 degrades the “passenger” strand. The activated RNA-induced silencing complex is directed specifically to recognize a target by intermolecular base pairing throughout a single-stranded “guide” RNA molecule.30,31 Argonaute-2 then selectively hunts down and degrades the mRNA complementary to the antisense strand,32 and endonucleolytic cleavage occurs between bases 10 and 11 relative to the 5′ end of the antisense siRNA strand33–36 (Figure 1A). This activated complex can propagate gene silencing further, destroying additional mRNA targets. This effect may last for 3–7 days in rapidly dividing cells and for many weeks in nondividing cells.37 Translating this into practical terms, siRNA could be produced synthetically and directly introduced into target cells, thereby bypassing Dicer mechanics. Other forms of siRNA could be discussed, such as micro RNA (miRNA),38 short hairpin RNA,39,40 and Piwi-interacting RNA.41 However, siRNA is the most commonly used for RNAi in therapeutic applications.
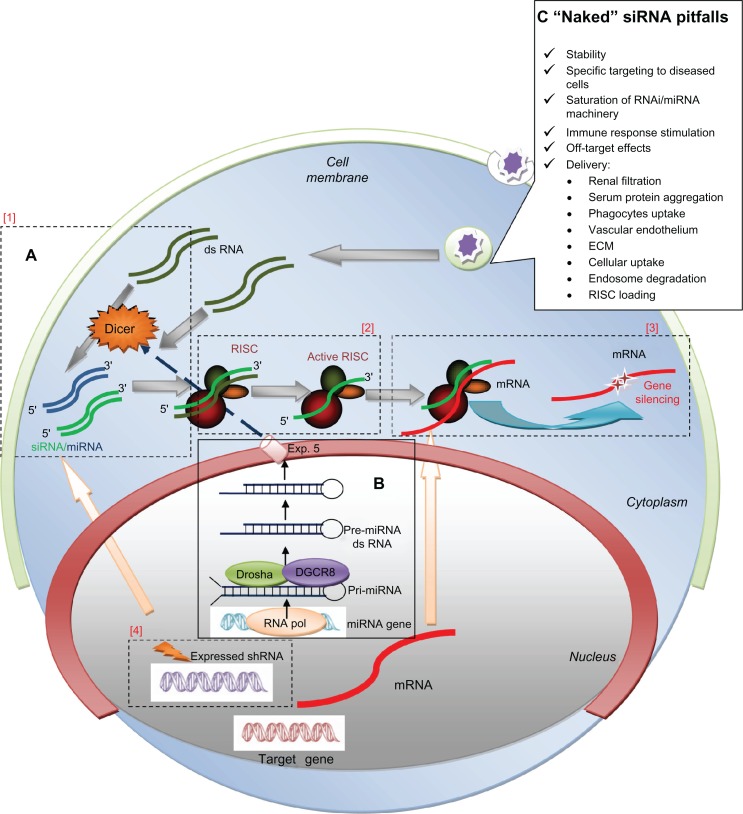
Figure 1.
RNAi/miRNA pathway schematization and major challenges for naked siRNA delivery in vivo. (A) Schematization of RNA interference (RNAi): non-translated double-stranded RNA (dsRNA) molecules called small interfering RNA (siRNA), of exogenous or endogenous origin, post-transcriptional regulate gene-expression through a sequence specific degradation of target messenger RNA (mRNA). [1] Longer siRNA molecules (dark green) are cleaved by the nuclease Dicer and [2] incorporated into a multiprotein RNA-inducing silencing complex (RISC). [2] The duplex RNA is unwound leaving the anti-sense strand (light green). [3] to guide RISC to complementary mRNA (red) for subsequent endonucleolytic cleavage and gene silencing. [4] Short hairpin RNAs (shRNA) (violet) are sequences of RNA encoded by specific genes; they are introduced into the nucleus, transcribed and transported into the cytoplasm where they follow the same fate of siRNA. (B) miRNA processing: microRNA (blue) are considered as the “endogenous substrate” of the RNAi machinery. They are trascribed by RNA-Pol III in long primary transcripts (pri-miR), then processed within the nucleus into precursor miRNA (pre-miRNA) by the RNase III enzyme Drosha-DGCR8. Pre-miRNA hairpins are exported from the nucleus in a process involving the nucleo-cytoplasmic shuttle Exportin-5 (Exp.5). In cytoplasm, the pre-miRNA hairpin is cleaved by Dicer and loaded into RISC as for siRNA. miRNAs often share only partial complementarity with target mRNAs, usually in the 3′UTR, acting mainly as a translational repressors. (C) “Naked” siRNA pitfalls. In the box are reported the major obstacles for therapeutic efficacy of siRNA without modifications (“naked”). See text for details.
In certain cases, only partial complementarity can occur between siRNA and its target mRNA, leading to suppression of translation or destabilization of transcripts. This phenomenon usually causes the so-called off-target effects of siRNA (see later) and mimics interactions with target sites of another class of regulatory small RNA, ie, miRNA. These molecules are thought to be the endogenous substrate for the RNAi machinery, because endogenous siRNA has not been found in mammalian systems. miRNA molecules have been identified in mammals as well as in many other organisms. They derive from long primary transcripts (pri-miRNA), which are processed within the nucleus into precursor miRNA (pre-miRNA, 60–70 bp hairpins) by the RNase III enzyme, Drosha-DGCR8 (Figure 1B). They are then exported to the cytoplasm by Exportin-5, where they are further processed by the Dicer enzyme which removes the loop, and one of the two strands is loaded into the RNA-induced silencing complex.38
Unlike siRNAs, mature miRNAs often share only par tial complementarity with sequences of target mRNAs, usually in the three prime untranslated regions. Therefore, the primary mechanism of action is translational repression, although miRNA molecules with full sequence complementarity can degrade mRNA, so it could be said that while siRNAs are able to control single gene expression fully, microRNAs can moderate the expression of gene networks.38
The short hairpin RNA approach is based on nuclear delivery of a gene that encodes for desired short hairpin RNAs. These are transcribed, transported to the cytoplasm via the miRNA machinery, and processed into siRNAs by Dicer, as mentioned earlier. The need for the short hairpin RNA gene to enter the nucleus adds complexity to the delivery and reduces the potency of siRNA gene silencing. On the other hand, the potential advantage of DNA-based RNAi is its stable introduction into cells as gene therapy, bypassing transient effects, so avoiding repeated or continuous administration in the clinical setting.39,40
Another variation of siRNA-based therapy is administration of longer precursors processed by Dicer endonuclease. Recent reports indicate that these precursors are several nucleotides longer and are much more powerful than the siRNAs widely used for inducing target gene silencing.42 This is probably because direct coupling of Dicer processing and the Argonaute-2 loading mechanism of siRNA is much more efficient.42
RNAi and cancer: therapeutic implications
Cancer is a disease that is often characterized by mutations in multiple signal transduction pathways, leading to uncontrolled cell proliferation. It is usually difficult to identify the key genes governing cell proliferation or survival which should be targeted to induce cell death. RNAi technology can be useful for cancer therapy because of its high efficacy and specificity in downregulating gene expression, and it is clearly important to understand the role of altered genes in the development of cancer. It must be stressed that blockage of a single gene is not sufficient to cure or control most cancers. One of the major likely benefits of the RNAi technique in the treatment of cancer is the feasibility of developing combination therapeutic RNAi approaches to inhibit multiple oncogenes or genes of proteins involved in tumorigenesis.43 A correct approach could involve hitting multiple pathways simultaneously and/ or combining gene silencing with other therapies.
Many in vitro and in vivo studies have evaluated the silencing of genes involved in pathways that drive cancer, such as oncogenesis, apoptosis, cell cycle regulation, cell senescence, tumor-host interaction, and resistance to conventional therapies.9,43,44 In addition, because miRNAs may work as both tumor suppressors and oncogenes, these molecules could also be investigated as therapeutic targets, in that they could be knocked down when expressed in excess or replaced in cells when they have been pathologically reduced or lost.45 Deregulated miRNA could also serve as a helpful biomarker for diagnosis and monitoring of disease.46 Biomarkers are easily and objectively measurable biological characteristics which can be used as indicators of normal and pathological processes. Notably, miRNAs can be detected in body fluids, such as blood and cerebrospinal fluid, enabling noninvasive and early detection of disease. A further example of the potential applications of miRNA is expression profiling in order to distinguish cancer from normal tissue and to classify, eg, cancer subtypes or the origin of metastatic cancer tissue.46
siRNA in the clinic
Difficulties in therapeutic application
As discussed, siRNAs hold promise as therapeutics in view of their ability to silence any gene with a known sequence. However, effective and well controlled in vivo delivery is challenging, and presently limits the use of RNAi in the clinic27 (see Table 1). In fact, these molecules, without any other modifications (“naked”), encounter several obstacles that reduce their therapeutic efficacy (Figure 1C). To be efficacious, they need to reach the cytoplasm of the cell in sufficient amounts to enable sustained target inhibition.47,48 However, naked siRNAs are highly unstable intravascularly, with a short half-life due to their susceptibility to serum RNAse A-type nucleases and rapid renal clearance.47–49 Moreover, unmodified siRNA molecules are not able to enter most cells readily because of their size (13 kDa) and highly polyanionic nature.47,48,50 Once in the cells, their effects are transient and diluted at each cell division,51 so repeated dosing is required to achieve long-term therapeutic effects. Unmodified siRNA molecules also have potential toxicities,52,53 via three main mechanisms, ie, saturation of RNAi machinery and competition within the miRNA pathway, stimulation of the immune response, and off-target effects.
Table 1.
Problems of Naked siRNA for clinical applications
|
Naked siRNA
| ||
|---|---|---|
| Problems | Explanation | Solution |
| Short half life | Serum nuclease susceptibility | Local or topical administration |
| Rapid renal clearance | Chemical modification of the sugars or the bases of oligoribonucleotides for stabilization | |
| Phagocyte uptake | Nanoparticle carriers | |
| Reduced uptake by cells | Anionic nature (charge obstacle) | Conjugation with nanoparticles |
| Too large to pass membrane | ||
| Transient effect | Dilution of siRNA concentration | shRNA for integration gene in cell genome |
| Toxic effect | ||
| • Saturation of RNAi machinery | Reduced accessibility to miRNA | Correct dosing and targeting |
| • Immune response stimulation | TLR activation and type I IFN response | 2′OH methylation modification use of nanoparticles |
| Non TLR mediated innate immune response | Chemical modification (2′OH methylation modification) | |
| • Off target effect | miRNA like off target silencing | 2′OH methylation – screening siRNA effect in vitro |
Abbreviations: TLR, Toll-Like Receptor; IFN, Interferon.
Saturation of RNAi machinery and competition in miRNA pathway
RNAi is an important regulatory mechanism in the cell which could be perturbed by exogenous introduction of dsRNA. The components of the cellular machinery involved in gene silencing could be less accessible to miRNAs entering the natural cellular pathway.47,54,55 Intact RNAi machinery is clearly essential for mammalian cells, as suggested by the early embryonic lethality of Dicer knockouts.
Stimulation of immune response
Some of the sequence motifs in siRNA molecules could have the ability to induce the innate immune response. The innate immune response is mediated principally by type I interferon and proinflammatory cytokines,56–58 and can be triggered in different ways, ie, mediated or not mediated by the Toll-like receptor (TLR). Briefly, when nucleic acids enter endosomal and lysosomal compartments, TLR-3, 7, and 8 are primed and activate interferon-alpha and inflammatory cytokines via nuclear translocation of the nuclear factor k-light chain enhancer of activated B cells.59 The innate immune response that is not mediated by TLR is triggered principally by cytoplasmic RNA sensors, including the retinoic acid-inducible gene 1 and dsRNA-binding protein kinase. This mechanism can lead to activation of interferon-beta and other inflammatory mediators.
Off-target effects
siRNA was initially described to be specific for the target gene. However, early genome-wide monitoring of gene activity in siRNA-treated cells disproved its almost ideal specificity.60,61 SiRNA shows miRNA-like off-target silencing of a large number of unintended transcripts with partial identity to its sequence.60,61 This mechanism has unpredictable cellular consequences, with important toxic phenotypic effects. Specific research efforts have been made to understand better and control these undesirable off-target effects and their long-term implications, and even more so now, considering the possible clinical reality of siRNA.47,62 These difficulties are likely to be less pronounced when therapy is localized to the more accessible organs or tissues, eg, eye, skin, and mucus membranes. In these contexts, siRNA therapy has higher bioavailability and may have fewer of the adverse effects associated with nonspecific siRNA delivery.
The ideal route and obstacles which can be encountered by molecules introduced into the human body through systemic delivery are still a matter of debate. Much research effort using in vivo tracking and fluorescent methods is presently focused on these issues. It is generally accepted that, after intravenous injection, the molecules travel throughout the whole body, navigate the circulatory system, in which they need to avoid renal filtration, are taken up by phagocytes (both in the bloodstream and in the extracellular matrix tissue) and aggregate with serum proteins (Figure 2).50 They have to cross biological barriers, ie, pass into the bloodstream and across vascular endothelial walls to be able to reach the target organ or tissue. Specifically, it has been reported that molecules larger than 5 nm in diameter do not readily cross the capillary endothelium, and therefore remain in the body if they are not cleared. However, larger molecules up to 200 nm in diameter are able to enter some tissues, including the spleen, liver, and several types of tumor, due to their different vasculature. The molecules must also diffuse through the extracellular matrix, which consists of a dense network of polysaccharides and fibrous proteins. The extracellular matrix is also rich in macrophages, which can obstruct transport of macromolecules and nanoparticles. The siRNA complex must then be taken up by target cells, at which point they have to evade the endosome system to reach the cytoplasm.63 Through a complex system of cell compartments with decreasing pH, the molecules can reach lysosomes to be degraded. Finally, to be effective, siRNA must be released from its carrier to reach the cellular machinery.50
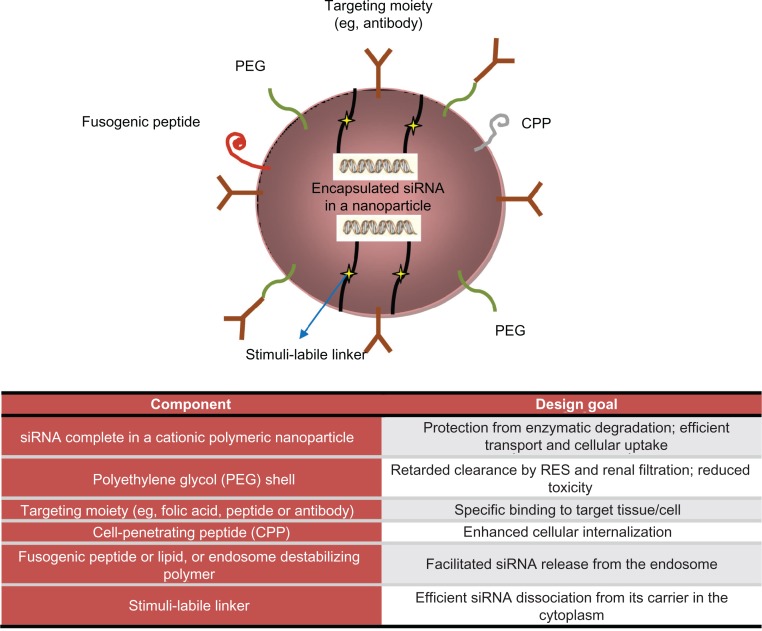
Figure 2.
Encapsulation technologies for siRNA delivery in vivo: nanoparticles strategies and advantages.
Readapted from Shim et al.165
Strategies for siRNA delivery: advantages and current limitations
Early delivery strategies and advent of nanotechnology
Early on, siRNA therapeutics were administered locally using naked or chemically modified siRNA.64 As already described, the complexity of physiological interactions, both biochemical and mechanical, immediately signaled the necessity to protect gene material and guide it to its target site of action. Preliminary approaches to protect siRNA arose from DNA transfection techniques. Generally speaking, these protective strategies can be divided in three categories, ie, mechanical, chemical, and biological. Mechanical approaches seek to enhance permeability of biological barriers, and in this way allow gene material to reach the cellular environment. The main mechanical permeabilization techniques are sonoporation and electroporation. Chemical and biological approaches, on the other hand, aim to transport genetic cargo through biological systems within a “Trojan horse” system. In this context, viral vectors provide the most efficient delivery, but their use is limited due to the host immune response and the inner complexities of this method. State-of-the art chemical approaches are the most promising, as a result of ever-increasing insight into techniques of synthesis and the biological behavior of nonbiological structures. Observations from clinically approved nanoparticle formulations suggest that lipidic carriers (Doxil®) and covalent coupling with albumin (Abraxane®)65 are viable strategies for siRNA delivery. However, bioavailability remains an unresolved issue because these vectors are unable to stabilize and protect siRNA in the biological environment.
Nanotechnology has been a most promising spinoff in molecular medicine. In this field, technical advances have suddenly accelerated towards more refined fabrication techniques and new materials with unique and innovative features.66,67 Starting from this technological platform, new possibilities and different points of view are coming to light which may resolve the present issues in drug delivery and remove the barriers to discovery of the “magic bullet” for cancer.68,69,168,169 Thus, a concrete opportunity to change current treatment regimens in a substantial way is offered by nanoparticles, which are nanostructured entities with tunable size, shape, surface, and biological behavior. The impact of nanostructured therapeutics in terms of efficacy and safety is promising to be revolutionary.
The dawn of nanotechnology, seen as the manipulation of matter on a nanometric scale, allows the design and fabrication of novel devices with uniquely tailored physicochemical properties and enormous therapeutic and diagnostic potential. Over the past two decades, there has been a steady rise in the number of commercially available nanoparticle therapeutics.69 The first example of a nanostructure with biological application was the lipidic vesicle (liposome), a spherical self-assembled layer of amphiphilic lipidic molecules of nanometric size. Liposomes became used as drug delivery systems, because of their circumscribed inner volume which can be used to contain and transport a drug to its target site of action. Chemical surface modifications, such as poly(ethylene) glycol (PEG), have been developed to increase bioavailability. Limiting factors to their successful use as carriers remain, especially concerning their stability in biological environments and their ability to transport drugs across biological barriers. In the following sections, we briefly highlight the advantages of nanoparticles which make them suitable for integration into siRNA technology. A broad overview is also provided of almost all delivery strategies that have been attempted to date, underlining the eventual limitations and pitfalls.
Nanoparticles
Nanoparticles are physical entities with a characteristic length of 1–100 nm. They can be considered as a unit in term of physicochemical properties. In nanotechnology, the synthesis and characterization of these objects is a broad field in constant development, heading towards “smart” structures with superior capabilities in various scientific and technological contexts, including catalysis, energy conversion, sensors, and actuators.70,71 The most promising and rapidly growing applications of nanoparticles are directly linked to biomedical issues, with cellular trafficking and accurate in vitro and in vivo tracking of biological systems being two examples of the new frontiers opened up for biological study by the application of luminescent inorganic nanoparticles.72
Nanoparticles can be categorized as organic or inorganic according to the bulk constituent materials in their structure. Classifications based on size, shape, surface, structure, and chemical behavior can be devised. Overall, what is particularly attractive about nanostructures is their surface to volume ratio. Nanosized entities enable a very limited volume to provide an enormous surface area for transport, chemical reactions, and interaction with biological systems. These features, combined with enhanced permeability to biological barriers, make nanoparticles the basis of nanomedicine and a powerful and promising tool in the design of new diagnostic and therapeutic devices.
Interest in nanoparticles has consistently grown in every branch of medicine. It has been driven by the potential of nanoparticles to achieve a high therapeutic index and corresponding clinical success with improved patient compliance and reduced side effects.73 Nanoparticle-based drug delivery systems play an important role in the development and future applications of new pharmaceutical formulations. They may be capable of improving bioavailability and reducing the frequency and dosage needed for currently used drugs.74,167 With this novel type of drug formulation, in which nanoparticles act as drug carriers, it may be possible to: improve drug stability and carrier capacity; target drug molecules to a specific diseased cell or tissue, avoiding toxicity to normal cells; confine release of the drug to the site of interest; allow efficient permeation of epithelial and endothelial barriers; and combine therapies with codelivery of two or more drugs.68,75–77 The latter could involve incorporation of a wide range of molecules, including both hydrophilic and hydrophobic substances, thereby improving the delivery of poorly water-soluble drugs.
Residence time in the circulation, maximal tolerated dose, and selectivity are the most important factors to consider when determining the optimal dosage.73 In drug delivery systems, the choice of bulk material constituent for the carrier is a key point, and tailored biological behavior (bioactivity, biocompatibility, biodegradability) must be fulfilled, with an improved payload capacity. It is important that the nanomaterial carrier is able to be eliminated harmlessly from the body in a reasonable period of time after releasing its cargo, and, in case of theranostic nanoparticles, accomplishing a desired diagnostic function.78 The drug could be adsorbed, dissolved, or dispersed throughout the matrix of the nanoparticles, or, alternatively, it can be covalently attached to the surface or inside the matrix. In the body, the drug loaded in nanoparticles is usually released from the matrix by diffusion, swelling, erosion, or degradation. Two kinds of targeting strategies are possible, ie, passive targeting and active targeting. Passive targeting does not require surface modification of the nanoparticles, and is achieved using the unique pathophysiology of diseased cells and controlling particle mobility through human tissues, according to particle size and route of administration. On the other hand, active targeting can be achieved by surface modification of nanoparticles and their functionalization with ligands or antibodies that can recognize and bind to complementary molecules or receptors found on the surface of specific cells.79
A further advance in drug delivery is represented by the multistage delivery system,80 which provides more accurate spatiotemporal targeting with efficient cargo release. Nanoparticles suitable for multistage drug delivery systems are hybrid nanostructures (for instance, with a core-shell structure) designed to reach injured tissues in a selective manner.81,82 Each part of the hybrid nanoparticle responds differently in the biological environment. These features, combined with conventional targeting against upregulated cell surface antigens, allow precise spatiotemporal drug delivery in selected areas of the host system. Furthermore, in the design of nanocarriers, it is possible to achieve stimuli-responsive behavior using pH-sensitive and/or temperature-sensitive materials.83 These carriers hold promise for realizing effective spatiotemporal delivery of drugs. In this sense, nanoparticles are designed to be part of a delivery system and it is necessary to consider the overall behavior of such systems, ie, carrier, payload, and host system properties, as well as mutual interactions. In a gene delivery context, a key feature of the carrier is the ability to protect the gene payload until it reaches the target site, with no modification in molecular structure of the gene or its biochemical activity.
This review explores strategies for siRNA delivery by nanoparticles. The pursuit of gene delivery has been undertaken using viral-based84 and nonviral-based85,86 vectors. These topics have already been extensively reviewed,87,88 so are not discussed in depth in this paper. Nanoparticle features in drug delivery are likely to overcome a number of issues concerning therapeutic applications of siRNA. The challenge of effective and nontoxic delivery is a crucial point and remains the most significant barrier to therapeutic application of siRNA technology.89 Moreover, nanoparticles can be engineered to have controlled release of functional siRNA. As discussed above, slow, sustained, and controlled release could be helpful for decreasing the frequency of treatment and lead to more effective therapies, especially in siRNA therapeutics.
In order to achieve effective siRNA delivery, several strategies have been studied. Leaving apart the viral vector delivery setup, we consider here the chemical modifications of siRNA, ligand-based (targeted or conjugated) siRNA, polymers, cationic and neutral liposomes, sensu stricto nanoparticles, and combined approaches. A short description and a few examples of each of these strategies is presented (see Table 2), and a more detailed revision can be found elsewhere.42
Table 2.
Delivery strategies for siRNA. Advantages and pitfalls
|
Modified siRNA (delivery strategies)
| ||
|---|---|---|
| Modification | Advantages | Pitfalls |
| Chemical modifications (<10 nm) | ||
• Sense and antisense strand
|
Enhanced serum stability, resistance to endonuclease Reduction of innate immune response stimulation |
Impaired biological activity and sometimes toxicity exacerbation |
• 3′ or 5′ modification in the sense strand
|
Enhanced of stabilization Prevention of protein absorption Enhanced interaction with cellular membrane (positive charge) Reduction of aggregations among particles Enhanced gene target efficacy in vivo Reduction of opsonization and phagocytosis Reduction sequester in RES (seticulo endothelial system) Cross cell membrane |
Impaired biological activity and sometimes toxicity exacerbation Pharmacokinetic and bio-distribution Immunogenicity High costs |
|
Enhanced stability Enhanced binding with serum albumin Enhanced bio-distribution in some organs and tissues (eg, liver) Enhanced cellular uptake (eg, LDL/HDL receptors) Enhanced gene silencing in vivo Enhanced delivery to some cell tumors reduction of immune stimulation Tumor specific delivery |
|
| Polymers (100–300 nm) | ||
| • Cationic polymers (Synthetic)
(Natural)
|
Stabilization Enhanced nuclease resistance Stimulation of non-specific endocytosis Endosomal escape (“proton-sponge-effect”) Functionalization of the corona Enhanced time in bloodstream Reduction on non specific bio-distribution Enhanced targeting when conjugated with targeting ligand |
Cytotoxicity (necrosis and apoptosis) |
| Liposomes | ||
• Cationic lipids (100–300 nm)
• Neutral nano-liposomes (30–40 nm; <200 nm)
|
Enhanced stabilization by electrostatic interactions Versatility and flexibility in structure Protection from nuclease Enhanced uptake by cells via endosomal pathway Enhanced siRNA half-life Efficient in vivo siRNA delivery Down-regulation of target genes – Inhibition of tumor growth in mouse models |
Short half-life in serum Lack of tissue specificity Rapid liver clearance (RES sequester) Reduced access in other tissue Cell toxicity Induction of type 1 and 2 – IFN response Dose-dependent toxicity Pulmonary inflammation |
| Nanoparticles, microspheres and hydrogels | ||
• Inorganic NP
• Organic NP
• Liposomes SLN |
Efficient target gene silencing Enhanced serum stability Minimal levels of cytotoxicity Enhanced targeting when conjugated with targeting ligand No immunotoxicity Tumoral specific delivery for EPR (Enhanced Permeability and Retention) effect |
RES clearance Immune system stimulation (opsonization) Hemolysis, thrombogenicity, complement activation with consequent altered biodistribution and potential toxicity. |
Abbreviations: IFN, Interferon; NP, Nanoparticles; RES, Reticuloendothelial System; SLN, Solid Lipid Nanoparticles.
Chemical modifications to siRNA
Various chemical modifications have been introduced to increase the in vivo metabolic stability of siRNA molecules less than 10 nm in size. Examples of such modifications and their advantages (without affecting the efficiency of RNAi) are listed below and have been reviewed extensively elsewhere:47,48,62,90–93
2′-O-Methyl modifications in the ribose structure of selected nucleotides within both sense and antisense strands
Introduction of phosphorothioate backbone linkages at the 3′ end of the RNA strands
Alternative 2′ sugar modifications (eg, fluorine substitution).
These chemical modifications, added to the sugars, backbone, or bases of dsRNA, improve intravascular stabilization and are able to reduce activation of the innate immune response, without significant loss of RNAi activity.51,59
Ligand-based targeting molecules
Among the chemical modifications, 3′ or 5′ modifications of siRNA deserve separate consideration. These can be useful for improving resistance to degradation, and also for introducing targeting or conjugating ligands less than 10 nm in size, such as peptides and aptamers. Indeed, the smallest siRNA nanoparticles derive from direct conjugation of small molecules, peptides, or polymers to the sense strand of siRNA. These modifications of the sense strand seem not to affect mRNA degradation by siRNA. Based on the additional features attributed to siRNA molecules, these further modifications can be classified as ligand-targeted (affecting target specificity) or ligand-conjugated (mostly affecting stability).
Ligand-targeted siRNAs
Terminal modification (5′ or 3′) of siRNA molecules using cholesterol is a useful strategy for increasing their stability and cellular uptake. In particular, it increases binding to serum albumin, with consequent improved biodistribution in certain target tissues, eg, the liver. The improvement in cellular uptake is mediated by in vivo interaction and incorporation into low-density and high-density lipoproteins. Cholesterol-modified siRNA are capable of silencing apolipoprotein B targets in the mouse liver and jejunum, and of ultimately reducing total cholesterol levels.18,94 However, although this type of chemical modification has improved siRNA delivery to tissues, it is often associated with impaired biological activity and increased toxicity.62,92,93 In addition, siRNA can be conjugated with other lipid-like molecules, such as long-chain or medium-chain fatty acids and bile salt derivatives. These interact with high-density and low-density lipoprotein receptors, enhancing delivery into the liver and gene silencing in vivo. Moreover, albumin serves as a primary carrier for siRNA conjugated to medium-chain fatty acids.42
One particular modification is that achieved by mipomersen, a 2′-O-(2-methoxyethyl)-modified single-stranded RNA molecule targeted to apolipoprotein B, frequently implicated in cardiovascular disease. Many promising Phase II trials have been reported in this regard, with encouraging safety and efficacy results.95 A Phase III trial assessing the efficacy of mipomersen is currently ongoing in patients with a family history of hypercholesterolemia.
The folate receptor is a membrane glycoprotein over-expressed in several human tumors, including ovarian, colorectal, and breast cancer, but is minimally present in normal tissues. This feature makes it an attractive target for drug delivery. Folate-mediated targeting has several advantages, including the small size of folic acid, its lack of immunogenicity, convenient availability, and easy chemical conjugation.42,96
Like the folate receptor, the transferrin receptor is a glycoprotein frequently overexpressed by tumor cells. The intracellular routing of transferrin receptor-mediated endocytosis has been well characterized, and this molecule is easily available. For this reasons, transferrin receptor-mediated delivery has been extensively explored in a variety of targets, including tumors, endothelial cells, and the brain.42 Nanoparticles are usually conjugated chemically to either transferrin itself or to antitransferrin receptor antibodies.42,52,97
Antibodies have also been used extensively in the treatment of cancer. Because of their high specificity and affinity for cancer cell antigens, they have the capacity to induce an immune response against target cancer cells. Antibodies have also been recognized as “delivery molecules”. Several studies have demonstrated antibody-targeted delivery of drugs and nucleic acids. Antibodies or their fragments can be used as targeting agents for nanoparticles. Among the successful in vivo demonstrations, are antibody-protamine fusions that bind siRNA.98 Several problems remain to be resolved for effective application of this strategy, in particular the potential for inducing immunogenicity and the high cost of the antibodies.42
Along with antibodies, aptamers99 have been used for site-specific delivery of siRNA because of their high affinity and specificity for targets, with the advantages of a much smaller size (<15 kDa versus 150 kDa for antibodies). For instance, prostate-specific membrane antigen is a cell surface receptor usually detected at abnormal levels in prostate cancer cells and in tumor vascular endothelium, and has been proposed for targeting siRNA to these sites. Delivery of siRNA mediated by aptamers is promising, albeit limited by some challenges that need to be investigated further, in particular, the pharmacokinetics and biodistribution of these molecules.42
Ligand-conjugated siRNAs
To increase in vivo transfer, siRNA can be conjugated with other molecules, eg, peptides or PEG, which are able to overcome steric hindrance, or linked with drugs or nucleic acids and nanoparticles. Cell-penetrating peptides, protein transduction domains, and membrane translocation sequences are counted among the peptides.42 For example, cell-penetrating peptides are able to cross the cell membrane, with the advantages of increased target gene knockdown in vitro and in vivo.100,101 In the case of PEG, introduction of sugar molecules, eg, cyclodextrin,97 and hyaluronic acid102–104 is the most common approach for preventing drug aggregation because of the surface charge of siRNA or other loaded drugs, which typically have a net positive charge.105 The benefits of this charge neutralization are increased circulation time for the compound and enhanced targeting through limiting nonspecific interactions between the positively charged siRNA drug and the negatively charged cell membrane. Overall, these mechanisms result in increased target gene knockdown in vitro and in vivo.50
Polymers are organic macromolecules that protect RNA from degradation and facilitate its sustained delivery to tissues. Many different types of polymers, varying in size, chemistry, and pharmacological properties, can be considered. Cationic polymers62,87,88,106–109 can be useful as transfection agents, given that they can bind one or more large nucleic acids, package them reversibly into stabilized nanoparticles, and protect them against a degrading bioenvironment. These vectors avoid enzymatic degradation by nucleic acids by forming condensed complexes along with targeted delivery to cells and tissue.110 Moreover, these compounds have demonstrated an ability to stimulate nonspecific endocytosis. Another useful feature for siRNA delivery is endosomal escape, whereby certain molecules, eg, polyethylenimine and cyclodextrin, lead to local accumulation of ions via a mechanism called the “proton-sponge” effect, and as a consequence, the osmotic pressure generated bursts the endosomal compartment in which they are contained. Cationic polymers are generally divided into synthetic and natural polymers, with a linear or branched structure. Synthetic polymers include branched or linear polyethylenimine,111 poly-L-lysine, and cyclodextrin-based polycations. On the other side, natural cationic polymers include chitosan, atelocollagen, and cationic polypeptides. Another advantage of cationic polymer-based delivery is the ease with which siRNA and polymer complexes can be formulated due to their opposite charges. In fact, cationic polymers usually form a complex with negatively charged siRNA upon simple mixing. Despite these advantages, appreciable cytotoxicity (both necrosis and apoptosis) is a major problem with the use of these compounds.112 However, cationic vectors with diminished cytotoxicity and enhanced efficacy are rapidly emerging as delivery systems of choice.113,114 A possible application of cationic polymers is their conjugation in more complex compounds such as polymeric nanoparticles. These molecules can be defined by the morphology and polymer composition in their core and corona73,110 (Figure 2). The therapeutic load (in this case siRNA) is usually conjugated to the surface of the nanoparticle or encapsulated and protected inside the core. Delivery systems composed of these compounds can be engineered to enable controlled or triggered release of therapeutic molecules.73 Furthermore, the nanoparticle surface may be functionalized by various methods to form a corona, which can provide increased residence time in blood, reduced nonspecific distribution, and in some cases, target tissues or specific cell surface antigens with a targeting ligand. For example, PEG allows steric stabilization or prevention of protein absorption. The final result is avoidance of opsonization and rapid clearance from bloodstream due to phagocytosis in the liver or spleen. As described, this delivery strategy combines the protective effect and loading capacity of the polymer with the specificity of an active targeting strategy through chemical molecular conjugation. During the past few years, several papers have reported delivery of nucleic acids mediated by cationic polymer carriers113,114 and described the generation of precise polymers, site-specific conjugation strategies, and multifunctional conjugates for nucleic acid transport.115,116
Lipid vectors
Also important in the field of delivery systems based on organic molecule carriers are lipid vectors, such as liposomes and cationic lipids.
Liposomes
Liposomes are compounds characterized by a phospholipid bilayer and an aqueous core. They can be created from single or multiple types of lipid. Unilamellar and multilamellar liposomes are commonly used as vehicles for delivery of pharmaceuticals. Liposomes interact with siRNAs to form complexes stabilized by electrostatic interactions.48,92 These are the delivery methods that are currently most mature and closest to clinical application, given that the platform has been adopted in clinical trials performed by several leading RNAi companies.117,118 However, some disadvantages must be overcome before they can be used in vivo. Liposomes tend to accumulate in the reticuloendothelial system, leading to rapid clearance by the liver and a short half-life in serum, so require either continuous infusion or frequent administration. Liposomes also lack target tissue specificity and have reduced access to other tissues. A possible approach to circumvent this problem is to develop sustained-release polymer formulations.107 For example, one group of polar head units can create the outer surface of the nanocomplex, while the other group of polar head units faces the interior hydrophilic core, which holds the nucleic acid payload (Figure 2). It is also possible to construct amorphous structures with liposomes, in which lipids and nucleic acids alternate with each other. The additional flexibility of these molecules can optimize the physical and chemical properties of the nanoparticle.
Neutral nanoliposomes
To avoid the potential toxicity associated with other delivery systems,112 neutral liposomes 30–40 nm in diameter which encapsulate siRNA are commonly being used. Unilamellar liposomes, such as dioleoyl phosphatidylcholine,119,120 with a hydrophilic core and a hydrophobic surface are able to protect siRNA from degradation by surrounding endonucleases and can enhance internalization through membrane fusion or receptor-mediated endocytosis.121
Cationic lipids
Cationic lipids 100–300 nm in size are able to protect siRNA from degradation by nucleases and increase the circulating half-life and uptake by cells. Several cationic lipids have been synthesized for formulation with siRNA, and have been evaluated in preclinical studies or animal models. However, they show significant cell toxicity and elicit hypersensitivity reactions in vivo. Liposomes in general and cationic lipids [DOTAP and DOTMA (N-[1-(2,3-dioleyloxy) propyl]-N, N, N-trimethyl ammonium chloride] in particular are able to induce strong type I and II interferon responses.122 They show dose-dependent toxicity, and pulmonary inflammation can arise as a result of reactive oxygen intermediates.123 Chemical additives could improve their formulation and reduce cell toxicity, thus making liposomes safer. In particular, liposomal nanoparticles comprise electrostatic complexes of nucleic acids and cationic lipids, such as dioleoyl trimethylammonium propane and N,N-dimethylaminopropane carbamoyl cholesterol.
A modern class of biodegradable solid lipid nanoparticles has also been developed. These nanoparticles are used to incorporate various drugs or imaging agents, with the benefit of using physiological and nontoxic lipids that remain solid at body temperature.124,125 Stable nucleic acid lipid particles (SNALPs) are a major advance in lipid-based siRNA delivery. A proof-of-concept study of the effectiveness of SNALP-siRNA therapeutics has been reported by Geisbert et al.126 In this recent work, siRNA was targeted to interfere with the expression of three Ebola virus proteins. This strategy was used successfully to protect nonhuman primates from lethal Ebola virus infection.
In the SNALP-siRNA panorama, it is worth mentioning a study by Morrissey et al17 which suggested that hepatitis B virus replication could be inhibited by delivery of an siRNA-SNALP complex targeting hepatitis B virus RNA. Finally, in very recent work by Lobovkina et al126 solid lipid nanoparticles were developed for sustained in vivo siRNA delivery in a mouse model. Even in this case, preparing nanoparticles from physiological lipids resulted in excellent biocompatibility, minimal toxicity, and less cost when compared with polymeric carriers.127
Nanoparticles and microspheres have also been developed as gene delivery vehicles. These are promising strategies because they afford improved siRNA delivery and stability, with minimal toxicity in animal models.47,108 They can be engineered to obtain target-specific delivery, for example, by tagging nanoparticle-siRNA oligonucleotide complexes with antibodies that bind the desired target cells. On the other hand, direct injection of synthetic oligonucleotides into solid tumors can be done for some neoplasms, such as mesothelioma (by intrapleural injection), ocular tumors, brain tumors, and sarcomas, and could reduce or eliminate off-target effects.62,106,108 Some companies are exploring these encapsulation technologies. Using this approach, oligonucleotides are sequestered in various kinds of nanoparticles to protect them from degradation and to direct them to appropriate tissues. The idea behind such an approach is that the nanoparticle is seen merely as a carrier, and thus the functions of the vector are to protect, stabilize, and transport the gene material, with the minimum of interaction. Here we give an overview of the main nanoparticles proposed for siRNA delivery and investigation, starting from inorganic crystals, noble metal nanoparticles, and other nanostructures (see Table 3).
Table 3.
Nanoparticles in siRNA-based delivery
| In vitro | In vivo | Clinical | Refs | |
|---|---|---|---|---|
| Nanoparticles in siRNA delivery and investigation | ||||
| Polymer | Yes | Yes | Yes | 28,83,105,128,129 |
| Liposome | Yes | Yes | Yes | 117,130,131,132,133 |
| Quantum dots (QDs) | Yes | Yes | No | 134,135,136 |
| Gold NP | Yes | No | No | 137,138,139,140 |
| Iron oxide | Yes | Yes | No | 141,142 |
| Silica | Yes | No | No | 143 |
| Porous silicon | Yes | Yes | No | 144 |
| Carbon nanotubes (CNT) | Yes | No | No | 145 |
Quantum dots are semiconducting inorganic crystals with superior photostability and tunable optical properties for an extensive selection of nonoverlapping colors.134 They have no bioactivity, and in certain cases, depending on the bulk material used, have no toxicity but a low payload capacity. These features make quantum dots basically useless in designing delivery systems, but they are a powerful tool in cancer targeting, imaging in living animals, and investigation of pathophysiology in tumor tissue.135
Incorporation of siRNA into gold nanoparticles was first accomplished by Oishi et al.137 Further advances in the use of gold particles as carriers have included creation of a supramolecular structure in a layer-by-layer assembly138 and particle surface chemical modification.139,140 Even though the reduced size provides a large surface area for the loading of siRNA-nanoparticle conjugates, chemical modification of both the carrier surface and the transported drug is required. To overcome such problems, new delivery strategies based on porous silicon80 have been developed.
New combination approaches
Multistage delivery systems show promise for meeting the challenges of targeting and overcoming biological barriers. Numerous, chemically different, core-shell nanoparticles with cationic cores and variable shells have been synthesized and tested for intracellular siRNA delivery. In general, this strategy enables tools for nanoparticle delivery to be equipped with the necessary targeting moieties, such as antibodies, aptamers, and small peptides, for direct delivery and improved specificity. These nanoparticles have been engineered to act in a depot manner, resulting in slow, sustained, and targeted release of siRNA. The majority of biodegradable formulations of this kind have used polymeric materials in which siRNA is incorporated in a polymeric core.146–149
Researchers have recently constrained siRNA between a cationic core composed of DOTAP and an outer lipid bilayer of 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-(polyethylene glycol-2000) and egg phosphatidylcholine obtaining a bloodstream circulation time of up to 20 hours after injection.150
A novel approach has been proposed by Tanaka et al144 involving a multistage delivery system composed of two biodegradable and biocompatible carriers. The first-stage carriers are mesoporous, microscale, biodegradable silicon particles that enable loading and release of the second-stage nanocarriers, ie, dioleoyl phosphatidylcholine nanoliposomal siRNA. In vivo mouse models provided the first evidence that single administration of multistage siRNA-dioleoyl phosphatidylcholine delivery resulted in sustained in vivo gene silencing for 3 weeks, with a significant antitumor effect in two orthotopic mouse models of ovarian cancer, and no observable concurrent toxicity.
Current limitations of nanoparticle-siRNA delivery
Nanoparticle-based drug delivery still has some limitations, and the barriers and obstacles encountered by nanoparticle-siRNA complexes are schematized in Figure 3 (adapted from Whitehead50). First of all, these vehicles must reach and enter target cells. The biological barriers encountered are the mucosa and the cellular and humoral arms of the immune system. These molecules tend to be sequestered by negatively charged serum proteins in the bloodstream. Moreover, immune cells can collect them via opsonization.
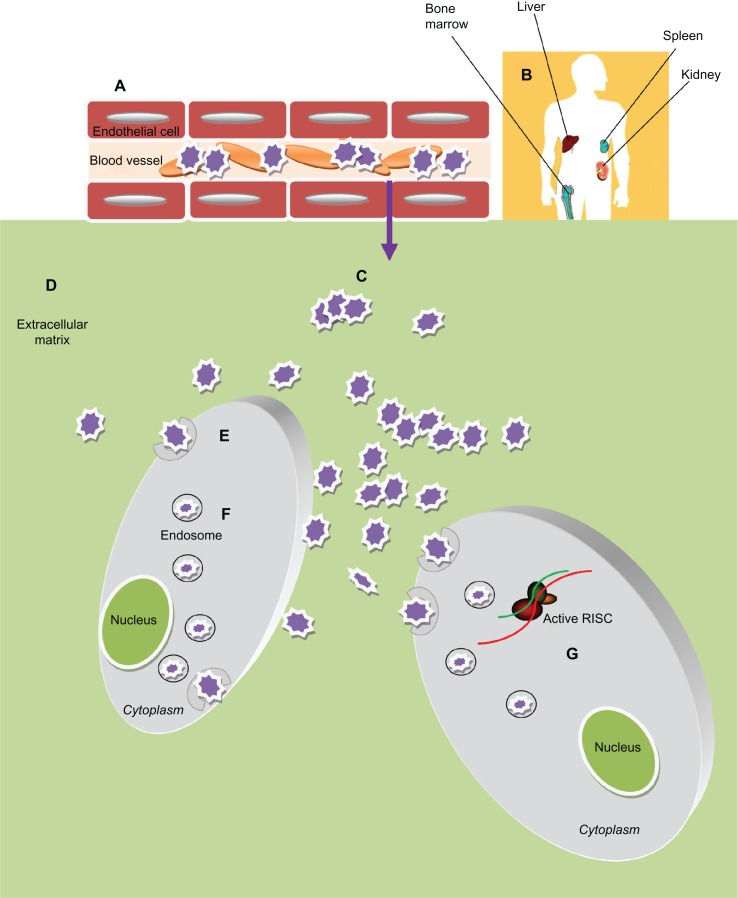
Figure 3.
Obstacles of Nanoparticles-based siRNA delivery in vivo. After administration into blood circulation the siRNA-nanoparticles (A) must avoid rapid degradation by plasma components (eg, cellular and humoral arm of the immune system) and sequester by negatively charged serum protein. (B) Then they need to escape renal filtration and/or clearance by the reticuloendothelial system (RES). (C) To reach the target cells they must overcome the capillary endothelium through an extravasation process and (D) overcome the extracellular matrix (ECM): a dense network of polysaccharides and fibrous proteins, rich in macrophages, which can obstacle the transport of nanoparticles. (E) Furthermore these particles must be taken up into the cells, usually bound to cellular receptors and transported into the cytoplasm through a receptor mediated endocytosis process. (F) Inside the cells the particles need to escape the endosome; (G) thus unpackage and release the siRNA to the RNA interference (RNAi) machinery.
Nanoparticles can be engineered in terms of size, surface functionalization, and core structures151 to overcome these barriers. Indeed, addition of PEG or other hydrophilic conjugates to the surface of a delivery vehicle can reduce protein sequestration, help in evading the immune system, provide steric stabilization, and protect against the effects of the surrounding microenviroment. These nanocomplexes generally undergo rapid clearance.152 The reticuloendothelial system in the liver, spleen, lung, and bone marrow is able to trap intravenously injected particles >100 nm in diameter, leading to their degradation by activated monocytes and macrophages. On the other hand, particles with a diameter <4 nm usually undergo rapid renal clearance. Thus, the optimal diameter of nanoparticles for intratumoral delivery, avoidance of the reticuloendothelial system, and renal clearance should be in the range of 5–100 nm.
Some tumor characteristics can be exploited for drug delivery. The enhanced permeability and retention effect takes advantage of the abnormal neovascularization typical of many cancers.73 This effect is often found in tumors as well as at sites of inflammation and body tissue with other types of disease. The blood vessels are either disrupted or not fully formed, allowing molecules of a certain size to be retained to a greater extent in abnormal tissues than in healthy ones. It has been shown that therapeutic nanoparticles can accumulate in specific target tissues as a result of this effect, potentially allowing for drug release over a longer period of time than for the same drug administered as a conventional preparation.
Conversely, limitations can arise from the intricate tumor microenvironment and interfere with achieving therapeutic concentrations of these molecules. The higher interstitial fluid pressure found in solid tumor tissue could prevent diffusion of nanoparticles.153,154 Strategies adopted to overcome this issue could include normalizing the tumor vasculature (eg, with antiangiogenic therapy) and using nanoparticles with a diameter larger than that of normal vessel fenestrations. The molecules must also face the extracellular matrix, and often the desmoplastic process in cancer creates difficulties in this respect.
Once inside the target cells, the nanoparticles must enter the intracellular trafficking pathway. Many nanoparticles enter cells through endocytosis, and the endosomal vesicles containing RNAi must escape the late endosomal phase in the lysosome that degrades RNA. Fusogenic lipids, fusogenic peptides, photosensitive molecules, pH-sensitive lipoplexes, and pH-sensitive polyplexes are some of the mechanisms used to improve endosomal escape.155
Nanoparticles have often been reported to have systemic toxicity, mostly in the liver.144 Hemolysis, thrombogenicity, and complement activation can also occur as a consequence of certain physical or chemical features of nanoparticles, eg, charge and size, leading to altered biodistribution and potential toxicity.156 Furthermore, most of the current methods used for RNAi delivery require frequent injections119,120 which could represent a great hindrance to their clinical application due to decreased patient compliance.
In the oncology field, given the heterogeneity of tumors, development of resistance seems inevitable. Furthermore, some cancers could have inherent resistance to some types of RNAi, owing to factors such as ethnicity, somatic mutations, altered RNAi processing machinery (Dicer, Drosha, RNA-induced silencing complex), and germline single nucleotide polymorphisms.27
Ongoing clinical trials of siRNA in cancer
The clinical utility of RNAi is still a matter of debate. There has been increasing interest in harnessing this versatile and multifaceted mechanism as a novel pharmacological approach to the treatment of human disease. As has already happened in other developing human therapeutic fields, including gene and antibody therapy, there has been a cooling off after initial enthusiasm. This was driven by a realistic understanding of the various complex milestones that needed to be reached for efficient RNAi-based therapy, before eventual approval for use in human therapy. This trend is now reversing as a result of the excitement generated by the advent of nanomedicine as a potential way of overcoming the challenges in this field. In recent years, there has been an increase in the numbers of preclinical and clinical RNAi-based trials being undertaken. Early proof-of-principle studies in animal models have strengthened the usefulness of RNAi as specific and powerful inhibitors of gene expression without significant toxicity. Translation of animal studies has in some cases progressed to early human trials.
In particular, the research interest is strongest when the aim is targeting genes with single nucleotide polymorphism mutations, as found in dominant-negative disorders, genes specific for pathogenic tumor cells, and genes that are critical for mediating the pathology of various other diseases. In these cases, RNAi could represent the only entities providing an opportunity for a potent and specific approach.157
The relevant clinical trials have addressed many kinds of disease, including retinal degeneration, dominantly inherited brain and skin diseases, viral infections, respiratory disorders, cancer, and metabolic disease). However, large-scale Phase III clinical trials and regulatory approvals remain distant. Use of RNAi for respiratory tract and neurological disorders, metabolic disease (such as hypercholesterolemia), and hepatic cancer has been widely revised in recent times.158 Most of the more advanced clinical trials focus on the treatment of age-related macular degeneration, which is a leading cause of blindness arising from excessive growth of blood vessels and their rupture within the cornea. Of note, a recent study has showed that the efficacy of antivascular endothelial growth factor siRNA in the eye is not due to specific gene silencing, but is actually caused by nonspecific stimulation of the TLR-3 pathway, which can reduce angiogenesis.59
Another important success has been achieved with a formulation targeting the nucleocapsid (N) gene of respiratory syncytial virus, a major cause of respiratory disease in infants and young children. An RSV01 formulation (ALN-RSV01) has completed Phase I investigations and was found to be well tolerated in healthy adults. In July 2009, the complete data from a Phase II study in adult lung transplant patients naturally infected with RSV were reported, documenting the significant antiviral efficacy, safety, and tolerability of this formulation. At this time, a Phase IIb study of ALN-RSV01 for the treatment of respiratory syncytial virus infection is ongoing in adult lung transplant patients. This study aims to repeat and extend the results already seen in this patient population.13,159
Another successful study is one of the siRNA, TD101.160,161 In this prospective, double-blind, split-body, vehicle-controlled, dose-escalation trial, TD101 was administered over a 17-week period to a single patient affected by pachyonychia congenita. This molecule specifically and potently targets keratin 6a N171K mutant mRNA without affecting wild-type keratin 6a mRNA. No adverse events were reported during the trial or during a 3-month washout period. This study was an example of siRNA application in a clinical setting to target a mutant gene or a genetic disorder, and the first to use siRNA targeted to human skin.
Recent reports have discussed other RNAi clinical trials in detail.43,157,158,162 Various clinical trials are in progress, involving many companies.163 We were able to find further as yet unpublished clinical trials registered at http://www.clinicaltrials.gov. Using the keyword “siRNA”, 24 trials were identified and are reported in Table 4. Among these, nine contained the keywords “siRNA and cancer therapy”. Although many hindrances remain for applying these technologies in the treatment of cancer, early clinical results are exciting, and suggest that we are moving forward quickly and also highlight how far we have already come.
Table 4.
Ongoing clinical trials in cancer and other diseases
| Clinical setting | Product | Targeted gene | Location | Disease | Admin | Phase | Stage |
|---|---|---|---|---|---|---|---|
| Cancer | |||||||
| 1 | Proteasome siRNA and tumor antigen RNA-transfected dendritic cells | Immunoproteasome | USA | Melanoma | Vaccination | I | Recruiting |
| 2 | SV40 | Tyrosine kinase | Israel | CML | – | – | Completed |
| 3 | CALAA-01 | Ribonucleotide reductase (M2 subunit) | USA | Cancers | IV | I | Recruiting |
| 4 | Atu027 | An siRNA | Germany | Advanced solid cancer | IV | I | Recruiting |
| 5 | TKM-080301 | PLK1 gene product | USA | MCRC(liver); MPC(liver); MGC(liver); MBC(liver); MOC(liver) | Intra-arterial | I | Recruiting |
| 6 | AHR siRNA | Aromatic hydrocarbon receptor | Taiwan | Neuroblastoma | – | – | Completed |
| 7 | siG12D LODER | – | Israel | PD-Adk; PC | – | I | Recruiting |
| 8 | B4GALNT | Taiwan | Neuroblastoma | – | – | Completed | |
| Others | |||||||
| 1 | AGN211745 siRNA027 |
VEGFR receptor-1 | USA | CNV-AMD | IVT | I/II | Completed* |
| 2 | AGN 211745; ranibizumab | VEGFR receptor-1 | USA | AMD | IVT | II | Terminated |
| 3 | td101 | Keratins, K6a | USA | Pachyonychia C | Intradermal | I | Completed |
| 4 | QPI-1007 | Caspase 2 | USA | Optic atrophy; N-AAION | IVT | I | Recruiting |
| 5 | Bevasiranib | VEGF | USA | DME | IVT | II | Completed |
| 6 | PRO-040201; placebo | – | USA | Hypercholesterolemia | – | I | Terminated |
| 7 | – | IL-10 | Taiwan | Preeclampsia | – | – | Terminated |
| 8 | SYL1001 | – | Spain | Ocular pain; dry eye | Topical | I | Recruiting |
| 9 | Bevasiranib; ranibizumab | VEGF | – | ARMD | IVT | III | Withdrawn |
| 10 | I5NP; placebo | p53 | USA | Injury of kidney Ac renal failure |
IV | I | Completed |
| 11 | I5NP; saline | p53 | USA | Delayed graft function; other complication of kidney transplant | IV | I/II | Recruiting |
| 12 | SYL040012 | β2adrenergic receptor | Spain | Glaucoma; ocular Hypertension |
Local | I/II | Recruiting |
| 13 | Simvastatine | Keratin (K6a-K17) | Israel | Pachyonychia C | Topical | I | Not yet recruiting |
| 14 | TBX3 | TBX3 | USA | Human ES cell differentiation | – | – | Unknown |
| 15 | Bevasiranib | VEGF | USA | MD | IVT | II | Completed |
| 16 | PF-04523655; PF-04523655 and ranibizumab; ranibizumab; PF-04523655 | – | USA | Choroidal Neovasculite Diabetic retinopathy DME |
IVT | II | Not yet recruit |
Abbreviations: MCRC, metastatic colorectal cancer; MPC, metastatic pancreatic cancer; MGC, metastatic Gastric cancer; MBC, Metastatic Breast cancer; MOC, Metastatic Ovarian Cancer; PD-ADK, Pancreatic Ductal Adenocarcinoma; PC, Pancreatic Carcinoma; Pachyonychia C, Pachyonychia Congenita; N-AAION, Non-Arteric Anterior Ischemic Optic Neuropathy; DME, Diabetic Macular Edema; ARMD, Age Related Macular Degeneration; MD, Macular Degeneration; CML, Chronic Myeloid Leukemia; AMD-CNV, Age-Related Macular Degeneration- Choroidal Neovascularization; AMD, Age-Related Macular Degeneration; IVT, Intravitreal Therapy.
An example of the clinical feasibility of siRNA application for cancer therapy is represented by chronic myeloid leukemia. In more than 90% of cases, this disease is caused by a chromosomal aberration, ie, the so-called Philadelphia chromosome. This abnormality is due to fusion between the Abelson (Abl) tyrosine kinase gene at chromosome 9 and the breakpoint cluster (Bcr) gene at chromosome 22, resulting in the chimeric oncogene Bcr-Abl and a constitutively active Bcr-Abl tyrosine kinase implicated in the pathogenesis of chronic myeloid leukemia. On the basis of these observations, an attempt was made to exploit RNAi for inhibition of tumor-specific genes to hit cancer cells in a selective manner. To this end, the Bcr-Abl clinical trial addressed safety and efficacy issues that have now been published.162,164,165 This clinical trial treated only one patient with recurrent Philadelphia chromosome-positive chronic myeloid leukemia by systemic administration of a nonvirally delivered synthetic siRNA against Bcr-Abl. However, effective knockdown of the target gene in circulating leukemic cells was difficult to assess due to concomitant treatment.162 The findings of this study imply that the clinical application of synthetic siRNA is feasible and safe, and has real potential for a genetic-based therapy using a synthetic nonviral carrier.
In 2010, Davis et al28 undertook the first proof-of-principle study of encapsulation technology in human patients. In this small Phase I clinical trial, nanoparticles packed with siRNA were administered systemically on days 1, 3, 8, and 10 every 21 days via a 30-minute intravenous infusion to patients with melanoma refractory to standard of care. This new “drug” was able to deliver its cargo to melanoma cells and to silence the targeted gene.
The nanoparticles used in this study are a paradigm of all that has been discussed to this point, in that they are comprised of separate components encapsulating RNA, targeting specific cell types, favoring stability, and avoiding aggregation, each of which is highly engineered and optimized. These nanoparticles contain a cyclodextrin-based polymer, a human transferrin protein to engage transferrin receptors on the surface of the cancer cells, PEG to promote stability of the nanoparticles in biological fluids, and siRNA. The results provide the first evidence of the importance of RNAi in humans, indicating that siRNA can be used as a gene-specific therapeutic agent.28
Conclusion and future prospects
In just over a decade, RNAi technology has progressed rapidly from an academic discovery to a potential new class of treatment for human disease. Initial observations that were useful for studying gene function in worms were quickly translated to other organisms, and in particular to mammals, revealing the potential clinical applications of siRNA, including an ability to induce potent, persistent, and specific silencing of a wide range of genetic targets. Early excitement was soon dampened by the hurdles and concerns about full exploitation of the power of the RNAi pathway. These include the need to: identify strategies to select potent siRNA with specificity for the target gene, thus minimizing off-target effects; reduce immune stimulation and competition with cellular RNAi components; and enhance effective in vivo delivery to the appropriate cells or tissues by manipulating biopharmaceutical properties. Many of these issues have already been addressed, and others are being addressed at the moment.
The advent of nanomedicine has represented a milestone, contributing strongly towards a solution to problems often encountered in the clinical setting. Recently, nanoparticle-based delivery systems have been shown to have potent RNAi effects after systemic administration. However, their delivery remains one of the most significant obstacles to the widespread use of RNAi therapeutics in the clinical setting, so further progress in this area is needed. To this end, strategies encompassing multiple delivery technologies including novel conjugations and formulations are being explored. Looking forward, synthetic nanoparticles will likely have a fundamental role in the systemic application of siRNA in the clinic. The conjugation of tissue-specific ligands with these particles will enable targeting, and support in vivo biodistribution and delivery.
Tremendous progress has made in exploiting the biological and physical features of cancer to enhance the use of nanoparticles in this disease. The ability of nanoparticles to overcome biological barriers, recognize cancerous versus normal tissue, and deliver therapeutic RNAi is a key determinant of the therapeutic index. In this regard, researchers must also take into account the practical considerations of patient compliance with repeated dosing. Due to the polygenic nature of cancer, the efficacy and specificity of antitumor treatment will probably be further enhanced by using a combination approach of siRNA with chemotherapy, radiotherapy, photodynamic therapy, and/or immunotherapy. It is anticipated that siRNA and nanotechnology will lead the next wave of drug development. In the future, nanoparticles containing siRNA will gain importance as a conventional treatment for cancer and other human diseases.
Acknowledgments
This work was supported in part by the Consorzio Sapienza Innovazione, Associazione Italiana Ricerca Cancro, Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR), Fondo per gli Investimenti della Ricerca di Base (FIRB), Progetti di Ricerca di Interesse Nazionale (PRIN) projects, the Istituto Italiano di Tecnologia (IIT) and European Framework Program 7 (EU-FP7) Initial Training Network (ITN) project n. 238186.
Footnotes
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
- 1.Druker B, Guilhot F, O’Brian S, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 2.Goldenberg MM. Trastuzumab, a recombinant DNA derived humanized monoclonal antibody, a novel agent for the treatment of metastatic breast cancer. Clin Ther. 1999;2:309–318. doi: 10.1016/S0149-2918(00)88288-0. [DOI] [PubMed] [Google Scholar]
- 3.Peterson C. Drug therapy of cancer. Eur J Clin Pharmacol. 2011;67:437–447. doi: 10.1007/s00228-011-1011-x. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert W. The RNA world. Nature. 1986;319:216. [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. Gene expression: RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 8.Wall NR, Shi Y. Small RNA: can RNA interference be exploited for therapy? Lancet. 2003;362:1401–1403. doi: 10.1016/S0140-6736(03)14637-5. [DOI] [PubMed] [Google Scholar]
- 9.Devi GR. siRNA-based approaches in cancer therapy. Cancer Gene Ther. 2006;13:819–829. doi: 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]
- 10.Abdelrahim M, Safe S, Baker C, Abudayyeh C. RNAi and cancer: implications and applications. J RNAi Gene Silencing. 2006;2:136–145. [PMC free article] [PubMed] [Google Scholar]
- 11.Gartel AL, Kandel ES. RNA interference in cancer. Biomol Eng. 2006;23:17–34. doi: 10.1016/j.bioeng.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Grimm D, Kay MA. RNAi and gene therapy: a mutual attraction. Hematol Am Soc Hematol Educ Program. 2007:473–481. doi: 10.1182/asheducation-2007.1.473. [DOI] [PubMed] [Google Scholar]
- 13.DeVincenzo J, Lambkin-Williams R, Wilkinson T, et al. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2010;107:8800–8805. doi: 10.1073/pnas.0912186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanya S, Kim SS, Manjunath N, Shankar P. RNA interference-based therapeutics for human immunodeficiency virus HIV-1 treatment: synthetic siRNA or vector-based shRNA? Expert Opin Biol Ther. 2010;2:201–213. doi: 10.1517/14712590903448158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Cheng G, Mahato RI. RNAi for treating hepatitis B viral infection. Pharm Res. 2008;1:72–86. doi: 10.1007/s11095-007-9504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashfaq UA, Yousaf MZ, Aslam M, Ejaz R, Jahan S, Ullah O. siRNAs: potential therapeutic agents against hepatitis C virus. Virol J. 2011;8:276. doi: 10.1186/1743-422X-8-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrissey DV, Lockridge JA, Shaw L, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 18.DiFiglia M, Sena-Esteves M, Chase K, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci U S A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farah MH. RNAi silencing in mouse models of neurodegenerative diseases. Curr Drug Deliv. 2007;4:161–167. doi: 10.2174/156720107780362276. [DOI] [PubMed] [Google Scholar]
- 20.Li T, Koshy S, Folkesson HG. RNA interference for CFTR attenuates lung fluid absorption at birth in rats. Respir Res. 2008;9:55. doi: 10.1186/1465-9921-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courties G, Presumey J, Duroux-Richard I, Jorgensen C, Apparailly F. RNA interference-based gene therapy for successful treatment of rheumatoid arthritis. Expert Opin Biol Ther. 2009;9:535–538. doi: 10.1517/14712590902926089. [DOI] [PubMed] [Google Scholar]
- 22.Martinez LA, Naguibneva I, Lehrmann H, et al. Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc Natl Acad Sci U S A. 2002;99:14849–14854. doi: 10.1073/pnas.222406899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pai SI, Lin YY, Macaes B, Meneshian A, Hung CF, Wu TC. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13:464–477. doi: 10.1038/sj.gt.3302694. [DOI] [PubMed] [Google Scholar]
- 24.Takeshita T, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689–696. doi: 10.1111/j.1349-7006.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whelan J. First clinical data on RNAi. Drug Discov Today. 2005;10:1014–1015. doi: 10.1016/S1359-6446(05)03547-6. [DOI] [PubMed] [Google Scholar]
- 26.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis ME, Zuckerman JE, Choi CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond SM. Dicing and slicing the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 30.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 31.Tolia NH, Joshua-Tor L. Slicer and the Argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 32.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 33.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 34.Dykxhoorn DM, Lieberman J. Knocking down disease with siRNAs. Cell. 2006;126:231–235. doi: 10.1016/j.cell.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 36.Lingel A, Sattler M. Novel modes of protein-RNA recognition in the RNAi pathway. Curr Opin Struct Biol. 2005;15:107–115. doi: 10.1016/j.sbi.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 40.Röther S, Meister G. Small RNAs derived from longer non-coding RNAs. Biochimie. 2011;93:1905–1915. doi: 10.1016/j.biochi.2011.07.032. [DOI] [PubMed] [Google Scholar]
- 41.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu B, Zhao X, Lee LJ, Lee RJ. Targeted delivery Systems for oligonucleotide therapeutics. AAPS J. 2009;11:195–203. doi: 10.1208/s12248-009-9096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen SH, Zhaori G. Potential clinical applications of siRNA technique: benefits and limitations. Eur J Clin Invest. 2011;41:221–232. doi: 10.1111/j.1365-2362.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- 44.Masiero M, Nardo G, Indraccolo S, Favaro E. RNA interference: implications for cancer treatment. Mol Aspects Med. 2007;1:143–166. doi: 10.1016/j.mam.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Baker M. Homing in on delivery. Nature. 2010;462:1225–1228. doi: 10.1038/4641225a. [DOI] [PubMed] [Google Scholar]
- 46.De Smaele E, Ferretti E, Gulino A. MicroRNAs as biomarkers for CNS cancer and other disorders. Brain Res. 2010;1338:100–111. doi: 10.1016/j.brainres.2010.03.103. [DOI] [PubMed] [Google Scholar]
- 47.Aagaard L, Rossi JJ. RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev. 2007;59:75–86. doi: 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X, Pan F, Holt CM, Lewis AL, Lu JR. Controlled delivery of antisense oligonucleotides: a brief review of current strategies. Expert Opin Drug Deliv. 2009;6:673–686. doi: 10.1517/17425240902992894. [DOI] [PubMed] [Google Scholar]
- 49.Bumcrot D, Manoharan M, Koteliansky V, Sah DWY. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;2:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;3:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 52.Heidel JD, Yu Z, Liu JY, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunitM2 siRNA. Proc Natl Acad Sci U S A. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desai AA, Schilsky RL, Young A, et al. A phase I study of antisense oligonucleotide GTI-2040 given by continuous intravenous infusion in patients with advanced solid tumors. Ann Oncol. 2005;6:958–965. doi: 10.1093/annonc/mdi178. [DOI] [PubMed] [Google Scholar]
- 54.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to over-saturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 55.Castanotto D, Sakurai K, Lingeman R, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35:5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hornung V, Guenthner-Biller M, Bourquin C, et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 57.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 58.Gantier MP, Williams BR. The response of mammalian cells to double-stranded RNA. Cytokine Growth Factor Rev. 2007;18:363–371. doi: 10.1016/j.cytogfr.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kleinman ME, Yamada K, Takeda A, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson AL, Bartz SR, Schelter J, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 61.Jackson AL, Burchard J, Schelter J, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1:347–355. [PubMed] [Google Scholar]
- 63.Oliveira S, van Rooy I, Kranenburg O, Storm G, Schiffelers R. Fusogenic peptides enhance endosomal escape improving siRNA-induced silencing of oncogenes. Int J Pharm. 2007;331:211–214. doi: 10.1016/j.ijpharm.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 64.Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J Intern Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miele E, Spinelli GP, Miele E, Tomao F, Tomao S. Albumin-bound formulation of paclitaxel (Abraxane ABI-007) in the treatment of breast cancer. Int J Nanomedicine. 2009;4:99–105. doi: 10.2147/ijn.s3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Angelis F, Gentile F, Mecarini F, et al. Breaking the diffusion limit with super-hydrophobic delivery of molecules to plasmonic nanofocusing SERS structures. Nat Photonics. 2011;5:682–687. [Google Scholar]
- 67.Pujia A, De Angelis F, Scumaci D, et al. Highly efficient human serum filtration with water-soluble nanoporous nanoparticles. Int J Nanomedicine. 2010;5:1005–1015. doi: 10.2147/IJN.S12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farokhzad OC, Langer R. Impact of nanotechnology on drug delivery. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 69.Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:61–71. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 70.Wong MS. Nanoparticles and catalysis. In: Astruc Didier., editor. Angewandte Chemie International Edition. Weindham, Germany: Wiley; 2008. [Google Scholar]
- 71.Knöpfel T, Akemann W. Nanobiotechnology: remote control of cells. Nat Nanotechnol. 2010;8:560–561. doi: 10.1038/nnano.2010.163. [DOI] [PubMed] [Google Scholar]
- 72.Michalet X, Pinaud FF, Bentolila LA, et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glass G. Pharmaceutical patent challenges – time for reassessment? Nat Rev Drug Discov. 2004;3:1057–1062. doi: 10.1038/nrd1581. [DOI] [PubMed] [Google Scholar]
- 75.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarrier as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 76.Weisleder R, Kimberly K, Yi Sun E, Shtaland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 77.Byrne JD, Betancourt T, Brannon-Peppas L. Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv Drug Deliv Rev. 2008;60:1615–1626. doi: 10.1016/j.addr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 78.Park Ji-Ho, Gu L, von Maltzahn G, Ruoslahti E, Bhatia SN, Sailor MJ. Biodegradable luminescent porous silicon nanoparticles for in vivo applications. Nat Mater. 2009;8:331–336. doi: 10.1038/nmat2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Janib SM, Moses AS, MacKay JA. Imaging and drug delivery using theranostic nanoparticles. Adv Drug Deliv Rev. 2010;62:1052–1063. doi: 10.1016/j.addr.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tasciotti E, Liu X, Bhavane R, et al. Mesoporous silicon particles as a multistage delivery system for imaging and therapeutic application. Nat Nanotechnol. 2008;3:152–157. doi: 10.1038/nnano.2008.34. [DOI] [PubMed] [Google Scholar]
- 81.Chan JM, Zhang L, Tong, et al. Spatiotemporal controlled delivery of nanoparticles to injured vasculature. Proc Natl Acad Sci U S A. 2010;107:2213–2218. doi: 10.1073/pnas.0914585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sengupta S, Eavarone D, Capila I, et al. Temporal targeting of tumor cells and neovasculature with a nanoscale delivery system. Nature. 2005;436:568–572. doi: 10.1038/nature03794. [DOI] [PubMed] [Google Scholar]
- 83.Bigall NC, Curcio A, Pernia Leal MP, et al. Magnetic nanocarriers with tunable pH dependence for controlled loading and release of cationic and anionic payloads. Adv Mater. 2011;23:5645–5650. doi: 10.1002/adma.201103505. [DOI] [PubMed] [Google Scholar]
- 84.Waehler R, Russell SJ, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8:573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109:259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- 86.Gao Y, Liu XL, Li XR. Research progress on siRNA delivery with nonviral carriers. Int J Nanomedicine. 2011;6:1017–1025. doi: 10.2147/IJN.S17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm Res. 2009;26:657–666. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- 88.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Ther. 2008;15:1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 89.Tokatlian T, Segura T. siRNA applications in nanomedicine. Nanomed Nanobiotechnol. 2010;2:305–315. doi: 10.1002/wnan.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Malik R, Roy L. Design and development of antisense drugs. Exp Opin Drug Discov. 2008;3:1189–1207. doi: 10.1517/17460441.3.10.1189. [DOI] [PubMed] [Google Scholar]
- 93.Chiarantini L, Cerasi A, Fraternale A, et al. Comparison of a novel delivery system for antisense peptide nucleid acids. J Control Release. 2005;109:24–36. doi: 10.1016/j.jconrel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 94.Soutschek J, Akinc A, Bramlage B, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 95.Raal FD, Santos RD, Blom DJ, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 96.Zhao X, Li H, Lee JR. Targeted drug delivery via folate receptors. Expert Opin Drug Deliv. 2008;5:309–319. doi: 10.1517/17425247.5.3.309. [DOI] [PubMed] [Google Scholar]
- 97.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 98.Song E, Zhu P, Lee SK, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 99.McNamara JO, 2nd, Andrechek ER, Wang Y, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 100.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 101.Hatakeyama H, Ito E, Akita H, et al. A pH-sensitive fusogenic peptide facilitates endosomal escape and greatly enhances the gene silencing of siRNA-containing nanoparticles in vitro and in vivo. J Control Release. 2009;139:127–132. doi: 10.1016/j.jconrel.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 102.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pirollo KF, Chang EH. Targeted delivery of small interfering RNA: approaching effective cancer therapies. Cancer Res. 2008;68:1247–1250. doi: 10.1158/0008-5472.CAN-07-5810. [DOI] [PubMed] [Google Scholar]
- 104.Lee H, Mok H, Lee S, Oh YK, Park TG. Target specific intracellular delivery of siRNA using degradable hyaluronic acid nanogels. J Control Release. 2007;119:245–252. doi: 10.1016/j.jconrel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 105.Rozema DB, Lewis DL, Wakefield DH, et al. Dynamic polyconjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci U S A. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zuhorn IS, Engberts JB, Hoeckstra D. Gene delivery by cationic lipid vectors: overcoming cellular barriers. Eur Biophys J. 2007;36:349–362. doi: 10.1007/s00249-006-0092-4. [DOI] [PubMed] [Google Scholar]
- 107.Chirila TV, Rakoczy PE, Garrett KL, Lou X, Constable IJ. The use of synthetic polymers for delivery of therapeutic antisense oligodeoxynucleotides. Biomaterials. 2002;23:321–342. doi: 10.1016/S0142-9612(01)00125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 109.Convertine AJ, Benoit DS, Duvall CL, Hoffman AS, Stayton PS. Development of a novel endosomolytic diblock copolymer for siRNA delivery. J Control Release. 2009;133:221–229. doi: 10.1016/j.jconrel.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wagner E. Polymers for siRNA delivery: inspired by viruses to be targeted, dynamic, and precise. Acc Chem Res. 2011;22 doi: 10.1021/ar2002232. [DOI] [PubMed] [Google Scholar]
- 111.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 112.Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61:850–862. doi: 10.1016/j.addr.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 113.Nimesh S, Gupta N, Chandra R. Cationic polymer based nanocarriers for delivery of therapeutic nucleic acids. J Biomed Nanotechnol. 2011;7:504–520. doi: 10.1166/jbn.2011.1313. [DOI] [PubMed] [Google Scholar]
- 114.Singha K, Namgung R, Kim WJ. Polymers in small-interfering RNA delivery. Nucleic Acid Ther. 2011;21:133–147. doi: 10.1089/nat.2011.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Troiber C, Wagner E. Nucleic acid carriers based on precise polymer conjugates. Bioconjug Chem. 2011;22:1737–1752. doi: 10.1021/bc200251r. [DOI] [PubMed] [Google Scholar]
- 116.Duncan R. Polymer therapeutics as nanomedicines: new perspectives. Curr Opin Biotechnol. 2011;22:492–501. doi: 10.1016/j.copbio.2011.05.507. [DOI] [PubMed] [Google Scholar]
- 117.Semple SC, Akinc A, Chen J, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 118.Guo P, Coban O, Snead NM, et al. Engineering RNA for targeted siRNA delivery and medical application. Adv Drug Deliv Rev. 2010;62:650–666. doi: 10.1016/j.addr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Merritt WM, Lin YG, Spannuth WA, et al. Effect of interleukin-8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100:359–372. doi: 10.1093/jnci/djn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Landen CN, Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 121.Dharmapuri S, Peruzzi D, Marra E, et al. Intratumor RNA interference of cell cycle genes slows down tumor progression. Gene Ther. 2011;18:727–733. doi: 10.1038/gt.2011.27. [DOI] [PubMed] [Google Scholar]
- 122.Ma Z, Li J, He F, Wilson A, Pitt B, Li S. Cationic lipids enhance siRNA-mediated interferon response in mice. Biochem Biophys Res Commun. 2005;330:755–759. doi: 10.1016/j.bbrc.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 123.Dokka S, Toledo D, Shi X, Castranova V, Rojanasakul Y. Oxygen radical-mediated pulmonary toxicity induced by some cationic liposomes. Pharm Res. 2000;17:52152–52155. doi: 10.1023/a:1007504613351. [DOI] [PubMed] [Google Scholar]
- 124.Mehnert W, Mäder K. Solid lipid nanoparticles: production, characterization and applications. Adv Drug Deliv Rev. 2001;47:165–196. doi: 10.1016/s0169-409x(01)00105-3. [DOI] [PubMed] [Google Scholar]
- 125.Gallarate M, Battaglia L, Peira E, Trotta M. Peptide-loaded solid lipid nanoparticles prepared through coacervation technique. International Journal of Chemical Engineering. 2011:1–6. [Google Scholar]
- 126.Geisbert TW, Lee AC, Robbins M, et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet. 2010;375:1896–1905. doi: 10.1016/S0140-6736(10)60357-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lobovkina T, Jacobson GB, Gonzalez-Gonzalez E, et al. In vivo sustained release of siRNA from solid lipid nanoparticles. ACS Nano. 2011;5:9977–9983. doi: 10.1021/nn203745n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. PEG conjugated VEGF siRNA for anti-angiogenic gene therapy. J Control Release. 2006;116:1239. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 129.Boussif O, Lezoualc’h F, Zanta MA, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zimmermann TS, Lee ACH, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 131.Akinc A, Goldberg M, Qin J, et al. Development of lipidoid-siRNA formulations for systemic delivery to the liver. Mol Ther. 2009;17:872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Akinc A, Zumbuehl A, Goldberg M, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vaishnaw AK, Gollob J, Gamba-Vitalo C, et al. A status report on RNAi therapeutics. Silence. 2010;1:14. doi: 10.1186/1758-907X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen AA, Derfus AM, Khetani SR, Bhatia SN. Quantum dots to monitor RNAi delivery and improve gene silencing. Nucleic Acids Res. 2005;33:190. doi: 10.1093/nar/gni188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gao X, Cui Y, Levenson RM, Chung LWK, Nie1 S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 136.Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. Proton-sponge coated quantum dots for siRNA delivery and intracellular imaging. J Am Chem Soc. 2008;130:9006–9012. doi: 10.1021/ja800086u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oishi M, Nakaogami J, Ishii T, Nagasaki Y. Smart PEGylated gold nanoparticles for the cytoplasmic delivery of siRNA to induce enhanced gene silencing Chem. Lett. 2006;35:1046–1047. [Google Scholar]
- 138.Elbakry A, Zaky A, Liebl R, Rachel R, Goepferich A, Breunig M. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009;5:2059–2064. doi: 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- 139.Lee JS, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Gold, poly(beta-amino ester) nanoparticles for small interfering RNA delivery. Nano Lett. 2009;6:2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lytton-Jean A, Langer R, Anderson DG. Five years of siRNA delivery: spotlight on gold nanoparticles. Small. 2011;7:1932–1937. doi: 10.1002/smll.201100761. [DOI] [PubMed] [Google Scholar]
- 141.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. In vivo imaging of siRNA delivery and silencing in tumors. Nat Med. 2007;3:372–377. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 142.Curcio A, Marotta R, Riedinger A, Palumberi D, Falqui A, Pellegrino T. Magnetic pH-responsive nanogels as multifunctional delivery tools for small interfering RNA (siRNA) and iron oxide nanoparticles (IONPs) Chem Commun (Camb) 2012;48:2400–2402. doi: 10.1039/c2cc17223b. [DOI] [PubMed] [Google Scholar]
- 143.Xia T, Kovochich M, Liong M, et al. Polyethyleneimine coating enhances the cellular uptake of mesoporous silica nanoparticles and allows safe delivery of siRNA and DNA constructs. ACS Nano. 2009;3:3273–3286. doi: 10.1021/nn900918w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tanaka T, Mangala LS, Vivas-Mejia PE, et al. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Res. 2010;70:3687–3696. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kam NW, Liu Z, Dai H. Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. J Am Chem Soc. 2005;127:12492–12493. doi: 10.1021/ja053962k. [DOI] [PubMed] [Google Scholar]
- 146.Lee S, Yang SC, Kao CY, Pierce RH, Murthy N. Solid polymeric microparticles enhance the delivery of siRNA to macrophages in vivo. Nucleic Acids Res. 2009;37:145. doi: 10.1093/nar/gkp758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jacobson GB, Shinde R, McCullough RL, et al. Nanoparticle formation of organic compounds with retained biological activity. J Pharm Sci. 2010;99:2750–2755. doi: 10.1002/jps.22035. [DOI] [PubMed] [Google Scholar]
- 148.Krebs MD, Jeon O, Alsberg E. Localized and sustained delivery of silencing RNA from macroscopic biopolymer hydrogels. J Am Chem Soc. 2009;131:9204–9206. doi: 10.1021/ja9037615. [DOI] [PubMed] [Google Scholar]
- 149.Patil Y, Panyam J. Polymeric nanoparticles for siRNA delivery and gene silencing. Int J Pharm. 2009;367:195–203. doi: 10.1016/j.ijpharm.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yagi N, Manabe I, Tottori T, et al. A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo. Cancer Res. 2009;69:6531–6538. doi: 10.1158/0008-5472.CAN-08-3945. [DOI] [PubMed] [Google Scholar]
- 151.Petros RA, DeSimone JM. Strategies in the design of nanoparticles for therapeutic applications. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 152.Longmire M, Choyke PL, Kobayashi H. Clearance properties of nanosized particles and molecules as imaging agents: considerations and caveats. Nanomedicine (Lond) 2008;5:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Decuzzi P, Causa F, Ferrari M, Netti PA. The effective dispersion of nanovectors within the tumor microvasculature. Ann Biomed Eng. 2006;4:633–641. doi: 10.1007/s10439-005-9072-6. [DOI] [PubMed] [Google Scholar]
- 154.Li L, Wilcox D, Zhao X, et al. Tumor vasculature is a key determinant for the efficiency of nanoparticle-mediated siRNA delivery. Gene Ther. 2011:1–6. doi: 10.1038/gt.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 156.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol Pharm. 2008;4:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Seyhan AA. RNAi: a potential new class of therapeutic for human genetic disease. Hum Genet. 2011;130:583–605. doi: 10.1007/s00439-011-0995-8. [DOI] [PubMed] [Google Scholar]
- 158.Davidson BL, McCray PB. Current prospects for RNA interference-based therapies. Nat Rev Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Alnylam Pharmaceuticals Alnylam achieves first human proof of concept for an RNAi therapeutic with GEMINI study. Available from: http://www.alnylam.com/Programs-and-Pipeline/Partner-Programs/index.php. Accessed April 17, 2012.
- 160.Leachman SA, Hickerson RP, Hull PR, et al. Therapeutic siRNAs for dominant genetic skin disorders including pachyonychia congenita. J Dermatol Sci. 2008;51:151–157. doi: 10.1016/j.jdermsci.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Leachman SA, Hickerson RP, Schwarz ME, et al. First-in-human mutation targeted siRNA phase Ib trial of an inherited skin disorder. Mol Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gondi C, Rao JS. Concepts in in-vivo siRNA delivery for cancer therapy. J Cell Physiol. 2009;220:285–291. doi: 10.1002/jcp.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ptasznik A, Nakata Y, Kalota A, Emerson SG, Gewirtz AM. Short interfering RNA (siRNA) targeting the Lyn kinase induces apoptosis in primary, and drug resistant, BCR-ABL1+ leukemia cells. Nat Med. 2004;10:1187–1189. doi: 10.1038/nm1127. [DOI] [PubMed] [Google Scholar]
- 164.Koldehoff M, Steckel NK, Beelen DW, Elmaagacli AH. Therapeutic application of small interfering RNA directed against bcr-abl transcripts to a patient with imatinib-resistant chronic myeloid leukaemia. Clin Exp Med. 2007;7:47–55. doi: 10.1007/s10238-007-0125-z. [DOI] [PubMed] [Google Scholar]
- 165.Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in-vivo. FEBS J. 2010;277:4814–4817. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- 166.Vaccari L, Canton D, Zaffaroni N, et al. Porous silicon as drug carrier for controlled delivery of doxorubicin anticancer agent. Micr Eng. 2006;83:1598–1601. [Google Scholar]
- 167.Gentile F, Tirinato L, Battista E, et al. Cells preferentially grow on rough substrates. Biomaterials. 2010;31:7205–7212. doi: 10.1016/j.biomaterials.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 168.Gaspari M, Ming-Cheng Cheng M, Terracciano R, et al. Nanoporous surfaces as harvesting agents for mass spectrometric analysis of peptides in human plasma. J Proteome Res. 2006;5:1261–1266. doi: 10.1021/pr050417+. [DOI] [PubMed] [Google Scholar]