Abstract
Infantile hypertrophic pyloric stenosis (IHPS), characterized by enlarged pyloric musculature and gastric-outlet obstruction, is associated with altered expression of neuronal nitric oxide synthase (nNOS). Here we have studied molecular mechanisms by which nNOS gene expression is altered in pyloric tissues of 16 infants with IHPS and 9 controls. A significant decreased expression of total nNOS mRNA was found by quantitative RT-PCR in IHPS after normalization against GAPDH, which predominantly affected exon 1c with a reduction of 88% compared with controls (P < 0.001). After normalization against the neuronal-specific gene PGP9.5, expression of exon 1c was still decreased (P < 0.001), whereas expression of exon 1f was increased significantly (P = 0.001), indicating a compensatory up-regulation of this nNOS mRNA variant. DNA samples of 16 IHPS patients and 81 controls were analyzed for nNOS exon 1c promoter mutations and single-nucleotide polymorphism (SNP). Sequencing of the 5′-flanking region of exon 1c revealed mutations in 3 of 16 IHPS tissues, whereas 81 controls showed the wild-type sequence exclusively. Carriers of the A allele of a previously uncharacterized nNOS exon 1c promoter SNP (-84G → A) had increased risk for development of IHPS (odds ratio, 8.0; 95% confidence interval, 2.5–25.6). Reporter gene assays revealed an unchanged promoter activity for mutations but a ≈30% decrease for the -84A SNP (P < 0.001). In summary, our findings indicate that genetic alterations in the nNOS exon 1c regulatory region influence expression of the nNOS gene and may contribute to the pathogenesis of IHPS.
Infantile hypertrophic pyloric stenosis (IHPS), with an incidence of 1–5 per 1,000 live births in whites and a marked preponderance of males to females (4:1), is the most frequent disorder requiring surgery in the first year of life (1). A genetic contribution toward the etiology of IHPS is well established with familiar aggregation, a concordance rate of 25–40% in monogenetic twins, a recurrence risk of 10% for males and 2% for females born after an affected child, and a ratio of risk of 18 for first-degree relatives compared with the general population (2).
IHPS is characterized by hypertrophy and hyperplasia of the circular muscle layer of the pylorus, leading to persistent vomiting 2–12 weeks after birth (3, 4). Defective pyloric relaxation and increased pyloric smooth muscle mass have been suggested to be responsible for gastric-outlet obstruction. In most cases, surgical pyloromyotomy is the treatment of choice, but conservative management has sometimes been used successfully (5, 6). Within a few months, hypertrophy resolves and no permanent functional or morphological abnormalities have been detected after healing (5, 7, 8).
Recent evidence suggested a reduced expression of neuronal nitric oxide synthase (nNOS) in IHPS, because the pyloric circular muscle layer shows decreased staining of nNOS-positive neurons (9) and decreased expression of nNOS mRNA normalized against GAPDH (10). Additionally, genetic-linkage analysis suggests the nNOS gene as a susceptibility locus for hereditary IHPS (11). In addition to decreased nNOS expression, other abnormalities have been described, such as a reduced amount of interstitial cells of Cajal (12) and a decreased expression of various neurotransmitters (for review see ref. 13).
NO, generated by nNOS, is a major inhibitory transmitter in the gut. It is involved in the intrinsic and extrinsic inhibitory innervation of the pyloric muscle (14), and genetic deletion of nNOSα in mice causes a phenotype closely resembling IHPS with delayed gastric emptying, enlargement of the stomach, and hypertrophy of the muscle layer of the antrum (15, 16). Furthermore, deletion of cGMP-dependent kinase I, the downstream target of the NO/cGMP signaling pathway, leads to a similar phenotype with pyloric stenosis, delayed gastric emptying, and thickened pyloric muscle (17). Additional evidence for an important role of the NO/cGMP/cGMP-dependent kinase signaling cascade in the pathogenesis of IHPS comes from observations in vascular and airway smooth muscles. In these cells, lack of NO results in proliferation and hypertrophy (18–20), suggesting an inhibitory effect of NO on smooth muscle cell growth beneath the well established role of NO in smooth muscle relaxation. Similar effects of NO have been observed in cardiomyocytes, in which signaling by cGMP-dependent kinase I acts as a negative regulator of cardiac myocyte hypertrophy (21), and in Xenopus brain, in which NO is an essential negative regulator of cell proliferation (22).
The major source for production of NO in the human gut is nNOS (23). The nNOS gene is one of the most complex genes known in terms of first-exon usage and alternative splicing (24). Nine distinct first-exon transcripts (exon 1a–1i), generated by alternative promoter usage, are present in the human gut, leading to nNOS mRNA variants with different 5′-untranslated regions and translational efficiencies (23–27). Cell- and site-specific expression patterns (23, 25) indicate a spatial and temporal regulation of nNOS expression under different physiological or pathological conditions.
Here, we present evidence for a specific decrease of nNOS exon 1c mRNA in neurons innervating the pyloric sphincter of patients with IHPS, whereas other transcripts were unaffected or up-regulated. We identified a previously uncharacterized single-nucleotide polymorphism (SNP) in the promoter of nNOS exon 1c (-84G → A) that increases the risk for development of IHPS substantially and alters promoter activity by ≈30%. Additionally, point mutations in the promoter region of nNOS exon 1c that may contribute to the pathogenesis of IHPS were found in a few patients.
Methods
Materials. The human neuroblastoma cell line TGW-nu-I was kindly provided by H. Esumi (National Cancer Center Research Institute East, Chiba, Japan) (28). All cell-culture reagents were obtained from Invitrogen. Primers were made by MWG Biotech (Ebersberg, Germany), and TaqMan probes were made by Applied Biosystems.
Patient Population and Tissue Samples. This study conformed to the Declaration of Helsinki and was approved by the institutional review board at the Université Libre de Bruxelles (Belgium). Sixteen patients with IHPS who underwent pyloromyotomy at the Department of Surgery (Université Libre de Bruxelles) were studied. In the IHPS group, the mean age was 5.2 ± 2.1 weeks (range, 2–9 weeks), and the male/female ratio was 2:1. Nine patients (seven boys and two girls) who underwent surgery as organ donor (n = 2), for pyloroplasty for gastroesophageal reflux disease (n = 5), and a postmortem specimen sampled with a delay of 24 h from an infant deceased presumably from sudden infant death syndrome (n = 1) served as controls. The mean age of the control group was 45.3 ± 47.0 weeks (range, 6–161 weeks). To study polymorphism of the promoter region of nNOS exon 1c, genomic DNA was isolated from leukocytes after obtaining informed consent from 72 control subjects (average age, 38.9 ± 13.1 years; range, 19–75 years; male/female ratio, 1.66:1), free of gastrointestinal disorders and with no history of IHPS, recruited at the outpatient clinic of the University Hospital of Munich (Klinikum rechts der Isar, Munich).
Tissue Preparation and RNA and DNA Isolation. Full-thickness pyloric muscle biopsy specimens were cleaned in PBS and immersed in RNAlater (Ambion, Austin, TX), and total RNA was isolated as described (29). Genomic DNA was isolated from pyloric tissue samples from 16 IHPS patients and 9 controls and lymphocytes from 72 blood donors by standard techniques (23).
Rapid Amplification of 5′ cDNA Ends (5′-RACE) PCR. To determine 5′-end mRNA variants of nNOS in the pyloric sphincter of patients with IHPS and controls, two independent RACE protocols were performed exactly as described (23).
Quantitative Real-Time RT-PCR. Real-time quantitative RT-PCR analyses were performed for the alternative first exons 1a–1i of human nNOS and exon 6/7 as a parameter for total nNOS mRNA expression exactly as described (25) by using the Prism 7700 Sequence Detection System (Applied Biosystems; for primer and probes see ref. 25). In addition, the neuronal inhibitory transmitter vasoactive intestinal peptide (VIP) was quantified by using intron-spanning primers and probes (for sequences see Table 2, which is published as supporting information on the PNAS web site). As endogenous references, the globally expressed housekeeping genes GAPDH, HPRT, and 18SRNA (primers and probes were purchased from Applied Biosystems) and the neuron-specific genes PGP9.5, Peripherin, and microtubule-associated protein 2 (MAP-2) were used (for primers and probes, see Table 2).
PCR. PCR amplification was carried out as described (23, 25). A 381-bp fragment containing the promoter of exon 1c (nucleotides -332 to +49) (25) was amplified by using a Pfu Turbo proofreading polymerase (Stratagene) and the primers P-332 (sense) and P+49 (antisense) (see Table 2). PCR products were analyzed by direct automated DNA sequencing with a Prism 377 DNA sequencer (Applied Biosystems).
Genotyping. Genotype analysis was performed by using the TaqMan 5′-nuclease assay (Applied Biosystems) with allele-specific fluorogenic oligonucleotide probes. Primers and probes for a newly identified G-to-A SNP of the nNOS exon 1c promoter at position -84 relative to the transcription start site (25) were designed by using primer express software (see Table 2). Allele-specific TaqMan PCR was performed by using the Prism 7700 Sequence Detection System, 30 ng of genomic DNA, 900 nM primers, 250 nM probes, and 12.5 μl of TaqMan Universal Master Mix in a 25-μl final reaction mixture (95°C, 10 min; 40 PCR cycles, 60°C, 1 min; 95°C, 15 sec). Signals were analyzed by sds 1.7 software (Applied Biosystems). The genotyping results were confirmed by direct sequencing of selected probes.
Plasmid Constructions. The human genomic 5′-flanking region of nNOS exon 1c was characterized by 5′-RACE PCR (25). PCR products containing the mutations and the G or A allele of the SNP within the promoter of exon 1c were blunt-end cloned into the SmaI site of the promoterless/enhancerless firefly luciferase reporter gene vector pGL3-basic (Promega). Construction of additional 5′ reporter constructs (pGL3-1938/+49, pGL3-48/+49) has been described (25). Integrity of all cloned sequences was confirmed by DNA sequencing.
Cell Culture, Transient Expression, and Reporter Gene Assays. TGW-nu-I cells were cultured and transiently cotransfected with the different nNOS exon 1c-pGL3 promoter constructs and the Renilla luciferase expression vector phRL-TK (Promega) to normalize for transfection efficiency and cell number as described (25). Forty-eight hours after transfection, cells were harvested, and firefly and Renilla luciferase activities were measured as described (23, 25). Data are presented as relative luciferase activity of firefly/Renilla luciferase.
Data Analysis. Unless otherwise indicated, all data were determined from three independent experiments, each done in triplicate, and expressed as mean values ± SD. Comparisons among data sets were made by using ANOVA, followed by Student's t test or Mann–Whitney rank sum test. For multiple testing, a Bonferroni correction of the P values was made. The χ2 or Fisher's exact test was used to compare the frequencies of the -84G/A SNP of the nNOS exon 1c promoter between IHPS cases and the control group. Logistic regression analysis was performed by using statxact 4.0.1 software (Cytel Software, Cambridge, MA) to determine the odds ratios with 95% confidence intervals for the risk of the occurrence of IHPS. Values of P < 0.05 (two-tailed) were considered to be statistically significant.
Results
Analysis of 5′-mRNA Variants of nNOS. Cloning of 5′-RACE PCR products from pyloric tissues of controls and patients with IHPS revealed expression of the already known alternative 5′-mRNA variants of nNOS, expressed in the human gut (23, 25). No novel 5′-mRNA forms were found. In addition, we could not detect the alternatively spliced untranslated leader exon AS in infantile pyloric specimens, which has been shown to alter translation of human nNOS (26).
Quantitative Expression Patterns of nNOS First-Exon mRNA Variants in Normal and Hypertrophic Pyloric Tissue Specimens. We analyzed mRNA expression of nine alternative first-exon variants of nNOS, present in the human gastrointestinal tract as determined by 5′-RACE PCR, by real-time quantitative RT-PCR in pyloromyotomy specimens of controls and patients with IHPS. As a parameter for total nNOS mRNA expression, a sequence between exons 6 and 7, encoding parts of the oxygenase domain that is essential for NOS activity, was amplified. To normalize nNOS mRNA expression, we used two different approaches. First, the globally expressed housekeeping genes GAPDH, HPRT, and 18SRNA were quantified to normalize nNOS mRNA expression for total pyloric tissue content. Second, the neuron-specific genes PGP9.5, MAP-2, and Peripherin were amplified to normalize nNOS mRNA expression in enteric neurons.
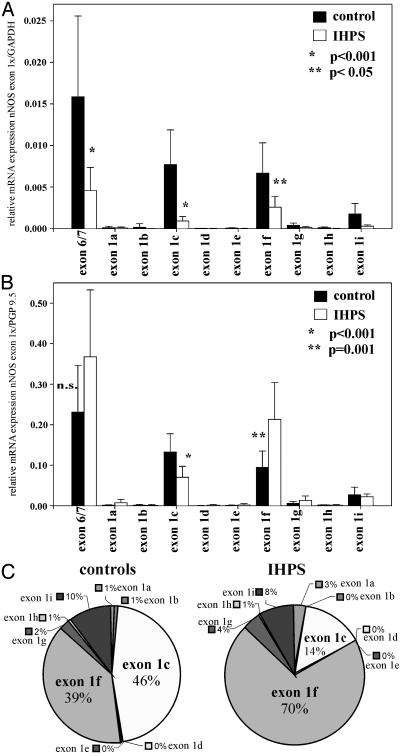
When normalized against GAPDH, pyloric tissues from control infants showed high expression levels of total nNOS mRNA and a characteristic pattern of nNOS first-exon variants (Fig. 1A). Exon 1c and 1f were the predominant forms showing ≈7.7 and 6.5 mRNA copies per 1,000 copies of GAPDH, respectively. In contrast, in infants with IHPS, the overall expression of nNOS mRNA was decreased significantly (P < 0.001; see Fig. 1A). This decrease predominantly affected exon 1c, with a reduction of ≈88% compared with control tissues (P < 0.001; 0.92 copies per 1,000 copies of GAPDH). Similar results were obtained when other ubiquitous housekeeping genes (HPRT and 18SRNA) were used as reference (data not shown). Taken together, these results suggest an undersupply of the thickened pyloric smooth muscle with NO in IHPS patients. Because nNOS is expressed in the human enteric nervous system but not in enteric smooth muscle cells (9, 30), the reduced abundance of nNOS mRNA transcripts normalized against ubiquitous housekeeping genes most likely reflects hypertrophy and hyperplasia of enteric smooth muscle cells leading to a relative decrease of enteric neurons in tissue specimen of IHPS patients when compared with controls. Therefore, expression of nNOS mRNA was assessed further by normalization against PGP9.5, a marker for general neural tissue (Fig. 1B), which has been used in several studies to normalize neuronal gene expression in the central and peripheral nervous system (e.g., ref. 31). Furthermore, it has been shown that PGP9.5 expression can be used as a reference to study gene expression in tissue specimens of IHPS patients and controls (30, 32). Therefore, PGP9.5 is a reliable internal standard for normalization of nNOS mRNA expression in enteric neurons of patients with IHPS. Interestingly, only exon 1c showed a significant reduced expression in IHPS tissues versus controls after normalization against the neuronal marker PGP9.5 (P < 0.001; Fig. 1B). In contrast, nNOS exon 1f expression was increased significantly in enteric neurons of patients with IHPS (P = 0.001; Fig. 1B), and total nNOS expression was unchanged when compared with controls. Similar results were obtained when two other general neuronal markers, Peripherin and MAP-2, were used for normalization of nNOS mRNA expression (see Fig. 4 A and B, which is published as supporting information on the PNAS web site).
Fig. 1.
Quantitative analysis of human nNOS mRNA expression by real-time RT-PCR in the pyloric sphincter of controls and patients with IHPS. A set of nine forward primers, specific for the nine alternative first exons of nNOS (exon 1a–1i), were used with a common exon 2-specific reverse primer and an internal exon 2-specific 6-carboxy-fluorescein-labeled TaqMan probe. As a parameter for total nNOS mRNA expression, a pair of primers specific for exons 6 and 7, present in all known nNOS cDNAs, were used with an exon 7-specific internal probe. Relative amounts of transcripts were calculated by using standard curves and dividing the expression levels of the different nNOS variants by the expression levels of the globally expressed GAPDH housekeeping gene (A) or the neuronal-specific gene PGP9.5 (B) measured in the same RNA preparation. Results shown are the mean ± SD of pyloric sphincter preparations of 9 controls and 16 IHPS patients. Individual cDNA samples were analyzed in triplicate with a given pair of primers. (C) Relative amounts of the different alternative first exons of nNOS normalized against GAPDH in controls and patients with IHPS as determined by quantitative RT-PCR.
Fig. 1C shows the relative amounts of the different alternative first-exon variants of nNOS. In controls, exon 1c and 1f are distributed almost equally (46% and 39%, respectively), but in patients with IHPS, there is a dramatic shift from exon 1c (14%) to exon 1f (70%).
To evaluate whether the major change in patients with IHPS is a general decrease in the total expression of neuronal proteins, we quantified mRNA levels of another inhibitory neurotransmitter, VIP, which is expressed in neurons of the myenteric plexus, and normalized them to PGP9.5, Peripherin, and MAP-2. When normalized against PGP9.5, Peripherin, and MAP-2, expression of the neuronal message, VIP, was statistically significantly increased in pyloric tissues from IHPS patients compared with controls (see Fig. 4C). These findings indicate that enteric neurons are able to up-regulate neuronal messages in patients with IHPS and are consistent with the observed increased expression of one specific nNOS mRNA variant in IHPS (nNOS exon 1f). Furthermore, these results suggest an adaptive increase of VIP to elevate inhibitory neuronal messages to improve pyloric sphincter relaxation. Therefore, it is unlikely that an overall decrease in the expression of neuronal proteins is the major change in IHPS. However, we cannot exclude the possibility that there is a deficit in mRNA expression of several other neuronal genes in IHPS.
To evaluate possible age- or disease-related changes of nNOS mRNA expression in the control group, we compared total and first-exon nNOS mRNA expression levels of young (n = 3; mean age, 9.7 ± 6.4 weeks; range, 6–17 weeks) and old (n = 4; mean age, 77.8 ± 55.8 weeks; range, 42–161 weeks) controls. In addition, we compared controls with gastrointestinal reflux disease (n = 5) and without gastrointestinal disease (n = 4). The different subgroups showed similar total nNOS mRNA levels and nNOS first-exon expression profiles, indicating that there are no age- or disease-related differences within the control group (data not shown).
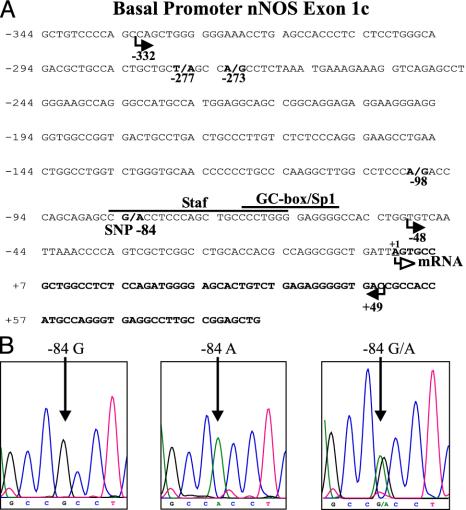
Analysis of the nNOS Exon 1c 5′-Flanking Region in Patients with IHPS and Controls. To further investigate decreased expression of nNOS exon 1c transcripts in patients with IHPS, we screened the 5′-flanking regulatory region of exon 1c by PCR and direct sequencing by using genomic DNA extracted from pyloric specimens. In addition, we used 72 genomic DNA samples of patients without gastrointestinal disorders. After direct sequencing of the PCR products, we found three different nucleotide-exchange mutations in the promoter region of exon 1c (-277T → A, -273A → G, or -98A → G) in pyloric specimens from three different IHPS patients (see Fig. 2A), whereas 9 control tissues and 72 control DNA blood samples showed the wild-type sequence exclusively. The mutated sequences were verified by three independent PCR and sequencing reactions to rule out PCR or sequencing errors.
Fig. 2.
Mutations and polymorphisms in the nNOS exon 1c promoter in patients with IHPS. (A) Nucleotide sequence of human nNOS exon 1c (bold) and in part of the 5′-flanking region including the minimal promoter. The transcription start site is indicated as +1. Cis-acting elements previously determined by gel-shift assays and site-directed mutagenesis (25) are indicated by the lines above the sequence. Mutations (-277T → A, -273A → G, and -98A → G) detected in DNA probes of IHPS patients and the -84G/A SNP are shown in bold and numbered below the sequence relative to the transcription start site (+1). Arrows below the nucleotide sequence depict the 5′-deletion sites of the different nNOS pGL3 promoter constructs used in reporter gene assays (Fig. 3). (B) DNA sequence analysis of the nNOS exon 1c -84G/A SNP. (Left) G/G homozygous DNA. (Center) A/A homozygous DNA. (Right) G/A heterozygous DNA. The position of nucleotide -84 is indicated with an arrow.
In addition to nucleotide-exchange mutations, we identified a previously uncharacterized SNP within the promoter region of exon 1c (-84G → A) (Fig. 2). Sixteen IHPS patients and 81 controls were analyzed for the -84 G/A genotype distribution (see Table 1). Comparison of genotype frequencies showed a significant difference between IHPS cases and controls (χ2 = 16.2, P = 0.0014, calculated by the χ2 test 3 × 2 contingency table, degrees of freedom = 2). The risk of IHPS for nNOS-exon 1c-84A carriers (both nNOS-exon 1c-84A homozygotes and nNOS-exon 1c-84G/A heterozygotes) was increased significantly, with an odds ratio of 8.0 (95% confidence interval = 2.5–25.6, P = 0.0004, calculated by Fisher's exact test; Table 1).
Table 1. Genotypic frequencies of the nNOS exon 1c promoter -84 G/A SNP in IHPS and controls.
| Genotype | Controls (n = 81) | IHPS (n = 16) | Odds ratio (95% confidence interval) | P value |
|---|---|---|---|---|
| G/G | 67 (82.72%) | 6 (37.5%) | — | — |
| G/A | 13 (16.05%) | 8 (50%) | 6.9 (G/A vs. G/G) (2.0-23.1) | 0.002 |
| A/A | 1 (1.23%) | 2 (12.5%) | 22.3 (A/A vs. G/G) (1.8-283.6) | 0.0271 |
| A carriers | 14 (17.28%) | 10 (62.5%) | 8.0 (A carriers vs. G/G) (2.5-25.6) | 0.0004 |
The statistical significance of differences between control subjects and patients was tested by Fisher's exact test.
Functional Promoter Activity of nNOS Exon 1c Promoter Polymorphism and Mutations in Patients with IHPS. We have shown previously that exon 1c of nNOS is driven by a separate promoter and demonstrated that transcription factors of the Sp and ZNF families play an essential role for basal promoter activity in TGW-nu-I neuroblastoma cells (25).
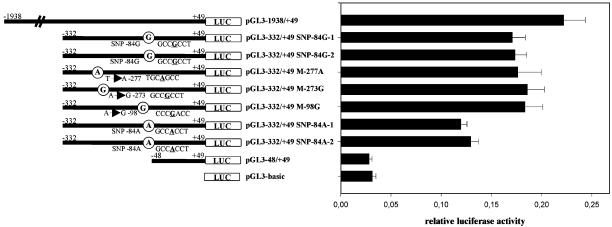
To examine the impact of the identified three mutations (nNOS-exon 1c -277T → A, -273A → G, and -98A → G) and the previously uncharacterized -84G/A SNP on nNOS exon 1c promoter activity in vitro, we performed reporter gene assays by using a -332/+49 construct of the basal nNOS exon 1c promoter, -332/+49 pGL3 constructs containing the different mutations (pGL3–332/+49 M-277A, pGL3–332/+49 M-273G, pGL3–332/+49 M-98G), and either the G or A allele of the -84 SNP (pGL3–332/+49 SNP-84G, pGL3–332/+49 SNP-84A) (see Fig. 3).
Fig. 3.
Functional analysis of mutations (-277T → A, -273A → G, and -98A → G) and the -84G/A SNP of the human nNOS exon 1c promoter in TGW-nu-I cells. The indicated DNA fragments containing the wild-type or mutant sequences of the nNOS exon 1c 5′-flanking region were ligated into promoterless/enhancerless firefly luciferase expression vector pGL3-basic. The different reporter gene constructs were transiently cotransfected with the herpes simplex virus thymidine kinase promoter-driven Renilla luciferase expression vector phRL-TK as internal control. pGL3-basic was used as a negative control. Data are expressed as means ± SD of three independent experiments in triplicate.
Because reporter gene assays were not feasible on isolated enteric neurons from the infantile pyloric sphincter, we transiently transfected TGW-nu-I cells with the different pGL3 plasmids. We showed previously that TGW-nu-I cells predominantly express nNOS exon 1c, 1f, and 1g; therefore, these cells show a similar nNOS first-exon expression profile as infantile pyloric sphincter specimens (see Fig. 1 A and B and ref. 25). After transfection of TGW-nu-I cells with promoter constructs containing the three different mutations (pGL3–332/+49 M-277A, pGL3–332/+49 M-273G, pGL3–332/+49 M-98G), we observed almost identical or slightly increased relative luciferase activities compared with wild-type pGL3–332/+49. In contrast, promoter constructs containing the A allele of the -84G/A SNP showed a significant decrease of ≈30% (P < 0.001) in normalized luciferase activity (see Fig. 3). These results indicate that the previously uncharacterized -84G/A promoter SNP has a major influence on the basal transcriptional activity of the nNOS exon 1c regulatory region.
Discussion
Our study demonstrates altered expression patterns of nNOS first-exon mRNA variants in patients with hypertrophic pyloric stenosis, together with mutations in the promoter region of nNOS exon 1c and a substantially increased risk to develop IHPS for infants carrying the A allele of a previously uncharacterized, identified exon 1c promoter SNP.
These data are consistent with previous studies presenting evidence for a major pathogenic role of the nNOS gene in IHPS (9–11). However, the causal mechanisms of changes of nNOS gene expression and therefore the molecular basis of the disease were not elucidated in these studies.
The pylorus seems to be normal at birth in most IHPS cases but deteriorates within 2–12 weeks after birth (4) and ultimately returns to normal after successful surgical or conservative treatment (5, 7, 8). Because the pyloric sphincter seems to serve no major digestive function in utero, a functional change occurs after birth, which could be accompanied by a change in gene expression as shown for several developmentally regulated genes (33). In patients with IHPS, the pyloric sphincter appears immature at the time of pyloromyotomy (34), suggesting that this change in gene expression is altered or delayed, resulting in smooth muscle hypertrophy, hyperplasia, and defective sphincter relaxation. The present study suggests a major role for the nNOS gene within this process. A reduction in total nNOS mRNA expression was observed at the time of pyloromyectomy in IHPS patients after normalization against the globally expressed housekeeping gene GAPDH, mainly caused by the severe decrease (88%) of nNOS exon 1c expression. Reduced total nNOS expression most likely results from hypertrophy and hyperplasia of pyloric smooth muscles (which express GAPDH but not nNOS), leading to a relative decrease of enteric neurons, which express nNOS. However, normalization of nNOS against GAPDH and thus total tissue content is useful, because it indicates that there is not enough nNOS present to produce sufficient amounts of NO to relax the thickened pyloric smooth muscle of IHPS patients. Therefore, the relative lack of nNOS in relation to increased total pyloric tissue volume seems to play an important role in defective sphincter relaxation in IHPS.
To evaluate nNOS expression in enteric neurons of the pyloric sphincter, we normalized against the general neuronal markers PGP9.5, Peripherin, and MAP-2 and observed no significant differences in total nNOS mRNA expression between IHPS patients and controls. In addition, we found a significantly decreased exon 1c and a significantly increased exon 1f expression. This reciprocal relationship between exons 1c and 1f containing nNOS transcripts in IHPS patients versus controls is of great interest, because it indicates a dynamic regulation of nNOS gene expression in the enteric nervous system. Exon 1c and 1f transcripts are the predominant forms in the pyloric sphincter. Therefore, up-regulation of nNOS exon 1f seems to be a regulatory compensatory mechanism in response to decreased nNOS exon 1c mRNA expression in IHPS.
Such regulatory mechanisms can be mediated by transcriptional or posttranscriptional control. We previously demonstrated that at least nine different first-exons of nNOS, driven by separate promoters, are present in the human gastrointestinal tract (23, 25). Transcriptional control by alternative usage of multiple promoters allows regulation of nNOS expression in different physiological and pathological conditions. Thus, up-regulation of nNOS exon 1f mRNA transcripts by activation of the nNOS exon 1f promoter could be a response of the organism to reduced exon 1c transcript abundance in IHPS. In addition, posttranscriptional regulation of nNOS may contribute to increased exon 1f and decreased exon 1c mRNA levels. For example, changes in mRNA stability could be involved but cannot be studied in pyloric sphincter specimens of IHPS patients and controls.
To evaluate the relative expression of other neuronal messages, we quantified VIP mRNA levels and observed a statistically significant increased mRNA expression in patients with IHPS compared with controls. This observation indicates a specific up-regulation of the inhibitory neurotransmitter VIP in enteric nerves of patients with IHPS and argues against a general decreased expression of neuronal messages in IHPS.
To gain insight into the molecular basis of decreased nNOS exon 1c transcripts, we screened its 5′-flanking region for mutations and polymorphisms that may be responsible for the observed reduced expression. We identified three independent mutations within the promoter region of exon 1c in three different patients and one SNP of the exon 1c promoter (-84G/A).
To evaluate the contribution of the -84G/A SNP for development of IHPS, we compared genotype frequencies between controls and IHPS patients. Carriers of the A allele of the -84G/A SNP have substantially increased risk for IHPS, with an odds ratio of 8.0, which could indicate that the -84G/A promoter SNP alters expression of nNOS exon 1c or is in linkage dysequilibrium with a functionally important sequence variant elsewhere in the nNOS transcription unit and therefore may serve as a informative marker for a functionally important genetic alteration. The observed correlation of the -84G/A SNP with an increased risk for development of IHPS is consistent with a report showing a strong correlation of a microsatellite polymorphism in the nNOS gene with a familiar form of IHPS (11). However, the -84G/A SNP does not account for all IHPS cases, and therefore other components of the NO-dependent signal transduction pathway or additional mechanisms and genes may be involved in the pathogenesis of IHPS (for a review see ref. 13). This is in accordance with recent observations suggesting a multifactorial etiology of IHPS (2).
To further test whether the previously uncharacterized nNOS exon 1c promoter mutations and the different alleles of the -84G/A SNP alter basal transcription of exon 1c, we constructed reporter gene plasmids carrying the different mutations (-277T → A, -273A → G, or -98A → G) and the G or A allele of the -84G/A SNP. Reporter gene assays in TGW-nu-I cells revealed a significant decreased transcription (≈30%) of exon 1c promoter plasmids carrying the A allele, compared with plasmids carrying the G allele. However, plasmids carrying the different mutations showed no altered transcriptional activity. This could be because of several reasons. (i) The mutations do not affect transcription of exon 1c, which seems to be unlikely, because none of 81 tested controls showed any mutation, indicating that genetic alterations in the nNOS exon 1c promoter specifically occur in patients with IHPS. (ii) The neuroblastoma cell line used in the study may not use exactly the same cis-regulatory DNA elements as enteric neurons for transcription of nNOS exon 1c despite similar nNOS first-exon expression profiles. (iii) Elements that are not present within the used nNOS exon 1c reporter gene construct may influence transcription. (iv) Chromatin structure and acetylation state have been shown to play an important role in regulation of gene expression, and therefore endogenous nNOS expression may be regulated at multiple levels that cannot be assessed completely by reporter gene assays (25).
Decreased transcriptional activity of nNOS exon 1c promoter constructs carrying the A allele of the -84G/A SNP may be caused by disruption of a Staf transcription factor-binding site within the exon 1c basal promoter (see Fig. 2 A). We have shown previously that the minimal exon 1c promoter is localized to 44 bp, with a cooperative binding of several transcription factors of the ZNF and Sp families (25). ZNF 76 and 143 bind to the Staf consensus-responsive DNA element in the exon 1c promoter and activate transcription of exon 1c in Drosophila Schneider cells on their own (25). Mutation of the Staf cis-acting element results in an ≈53% decreased transcriptional activity of the nNOS exon 1c promoter in TGW-nu-I cells (25). Therefore, the A allele of the -84G/A SNP may reduce DNA-binding affinity of ZNF 76 and 143 to the Staf element and thus impairs transcription of nNOS exon 1c. Taken together, these results indicate that transcriptional regulation of exon 1c is an important component of decreased abundance of nNOS exon 1c transcripts.
IHPS is diagnosed and treated by pyloromyotomy when hypertrophy and hyperplasia of the pyloric sphincter is already apparent. Therefore, it is not possible to study the unknown initial events in the pathogenesis of IHPS. We suggest that the profound reduced expression of one of the predominant nNOS transcripts (exon 1c) leads to transiently decreased total nNOS expression and impaired NO production, resulting in hypertrophy and hyperplasia of pyloric sphincter smooth muscle cells and defective sphincter relaxation. Such effects of NO have been reported for the cardiovascular system, because lack of NO or blockade of the NO/cGMP/cGMP-dependent kinase signaling cascade results in hypertrophy and proliferation of smooth muscle cells (18–20). As a reaction to decreased nNOS exon 1c transcript abundance, most likely transcriptional regulation by alternative promoter usage up-regulates an alternative first-exon variant of nNOS (exon 1f) in an attempt to increase nNOS expression and normalize pyloric sphincter function. Therefore, the intriguing diversity of the nNOS gene, providing many possibilities for a dynamic transcriptional and posttranscriptional regulation, seems to be able to overcome IHPS. However, because this adaptive process may take a while at a period when adequate nutrition is essential for optimal infantile development, pyloromyotomy currently remains the best therapeutic option for most IHPS patients.
Supplementary Material
Acknowledgments
We thank Dr. H. Esumi (National Cancer Center Research Institute East, Chiba, Japan) for providing TGW-nu-I cells, U. Goetz and M. Werb for excellent technical assistance, and R. Rad for statistical assistance. This study was supported by Deutsche Forschungsgemeinschaft Sonderforschungsbereich Grant 391 C5 and Kommission für Klinische Forschung Technical University of Munich Grant F47-01 (to D.S.). J.-M.V. is a Senior Research Associate of the National Fund for Scientific Research (Belgium) and is supported by grants from Fonds de la Recherche Scientifique Médicale, Fondation Médicale Reine Elisabeth, and Fondation Universitaire David et Alice Van Buuren (Belgium).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IHPS, infantile hypertrophic pyloric stenosis; nNOS, neuronal nitric oxide synthase; SNP, single-nucleotide polymorphism; 5′-RACE, rapid amplification of 5′ cDNA ends; VIP, vasoactive intestinal peptide; MAP-2, microtubule-associated protein 2.
References
- 1.Rasmussen, L., Green, A. & Hansen, L. P. (1989) Int. J. Epidemiol. 18, 413-417. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell, L. E. & Risch, N. (1993) Am. J. Dis. Child. 147, 1203-1211. [DOI] [PubMed] [Google Scholar]
- 3.Oue, T. & Puri, P. (1999) Pediatr. Res. 45, 853-857. [DOI] [PubMed] [Google Scholar]
- 4.Rollins, M. D., Shields, M. D., Quinn, R. J. & Wooldridge, M. A. (1989) Arch. Dis. Child. 64, 138-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludtke, F. E., Bertus, M., Voth, E., Michalski, S. & Lepsien, G. (1994) J. Pediatr. Surg. 29, 523-526. [DOI] [PubMed] [Google Scholar]
- 6.Kawahara, H., Imura, K., Nishikawa, M., Yagi, M. & Kubota, A. (2002) Arch. Dis. Child. 87, 71-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanderwinden, J. M., Liu, H., Menu, R., Conreur, J. L., De Laet, M. H. & Vanderhaeghen, J. J. (1996) J. Pediatr. Surg. 31, 1530-1534. [DOI] [PubMed] [Google Scholar]
- 8.Okorie, N. M., Dickson, J. A., Carver, R. A. & Steiner, G. M. (1988) Arch. Dis. Child. 63, 1339-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanderwinden, J. M., Mailleux, P., Schiffmann, S. N., Vanderhaeghen, J. J. & De-Laet, M. H. (1992) N. Engl. J. Med. 327, 511-515. [DOI] [PubMed] [Google Scholar]
- 10.Kusafuka, T. & Puri, P. (1997) Pediatr. Surg. Int. 12, 576-579. [DOI] [PubMed] [Google Scholar]
- 11.Chung, E., Curtis, D., Chen, G., Marsden, P. A., Twells, R., Xu, W. & Gardiner, M. (1996) Am. J. Hum. Genet. 58, 363-370. [PMC free article] [PubMed] [Google Scholar]
- 12.Vanderwinden, J. M., Liu, H., De Laet, M. H. & Vanderhaeghen, J. J. (1996) Gastroenterology 111, 279-288. [DOI] [PubMed] [Google Scholar]
- 13.Ohshiro, K. & Puri, P. (1998) Pediatr. Surg. Int. 13, 243-252. [DOI] [PubMed] [Google Scholar]
- 14.Tomita, R., Tanjoh, K., Fujisaki, S. & Fukuzawa, M. (1999) Hepatogastroenterology 46, 2999-3003. [PubMed] [Google Scholar]
- 15.Huang, P. L., Dawson, T. M., Bredt, D. S., Snyder, S. H. & Fishman, M. C. (1993) Cell 75, 1273-1286. [DOI] [PubMed] [Google Scholar]
- 16.Mashimo, H., Kjellin, A. & Goyal, R. K. (2000) Gastroenterology 119, 766-773. [DOI] [PubMed] [Google Scholar]
- 17.Pfeifer, A., Klatt, P., Massberg, S., Ny, L., Sausbier, M., Hirneiss, C., Wang, G. X., Korth, M., Aszodi, A., Andersson, K. E., et al. (1998) EMBO J. 17, 3045-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer, P. M., Buga, G. M. & Ignarro, L. J. (2001) Proc. Natl. Acad. Sci. USA 98, 12802-12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamad, A. M. & Knox, A. J. (2001) FEBS Lett. 506, 91-96. [DOI] [PubMed] [Google Scholar]
- 20.Ueno, H., Kanellakis, P., Agrotis, A. & Bobik, A. (2000) Hypertension 36, 89-96. [DOI] [PubMed] [Google Scholar]
- 21.Wollert, K. C., Fiedler, B., Gambaryan, S., Smolenski, A., Heineke, J., Butt, E., Trautwein, C., Lohmann, S. M. & Drexler, H. (2002) Hypertension 39, 87-92. [DOI] [PubMed] [Google Scholar]
- 22.Peunova, N., Scheinker, V., Cline, H. & Enikolopov, G. (2001) J. Neurosci. 21, 8809-8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saur, D., Paehge, H., Schusdziarra, V. & Allescher, H. D. (2000) Gastroenterology 118, 849-858. [DOI] [PubMed] [Google Scholar]
- 24.Wang, Y., Newton, D. C., Robb, G. B., Kau, C. L., Miller, T. L., Cheung, A. H., Hall, A. V., VanDamme, S., Wilcox, J. N. & Marsden, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 12150-12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saur, D., Seidler, B., Paehge, H., Schusdziarra, V. & Allescher, H. D. (2002) J. Biol. Chem. 277, 25798-25814. [DOI] [PubMed] [Google Scholar]
- 26.Newton, D. C., Bevan, S. C., Choi, S., Robb, G. B., Millar, A., Wang, Y. & Marsden, P. A. (2003) J. Biol. Chem. 278, 636-644. [DOI] [PubMed] [Google Scholar]
- 27.Xie, J., Roddy, P., Rife, T., Murad, F. & Young, A. P. (1995) Proc. Natl. Acad. Sci. USA 92, 1242-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogura, T., Nakayama, K., Fujisawa, H. & Esumi, H. (1996) Neurosci. Lett. 204, 89-92. [DOI] [PubMed] [Google Scholar]
- 29.Saur, D., Neuhuber, W. L., Gengenbach, B., Huber, A., Schusdziarra, V. & Allescher, H. D. (2002) Am. J. Physiol. 282, G349-G358. [DOI] [PubMed] [Google Scholar]
- 30.Gentile, C., Romeo, C., Impellizzeri, P., Turiaco, N., Esposito, M., Di Mauro, D. & Mondello, M. R. (1998) Pediatr. Surg. Int. 14, 45-50. [DOI] [PubMed] [Google Scholar]
- 31.Golder, M., Burleigh, D. E., Belai, A., Ghali, L., Ashby, D., Lunniss, P. J., Navsaria, H. A. & Williams, N. S. (2003) Lancet 361, 1945-1951. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, H., Miyahara, K., Yamataka, A., Lane, G. J., Sueyoshi, N. & Miyano, T. (2001) J. Pediatr. Surg. 36, 1277-1279. [DOI] [PubMed] [Google Scholar]
- 33.Montgomery, R. K., Mulberg, A. E. & Grand, R. J. (1999) Gastroenterology 116, 702-731. [DOI] [PubMed] [Google Scholar]
- 34.Guarino, N., Shima, H. & Puri, P. (2000) Pediatr. Surg. Int. 16, 282-284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.