Abstract
Human endogenous retroviruses (HERVs) are a potential source of genetic diversity in the human genome. Although many of these elements have been inactivated over time by the accumulation of deleterious mutations or internal recombination leading to solo-LTR formation, several members of the HERV-K family have been identified that remain nearly intact and probably represent recent integration events. To determine whether HERV-K elements have caused recent changes in the human genome, we have undertaken a study of the level of HERV-K polymorphism that exists in the human population. By using a high-resolution unblotting technique, we analyzed 13 human-specific HERV-K elements in 18 individuals. We found that solo LTRs have formed at five of these loci. These results enable the estimation of HERV solo-LTR formation in the human genome and indicate that these events occur much more frequently than described in inbred mice. Detailed sequence analysis of one provirus shows that solo-LTR formation occurred at least three separate times in recent history. An unoccupied preintegration site also was present at this locus in two individuals, indicating that although the age of this provirus is estimated to be ≈1.2 million years, it has not yet become fixed in the human population.
At least 8% of the human genome is made up of endogenous retroviruses and related sequences, which form ≈200 distinct groups and subgroups (1–3). Most of these elements represent ancient retroviral infections, as evidenced by their wide distribution in primate species, and no infectious counterparts of human endogenous retroviruses (HERVs) are known to exist today. Many HERV elements have been found to be at identical sites in both Old World monkeys and apes, which diverged ≈25 million years ago (4, 5), implying that the virus that gave rise to them existed at least that long ago. As a consequence of their long residence in the genome, most HERVs have acquired numerous mutations in their coding sequences, rendering them incapable of further colonization of the genome by either reinfection or intracellular retrotransposition. More ancient proviruses have more divergent LTRs, which are identical at the time of integration and acquire mutations over time (6). Inactivation of HERV elements can occur also by solo-LTR formation, which is the result of homologous recombination between the two LTRs flanking the provirus and the subsequent deletion of the internal sequence. The vast majority of HERV elements in the human genome exist as solo LTRs (7).
The elements in our genome that appear to be the most recently active belong to the HERV-K family. The oldest members of this family entered the genome before the Old World monkey–ape divergence, but HERV-K elements have undergone several periods of expansion throughout primate evolution (7, 8). For example, there are many HERV-K elements that are present in the African great apes and humans but not in orangutans and the lesser apes, indicating a relatively recent integration time of 8–15 million years (4, 9). There are also a number of elements that are unique to humans, implying still more recent activity (5, 9, 10; see also ref. 32). Two HERV-K elements, one of which was completely intact, were identified in a genomic library (11) and found to be polymorphic in the human population. This observation indicates that HERV-K family members were active in very recent evolutionary time, and the possibility exists that they may still be capable of causing changes in our genome by further expansion due to new integration events or loss due to solo-LTR formation.
To examine this possibility, we studied of the level of HERV-K polymorphism in the human population by using a high-resolution unblotting technique. A previous study had used Southern blot analysis to search for HERV-K content differences that might exist in the human population (12). Genomic DNA samples from 37 Caucasian and 4 Chinese individuals were examined, and only one polymorphism was found, but it was not characterized. Another study demonstrated a genetic variation due to the presence of a HERV solo LTR instead of its full-length counterpart in a small number of individuals (13). In the present study, we analyzed 13 human-specific HERV-K elements in individuals and found that solo LTRs have formed at five of these loci, suggesting a relatively high frequency of occurrence. Furthermore, an unoccupied preintegration site was found to exist in some individuals for two of these elements, indicating that they have not yet reached fixation. One of these loci exhibits a particularly high level of polymorphism because two alleles of the full-length element and three different solo-LTR alleles were identified in the sampled individuals.
Materials and Methods
DNA Samples. Human genomic DNA samples were isolated by using the QIAamp DNA blood maxi kit (Qiagen, Valencia, CA) from either whole blood (5 ml) drawn from healthy individuals or from the following lymphoblast cell lines: Biaka, GM10469A; Mbuti, GM010492A; Maya, GM10975; Quecha, GM11197; Khmer, GM11377; Druze, GM11522; Ami, GM13607; and Adygei, GM13619 (Human Diversity Collection, Coriell Cell Repositories, Camden, NJ).
Unblotting. Unblotting, or hybridization in dried agarose, is described in ref. 14. Briefly, genomic DNA digested with Ase I was electrophoresed in an 0.8% agarose gel and stained with ethidium bromide. The DNA in the gel was then denatured and neutralized. After drying, the gel was hybridized overnight at 55°C with a 5′-end 32P-labeled oligonucleotide probe corresponding to positions 1069–1091 of HERV-K10 at 2 × 106 cpm/ml. The dried gel was then washed, briefly air dried, and exposed to BioMax MS (Kodak) maximum-sensitivity film for 5 days by using an intensifying screen at -70°C.
PCR Analysis. PCRs contained 200 ng of genomic DNA, 1.5–3.5 mM MgCl2, 50 μM each dNTP, 0.2 μM each primer, and 2.5 units of Taq DNA polymerase (Sigma). Primers and conditions used for each reaction are available on request.
Divergence Date Calculations. Calculations of integration time and divergence were based on the average nucleotide substitution rate of 19 full-length HERV-K loci in the human genome, 3.77 ± 1.33 × 10-9 substitutions per site for each year, estimated by comparing the human sequences with the corresponding chimpanzee loci (J.F.H. and J.M.C, unpublished data). Genetic distances between LTRs of each provirus were calculated by using the Kimura two-parameter estimate, which corrects for the occurrence of multiple mutations at the same site, back mutations, and convergent substitutions.
Estimation of Frequency of Solo-LTR Formation. A total of seven instances of solo-LTR formation was detected by this analysis. To calculate the rate of solo-LTR formation, this value was multiplied by 20,000, which was derived from the probability that a neutral allele will be fixed in randomly mating population 1/2N, where N is the effective population size, commonly10,000 for humans (15). Because the solo LTRs in this study had not yet reached fixation, however, the calculation for each element was adjusted by factoring in its allele frequency, which corresponds to its current probability of becoming fixed. For example, the frequency of the solo-LTR allele at the 12q14 locus is 0.44, which is also the probability that it will reach fixation. Therefore, 20,000 is multiplied by this probability, and each locus is then treated in the same manner and summed to give the extrapolated total number of events. The resulting value was divided by the total length of residence in the genome for all 13 human-specific HERV-K elements (estimated to be ≈23.0 million years, or ≈1.15 million generations, given the 20-years generation time commonly used for ancestors). This calculation results in a frequency of ≈0.002 solo LTRs formed per site for each generation.
Results
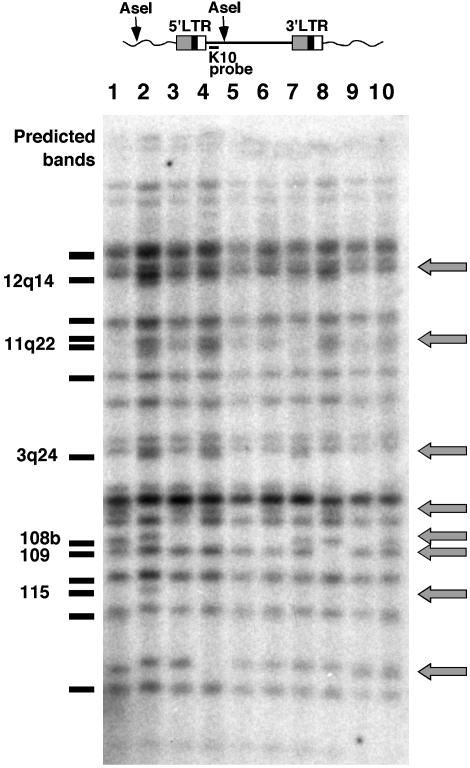
Detection of HERV-K Polymorphisms By Using High-Resolution Unblotting. To examine HERV-K polymorphism in the human genome at a higher resolution than had been possible before, we initially examined genomic DNA samples from 10 individuals for their content of human-specific proviruses. For this purpose, a high-resolution hybridization (unblotting) strategy was developed by using an oligonucleotide probe (K10) derived from the sequence of HERV-K10 (16) between the 5′LTR and the start of gag. This probe was used to detect 5′ junction fragments in DNA cleaved with Ase I, which cleaves the proviruses in the database 3′ of the probe site and, frequently, elsewhere in human DNA (Fig. 1 Upper). Each HERV-K element detected by the probe should give rise to a unique band, the size of which depends on its proximity to an upstream Ase I site in the genomic flanking DNA. When this analysis was performed, ≈24 bands were observed in each DNA sample (Fig. 1 Lower).
Fig. 1.
Detection of HERV-K polymorphisms. (Upper) The position of the K10 oligonucleotide probe and putative Ase I sites relative to a typical HERV-K element. (Lower) Unblotting of a set of human genomic DNA samples digested with Ase I and probed with K10. Gray arrows indicate bands that are polymorphic within the sample. Bands predicted from the genomic database are plotted to the left of the blot, and the bands that appear to be polymorphic are indicated. Five of these elements have been identified: HERV-K108, 109, and 115 (9, 11); and HERV-K11q22 and 12q14 (29). The HERV-K element 3q24 is not full-length and was identified in the clone with GenBank accession no. AC069410 (Human Bacterial Artificial Chromosome Library, Roswell Park Cancer Institute, Buffalo, NY). The band marked 108b is the size expected for the central LTR of the tandem form of HERV-K 108 (20) (see Fig. 2).
We used a blast search of the human genome sequence (17) to predict the number of elements that contained the probe sequence and the length of the Ase I fragment for each element (Fig. 1, left of blot). The overall pattern of the predicted bands was quite similar to the observed pattern, and most of the proviruses could be identified from the size of the Ase I fragments. We identified 19 full-length HERV-K elements and related fragments that contained from zero to two mismatches with the K10 probe sequence, implying that approximately five proviruses are not in the database. One of the missing proviruses corresponds to the polymorphic HERV-K115 identified recently, whose Ase I fragment size could also be predicted (11).
Quite unexpectedly, we found a number of proviruses that were present in some individuals and not others within the small analyzed sample (Fig. 1, arrows). These polymorphisms could represent recently integrated elements that are present only in some individuals, or they could be indicative of element loss by solo-LTR formation or deletion, or polymorphisms in the probe or Ase I sites. Six polymorphic elements could be identified from the database analysis. Five of these elements represent full-length HERV-K proviruses. The only polymorphic element that is not full-length, HERV-K3q24, represents a previously uncharacterized HERV-K-type element and is described elsewhere (18). All six of the polymorphic elements identified in this study were found to be human-specific (ref. 10 and J.F.H. and J.M.C., unpublished results). The estimated integration times, based on LTR divergences, for these elements are given in Table 1.
Table 1. Polymorphic HERV-K loci in the human genome.
| HERV-K | No. of differences between LTRs | Integration time (mya)* |
|---|---|---|
| 11q22 | 4 | 0.81-2.3 |
| 12q14 | 4 | 0.81-2.3 |
| 109 | 3 | 0.61-1.8 |
| 1p31† | 3 | 0.82-2.4‡ |
| 115 | 5 | 1.0-2.9§ |
| 108 | 6 | 1.2-3.5 |
mya, million years ago.
Calculations based on the average nucleotide substitution rate of HERV-K loci of 3.77 × 10-9 ± 1.33
This element was not located in the blot but was found by PCR analysis to be polymorphic
Estimated from an alignment of 715 bp, instead of the full-length 968, because of missing sequence data in the 5′ LTR
Integration time estimate taken from ref. 12
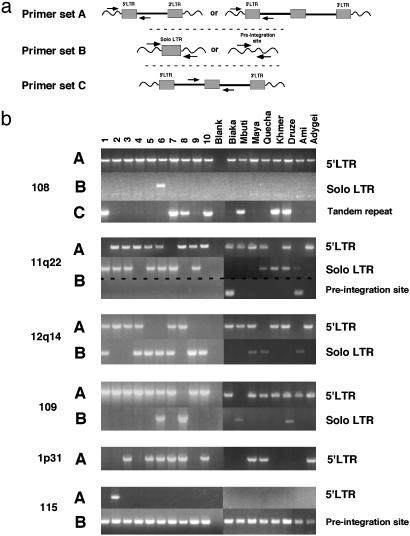
Most HERV-K Polymorphisms Detected Are Because of the Presence of Solo LTRs. To determine the nature of the polymorphisms at these and other human-specific loci, PCR primers were designed based on the flanking sequence information (Fig. 2a). Eight additional samples representing long-isolated, diverse populations were included in the PCR analysis to determine the extent of polymorphism at these loci among more diverse human populations.
Fig. 2.
Detection of HERV-K solo LTRs. Two primer sets were used for each polymorphic HERV-K locus identified in the unblotting analysis, labeled A and B. A similar PCR strategy was used to probe for polymorphisms at the nine other known human-specific HERV-K loci (data not shown), and only HERVK1p31 was found to be polymorphic. Primer set A was used to amplify the 5′ LTR of the full-length element, and primer set B was used to amplify the corresponding solo LTR or unoccupied preintegration site at each locus. Samples 1–10 are the same as in Fig. 1, and the remaining samples are labeled according to the population of origin of their donors. The primer set used in each reaction is indicated to the left of the unblots, and the identity of each product, confirmed by sequence analysis, is indicated the right of the unblots.
Unblotting analysis of these samples indicated that they did not contain any polymorphisms that were not observed in the initial analysis (data not shown).
A polymorphism at the HERV-K108 locus, also known as HERV-K(HML-2.HOM), was characterized (19) and found to comprise different forms: a tandemly duplicated copy with three full-length LTRs and a single full-length element. Our unblotting analysis was consistent with the existence of these forms. By using PCR primers that amplify the tandem provirus specifically (19), we confirmed the results obtained in the unblotting analysis (Fig. 2b, 108 B). Additionally, a solo LTR, which was not found in the study, was detected in one individual. Interestingly, the other product of the unequal homologous recombination event proposed to account for the appearance of the tandemly duplicated copy of HERV-K108 is predicted to be a solo LTR. Therefore, this solo LTR might not be the result of a recombination event between the two LTRs of one element, but may represent the reciprocal product of the generation of the duplicated allele.
Solo LTRs were detected at the three other polymorphic HERV-K loci 11q22, 12q14, and 109, with allele frequencies of 0.39, 0.44, and 0.17, respectively (Fig. 2b). One of these elements, HERV-K11q22, was also insertionally polymorphic, as evidenced by the amplification of the unoccupied preintegration site in two unrelated individuals. Another human-specific HERV-K element, HERV-K1p31, which was not identified in the unblotting analysis because it contains a deletion spanning the region from which the K10 probe is derived, was found to be polymorphic by PCR analysis (Fig. 2b). Although PCRs to detect the corresponding solo LTR by using primers located in the sequences flanking this element were inconclusive because of its location in a region of the genome rich in repetitive elements, a solo-LTR variant at this locus was found in the sequence database (GenBank accession no. AC053498), indicating that the solo-LTR allele exists in some fraction of the population.
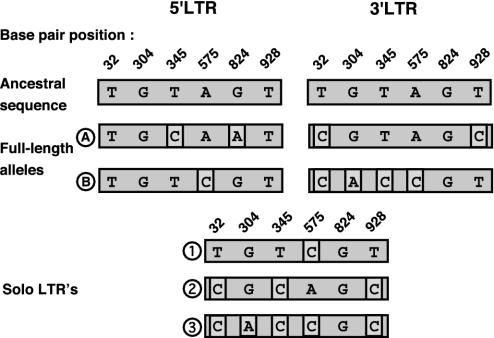
Solo LTRs are formed as the result of the recombination between the two LTRs of a full-length element. If enough mutations had occurred in the two LTRs of this element before solo-LTR formation, it would be possible to identify recombination crossover points and determine whether solo-LTR formation had occurred at this locus more than one time. To examine this possibility, the 5′ and 3′ LTRs of the full-length element and the corresponding solo LTRs were sequenced in individuals heterozygous for these alleles. Evidence for multiple solo-LTR formation events was found at one locus, HERVK11q22. In total, there were six sites along the length of the 968-bp LTR sequence at which the 5′ and 3′ LTRs differed from each other (Fig. 3). Surprisingly, there were two distinct alleles of the full-length element identified in these individuals that could be differentiated at seven polymorphic sites, indicating a divergence date of ≈0.7–2.0 million years ago. Three different crossover patterns were evident by examining the solo-LTR sequences. These crossover patterns were quite complex and may indicate the occurrence of further recombination and/or gene conversion events after the solo-LTR formation. However, no substitutions were evident in the solo-LTR sequences that were not already present in either the 5′ or 3′ LTR of the corresponding full-length element, indicating that the recombination events took place relatively recently (<580,000 years ago).
Fig. 3.
Multiple alleles of HERV-K11q22 and its solo LTRs. The 5′ and 3′ LTRs of the full-length element and the solo LTR contain six polymorphic sites in the 968 base pairs sequenced in the five heterozygous individuals studied. The base pair positions and the identities of these sites are indicated. The ancestral states were determined from character state reconstructions (304, 824, and 928) or assigned by homology to other, closely related HERV-K loci (32, 345, and 575), in cases of ambiguity. There are two alleles (A and B) of the full-length element and three alleles (1, 2, and 3) of the solo LTR. The polymorphic sites that differ from the inferred ancestral sequence are shown in boxes. Of the five heterozygous individuals, two had full-length element A and solo-LTR 2. The remaining individuals each had one of the following combinations: A and solo-LTR 1, B and solo-LTR 1, and B and solo-LTR 3.
High Frequency of Solo-LTR Formation in the HERV-K Family in Humans. Solo-LTR formation occurred at no fewer than 5 of the 13 human-specific full-length HERV-K elements, and it occurred at least three times at one of these loci for a total of seven or more independent instances of solo-LTR formation. To calculate the frequency of these events, the total length of residence in the genome for all 13 human-specific HERV-K elements was estimated to be ≈23.0 million years, or ≈1.15 million generations. Taking into account the probability that a newly formed solo LTR would not be lost because of random genetic drift (15), we obtain a frequency of ≈0.002 solo LTRs formed per site for each generation.
Discussion
By using a high-resolution hybridization method, we have found that the HERV-K family of endogenous retroviruses is quite polymorphic in the human population. Of the ≈24 proviruses detected by our probe, only the more recently integrated elements, namely those that were found to be human-specific, contain polymorphic loci. Four such human-specific elements, including two that were identified in ref. 11, are apparently not yet fixed in the human population, as evidenced by the presence of unoccupied preintegration sites in the genomes of some individuals. One of these elements, HERV-K115, is very rare in the population and, therefore, most likely represents a more recent integration event. The remaining polymorphic elements have undergone solo-LTR formation at some point after their integration, and these solo-LTR alleles exist at different frequencies in the human population, perhaps reflecting their time of formation. The time that it would take for a neutral allele to reach fixation in the human population is ≈800,000 years, or 4N generations, given an effective population size of 10,000 and a generation time of 20 years (20). Therefore, in the absence of selection, a solo LTR that has formed within the last 800,000 years will not yet be fixed.
Analysis of the structure of the proviruses and their solo LTRs can provide some insight into the evolutionary processes affecting them. At least six different alleles have been maintained at the HERV-K11q22 locus: two full-length variants, three solo LTRs, and the unoccupied preintegration site. Given the estimated age (1.2 million years) of this provirus, the maintenance of the ancestral state, or the unoccupied preintegration site, at this locus is unexpected. This high level of genetic heterogeneity could be indicative of balancing selection acting to maintain heterozygosity. No function has been attributed to this particular HERV element; therefore, mutations at this locus might be considered neutral. Functional neutrality is only an assumption, however, and has not been tested in any way. If selection is not acting directly on the HERV sequence, the high level of heterogeneity could be due to a hitchhiking effect with a nearby gene under selective pressure. However, a search of the human genome database revealed no obvious candidates in the vicinity of the provirus. The distribution of the two full-length alleles did not reveal any regional specificity, indicating that they originated either before or near to the time of the emergence of modern humans from Africa.
The predicted date of origin (≈1–2 million years ago) of the full-length alleles at HERV-K11q22 is relatively ancient, whereas all three solo LTRs seem to have formed more recently (<580,000 years ago). Consistent with this range, neither a HERV-K11q22 solo LTR nor a 12q14 solo LTR were found in either tested African pygmy sample (Biaka and Mbuti), even though the alleles are quite common among the remaining individuals. This distribution may indicate that these loci were formed close to or after the time of the emergence of modern humans from Africa, which began ≈200,000 years ago (21). This conclusion is speculative, of course, given the small set of analyzed samples.
An alternative interpretation of the divergence date of the two full-length alleles is that they are indicative of a more ancient origin of human genetic history than implied by the “Out of Africa” model, which argues for a complete replacement of ancient populations 100,000–200,000 years ago. Similar divergence dates of >1 million years ago have been found in studies examining large, noncoding regions of the genome (22, 23). The widespread distribution of the more recently formed solo-LTR alleles does not necessarily refute this hypothesis, but it suggests instead a more complicated scenario than either the strictly multiregional or recent replacement models of human evolution allow. The pattern may be indicative of multiple migrations from Africa accompanied by a high level of gene flow among populations throughout human history (24).
This study examines HERV loci systematically for evidence of polymorphism in the human genome. We have found that some of these loci undergo solo-LTR formation in the germ-line at a relatively high frequency. One estimate of the frequency of solo-LTR formation comes from studies of the DBA strain of mice, which have a characteristic coat-color mutation that was generated originally by the integration of a murine leukemia virus. Solo-LTR formation at this locus causes a reversion of the phenotype; consequently, the frequency of such events could be estimated directly and was found to be 4.5 × 10-6 times per gamete at this locus in mice (25). This estimate, which is ≈450 times less frequent than the HERV-K estimate, is in agreement with the solo-LTR formation frequency calculated in a larger study of 103 murine leukemia virus loci in recombinant inbred strains of mice (26) as well as with the observation that solo-LTR loci make up a small fraction (≈10%) of total proviral loci in these genomes (27). Whether this discrepancy is due to a large difference in rate of solo-LTR formation from one site to another (due to structural differences between murine leukemia virus and HERV-K) or a difference from one species to another remains to be seen.
Both studies in mice differ in approach from the present analysis. The system used to study DBA mice allowed for the measurement of the phenomenon of solo-LTR formation as it occurs because there is a distinct and readily observable phenotype associated with deletion of the provirus, but it was restricted to analysis of one locus, whereas the recombinant inbred-strain studies observe solo-LTR formation over a relatively short amount of time (≈7,000 generations). In the human case, solo-LTR formation can be viewed only from an evolutionary perspective. Therefore, only events that have survived in the genome for hundreds of thousands of years and over one million generations are taken into account. The rate calculation in the human genome assumes selective neutrality of the newly formed solo-LTR allele; therefore, it would be an overestimate if this assumption were not true and fixation of solo LTRs were influenced by selection. Given the preponderance of solo LTRs in the human genome compared with their full-length proviral counterparts (7), selection may have played a role in reducing the proviral number in the genome if such events were beneficial to ancestors, either by reducing the expression of retroviral proteins, by decreasing the frequency of reinsertion events (28), or, as in the mouse model, by altering the expression of cellular genes.
Acknowledgments
We thank Tara Love for assistance with sample collection. This work was supported by National Cancer Institute Research Grant R01CA89441. J.M.C. was a research professor of the American Cancer Society, and J.F.H. was supported in part by National Cancer Institute Training Grant CA5441.
Abbreviation: HERV, human endogenous retrovirus.
References
- 1.Jurka, J. (2000) Trends Genet. 16, 418-420. [DOI] [PubMed] [Google Scholar]
- 2.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 3.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 4.Johnson, W. E. & Coffin, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medstrand, P. & Mager, D. L. (1998) J. Virol. 72, 9782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dangel, A. W., Baker, B. J., Mendoza, A. R. & Yu, C. Y. (1995) Immunogenetics 42, 41-52. [DOI] [PubMed] [Google Scholar]
- 7.Sverdlov, E. D. (1998) FEBS Lett. 428, 1-6. [DOI] [PubMed] [Google Scholar]
- 8.Sverdlov, E. D. (2000) BioEssays 22, 161-171. [DOI] [PubMed] [Google Scholar]
- 9.Barbulescu, M., Turner, G., Seaman, M. I., Deinard, A. S., Kidd, K. K. & Lenz, J. (1999) Curr. Biol. 9, 861-868. [DOI] [PubMed] [Google Scholar]
- 10.Buzdin, A., Ustyugova, S., Khodosevich, K., Mamedov, I., Lebedev, Y., Hunsmann, G. & Sverdlov, E. (2003) Genomics 81, 149-156. [DOI] [PubMed] [Google Scholar]
- 11.Turner, G., Barbulescu, M., Su, M., Jensen-Seaman, M. I., Kidd, K. K. & Lenz, J. (2001) Curr. Biol. 11, 1531-1535. [DOI] [PubMed] [Google Scholar]
- 12.Steinhuber, S., Brack, M., Hunsmann, G., Schwelberger, H., Dierich, M. P. & Vogetseder, W. (1995) Hum. Genet. 96, 188-192. [DOI] [PubMed] [Google Scholar]
- 13.Mager, D. L. & Goodchild, N. L. (1989) Am. J. Hum. Genet. 45, 848-854. [PMC free article] [PubMed] [Google Scholar]
- 14.Stoye, J. P., Frankel, W. N. & Coffin, J. M. (1991) Technique (Philadelphia) 3, 123-128. [Google Scholar]
- 15.Takahata, N. (1993) Mol. Biol. Evol. 10, 2-22. [DOI] [PubMed] [Google Scholar]
- 16.Ono, M. (1986) J. Virol. 58, 937-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, J. F. & Coffin, J. M. (2002) Genomics 80, 453-435. [PubMed] [Google Scholar]
- 19.Reus, K., Mayer, J., Sauter, M., Scherer, D., Muller-Lantzsch, N. & Meese, E. (2001) Genomics 72, 314-320. [DOI] [PubMed] [Google Scholar]
- 20.Kimura, M. (1983) The Neutral Theory of Molecular Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 21.Ruvolo, M. (1996) Mol. Phylogenet. Evol. 5, 202-219. [DOI] [PubMed] [Google Scholar]
- 22.Yu, N., Zhao, Z., Fu, Y. X., Sambuughin, N., Ramsay, M., Jenkins, T., Leskinen, E., Patthy, L., Jorde, L. B., Kuromori, T. & Li, W. H. (2001) Mol. Biol. Evol. 18, 214-222. [DOI] [PubMed] [Google Scholar]
- 23.Zhao, Z., Jin, L., Fu, Y. X., Ramsay, M., Jenkins, T., Leskinen, E., Pamilo, P., Trexler, M., Patthy, L., Jorde, L. B., Ramos-Onsins, S., Yu, N. & Li, W. H. (2000) Proc. Natl. Acad. Sci. USA 97, 11354-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Templeton, A. (2002) Nature 416, 45-51. [DOI] [PubMed] [Google Scholar]
- 25.Seperack, P. K., Strobel, M. C., Corrow, D. J., Jenkins, N. A. & Copeland, N. G. (1988) Proc. Natl. Acad. Sci. USA 85, 189-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankel, W. N., Stoye, J. P., Taylor, B. A. & Coffin, J. M. (1990) Genetics 124, 221-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankel, W. N. & Coffin, J. M. (1994) Mamm. Genome 5, 275-281. [DOI] [PubMed] [Google Scholar]
- 28.Boeke, J. D. & Stoye, J. P. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 343-436. [PubMed]
- 29.Hughes, J. F. & Coffin, J. M. (2001) Nat. Genet. 29, 487-489. [DOI] [PubMed] [Google Scholar]