Abstract
The unique features of mtDNA, together with the lack of a wide range of mouse cell mtDNA mutants, have hampered the creation of mtDNA mutant mice. To overcome these barriers mitochondrial defects were created by introducing mitochondria from different mouse species into Mus musculus domesticus (Mm) mtDNA-less (ρ0) L cells. Introduction of the closely related Mus spretus (Ms) or the more divergent Mus dunni (Md) mitochondria resulted in xenocybrids exhibiting grossly normal respiratory function, but mild metabolic deficiencies, with 2- and 2.5-fold increases in lactate production compared with controls. The transfer of this model from in vitro to in vivo studies was achieved by introducing Ms and Md mitochondria into rhodamine-6G-treated Mm mouse embryonic stem (ES) cells. The resultant xenocybrid ES cells remained pluripotent, and live-born chimerae were produced from both Ms and Md xenocybrid ES cells. Founder chimeric females (G0) were mated with successful germ-line transmission of Ms or Md mtDNA to homoplasmic G1 offspring. These xenocybrid models represent the first viable transmitochondrial mice with homoplasmic replacement of endogenous mtDNA and confirm the feasibility of producing mitochondrial defects in mice by using a xenomitochondrial approach.
The creation of in vitro and in vivo mouse models of mtDNA diseases is of great current interest. However, the unique features of mitochondrial genetics, including the high cellular mtDNA copy number and the lack of demonstrated mechanisms of recombination, have presented technical barriers to producing targeted mtDNA mutant mice.
Various attempts have been made to introduce exogenous DNA into mitochondria. One approach that uses a peptide nucleic acid linked to a mitochondrial target peptide leader sequence has proved promising (1, 2). Similarly, electroporation of plasmid DNA into isolated mitochondria was successful but has not proceeded further (3-5).
Several techniques have been used successfully to introduce genetically distinct mtDNA molecules into the mouse female germ line (see ref. 6 for review). Heteroplasmic mice have been created by fusion of cytoplasts generated from NZB/BINJ mouse ova with single-cell C57BL/6 (B6) or BALB/c zygotes (7-9), by fusion of membrane-bound karyoplasts which contain a zygote nucleus and a portion of the oocyte cytoplasm with enucleated eggs (10, 11), and by microinjection of Mus spretus (Ms) somatic cell mitochondria into Mus musculus domesticus (Mm) zygotes (12, 13). Fusion of cytoplasts heteroplasmic for a 4,696-bp mtDNA deletion to pronucleus-stage zygotes has also been used to create the first mouse model of mtDNA disease (14).
Fusion of cytoplasts to undifferentiated mouse female embryonic stem (ES) cells has also been used to introduce one of the few characterized mouse mtDNA mutants [a 16S rRNA mutation resulting in chloramphenicol resistance (CAPR)] into chimeric animals (15-17) and the mouse female germ line (17). Mice homoplasmic for the CAPR mutation presented with myopathy, dilated cardiomyopathy, and perinatal or in utero lethality. The successful creation of the CAPR mouse has validated the ES cell approach to producing transmitochondrial mice.
A different approach to producing oxidative phosphorylation (OXPHOS) defects was established where human mtDNA-less (ρ0) cells could be repopulated with closely related primate mtDNAs (18). The resulting “xenomitochondrial cybrids” (xenocybrids) had OXPHOS enzyme complexes composed of subunit proteins encoded by both human nuclear genes and primate mtDNA genes. These xenocybrids could replicate and transcribe the foreign mtDNAs but showed defective respiratory chain complex I with preserved function of the other complexes (19).
Similar defects were created in mouse cells by using the xenocybrid system, with the numerous extant Murinae species providing a wide variety of possible mtDNA donors. In xenocybrids generated by introducing Rattus norvegicus mtDNA into a Mm ρ0 cell line, severe multiple respiratory-chain defects resulted, with reduced complex I, III, and IV activities (20-22).
Here we describe the creation of xenocybrid ES cell lines and xenomitochondrial chimeric mice. Germ-line transmission was achieved by using both Ms and Mus dunni (Md) xenocybrid ES cells, resulting in the first homoplasmic xenomitochondrial mice. By using other xenocybrids, mice with OXPHOS defects of varying severity can be generated to model the wide range of clinical phenotypes seen in humans.
Methods
Cell Lines and Culture Conditions. All cells were grown in RPMI 1640 medium supplemented with 10% FBS (Invitrogen), 4.5 mg/ml glucose, 50 μg/ml uridine, and 1 mM pyruvate (RPMI/GUP medium) at 37°C and 5% CO2/95% air unless otherwise specified.
To minimize the chance of using cell lines with fixed mtDNA mutations, primary fibroblast lines were created from a 2-day-old laboratory mouse (Mm, CBA × B6 F1 cross) and a 5-week-old Ms mouse as described (20). Cells used for fusions were passage 5 or lower. An Md primary fibroblast cell line, designated III8C, was obtained from American Type Culture Collection. The mouse ρ0 cell clone LMEB3 was derived from the parental line LMTK- by exposure to ethidium bromide (23).
Production of Xenomitochondrial L Cell Cybrids. Mouse L cell cybrids were produced by enucleation of mitochondrial donor cells and fusion of the cytoplasts with mouse ρ0 cells followed by selection for respiratory-competent transformants. Procedures were described in detail (20). Cells used as mitochondrial donors included the Mm, Ms, and Md primary fibroblast lines described above.
In brief, ≈5 × 106 cells were harvested and resuspended in sterile 50-ml polysulfone centrifuge tubes (Nalge) in a mixture of 10 ml of RPMI/GUP medium, 10 ml of Percoll (Amersham Pharmacia Biotech), 1× penicillin/streptomycin, and 20 μg/ml cytochalasin B (from a 2 mg/ml stock in DMSO). Cells were centrifuged at 19,000 rpm (44,000 × gav) for 70 min at 37°C, and the cytoplast/karyoplast mixture was collected from the medium/Percoll interface. Previous investigation of this mixture had shown that >90% of cells were enucleated (24). The mixture was diluted in 15 ml of RPMI/GUP medium and centrifuged at 5,000 × g for 3 min to remove excess Percoll before being combined with 2 × 106 mouse ρ0 cells and centrifuged at 10,000 × g for 10 min. The supernatant was removed by aspiration, and the pellet was overlaid with polyethylene glycol (Sigma) for 1 min. The polyethylene glycol was then removed, and the cells were gently resuspended and plated at 104 and 105 cells per 100-mm dish in RPMI/GUP medium. After 24 h the medium was replaced with select medium: RPMI 1640 medium supplemented with 5% dialyzed FBS (Invitrogen), 4.5 mg/ml glucose, and 50 μg/ml BrdUrd (Sigma). After 7-10 days cybrid clones were isolated by using cloning cylinders, expanded, and viably frozen. Three independent clones were frozen from each fusion experiment, and cybrids were used in experiments from passages 5-10.
Production of Xenomitochondrial ES Cell Cybrids. All ES cells were grown in ES medium (DMEM; Invitrogen) supplemented with 15% ES cell grade FCS (Invitrogen), 1× nonessential amino acids (Invitrogen), 103 units/ml leukemia inhibitory factor (Amrad, Melbourne), 100 μM 2-mercaptoethanol, and 1× penicillin/streptomycin at 37°C and 5% CO2/95% air. ES cells were cultured on STO feeder cells (gift of K. Fowler, Murdoch Institute, Melbourne) mitotically inactivated by treatment with 10 μg/ml mitomycin c for 3 h. The mouse female ES cell line CC9.3.1 was derived from a 129SvEv-Gpi1c (129S6) embryo (gift of A. Bradley, Baylor College of Medicine, Houston) and was shown to have the potential to contribute to the female germ line and produce normal fertile females (17).
CC9.3.1 ES cells were treated with 1 μg/ml rhodamine-6G (from a 500 μg/ml stock dissolved in 2% ethanol) in ES medium for 72 h with replacement of medium at 24-h intervals. During the treatment period the ES medium was supplemented with 50 μg/ml uridine and 1 mM pyruvate. After treatment, ES cells were cultured in normal ES medium (no rhodamine-6G) for 2 h before cytoplast fusion.
Enucleated Ms or Md xenocybrid fibroblasts (≈2 × 106) were combined with 1 × 106 rhodamine-6G-treated ES cells in sterile 50-ml Nalgene tubes in 20 ml of ES medium. Fusions were performed as described above for fibroblasts. The fusion mixtures were plated onto mitomycin c-treated STO feeder cells at three densities (1.5 × 105, 2.5 × 105, and 6 × 105 ES cells per plating) in ES cell media with 1× hypoxanthine/aminopterin/thymidine (HAT). The HAT selection removed residual nucleated L cells, which were TK-. Colonies appeared after 4-5 days and were picked after 7 days for further expansion and genotyping.
Extraction of Total DNA from Cultured Cells and Mouse Tissue. Approximately 1 × 106 cells or 0.5 g of mouse tissue were digested with 100 μg/ml proteinase K (Invitrogen) in STE buffer (100 mM NaCl/25 mM EDTA/10 mM Tris·HCl, pH 8.0) with 0.5% SDS for 4 h at 55°C, followed by digestion with 40 μg/ml RNase A (Invitrogen) for 1 h at 37°C. DNA was extracted by using phenol/chloroform/isoamyl alcohol (25:24:1) followed by ethanol precipitation. DNA pellets were resuspended to 200 μg/ml in TE (1 mM EDTA/10 mM Tris·HCl, pH 8.0).
mtDNA PCR Amplification. A 461-bp mtDNA fragment containing part of the D loop and the tRNA-Phe gene was generated by using 1.0 unit of Taq polymerase (Invitrogen) and 35 cycles of 94°C for 30 sec, 55°C for 1 min, and 72°C for 1 min (20). Primers used for amplification were designed to amplify murid species diverging from Mus musculus to R. norvegicus as described (20): forward primer, 5′-ctc aac ata gcc gtc aag gc-3′ [representing nucleotides 15934-15953 (25)]; reverse primer, 5′-acc aaa cct ttg tgt tta tgg g-3′ (representing nucleotides 80-59).
The complete cytochrome b gene was amplified by using the forward primer 5′-cga agc ttg ata tga aaa acc atc gtt g-3′ and the reverse primer 5′-tct tca ttt ywg gtt tac aag ac-3′ with 1.0 unit of Taq polymerase (Invitrogen) and 35 cycles of 94°C for 30 sec, 55°C for 1 min, and 72°C for 1 min (26).
Sequencing of mouse mtDNA was performed by using the primers described for PCR amplification and the primers UMMZ12 5′-rta dgg gtg raa tgg rat ttt wtc-3′ and UMMZ13 5′-cay gaa wca ggv tca aay aay cc-3′ for the cytochrome b gene (26). PCR templates were column-purified (Wizard columns, Promega), and sequencing reactions used a BigDye terminator cycle-sequencing kit (Applied Biosystems) as per manufacturer's instructions. Reactions were analyzed by using an ABI 377 automated sequencer and sequencing analysis software (Applied Biosystems).
mtDNA Genotyping by Restriction Enzyme Analysis, Southern Blot Analysis, and DNA Sequencing. Genotyping of mouse cells was performed by restriction enzyme analysis of the 461-bp mtDNA D loop PCR amplicon. Mm and Ms derived amplicons were digested with either 10 units of StyI (NEB, Beverly, MA) or 10 units of Bsu36I (NEB) at 37°C for 4 h. Mm- and Md-derived amplicons were digested with 10 units of SpeI or 10 units of ScaI at 37°C for 4 h.
Total DNA was isolated from Ms:Mm and Md:Mm chimeric founders and putative homoplasmic offspring (agouti coat color) of female chimeras. Similarly, total DNA was isolated from tissues of Ms and Mm control mice or Md cells. For total DNA samples, 10 μg were digested with Bsu36I and XhoI (Ms:Mm analyses) or Bsu36I and EcoRI (Md:Mm analyses), loaded onto 0.8% agarose gels, and transferred to nylon membranes.
For Southern blot analysis, a 193-bp fragment corresponding to a region within the genes encoding ATP synthase 8 and ATP synthase 6 was used to probe mtDNA. The fragment was amplified by PCR with Ms primary kidney cell DNA and inserted into pGEM-T Easy Cloning vector (Promega). Primers were selected based on homology to known Mm, Ms, and Md mtDNA sequence within the amplified area (unpublished sequence data). The forward primer sequence was 5′-ggc acc ttc acc aaa atc ac-3′, representing nucleotides 7870-7889, and the reverse primer sequence was 5′-gga aag aat gga gac ggt tg-3′, representing nucleotides 8062-8043 (GenBank accession nos. J01420 and AY172335). Amplification was performed with 100 ng of DNA, 1 unit of Taq polymerase (Invitrogen), 1.5 mM MgCl2, 0.25 μM each primer, 0.20 μM dNTPs, and 40 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec. Plasmid DNA containing the mtDNA fragment was submitted for automatic sequencing to the URMC Functional Genomics Center (Rochester, NY). To generate probe template, an amplicon was produced by using 1 ng of plasmid DNA along with the primers and PCR conditions described above. Probes were labeled with [32P]dCTP by using the Random Primers DNA Labeling Kit (Invitrogen) and 100 ng of purified amplicon. The probe annealing and washing were done by using standard conditions.
Lactate Measurement. Lactate was measured in medium with a commercial kit (Sigma); 1 × 106 cells were harvested into 15-ml tubes, resuspended in 1 ml of RPMI/GUP medium, and incubated at 37°C in 5% CO2/95% air for up to 66 h. Ten microliters of medium were removed at various intervals and combined with 500 μl of reaction mixture (0.2 M glycine and hydrazine, pH 9.2/33 units/ml lactate dehydrogenase/3.3 mg/ml NAD). Reactions were incubated at 37°C for 15 min before spectrophotometric determination of NAD reduction by measuring absorbance at 340 nm. Measurements were compared with a lactate standard and standardized per cell.
Mitochondrial Isolation and Polarography. Mitochondrial isolation and polarography using freshly isolated mitochondria were performed as described (24). Cultures were expanded to ≈109 cells by seeding 2 × 107 cells into roller bottles (Corning) in 300 ml of RPMI/GUP medium, expanded after 4 or 5 days to 1,000 ml and then harvested after another 4 days.
OXPHOS Enzymology. Frozen mitochondrial aliquots were thawed and diluted to 2 μg/μl in mitochondrial isolation buffer and used to determine mitochondrial respiratory chain activity. Complex I (NADH:ubiquinone oxidoreductase, EC 1.6.5.3), complex II (succinate:ubiquinone oxidoreductase, EC 1.3.5.1), complex II+III (succinate:cytochrome c oxidoreductase), and complex IV (ferrocytochrome c:oxygen oxidoreductase or cytochrome oxidase, EC 1.9.3.1) activities were measured spectrophotometrically as described (24). Rotenone (5 μg) was also added to the complex I reaction mixture of duplicate reactions to measure complex I rotenone-insensitive activity.
Production of Transmitochondrial Mice. Procedures used for the collection, culture, and manipulation of B6 blastocysts were described in detail (27-29). Blastocysts were recovered from uteri of mated females (Taconic Farms) on day 3.5 (72 h after human chorionic gonadotropin) and maintained in microdrop cultures at 37°C in an atmosphere of 5% CO2/5% O2/90% N2 until ES cells were readied for injection. For injections, blastocysts were placed in either M16 or BMOC3 medium supplemented with Hepes (29). Injected blastocysts were transferred to the oviducts of D1 pseudopregnant ICR recipient mice. Resulting progeny were initially identified by coat color chimerism, and all founders were genotyped by using species-specific PCR analyses. Dead or euthanized animals were dissected, and DNA was extracted from various tissues as described above. PCR and Southern blotting were performed to confirm mitochondrial genotypes and tissue distribution.
Founder chimeric (heteroplasmic for Ms or Md mitochondria) females (G0) were mated to control B6 males (homoplasmic for Mm mitochondria). Tail biopsies from first-generation offspring (G1) were taken, and DNA extraction, PCR, and representative Southern blot analyses were performed for confirmation of genotype.
All mice were maintained in a specific pathogen-free barrier facility and were anesthetized by Avertin (2,2,2-tribromoethanol) injection during any surgical procedures. Mice were euthanized by CO2 inhalation or by Avertin sedation and cervical dislocation. All procedures adhered to the American Veterinary Medical Association Guide.
Results
Genetic Comparison of Murid Species. Because sequence data for mtDNA genes were lacking for the mouse species used, we sequenced the cytochrome b gene in all three species to verify that increasing amino acid divergence exists. The cytochrome b sequence of the Mm primary culture was identical with the published sequence (25), whereas the Ms and Md sequences showed 91% and 87% identity, respectively. Translation of these sequences revealed a high degree of conservation. The Ms amino acid sequence exhibited 99% identity with the Mm cytochrome b apoprotein (4 aa replacements), whereas the Md sequence showed 98% identity (7 aa replacements; ref. 30).
Production of Transmitochondrial L Cell Cybrids. Control cybrids produced by fusion of the ρ0 cell clone LMEB3 with enucleated Mm primary fibroblasts were obtained at a frequency of about one per 5 × 103 ρ0 cells used. Similar cybrid frequencies were obtained in fusions of LMEB3 with enucleated Ms and Md primary fibroblast cells. All xenomitochondrial fusion cybrids were genotyped by restriction enzyme digestion and sequencing of a mitochondrial D loop PCR amplicon. The transfer of mtDNA from Ms primary fibroblasts to the LMEB3 cell line was confirmed by digestion of the D loop PCR amplicon from the Ms xenocybrids with StyI (cleaves Ms but not Mm) and Bsu36I (cleaves Mm but not Ms) (not shown). Similar identification was performed for Md xenocybrid cybrids with ScaI (cleaves Mm but not Md) and SpeI (cleaves Md but not Mm) (not shown).
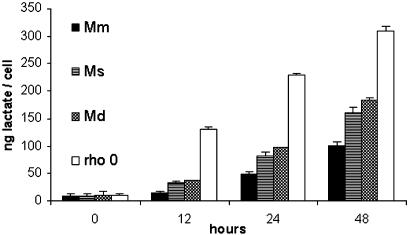
Lactate Measurement. Lactate production was measured as a marker of cell metabolism in the Mm control cybrid, the Ms and Md xenocybrids, and the ρ0 cell line LMEB3. Cell lactate is increased when mitochondrial oxidative ATP production is compromised. The Ms and Md xenocybrids showed significant 2- and 2.5-fold increases in lactate production compared with the control cybrid after 12 h (Fig. 1). The ρ0 cell line LMEB3, which has no oxidative ATP production, produced nine times more lactate than the control cell line after 12 h.
Fig. 1.
Lactate production in xenomitochondrial cybrids and ρ0 cells. Lactate production in the LMEB3 ρ0 cell line (rho 0), without oxidative ATP production, was nine times greater than in the control Mm after 12 h. Mild respiratory defects were indicated by 2- and 2.5-fold increases in lactate production in Ms and Md xenomitochondrial cybrids after 12 h.
Polarographic and Enzymological Measurement of OXPHOS in the Rodent Xenocybrids. Comparison of the mean specific respiratory rates determined by polarographic analysis from three independent experiments revealed that the Ms and Md xenocybrids have similar respiratory capacity to the control (data not shown). The mean activity of OXPHOS complexes I, II, II+III, and IV were also measured in triplicate by using three independent mitochondrial isolates for each xenocybrid. No significant differences were found in the specific activity of the enzyme complexes compared with the control (data not shown).
Production of Xenomitochondrial ES Cell Cybrids and Mice. Xenocybrid ES cells were produced by fusion of the Mm CC9.3.1 ES cell line (pretreated with rhodamine-6G) with cytoplasts from enucleated Ms or Md cells and obtained at a frequency of about one per 5 × 104 ES cells used. Both fusions resulted in xenocybrid ES cell clones with undifferentiated morphology and growth characteristics similar to the parental cell line.
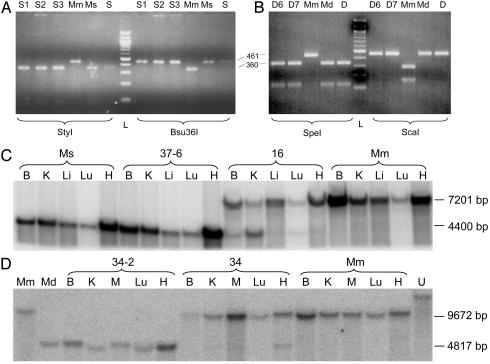
Xenocybrid ES cell cybrid clones were genotyped by restriction digest of a PCR amplicon containing a region of the mitochondrial D loop as per the LMEB3 fibroblast fusions. Three Ms ES cell xenocybrid clones were genotyped by digestion with StyI and Bsu36I (Fig. 2A), with each clone homoplasmic for Ms mtDNA. Similarly digestion with SpeI and ScaI revealed two Md ES cell xenocybrid clones homoplasmic for Md mtDNA (Fig. 2B).
Fig. 2.
Genotyping of Ms and Md xenomitochondrial ES cell clones and mice by restriction digest and Southern blot analysis. (A and B) A 461-bp PCR amplicon containing a region of the mtDNA D loop was digested with StyI, Bsu36I, SpeI, or ScaI. (A) Ms mtDNA was detected in three Ms xenomitochondrial ES cell clones (S1, S2, and S3), the Ms xenocybrid (MS), and MS primary fibroblasts (S) by a StyI restriction site (control Mm mtDNA s not cleaved by StyI). Conversely, Mm but not Ms mtDNA is cleaved by Bsu36I. The three Ms xenocybrid ES cell clones are homoplasmic for Ms mtDNA. (B) Md mtDNA was detected in two Md xenomitochondrial ES cell clones (D6 and D7), the Md xenocybrid (Md), and Md primary fibroblasts (D) by a SpeI restriction site (control Mm mtDNA is not cleaved by SpeI). Conversely, Mm but not Md mtDNA is cleaved by ScaI. The two Md xenocybrid ES cell clones are homoplasmic for Md mtDNA. (C) Hybridization of Ms and Mm mtDNA visualizing a 4,400-bp fragment and 7,201-bp fragment, respectively. Mouse #37-6 was a Ms homoplasmic offspring of founder dam S2 #37. Mouse #16 was a female founder displaying 20% chimerism by coat color. Ms and Mm, tissue from control mice. (D) Hybridization of Md and Mm mtDNA visualizing 4,817-bp and 9,672-bp fragments, respectively. Mouse #34-2 was the Md homoplasmic offspring of founder female #34 (80% coat-color chimera). Md and Mm were control cells or mouse tissue. B, brain; K, kidney; M, muscle; Li, liver; Lu, lung; H, heart; U, uncut DNA from Mm ES cells; L, DNA ladder.
Two ES cell clones for each xenocybrid were used for blastocyst injections. ES xenocybrid clones S2 and S3 (Ms) and D6 and D7 (Md) were injected into B6 blastocysts, and chimeric heteroplasmic founder mice were obtained (Table 1). Coat color (fur pigmentation) should reflect the source of nuclear genes, associated with either the host blastocyst (black) or the ES cell lines (agouti). However, mtDNA and nuclear DNA should be associated in maternal lineages and dissociated in paternal lineages. Indeed, this was confirmed in analyzed G1 offspring. Survival of newborn mice was low, but it was similar to yields in unrelated studies housed in the same room during the same period and appeared to reflect environmental perturbations. Health monitoring, including comprehensive necropsies of representative mice, did not identify murine pathogens or disease.
Table 1. Generation of chimeric heteroplasmic founder mice.
| ES cells | Clone | Ova injected | Founders | Weaned | Chimeras |
|---|---|---|---|---|---|
| Ms xenocybrid | S2 | 227 | 66 | 42 | 13/61 |
| S3 | 194 | 28 | 14 | 5/26 | |
| Md xenocybrid | D6 | 169 | 43 | 30 | 7/38 |
| D7 | 546 | 102 | 53 | 22/96 |
Chimeric heteroplasmic founder mice generated from different ES xenomitochondrial cybrid clones. All ES cell clones used were homoplasmic by restriction digest analysis.
Founder Ms:Mm xenocybrid mice (n = 18) were generated from ES cell clones S2 and S3. One male Ms xenocybrid mouse, generated from Ms ES xenocybrid clone S2 and determined to be ≈80% chimeric by coat color, was killed, and DNA was extracted from various tissues. Semiquantitative genotyping by PCR and restriction digest revealed the presence of a high percentage (≈50%) of Ms mtDNA in testes and spleen and lower percentages (≈5%) in kidney, skin, and lung, whereas it did not detect Ms mtDNA in heart, intestine, liver, and muscle (not shown).
Seven Ms xenocybrid females (1-20% coat-color chimeras) were mated with B6 males (Table 2). Breeding of the Ms xenocybrid female #37 (1% coat-color chimera) resulted in the production of 20 offspring, including one homoplasmic Ms xenocybrid male (100% agouti coat color). DNA was isolated and purified from tissue samples from this mouse, and subsequent PCR and Southern blot analyses confirmed homoplasmic germ-line transmission of Ms mtDNA to this pup (Fig. 2C).
Table 2. Breeding chimeric founder females.
| Clone | ♀ (% chimerism) | Pups | Germ-line transmission |
|---|---|---|---|
| Ms (clone S2) | 13 (20) | 19 | 0/19 |
| 26 (15) | 54 | 0/47 | |
| 33 (5) | 10 | 0/5 | |
| 37 (1) | 20 | 1/15 (♂) | |
| Ms (clone S3) | 2 (10) | 46 | 0/35 |
| 16 (20) | 29* | 0/27 | |
| 17 (5) | 9 | 0/9 | |
| Md (clone D6) | 1 (20) | 48 | 0/48 |
| 27 (20) | 33 | 0/33 | |
| Md (clone D7) | 1 (90) | 31 | 0/26 |
| 2 (10) | 32 | 0/29 | |
| 34 (80) | 34 | 1/33 (♂) | |
| 35 (80) | — | — | |
| 55 (90) | 31 | 3/23 (2♀/1♂)† | |
| 62 (95) | 57 | 0/38 | |
| 63 (85) | 59 | 1/46 (♂) | |
| 87 (85) | 17 | 0/14 | |
| 89 (80) | 8 | 0/7 |
Weaned chimeric founder females (heteroplasmic for either Ms or Md mitochondria) were bred with B6 males to generate homoplasmic offspring. Breeding of the Ms chimeric founder female S2 #37 (generated from the S2 clone) resulted in the germ-line transmission of Ms mtDNA in one male pup. This pup was homoplasmic for Ms mtDNA. Similarly, breeding of Md chimeric founder females D7 #34 and #55 (generated from the D7 clone) to B6 males resulted in the germ-line transmission of Md mtDNA to one male pup each that was homoplasmic for Md mtDNA. The three homoplasmic xenomitochondrial offspring had agouti fur pigmentation; all other offspring from female lineages bred to control B6 males had black fur.
Chimera S3 #16 died at 5 weeks of age. Ovaries were harvested and transferred to nude female recipients to attempt lineage rescue
Pups from chimera D7 #55 died or were killed within 24 h after birth
Founder Md:Mm chimeras (n = 29) were generated from ES cell clones D6 and D7. One male Md xenocybrid mouse, generated from Md ES xenocybrid clone D6 and determined to be ≈20% chimeric by coat color, was euthanized, and DNA was extracted from various tissues. Semiquantitative genotyping by PCR and restriction digest revealed ≈10-20% Md mtDNA with variability between brain, heart, muscle, intestine, lung, skin, and spleen, whereas it did not detect Md mtDNA in kidney, liver, pancreas, and testis (not shown).
Eleven Md xenocybrid females (10-95% coat-color chimerism) were also mated with B6 males (Table 2). Breeding of the Md xenocybrid female #34 (80% coat-color chimera) resulted in the production of 34 offspring, including one homoplasmic Md xenocybrid male (100% agouti coat color). DNA was purified from tissue samples; subsequent PCR and Southern blot analyses confirmed homoplasmic germ-line transmission of Md mtDNA (Fig. 2D). As observed with the Ms homoplasmic male, this male was fertile. Md xenocybrid female #63 (85% coat-color chimera) produced 46 offspring, including one homoplasmic Md xenocybrid male (100% agouti coat color) in her sixth litter. Md xenocybrid female #55 produced 31 offspring in three litters, although pups died or were killed within 24 h after birth. Three of 21 analyzed pups were homoplasmic for Md mitochondria.
Representative founder chimeric males derived from clone S2 or D6 were bred to two or more B6 females for a minimum of 2 months, at which time females were necropsied to establish male infertility [males: S2, #11 (20% coat-color chimera) and #12 (80%); D6, #13 (20%), #30 (20%), and #31 (20%)]. In contrast to males derived from these two clones, bred D7 males [#8 (20%), #49 (60%), and #60 (99%)] proved fertile with dead neonates or black pups obtained; Mm but not Ms or Md mtDNA was identified by PCR (not shown). Two homoplasmic male offspring of females S2 #37 and D7 #34 were bred and proved fertile with both agouti and black G2 pups obtained; PCR analyses indicated both an absence of Ms or Md mtDNA and the presence of Mm mtDNA (not shown).
Discussion
By introducing Ms and Md mtDNA into a female ES cell line, two species of xenocybrid chimeric mice were created; and germ-line transmission of the introduced mitochondrial backgrounds was identified in four separate lineages. Successful germ-line transmission of these mtDNAs was also achieved, resulting in the first homoplasmic xenomitochondrial mice. The first homoplasmic transmitochondrial mice were produced with a CAPR marker (17). Such mice showed a perinatal lethal phenotype, and the “proof-of-principle” mice produced in the same article by using an NZB strain mtDNA curiously showed a heteroplasmic genotype. From that work, it may be inferred that, without the CAPR marker, homoplasmic replacement of mtDNAs is not possible with the rhodamine-6G method. Our results differ by using a more divergent, interspecific mtDNA transfer. Further, the normal development and aging of the Ms xenocybrid mouse demonstrates that this homoplasmic replacement and rhodamine-6G manipulations are not the cause of the severe phenotype in the CAPR mouse.
In xenocybrid fibroblasts the severity of the OXPHOS defect and the viability of xenocybrid fusion depend on evolutionary divergence. The Ms and Md mtDNAs (divergence from Mm ≈2 and 4 million years before the present; refs. 31-33) are readily maintained when introduced into Mm ρ0 cells, with the resulting xenocybrids exhibiting normal OXPHOS. However, a slight increase in cellular lactate production (indicating an increase in glycolytic ATP generation because of subtle respiratory defects) was detectable in Ms (2-fold increase) and Md (2.5-fold increase) xenocybrid fibroblasts.
In a fashion similar to the xenocybrid fusion experiments using LMEB3 ρ0 fibroblasts, the viability of xenocybrid ES fusion also depended on evolutionary divergence. The Ms and Md mtDNAs were readily maintained when introduced into rhodamine-6G-pretreated Mm ES cells, with the resulting ES xenocybrids remaining undifferentiated and pluripotent. R. norvegicus mtDNA, which is replicated, transcribed, and translated efficiently in ρ0 fibroblasts, is not readily maintained in rhodamine-6G-pretreated ES cells. Rat:mouse xenocybrids were not viable, with ES clones either differentiating into fibroblast-type cells or repopulating with Mm mtDNA (data not shown). The nuclear-mitochondrial protein interaction between the Mm and rat OXPHOS subunits creates a severe respiratory defect (20) that appears to be too great for ES cell viability, contrary to an earlier claim that Rattus xenocybrids could be used to produce a mouse model (22).
The successful creation of homoplasmic Ms and Md xenocybrid mice confirms not only the viability of totally repopulating ES cells with mtDNA of a different species but also the ability to transfer these mtDNAs through the female germ line. Both the Ms and Md homoplasmic mice appear to have normal physiology and development, although pathological symptoms possibly may arise with age. Because we have obtained limited numbers of live homoplasmic mice to date, phenotypic and pathological studies have not been undertaken to examine what effects the subtle OXPHOS defects seen in the fibroblast cybrid studies may have in vivo. Xenomitochondrial mice made by using other species that are further removed from Mm may result in more overt phenotypes.
Currently only two mtDNA mouse models exist: the mtDNA deletion (14) and the 16S CAPR (15-17) mutations. Both of these models are highly informative and have provided important previously uninvestigated information on mitochondrial dysfunction. However, anomalies exist between their genetic features and those seen in human mtDNA disease. The phenotypes and inheritance patterns in the mtDNA deletion model are different from those seen in patients with human mtDNA rearrangements. In humans most deletion mutations are spontaneous, and patients with a very high percentage of deleted mtDNAs are severely affected in childhood. The 16S CAPR mutation results in a perinatal lethal phenotype in the homoplasmic mouse, whereas in humans many base substitution mutations in protein synthesis genes are usually compatible with maturation to at least late childhood.
Other models of human mtDNA disease are required; and because a paucity of known mouse mutants in cell culture exists, the xenocybrid system provides an alternative technology to creating mitochondrial defects. The homoplasmic Ms and Md xenocybrid mice described here prove that germ-line transmission of interspecies mtDNA is readily possible.
Germ-line transmission of introduced Ms and Md mtDNA in maternal lineages proved inefficient with the CC9.3.1 ES cell line. In studies where the same cell line was used (17), and in other reports where founder transmitochondrial mice were created by using ES cell transfer, demonstration of maternal germ-line transmission of a mutant/introduced mitochondrial background proved difficult. ES cell populations commonly used to produce chimeric mice are traditionally derived from male (XY) embryos, and certain developmental characteristics and efficiencies favor their usage (34, 35). Genetic and epigenetic changes were illustrated, in addition to varying percentages of abnormal karyotypes in cultured ES cells (35, 36). The potential for minor genetic changes or DNA methylation/imprinting concerns, coupled with the limited number of cell lines available, illustrates that additional female ES cell lineages are needed in further studies.
To accurately model human mitochondrial diseases a better understanding of the effects of human mtDNA mutations on not only OXPHOS but also cell death and reactive oxygen species generation is needed. The value of the xenomitochondrial approach is that the large range of evolutionary divergence in the Murinae subfamily can be used to create different cybrids with a range of OXPHOS defects. Other species of xenocybrid mice are currently being created, and appropriate models that mimic the clinical presentations and genetic features of human mitochondrial disease can be used for further study and possible therapeutic development. Mice with mildly defective OXPHOS, resulting in relatively subtle age-related pathology, will be of particular interest. By cross-breeding with nuclear gene models, such mice could then be used to directly test hypothetical involvement of OXPHOS dysfunction in the common neurodegenerative diseases of aging (37). A recent report showing that subtle neurological phenotypes can be ascertained in a nuclear-mitochondrial mismatch obtained by traditional back-crossing, by using the less divergent NZB mtDNA on a Mm nuclear background, augers favorably for our approach (38).
Acknowledgments
We thank D. Cotton for support; M. Chiotis for help with the mtDNA sequencing; C. A. Lerner, C. A. Ingraham, K. A. Bussey, and D. A. Dunn for mtDNA PCR genotyping; and R. L. Howell and C. L. Donegan for animal modeling and microinjection expertise. The CC9.3.1 cell line was a gift of A. Bradley (Baylor College of Medicine, Houston). This work was supported in part by Grant 145719 from the National Health and Medical Research Council of Australia (to I.A.T.) and by National Institutes of Health Grants RR16286 and DE12634 and institutional funds (to C.A.P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Mm, Mus musculus domesticus; Ms, Mus spretus; Md, Mus dunni; B6, C57BL/6; CAPR, chloramphenicol resistance; OXPHOS, oxidative phosphorylation; ES, embryonic stem.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AY193770 (M. dunni cytochrome b)].
References
- 1.Seibel, M., Bachmann, C., Schmiedel, J., Wilken, N., Wilde, F., Reichmann, H., Isaya, G., Seibel, P. & Pfeiler, D. (1999) Biol. Chem. Hoppe-Seyler 380, 961-967. [DOI] [PubMed] [Google Scholar]
- 2.Flierl, A., Jackson, C., Cottrell, B., Murdock, D., Seibel, P. & Wallace, D. C. (2003) Mol. Ther. 7, 550-557. [DOI] [PubMed] [Google Scholar]
- 3.Collombet, J. M., Wheeler, V. C., Vogel, F. & Coutelle, C. (1997) J. Biol. Chem. 272, 5342-5347. [DOI] [PubMed] [Google Scholar]
- 4.Pinkert, C. A. & Trounce, I. A. (2002) Methods 26, 348-357. [DOI] [PubMed] [Google Scholar]
- 5.Irwin, M. H., Parrino, V. & Pinkert, C. A. (2001) Adv. Reprod. 5, 59-66. [Google Scholar]
- 6.Wallace, D. C. (2001) Am. J. Med. Genet. 106, 71-93. [DOI] [PubMed] [Google Scholar]
- 7.Jenuth, J. P., Peterson, A. C., Fu, K. & Shoubridge, E. A. (1996) Nat. Genet. 14, 146-151. [DOI] [PubMed] [Google Scholar]
- 8.Jenuth, J. P., Peterson, A. C. & Shoubridge, E. A. (1997) Nat. Genet. 16, 93-95. [DOI] [PubMed] [Google Scholar]
- 9.Meirelles, F. V. & Smith, L. C. (1998) Genetics 148, 877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laipis, P. J. (1996) Methods Enzymol. 264, 345-357. [DOI] [PubMed] [Google Scholar]
- 11.Meirelles, F. V. & Smith, L. C. (1997) Genetics 145, 445-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irwin, M. H., Johnson, L. W. & Pinkert, C. A. (1999) Transgenic Res. 8, 119-123. [DOI] [PubMed] [Google Scholar]
- 13.Pinkert, C. A., Irwin, M. H., Johnson, L. W. & Moffatt, R. J. (1997) Transgenic Res. 6, 379-383. [DOI] [PubMed] [Google Scholar]
- 14.Inoue, K., Nakada, K., Ogura, A., Isobe, K., Yi, G., Nonaka, I. & Hayashi, J. I. (2000) Nat. Genet. 26, 176-181. [DOI] [PubMed] [Google Scholar]
- 15.Levy, S. E., Waymire, K. G., Kim, Y. L., MacGregor, G. R. & Wallace, D. C. (1999) Transgenic Res. 8, 137-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchington, D. R., Barlow, D. & Poulton, J. (1999) Nat. Med. 5, 957-960. [DOI] [PubMed] [Google Scholar]
- 17.Sligh, J. E., Levy, S. E., Waymire, K. G., Allard, P., Dillehay, D. L., Nusinowitz, S., Heckenlively, J. R., MacGregor, G. R. & Wallace, D. C. (2000) Proc. Natl. Acad. Sci. USA 97, 14461-14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenyon, L. & Moraes, C. T. (1997) Proc. Natl. Acad. Sci. USA 94, 9131-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrientos, A., Kenyon, L. & Moraes, C. T. (1998) J. Biol. Chem. 273, 14210-14217. [DOI] [PubMed] [Google Scholar]
- 20.McKenzie, M. & Trounce, I. (2000) J. Biol. Chem. 275, 31514-31519. [DOI] [PubMed] [Google Scholar]
- 21.Dey, R., Barrientos, A. & Moraes, C. T. (2000) J. Biol. Chem. 275, 31520-31527. [DOI] [PubMed] [Google Scholar]
- 22.Yamaoka, M., Isobe, K., Shitara, H., Yonekawa, H., Miyabayashi, S. & Hayashi, J.-I. (2000) Genetics 155, 301-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trounce, I., Schmiedel, J., Yen, H. C., Hosseini, S., Brown, M. D., Olson, J. J. & Wallace, D. C. (2000) Nucleic Acids Res. 28, 2164-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trounce, I., Kim, Y. L., Jun, A. S. & Wallace, D. C. (1996) Methods Enzymol. 264, 484-509. [DOI] [PubMed] [Google Scholar]
- 25.Bibb, M. J., Van Etten, R. A., Wright, C. T., Walberg, M. W. & Clayton, D. A. (1981) Cell 26, 167-180. [DOI] [PubMed] [Google Scholar]
- 26.Jansa, S. A., Goodman, S. M. & Tucker, P. K. (1999) Cladistics 15, 253-270. [DOI] [PubMed] [Google Scholar]
- 27.Pinkert, C. A., ed. (2002) Transgenic Animal Technology: A Laboratory Handbook (Academic, San Diego).
- 28.Pinkert, C. A., Irwin, M. H. & Moffatt, R. J. (1997) in Encyclopedia of Molecular Biology and Molecular Medicine, ed. Meyers, R. A. (VCH, New York), Vol. 6, pp. 63-74. [Google Scholar]
- 29.Polites, H. G. & Pinkert, C. A. (2002) in Transgenic Animal Technology: A Laboratory Handbook, ed. Pinkert, C. A. (Academic, San Diego), pp. 15-70.
- 30.McKenzie, M., Chiotis, M., Pinkert, C. A. & Trounce, I. A. (2003) Mol. Biol. Evol. 20, 1117-1124. [DOI] [PubMed] [Google Scholar]
- 31.Catzeflis, F. M., Dickerman, A. W., Michaux, J. & Kirsch, J. A. W. (1993) Mammal Phylogeny: Placentals (Springer, New York).
- 32.Bonhomme, F. & Guenet, J.-L. (1996) Genetic Variants and Strains of the Laboratory Mouse (Oxford Univ. Press, Oxford).
- 33.Silver, L. M. (1995) Mouse Genetics (Oxford Univ. Press, Oxford).
- 34.Nagy, A., Gertsenstein, M., Vintersten, K. & Behringer, R. (2003) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, New York).
- 35.Robertson, E. J. (1987) Teratocarcinomas and Embryonic Stem Cells: A Practical Approach (IRL, Oxford).
- 36.Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J. C. (1993) Proc. Natl. Acad. Sci. USA 90, 8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manfredi, G. & Beal, M. F. (2000) Brain Pathol. 10, 462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roubertoux, P. L., Sluyter, F., Carlier, M., Marcet, B., Maarouf-Veray, F., Cherif, C., Marican, C., Arrechi, P., Godin, F., Jamon, M., et al. (2003) Nat. Genet. 35, 65-69. [DOI] [PubMed] [Google Scholar]