Abstract
Coelenterazine is widely distributed among marine organisms, producing bioluminescence by calcium-insensitive oxidation mediated by Renilla luciferase (Rluc) and calcium-dependent oxidation mediated by the photoprotein aequorin. Despite its abundance in nature and wide use of both proteins as reporters of gene expression and signal transduction, little is known about mechanisms of coelenterazine transport and cell permeation. Interestingly, coelenterazine analogues share structural and physiochemical properties of compounds transported by the multidrug resistance MDR1 P-glycoprotein (Pgp). Herein, we report that living cells stably transfected with a codon-humanized Rluc show coelenterazine-mediated bioluminescence in a highly MDR1 Pgp-modulated manner. In Pgp-expressing Rluc cells, low baseline bioluminescence could be fully enhanced (reversed) to non-Pgp matched control levels with potent and selective Pgp inhibitors. Therefore, using coelenterazine and noninvasive bioluminescence imaging in vivo, we could directly monitor tumor-specific Pgp transport inhibition in living mice. While enabling molecular imaging and high-throughput screening of drug resistance pathways, these data also raise concern for the indiscriminate use of Rluc and aequorin as reporters in intact cells or transgenic animals, wherein Pgp-mediated alterations in coelenterazine permeability may impact results.
Bioluminescence, a common phenomenon particularly in marine ecosystems, is the emission of light by living organisms, representing a major form of communication and camouflage and a potential adaptation to oxidative stress (1). A substrate (luciferin) is oxidized by an enzyme (luciferase) in the presence of oxygen, yielding an excited intermediate that emits light on returning to the ground state (2, 3). Coelenterazine, the imidazolopyrazine luciferin, is widely distributed among coelenterates, fishes, squids, and shrimps (4, 5). Renilla luciferase (Rluc), purified from a bioluminescent soft coral known as sea pansy (Renilla reniformis), catalyzes the calcium-insensitive oxidation of coelenterazine (6). The gene has been cloned and sequenced (7) and used extensively in reporter gene assays in bacteria, yeast, plant, and mammalian cells (8) for bioluminescence resonance energy transfer studies and for high throughput drug screening (9-11). Rluc provides a kinetically rapid, alternative bioluminescence signal to firefly (Photinus pyralis) luciferase and, when coexpressed, enables dual reporter readouts.
A rapidly evolving area of biomedical research, use of bioluminescence as an optical marker for gene expression has been recently extended to noninvasive, real-time analysis of molecular events in intact cells and living animals (12-14). Although previous data suggested that Rluc could be used for reporter imaging in mice in vivo (15), limited utility and poor bioavailability of coelenterazine was reported in a mouse model of viral infection with an Rluc reporter strain of herpes simplex virus 1 (16). Interestingly, anabolic enzymes enabling de novo synthesis of coelenterazine are lacking in many bioluminescent organisms, and a dietary requirement for coelenterazine within the food chain has been demonstrated (17), implying mechanisms for absorption and retention of the compound. A hydrophobic heterocycle with protonated nitrogens (3), the structure of coelenterazine shares physicochemical features of many natural substrates of the ATP-binding cassette transporter superfamily, especially the multidrug resistance MDR1 P-glycoprotein (ABCB1; Pgp), a 170-kDa transmembrane protein encoded by the MDR1 gene (18-20). In cancer, overexpression of MDR1 confers cross-resistance to a broad array of natural product cytotoxic agents (21). Pgp putatively acts as an energy-dependent efflux transporter that efficiently pumps drugs out of cells or off the plasma membrane, resulting in decreased intracellular accumulation of anticancer drugs and other substrates (22, 23). In this study, we report that coelenterazine is a transport substrate recognized by human Pgp, thus enabling noninvasive bioluminescence imaging of Pgp transport activity in living animals.
Methods
Reagents. Native coelenterazine (Biotium, Hayward, CA) and five semisynthetic analogues, coelenterazine n, f, h, hcp, and cp (Molecular Probes), were dissolved in ethanol as 4.7 mM stock solutions. N-(4-[2-(1,2,3,4-tetrahydro-6,7-dimethoxy-2-isoquinolinyl)ethyl]-phenyl)-9,10-dihydro-5-methoxy-9-oxo-4-acridine carboxamide (GF120918, Elacridar; gift of Glaxo Wellcome), (3′-keto-Bmt′)-[Val-2]cyclosporin A (PSC 833, Valspodar; gift of Mallinckrodt), the anthranilamide derivative XR9576 (Tariquidar; gift of QLT), cyclosporin A, methotrexate, and cisplatin were prepared as stock solutions in DMSO. All other reagents were obtained from Sigma. The final concentration of DMSO or ethanol in cell buffers was 0.1%, which had no effect on the results (data not shown).
Cell Culture. Human epidermoid carcinoma drug-sensitive KB 3-1 cells and colchicine-selected KB 8-5-11 cells were transfected with a codon-humanized vector pRluc-N3 (BioSignal Packard, Montreal). Stable clones were isolated and maintained in DMEM supplemented with l-glutamine (1%), heat-inactivated FBS (10%), penicillin/streptomycin/fungizone (0.1%), and G418 (1 mg/ml) in a 5% CO2 incubator at 37°C. KB 8-5-11 and KB 8-5-11 Rluc cells were cultured in the presence of colchicine (100 ng/ml) to maintain expression of Pgp.
Western Blots. Pgp was detected in enriched membrane fractions from cell lines by using mAb C219 (Signet Laboratories, Dedham, MA) as described (24). Uniform loading was confirmed with anti-β-catenin Ab (BD Pharmingen).
Cellular Accumulation of 99mTc-Sestamibi. Transport function and modulation of MDR1 Pgp with GF120918 (800 nM) under steady-state conditions were assayed with 99mTc-Sestamibi as described (25). Cell-associated tracer is expressed as fmol/mg protein per nM, where cell content of Tc-Sestamibi (fmol) was normalized to mg of cell protein and extracellular concentration of tracer (nM).
Renilla Lysis Assay. Luciferase (R. reniformis) activity in cell lysates was detected according to the manufacturer's protocol (Promega). Cells were plated at a density of 2 × 105 cells per well into six-well plates. After 2 days, cells were lysed with lysis buffer (1×, 500 μl) and stored at -80°C until further use. For the assay, cell lysates (20 μl) were mixed with Rluc reagent (100 μl). After a 1-min incubation, luminescence was measured from each sample in a table-top beta counter (Beckman Coulter). Results were normalized to total protein as determined by BCA assay (Pierce).
Bioluminescence Imaging of Cell Lines. Cells were plated at a density of 5 × 104 cells per well into 24-well plates and grown to 80-100% confluency (≈2 days). Just before imaging, media were changed to a colorless solution containing (in mM): 2.7 KCl, 139 NaCl, 8.1 Na2HPO4. 7 H2O, 1.5 KH2PO4, 1.8 CaCl2, 1 MgCl2, and 5.5 d-glucose. Cells were preincubated for 15 min in the absence or presence of Pgp modulator, as indicated, after which coelenterazine (final concentration of 470 nM) was added directly to the cells. Bioluminescence imaging of Rluc was performed with the in vivo imaging system (Xenogen, Alameda, CA). Acquisition times depended on light output and are indicated in figure legends. After in vivo images were taken, cells were lysed to determine total protein and corrected for total protein in each well. Data analysis was performed as described (16).
Bioluminescence Imaging of Animals. Animal care and euthanasia were approved by the Washington University Medical School Animal Studies Committee. KB 3-1 and KB 3-1 Rluc cells (1 × 107) and KB 8-5-11 and KB 8-5-11 Rluc cells (1.5 × 107) were injected s.c. into flanks of 6-week-old male NCr nu/nu mice (Taconic Farms). After development of ≈5-mm tumors, the animals were pretreated with vehicle (0.5% hydroxy-propylmethylcellulose/1% Tween 80) on day 1 and with GF120918 (250 mg/kg of body weight in 0.5% hydroxypropyl methylcellulose/1% Tween 80) on day 2 by oral gavage 4 h before imaging. Mice were anesthetized with metofane or isoflurane before tail vein injection of coelenterazine (4 μg/g) formulated from an ethanol stock diluted in sodium phosphate buffer (50 mM). Bioluminescence imaging was performed on the in vivo imaging system at 2, 6, 8, and 11 min after injection as described (16) (acquisition time, 5 s; binning, 8; field of view, 10 cm; f/stop, 1). Anesthesia was maintained during imaging by nose cone delivery of 2.5% isoflurane. After imaging, animals were killed by cervical dislocation; tumors were then harvested and weighed.
Data Analysis. Corresponding grayscale photographs and color luciferase images were superimposed and analyzed with livingimage (Xenogen) and igor (WaveMetrics, Lake Oswego, OR) image analysis software. Signal intensities from regions of interest (ROI) were defined either with a grid template for cells grown in 24-well plates or manually for tumors. Light intensities of the ROIs were expressed as total flux (photons per s). Background total flux for tumors was defined from an ROI of the same size drawn over the thorax of each animal, and these data were subtracted from signal intensities measured over tumors. Data were reported as means ± SEM for the number of wells or animals as indicated in figure legends. Pairs were compared with Student's t test (26), and values of P ≤ 0.05 were considered to be significant.
Results
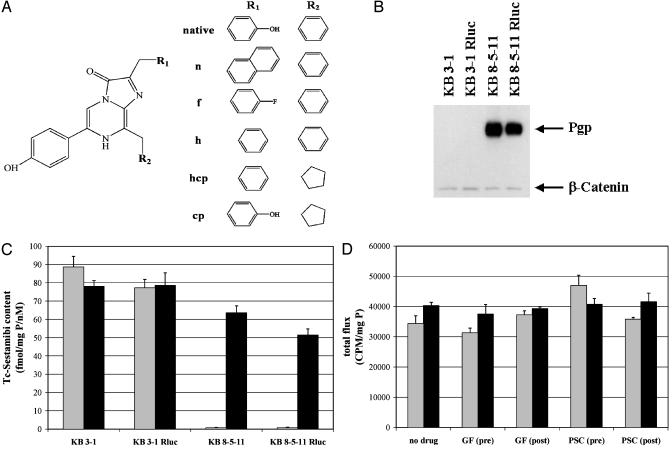
Pgp-Mediated Transport of Coelenterazine in Stably Transfected Rluc Cell Lines. To test the hypothesis that native coelenterazine (Fig. 1A) was transported by MDR1 Pgp, two human cell lines with different Pgp status were stably transfected with a codonhumanized vector encoding Rluc. Drug-sensitive KB 3-1 cells lack immunodetectable Pgp, and colchicine-selected KB 8-5-11 cells express high levels of Pgp. Western blotting confirmed that Pgp levels were not changed substantially between parental and Rluc transfected cell lines (Fig. 1B). To functionally compare Pgp expression levels in intact parental and transfected cell lines, cell tracer transport assays were performed with 99mTc-Sestamibi, a validated transport substrate for Pgp (27), in the absence or presence of GF120918 (800 nM), a potent and highly selective modulator (inhibitor) of Pgp transport function (28). As expected, 99mTc-Sestamibi accumulation was 100-fold less in MDR KB 8-5-11 cells compared with KB 3-1 cells, and in KB 8-5-11 cells only, tracer accumulation was enhanced by treatment with GF120918. These Pgp-mediated transport differences were identical between parental cells and cells stably transfected with Rluc (Fig. 1C). In addition, levels of Rluc enzyme activity were comparable in KB 3-1 Rluc and KB 8-5-11 Rluc cells as determined in cell lysates normalized to total cell protein (Fig. 1D). Furthermore, control experiments with cell lysates showed no effect of MDR modulators on Rluc-mediated coelenterazine oxidation whether performed with lysates containing freshly added modulator or with lysates from cells preincubated for 15 min with modulators before lysis. A low background signal was detected in lysates of nontransfected parental cells representing light emitted from enzyme-independent oxidation (autoluminescence) of coelenterazine.
Fig. 1.
Structures of coelenterazines and validation of the system. (A) Chemical structures of native coelenterazine and semisynthetic analogues. (B) Expression of Pgp (170 kDa) in KB cells as determined with mAb C219. Equal loading was confirmed with anti-β-catenin Ab. (C) Pgp function in KB cells verified with a tracer transport assay. Cells were incubated in buffer containing 99mTc-Sestamibi in the absence (gray) or presence (black) of GF120918 (800 nM). Each bar represents the mean ± SEM of four determinations. (D) Renilla lysis assays document comparable Rluc activity levels in transfected KB cells independent of the Pgp status. In vitro bioluminescence (cpm) corrected to total protein is plotted for the indicated cell lysates of KB 3-1 Rluc (gray) and KB 8-5-11 Rluc (black): lysates alone; lysates with GF120918 (300 nM) or PSC 833 (2 μM) (GF and PSC post); and lysates from cells preincubated with GF120918 (300 nM) or PSC 833 (2 μM) before lysis (GF and PSC pre).
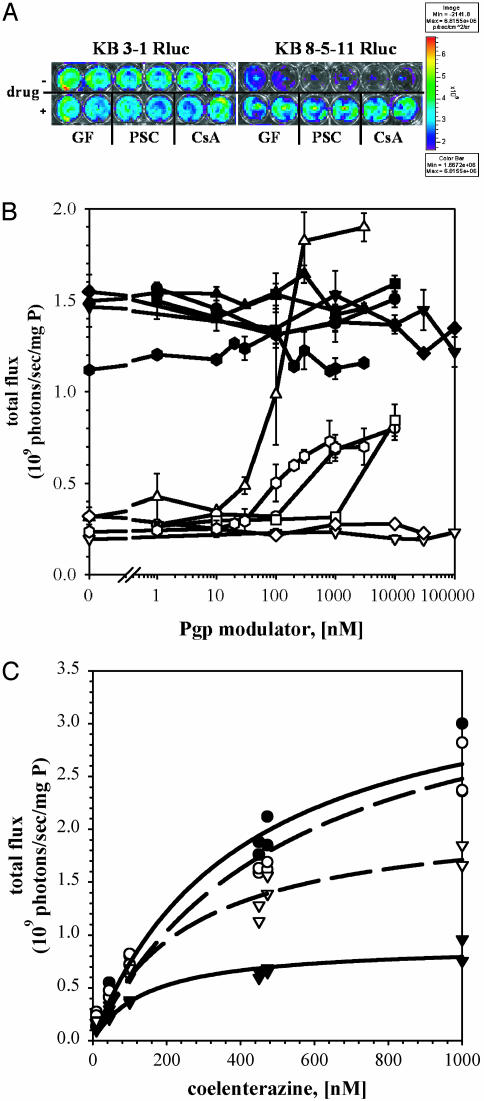
On addition of coelenterazine to the buffer, intact KB 3-1 Rluc cells showed high bioluminescence, whereas intact KB 8-5-11 Rluc cells showed low bioluminescence (Fig. 2A). Normalized to levels of total protein in each cell line, KB 3-1 Rluc cells showed a specific total flux of 1.4 × 109 photons per s per mg of protein, 4-fold greater than KB 8-5-11 Rluc cells. The signal in KB 3-1 Rluc cells was observed as early as 6 s after substrate addition (data not shown), remained stable for 30 min before declining from substrate depletion, and was 40-fold greater than autoluminescence from untransfected cells. In KB 8-5-11 Rluc cells, the low bioluminescence also was rapidly observed and remained stable for at least 30 min, but on treatment with saturating concentrations of the potent Pgp modulators GF120918 and PSC 833 or the classic Pgp modulator cyclosporin A (21, 29), they could be reversed to control levels. There were no modulator-induced changes in bioluminescence signals emitted from KB 3-1 Rluc cells (Fig. 2 A). Thus, a specific Pgp-mediated reduction in steady-state content of coelenterazine existed in KB 8-5-11 Rluc cells compared to KB 3-1 Rluc cells. Because Rluc is a cytosolic enzyme (8), the Pgp-mediated difference in bioluminescence in each cell line was consistent with either a Pgp-mediated outward translocation or a Pgp-mediated permeation barrier to coelenterazine at the level of the plasma membrane.
Fig. 2.
Bioluminescence imaging of intact KB Rluc cells. (A) KB 3-1 Rluc cells (Left) and KB 8-5-11 Rluc cells (Right) were imaged in an in vivo imaging system. The first row represents bioluminescence signal in the absence of drug, and the second row shows cells pretreated with GF120918 (300 nM), PSC 833 (2 μM), and cyclosporin A (5 μM) as indicated. Images show raw photon flux, uncorrected for mg of protein per well (exposure time 30 s, 6 min after the addition of 470 nM coelenterazine). (B) KB 3-1 Rluc (filled symbols) and KB 8-5-11 Rluc cells (open symbols) were imaged as above in the absence and after pretreatment with various concentrations of GF120918 (▵, ▴), XR9576 ( ,
,  ), PSC 833 (○, •), cyclosporin A (□, ▪), methotrexate (▿, ▾), and cisplatin (⋄, ♦). Total flux corrected for total cell protein is plotted versus drug concentrations (nM). Data represent the mean ± range of two independent determinations. (C) KB 3-1 Rluc (○, •) and KB 8-5-11 Rluc (▿, ▾) cells were imaged with increasing concentrations of coelenterazine in the absence (•, ▾; solid line) and in the presence of GF120918 (300 nM) (○, ▿; dashed line). Total flux normalized to total cell protein is plotted against coelenterazine concentration (nM). Lines represent Michaelis-Menten curves fit to the data.
), PSC 833 (○, •), cyclosporin A (□, ▪), methotrexate (▿, ▾), and cisplatin (⋄, ♦). Total flux corrected for total cell protein is plotted versus drug concentrations (nM). Data represent the mean ± range of two independent determinations. (C) KB 3-1 Rluc (○, •) and KB 8-5-11 Rluc (▿, ▾) cells were imaged with increasing concentrations of coelenterazine in the absence (•, ▾; solid line) and in the presence of GF120918 (300 nM) (○, ▿; dashed line). Total flux normalized to total cell protein is plotted against coelenterazine concentration (nM). Lines represent Michaelis-Menten curves fit to the data.
Effects of MDR Modulators on Coelenterazine Transport. To further characterize pharmacological inhibition, the two cell lines were tested by using the in vivo imaging system over a range of concentrations with known Pgp modulators (GF120918, PSC 833, XR9576, and cyclosporin A) and with drugs that have no effect on Pgp (methotrexate and cisplatin) (25). Bioluminescence signals were measured over drug concentrations ranging from 1 nM to 100 μM (as indicated in Fig. 2B) and were compared to background signals for untreated cells and cells in the presence of vehicle (DMSO) at the highest tested concentration. Bioluminescence output from KB 3-1 Rluc cells (Fig. 2B) was unaffected by all drugs over the concentration range tested. For this series of experiments, bioluminescence from KB 8-5-11 Rluc cells was one order of magnitude less than signals from KB 3-1 Rluc cells but could be completely reversed by GF120918 with an IC50 of 115 nM. Furthermore, the reversal agents XR9576 (30) (IC50 = 97 nM), PSC 833 (IC50 = 509 nM), and cyclosporin A (IC50 = 16 μM) also enhanced bioluminescence signals at concentrations consistent with the known potency rank order of these compounds, although in contrast to GF120918, full reversal was not observed. In addition, these IC50 values, right-shifted to higher concentrations in these highly Pgp-expressing KB 8-5-11 Rluc cells, matched data reported previously with other transport substrates of Pgp (25). As expected, methotrexate and cisplatin had no effect on light emission from both cell lines. Thus, coelenterazine and bioluminescence quantitatively reported the pharmacology of MDR reversal in cells in culture.
Bioluminescence signals from KB 3-1 Rluc cells, both in the absence and presence of GF120918 (300 nM), were readily detected with as little as 1 nM extracellular coelenterazine and increased in a concentration-dependent manner to 1 μM extracellular coelenterazine (Fig. 2C). In Pgp-expressing cells, there was evidence for a GF120918-reversible concentration-dependent saturation of bioluminescence with an EC50 of 257 nM coelenterazine. The plateau in bioluminescence signal observed in KB 8-5-11 Rluc cells implied that the capacity of Pgp to limit delivery of coelenterazine to cytosolic Rluc was not exceeded within the concentration range tested. Michaelis-Menten curve fits resulted in the following parameters (Km, Vmax): KB 3-1 Ruc, GF (-) (402 nM, 3. 7 × 109 total flux); KB 3-1 Rluc, GF (+) (537 nM, 3.8 × 109 total flux); KB 8-5-11 Rluc, GF (-) (147 nM, 9.1 × 108 total flux); KB 8-5-11 Rluc, GF (+) (257 nM, 2.2 × 109 total flux).
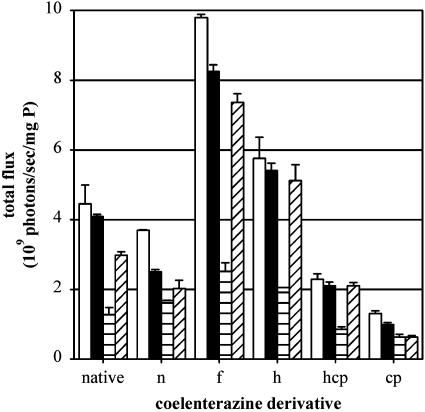
Coelenterazine Analogs. To characterize substrate specificity of imidazolopyrazine derivatives, additional semisynthetic coelenterazine analogues were analyzed. By using KB 3-1 Rluc and KB 8-5-11 Rluc cells, coelenterazine n, f, h, hcp, and cp (Fig. 1 A) were evaluated in the absence and presence of GF120918 (800 nM). In particular, coelenterazine f resulted in 1.5-fold more signal than native coelenterazine in a Pgp- and GF120918-dependent manner (Fig. 3). Overall, in addition to native coelenterazine, coelenterazines f, h, and hcp were transported by Pgp, whereas coelenterazines n and cp were not, showing smaller transport differences that were independent of GF120918.
Fig. 3.
Pgp-mediated transport of semisynthetic coelenterazine derivatives. KB 3-1 Rluc (white and black bars) and KB 8-5-11 Rluc (horizontal and diagonal lined bars) cells were imaged as described by using the indicated coelenterazine analogues in the absence (white and horizontal lined bars, respectively) and in the presence (black and diagonal lined bars, respectively) of 800 nM GF120918. Total flux was normalized by total cell protein. Bars represent the mean ± range of two determinations, representative of three independent experiments.
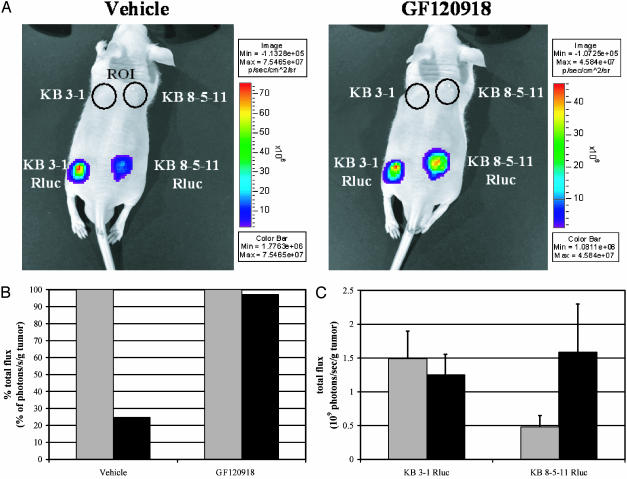
Bioluminescence Imaging of Pgp Transport Activity and Inhibition in Living Mice. We then sought to examine Pgp transport activity and the pharmacodynamics of inhibitors in vivo with noninvasive bioluminescence imaging. Tumor xenografts of KB 3-1 and KB 8-5-11 cells (control tumors) and KB 3-1 Rluc and KB 8-5-11 Rluc cells were grown in nu/nu mice and imaged with an in vivo imaging system and a charge-coupled device camera (Fig. 4A). Before tail vein administration of coelenterazine (4 μg/g) and imaging, mice were either first gavaged with vehicle, or the following day, the same mice were gavaged with GF120918 (250 mg/kg), thus enabling each mouse to serve as its own control (Fig. 4A). At baseline, KB 3-1 Rluc tumors showed a 4-fold higher bioluminescence signal in vivo than KB 8-5-11 Rluc tumors. In the presence of GF120918, bioluminescence signals of only KB 8-5-11 Rluc tumors increased, resulting in equilibration of bioluminescence signal emitted from both tumors. As expected, no bioluminescence was detected from nontransfected tumors. Variation in total bioluminescence signal from mouse to mouse could be accounted for by pharmacokinetic differences in injection efficiencies and substrate delivery within a given animal. Nonetheless, normalized to bioluminescence signals arising from KB 3-1 Rluc tumors, signals emitted from KB 8-5-11 Rluc tumors in vivo were consistently reduced by ≈75% (Fig. 4B). Overall, for all mice investigated (n = 5), bioluminescence signals (corrected to tumor weight) showed 3-fold lower values in the Pgp-expressing tumors in vivo and complete reversal on administration of GF120918 (Fig. 4C).
Fig. 4.
Imaging Pgp-modulated bioluminescence in living mice. (A) KB 3-1 and KB 3-1 Rluc cells (1 × 107) and KB 8-5-11 and KB 8-5-11 Rluc cells (1.5 × 107) were injected into nu/nu mice as indicated. Shown are representative pseudocolor images of the same mouse pretreated by gavage with vehicle alone on day 1 (Left) or with GF120918 (250 mg/kg) the following day (Right). Images on both days were taken 11 min after tail vein injection of coelenterazine (4 μg/g). Tumors of control cells are indicated with black circular ROIs. (B) Quantification of data obtained from mouse shown in A. The gray bar represents the total flux of KB 3-1 Rluc tumors, and the black bar depicts the signal of KB 8-5-11 Rluc tumors. Total flux is shown as a percent of signal emitted from control KB 3-1 Rluc tumors normalized to tumor weight. (C) Raw total flux signals normalized to tumor weight were averaged for all mice used in this series of experiments. The gray bar represents signal after vehicle alone (day 1), and the black bar depicts signal after treatment with GF120918 (day 2) for each tumor xenograft. Bars represent mean values ± SEM of five mice.
Discussion
We show that coelenterazine, the chromophoric substrate for Rluc, is avidly transported by MDR1 Pgp. While providing new opportunities to study Pgp transport activity in vivo, this finding dampens enthusiasm for the indiscriminate use of Rluc as a reporter gene in intact cells or transgenic animals where Pgp status is unknown or changes of Pgp expression may be overlooked under various experimental conditions. Importantly, aequorin, the calcium-sensitive photoprotein from the jellyfish Aequorea aequorea, has been widely used as a bioluminescent probe to monitor intracellular free calcium levels in signal transduction and contains coelenterazine as its chromophoric ligand (3, 31). Consequently, aequorin-dependent readout and calibration of free calcium determinations in intact cells may need to be reexamined in the context of Pgp expression. In this circumstance, use of the semisynthetic ligands coelenterazine cp and coelenterazine n, rather than native coelenterazine, may minimize contamination by Pgp-mediated modulation of bioluminescence in intact organisms and cells, but at the expense of an overall reduction in light emission. By comparison, pilot data indicate that d-luciferin, the substrate for firefly (P. pyralis) luciferase, is not transported by Pgp (data not shown).
Noninvasive imaging of ATP-binding cassette transporters in living animals and humans has previously been reported with radiolabeled transport substrates and provides dynamic information on MDR1 Pgp and MRP1 functions in vivo (21, 23). Selected radiopharmaceuticals have been validated in clinical studies for pretreatment assessment of Pgp-mediated MDR (32) and to directly visualize pharmacological modulation in patients (33). Imaging Pgp transport activity with bioluminescence offers an alternative biochemical readout to observe Pgp transport function in vitro and in living animals in vivo. Because Rluc resides in the cytosolic compartment and coelenterazine emits a single photon on oxidation, signals arising from Rluc transfected cells rapidly attain a steady state (within 6 s), representing the rate of Rluc enzyme activity. In effect, Pgp indirectly modulates intracellular Rluc enzyme kinetics by regulating the transmembrane translocation and availability of substrate. This fundamentally differs from readouts obtained with fluorescent probes such as rhodamine 123 and Hoechst 33342 (34, 35), BCECF-AM and SPQ (36), or activated fluorescent compounds such as calcein-AM (37) or radiotracer substrates (23, 38, 39) that interact with Pgp. These probes and their output signals are retained for significant times after exposure of the cells to the probes, or alternatively, the Pgp-mediated readout depends on washout kinetics over time. In contrast, once coelenterazine is oxidized to coelenteramide by Rluc, the compound is optically silent. Thus, on substrate depletion, bioluminescence signals return to baseline, thereby enabling noninvasive, repetitive interrogation of Pgp transport function over short time intervals in both cells and animals.
The low cost and convenience of bioluminescence provides a potential advantage for this technique over other approaches and also enables a platform readily adapted to high-throughput screens of compounds that interact with or inhibit Pgp. Furthermore, Rluc transgenic mice may provide opportunities to study regulation of Pgp transport activity and pharmacological inhibition in a variety of tissues within the intact animal with bioluminescence. In addition, discovery of coelenterazine as a natural product substrate of Pgp may provide new insight into marine ecosystems and the coelenterazine food chain within the oceans. It might be speculated that homologues of ATP-binding cassette transporters are involved in regulation of dietary absorption, transport, and biodistribution of coelenterazines in marine organisms and ultimately humans.
Acknowledgments
This work was supported by National Institutes of Health Grant P50 CA94056 and U.S. Department of Energy Grant 94ER61885.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MDR, multidrug resistance; Pgp, P-glycoprotein; Rluc, Renilla luciferase; GF120918, N-(4-[2-(1,2,3,4-tetrahydro-6,7-dimethoxy-2-isoquinolinyl)ethyl]-phenyl)-9,10-dihydro-5-methoxy-9-oxo-4-acridine carboxamide; PSC 833, (3′-keto-Bmt′)-[Val-2]cyclosporin A.
References
- 1.Herring, P. J. (2000) Philos. Trans. R. Soc. London B 355, 1273-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson, T. & Hastings, J. W. (1998) Annu. Rev. Cell Dev. Biol. 14, 197-230. [DOI] [PubMed] [Google Scholar]
- 3.Head, J., Inouye, S., Teranishi, K. & Shimomura, O. (2000) Nature 372-376. [DOI] [PubMed]
- 4.Jones, K., Hibbert, F. & Keenan, M. (1999) Trends Biotechnol. 17, 477-481. [DOI] [PubMed] [Google Scholar]
- 5.Hastings, J. (1996) Gene 173, 5-11. [DOI] [PubMed] [Google Scholar]
- 6.Rees, J. F., de Wergifosse, B., Noiset, O., Dubuisson, M., Janssens, B. & Thompson, E. M. (1998) J. Exp. Biol. 201, 1211-1221. [DOI] [PubMed] [Google Scholar]
- 7.Lorenz, W., Ro, M., Longiaru, M. & Cormier, M. (1991) Proc. Natl. Acad. Sci. USA 88, 4438-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz, W. W., Cormier, M. J., O'Kane, D. J., Hua, D., Escher, A. A. & Szalay, A. A. (1996) J. Biolumin. Chemilumin. 11, 31-37. [DOI] [PubMed] [Google Scholar]
- 9.Boute, N., Jockers, R. & Issad, T. (2002) Trends Pharmacol. Sci. 23, 351-354. [DOI] [PubMed] [Google Scholar]
- 10.Xu, Y., Kanauchi, A., von Arnim, A. G., Piston, D. W. & Johnson, C. H. (2003) Methods Enzymol. 360, 289-301. [DOI] [PubMed] [Google Scholar]
- 11.Issad, T., Boute, N. & Pernet, K. (2002) Biochem. Pharmacol. 64, 813-817. [DOI] [PubMed] [Google Scholar]
- 12.Contag, C. H. & Bachmann, M. H. (2002) Annu. Rev. Biomed. Eng. 4, 235-260. [DOI] [PubMed] [Google Scholar]
- 13.Contag, C. & Ross, B. (2002) J. Magn. Reson. 16, 378-387. [DOI] [PubMed] [Google Scholar]
- 14.Luker, G., Pica, C., Song, J., Luker, K. & Piwnica-Worms, D. (2003) Nat. Med. 9, 969-973. [DOI] [PubMed] [Google Scholar]
- 15.Bhaumik, S. & Gambhir, S. (2001) Proc. Natl. Acad. Sci. USA 99, 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luker, G., Bardill, J., Prior, J., Pica, C., Piwnica-Worms, D. & Leib, D. (2002) J. Virol. 76, 12149-12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haddock, S., Rivers, T. & Robison, B. (2001) Proc. Natl. Acad. Sci. USA 98, 11148-11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riordan, J. R. & Ling, V. (1985) Pharmacol. Ther. 28, 51-75. [DOI] [PubMed] [Google Scholar]
- 19.Ford, J. M. & Hait, W. N. (1990) Pharmacol. Rev. 42, 155-199. [PubMed] [Google Scholar]
- 20.Ambudkar, S., Dey, S., Hrycyna, C., Ramachandra, M., Pastan, I. & Gottesman, M. (1999) Annu. Rev. Pharmacol. Toxicol. 31, 361-398. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman, M., Fojo, T. & Bates, S. (2002) Nat. Rev. Cancer 2, 48-58. [DOI] [PubMed] [Google Scholar]
- 22.Sauna, Z. E., Smith, M. M., Muller, M., Kerr, K. M. & Ambudkar, S. V. (2001) J. Bioenerg. Biomembr. 33, 481-491. [DOI] [PubMed] [Google Scholar]
- 23.Sharma, V., Luker, G. & Piwnica-Worms, D. (2002) J. Magn. Reson. 16, 336-351. [DOI] [PubMed] [Google Scholar]
- 24.Rao, V., Dahlheimer, J., Bardgett, M., Snyder, A., Finch, R., Sartorelli, A. & Piwnica-Worms, D. (1999) Proc. Natl. Acad. Sci. USA 96, 3900-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piwnica-Worms, D., Rao, V., Kronauge, J. & Croop, J. (1995) Biochemistry 34, 12210-12220. [DOI] [PubMed] [Google Scholar]
- 26.Glantz, S. A. (1987) Primer of Biostatistics (McGraw-Hill, New York), 2nd Ed., p. 379.
- 27.Piwnica-Worms, D., Chiu, M., Budding, M., Kronauge, J., Kramer, R. & Croop, J. (1993) Cancer Res. 53, 977-984. [PubMed] [Google Scholar]
- 28.Hyafil, F., Vergely, C., Du Vignaud, P. & Grand-Perret, T. (1993) Cancer Res. 53, 4595-4602. [PubMed] [Google Scholar]
- 29.Sikic, B. I., Fisher, G. A., Lum, B. L., Halsey, J., Beketic-Oreskovic, L. & Chen, G. (1997) Cancer Chemother. Pharmacol. 40, Suppl., S13-S19. [DOI] [PubMed] [Google Scholar]
- 30.Mistry, P., Stewart, A., Dangerfield, W., Okiji, S., Liddle, C., Bootle, D., Plumb, J., Templeton, D. & Charlton, P. (2001) Cancer Res. 61, 749-758. [PubMed] [Google Scholar]
- 31.Sala-Newby, G., Badminton, M., Evans, W., George, C., Jones, H., Kendall, J., Ribeiro, A. & Campbell, A. (2000) Methods Enzymol. 305, 479-498. [DOI] [PubMed] [Google Scholar]
- 32.Ciarmiello, A., Del Vecchio, S., Silvestro, P., Potena, M. I., Carriero, M. V., Thomas, R., Botti, G., D'Aiuto, G. & Salvatore, M. (1998) J. Clin. Oncol. 16, 1677-1683. [DOI] [PubMed] [Google Scholar]
- 33.Agrawal, M., Abraham, J., Balis, F., Edgerly, M., Stein, W., Bates, S., Fojo, T. & Chen, C. (2003) Clin. Cancer Res. 9, 650-656. [PubMed] [Google Scholar]
- 34.Shapiro, A. B. & Ling, V. (1998) Acta Physiol. Scand. Suppl. 643, 227-234. [PubMed] [Google Scholar]
- 35.Sharom, F. J., Liu, R., Qu, Q. & Romsicki, Y. (2001) Semin. Cell Dev. Biol. 12, 257-265. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman, M. & Roepe, P. (1997) Biochemistry 36, 11153-11168. [DOI] [PubMed] [Google Scholar]
- 37.Homolya, L., Hollo, Z., Germann, U. A., Pastan, I., Gottesman, M. M. & Sarkadi, B. (1993) J. Biol. Chem. 268, 21493-21496. [PubMed] [Google Scholar]
- 38.Sharma, V. & Piwnica-Worms, D. (1999) Chem. Rev. 99, 2545-2560. [DOI] [PubMed] [Google Scholar]
- 39.Hendrikse, N., Franssen, E., van der Graaf, W., Vaalburg, W. & de Vries, E. (1999) Eur. J. Nucl. Med. 26, 283-293. [DOI] [PubMed] [Google Scholar]