Abstract
Background
This study is the first to investigate the Brazilian Amazonian Forest to identify new D-xylose-fermenting yeasts that might potentially be used in the production of ethanol from sugarcane bagasse hemicellulosic hydrolysates.
Methodology/Principal Findings
A total of 224 yeast strains were isolated from rotting wood samples collected in two Amazonian forest reserve sites. These samples were cultured in yeast nitrogen base (YNB)-D-xylose or YNB-xylan media. Candida tropicalis, Asterotremella humicola, Candida boidinii and Debaryomyces hansenii were the most frequently isolated yeasts. Among D-xylose-fermenting yeasts, six strains of Spathaspora passalidarum, two of Scheffersomyces stipitis, and representatives of five new species were identified. The new species included Candida amazonensis of the Scheffersomyces clade and Spathaspora sp. 1, Spathaspora sp. 2, Spathaspora sp. 3, and Candida sp. 1 of the Spathaspora clade. In fermentation assays using D-xylose (50 g/L) culture medium, S. passalidarum strains showed the highest ethanol yields (0.31 g/g to 0.37 g/g) and productivities (0.62 g/L·h to 0.75 g/L·h). Candida amazonensis exhibited a virtually complete D-xylose consumption and the highest xylitol yields (0.55 g/g to 0.59 g/g), with concentrations up to 25.2 g/L. The new Spathaspora species produced ethanol and/or xylitol in different concentrations as the main fermentation products. In sugarcane bagasse hemicellulosic fermentation assays, S. stipitis UFMG-XMD-15.2 generated the highest ethanol yield (0.34 g/g) and productivity (0.2 g/L·h), while the new species Spathaspora sp. 1 UFMG-XMD-16.2 and Spathaspora sp. 2 UFMG-XMD-23.2 were very good xylitol producers.
Conclusions/Significance
This study demonstrates the promise of using new D-xylose-fermenting yeast strains from the Brazilian Amazonian Forest for ethanol or xylitol production from sugarcane bagasse hemicellulosic hydrolysates.
Introduction
Growing environmental concerns over the use and depletion of non-renewable fuel sources, together with the rising price of oil and the instability of the oil market, have stimulated interest in optimizing fermentation processes for the large-scale production of alternative fuels such as ethanol [1]. The largest potential feedstock for ethanol is lignocellulosic biomass, which includes materials such as agricultural residues (corn stover, crop straws, sugarcane bagasse), herbaceous crops, short rotation woody crops, forestry residues, waste paper and other plant wastes [2].
Lignocellulosic biomass varies among plant species but generally consists of ∼25% lignin and ∼75% carbohydrate polymers (cellulose and hemicellulose). It is the largest known renewable carbohydrate source. The cellulosic and hemicellulosic portions of biomass can be separated from the lignin and depolymerized by hydrolysis to obtain their constituent sugars, mainly glucose from cellulose and D-xylose from hemicellulose [3]. As the major sugar in hemicellulose, D-xylose is the second most abundant sugar in lignocellulose [4]. The successful conversion of hemicellulose into fuel ethanol at high yields is the deciding factor for the economic viability of the process [5]. Thus, the efficient use of lignocellulosic biomass as a substrate for ethanol production requires effective utilization of D-xylose [4].
Yeasts that produce ethanol from D-xylose have been isolated from various locations, including tree exudates [6], wood-boring insects [7], [8], decaying wood [7], [9], rotten fruit and tree bark [10]. Known D-xylose-fermenting yeasts are principally from the species Scheffersomyces (Pichia) stipitis, Candida shehatae, C. lignosa, C. insectosa, C. tenuis, Pachysolen tannophilus [11]–[13], Spathaspora passalidarum [14] and S. arborariae [9], [15]. Among these naturally D-xylose-fermenting yeasts, S. stipitis and S. passalidarum are considered the best ethanol producers [11], [16]. Despite the existence of these microorganisms, it is still challenging to reach high yields of ethanol from pentose sugars on a large scale [17] because no microorganisms that robustly convert pentose sugars into ethanol at high yields while withstanding fermentation inhibitors have been identified [18].
According to Jeffries and Kurtzman [19], the identification of yeast strains that ferment hemicellulosic sugars will improve prospects for lignocellulosic ethanol production. The strains can be obtained by isolating them from the environment, by mutating and selecting strains in the laboratory [20] or by engineering strains of Saccharomyces cerevisiae that can ferment pentoses [21].
The Amazon basin sustains almost 60% of the world’s remaining tropical rainforest, with Brazilian Amazonia alone comprising ∼30% of the world’s current primary tropical rainforests. This environment plays crucial roles in biodiversity conservation, carbon storage, and regional hydrology and climate [22], [23]. Even considering all of the research that has been performed on biodiversity in the Amazonia to date, clearly much more research is needed to understand the enormous diversity and complexity of this region. Few studies have characterized the yeast diversity of Brazilian Amazonian environments [24]–[26]. Works related to yeast diversity in the region have identified a number of potential new species, but only one species, Candida amapae (Saccharomycopsis clade), from the region was characterized [25]. In this work, we studied yeast diversity in rotting wood collected from two Amazonian sites, focusing on the isolation of new D-xylose-fermenting yeasts that might potentially be used in the production of ethanol from sugarcane bagasse hemicellulosic hydrolysates.
Materials and Methods
Yeast Isolation
Yeasts were isolated from rotting wood samples collected in two sites of Amazonian Forest in the state of Roraima, in northern Brazil. These sites are maintained by Embrapa (Empresa Brasileira de Pesquisa Agropecuária)-Roraima for long-term experiments and are located in the municipalities of Mucajaí (2° 25′ 48′′N and 60° 55′ 11′′W) and São João da Baliza (00° 56′ 58′′N and 59° 54′ 41′′W). The collection sites were the experimental ecological reserve Serra da Prata (Mucajaí), belonging to Embrapa-Roraima, and the ecological reserves belonging to the owners of private landOsvaldo Antônio Sant’ana (Mucajaí) and José Lopes (São João da Baliza). All necessary permits were obtained from Embrapa-Roraima (collection permission obtained by Jerri E. Zilli) and from the owners of the private lands for the described field studies. The predominant vegetation in these sites is characterized as an Amazonian Forest biome. The climate is hot and humid, with an annual precipitation between 1,300 and 2,900 mm, and the average temperature ranges from 25.6 to 27.6°C. The Amazonian Forest comprises a continuum of nine main, floristically distinct vegetal formations, and 70% of it is occupied by upland forests that are characterized by their high richness and diversity of tree species [27]. The field collections were made according to the Brazilian diversity rules.
Forty decayed wood samples, 20 samples from each site, were collected in October 2009. Each sample was collected approximately 5 m from the other. The samples were stored in sterile plastic bags and transported under refrigeration to the laboratory within 24 h. One gram of each sample was placed, separately, in 125 mL Erlenmeyer flasks with 20 mL of sterile YNB-D-xylose medium (yeast nitrogen base, 6.7 g/L; D-xylose, 5 g/L; chloramphenicol, 0.2 g/L) or 20 mL sterile YNB-xylan medium (yeast nitrogen base, 6.7 g/L; xylan, 10 g/L; chloramphenicol. 0.2 g/L; pH 5.0±0.2). D-xylose, xylan and YNB solutions were sterilized separately. The flasks were incubated at 25°C on an orbital shaker at 150 rpm for 3–10 days. When growth was detected, 0.5 mL was transferred to a tube containing 5 mL sterile YNB-D-xylose or YNB-xylan media. The tubes were incubated on an orbital shaker as described above. After growth detection, one loopful of each tube was streaked on YNB-D-xylose or YNB-xylan agar media. The plates were incubated at 25°C until yeast colonies developed [9]. The different yeast morphotypes were purified by restreaking on yeast extract–malt extract agar (YMA – glucose, 10 g/L; peptone, 5 g/L; yeast extract, 3 g/L; malt extract, 3 g/L; agar, 20 g/L) and stored at −80°C or in liquid nitrogen for later identification.
Yeast Identification
The yeasts were preliminarily grouped according to various characteristics including their colony morphology and standard tests for growth on different carbon and nitrogen sources [28]. Physiology-based groupings were confirmed by PCR fingerprinting using the Intron Splice Site primer EI-1 (5′CTGGCTTGGTGTATGT) [29]. Yeast strains with identical DNA banding patterns were grouped and putatively considered to belong to the same species [30]. At least one representative strain from each EI-1 PCR group was subjected to sequence analysis of the D1/D2 region and internal transcribed spacer (ITS) domains of the large subunit of the rRNA gene as described below. Physiologically distinct strains with unique EI-1 PCR banding patterns were also selected for direct identification by sequencing of the D1/D2 region and ITS domains.
The D1/D2 and ITS domains were amplified by PCR directly from whole cells as described previously [31]. The amplified DNA was concentrated, cleaned and sequenced in a Mega-BACE™ 1000 automated sequencing system (Amersham Biosciences, USA). Potential new species were also sequenced using an ABI sequencer at the John P. Robarts Research Institute, London, Ontario, in Canada. The sequences were assembled, edited and aligned with the program MEGA5 [32]. Sequences of the new species isolated in this work were deposited in GenBank. The existing sequences for other yeasts were retrieved from GenBank. Phylogenetic placements of new species were based on maximum parsimony analysis of the sequences of the D1/D2 domains of the large subunit of the rRNA gene.
Screening of D-xylose-fermenting Yeasts
The ability to ferment D-xylose was tested in Durham tubes containing a 2% (w/v) solution of the sugar. The tubes were incubated at 25°C on an orbital shaker at 100 rpm for 25 days and observed daily for gas production. Candida shehatae CBS 5813, C. insectosa CBS 4286, C. lignosa CBS 4705 and S. stipitis NRRL Y-7124 were used as positive controls for D-xylose fermentation [9].
Yeasts showing the development of gas inside the Durham tubes were tested for fermentation in D-xylose culture medium (YPX: yeast extract-peptone-D-xylose medium) as described below. In addition, the yeast isolates identified by D1/D2 rRNA gene sequencing as belonging to the D-xylose-fermenting clades Spathaspora or Scheffersomyces were also tested for fermentation in YPX medium. Yeasts with the best D-xylose consumption and highest ethanol yields (Yp/s et) were assayed for their ability to ferment sugars in sugarcane bagasse hemicellulosic hydrolysate. Candida lignosa CBS 4705 and S. stipitis NRRL Y-7124 were used as positive controls.
Fermentation Assays
Inoculum preparation
Yeast inocula were prepared on yeast extract–malt extract agar (YMA – glucose, 10 g/L; peptone, 5 g/L; yeast extract, 3 g/L; malt extract, 3 g/L; agar 20 g/L) plates at 30°C for 24–48 h. Cells were cultured in 50 mL YPX liquid medium (yeast extract, 10 g/L; peptone, 20 g/L; D-xylose, 30 g/L) in 125 mL Erlenmeyer flasks at 30°C with continuous shaking (200 rpm) for 24 h at 30°C. D-xylose and yeast extract-peptone solutions were sterilized separately. Cells were recovered by centrifugation at 2,600×g for 20 min, washed twice and resuspended in the fermentation media to a final concentration of 0.5 g/L.
Medium composition and cultivation conditions
Batch fermentation experiments were carried out in 50 mL D-xylose culture medium (yeast extract, 10 g/L; peptone, 20 g/L; D-xylose, 50 g/L), pH 6.0, in 125 mL Erlenmeyer flasks incubated as described above for 48 h. Fermentation was monitored by taking samples at 0, 12, 24 and 48 h.
Sugarcane bagasse was supplied by Usina São Francisco (Sertãozinho, SP, Brazil). Hemicellulosic hydrolysate was prepared as described previously [33] in a 250 L stainless steel reactor loaded with sugarcane bagasse and sulfuric acid solution (100 mg acid/g dry matter). The reactor was operated with a solid/liquid ratio of 1∶10 at 121°C for 20 min. After hydrolysis, the resulting solid material was removed by filtration. The hemicellulosic hydrolysate was concentrated in a 30 L evaporator at 70±5°C to obtain a xylose concentration of about 70 g/L. To reduce inhibitors, a detoxification assay was performed as described in Alves et al. [34] by first raising the pH to 7.0 with calcium oxide and then decreasing it to pH 5.5 with phosphoric acid, adding active charcoal (2.5% w/v) and incubating at 200 rpm at 30°C for 1 h. The precipitates resulting after each procedure were removed by vacuum filtration. The sugar composition of the hydrolysate before autoclaving was 59 g/L D-xylose, 6.2 g/L glucose and 6.4 g/L L-arabinose. The treated hydrolysate was autoclaved at 111°C for 15 min and supplemented with yeast extract solution (3 g/L). Experiments were carried out in 250 mL flasks with 100 mL supplemented hemicellulosic hydrolysate. The average hydrolysate was at pH 5.1 and composed of 50.2 g/L D-xylose, 5.3 g/L glucose, 5.8 g/L L-arabinose, 3.2 g/L acetic acid, 0.01 g/L furfural, 0.01 g/L hydroxymethylfurfural, and 1.3 g/L total phenols. The flasks were incubated as described above for 96 h. Samples were taken at 0, 12, 24, 48, 72 and 96 h. Samples were stored at −20°C until analysis. All experiments were performed in duplicate.
Analytical Methods
Cell concentrations were determined by correlating optical density (OD) measurements taken with a Beckman DU 640B spectrophotometer at 600 nm with a previously constructed calibration curve (dry weight × optical density). After the cell concentration determined, samples were centrifuged at 2,600×g for 15 min, and the supernatant was diluted and filtered using a Sep-Pak C18 (Millipore) filter. Monosaccharides (glucose, D-xylose and L-arabinose), xylitol, glycerol, ethanol, and acetic acid levels were determined by HPLC (Waters 410, Milford, MA, USA) using a Bio-Rad Aminex HPX-87H (300×7.8 mm) column at 45°C with a sample injection volume of 20 µL, a Waters 410 refraction index, a mobile phase of 0.01 N H2SO4 and a flow rate of 0.6 mL/min.
Fermentation Parameter Calculation
The fermentation parameters Yp/s et (g/g, ethanol yield), Yp/s xy (g/g, xylitol yield), Qp (g/L·h, ethanol productivity), η (%, fermentation efficiency) and D-xylose and/or glucose consumption (%) were experimentally determined. Ethanol (Yp/s et, g/g) and xylitol (Yp/s xy, g/g) yields were calculated following the methods of Schmidell et al. [35]), which correlated ΔP produced ( ΔPethanol or ΔPxylitol) with ΔS consumed (substrate consumed to product obtained, derived by determining the total, initial and consumed substrate). The slope of the line through the origin provided the estimate of Yp/s et and Yp/s xy. Ethanol productivity (Qp, g/L·h) was determined by the ratio of ethanol concentration (g/L) to fermentation time (h). Conversion efficiency (η, %) was calculated as a percentage of the maximum theoretical ethanol yield (0.51 g ethanol/g D-xylose and/or glucose). D-xylose and/or glucose consumption (%) was determined as a percentage of the initial sugar concentration.
Results and Discussion
Yeast Isolation and Diversity
In this work, we studied the diversity of rotting-wood-associated yeasts from two Amazonian sites. A total of 224 yeast strains were isolated from rotting wood samples from the forest reserve sites of São João da Baliza (114 yeast strains) and Mucajaí (110 yeast strains). Of these strains, 118 were obtained following growth in YNB-D-xylose medium, and 106 were obtained from growth in YNB-xylan medium.
Table 1 shows the results of yeast species identification, the occurrence of each species by isolation site, the number of samples cultured in both media (YNB-D-xylose and xylan) and the results of the Durham tube fermentation tests. Of the 33 yeast species identified, 26 species were previously known and seven were new (Table S1). Eleven species were isolated from both isolation sites, whereas 22 species were observed in only one site. Sixteen of the 22 species were observed in the São João da Baliza forest reserve and the remaining six were observed in the Mucajaí forest reserve. Species in the genus Candida (namely 16 species related to the Candida glaebosa, Kurtzmaniella, Lindnera, Lodderomyces/Spathaspora, Metschnikowia, Ogataea, Wickerhamomyces and Yamadazyma clades were the most prevalent, followed by the genus Spathaspora, with four species. Candida tropicalis (Lodderomyces/Spathaspora clade) was the most frequently isolated yeast, occurring in 15 samples cultured on YNB-D-xylose medium and 13 samples in YNB-xylan medium, followed by Asterotremella humicola (eight samples on YNB-D-xylose medium and 10 in YNB-xylan medium) and Candida boidinii (Ogataea clade; 10 samples on YNB-D-xylose medium and seven samples on YNB-xylan medium). Strains of C. tropicalis have been reported in fruit, flowers, soil, water, and clinical specimens [36], and this species has already been shown to produce ethanol and, mainly, xylitol from D-xylose [37]–[40]. Strains belonging to the genus Asterotremella, including A. humicola, have been isolated from soil, plants and mushrooms [41]. Candida boidinii has been found with high regularity in the sap of many tree species in geographically distinct regions of the world. Specific substrates associated with this species are largely linked to its ability to assimilate the methanol produced in decaying plant tissues [36].
Table 1. Identification, occurrence and fermentation in Durham tube test of yeasts isolated in Amazonian forest reserves.
| Yeast species | Sampled medium | Fermentation in Durham tube test | |
| YNB-D-xylose (n = 40) | YNB-xylan (n = 40) | ||
| São João da Baliza Forest Reserve | |||
| Asterotremella humicola | 21 | 4 | − |
| Blastobotrys mokoenaii 3 | 1 | 1 | − |
| Candida amphixiae 3,5 | − | 1 | − |
| C. boidinii | 5 | 5 | − |
| C. gorgasii 2,3 | 1 | 1 | − |
| C. intermedia | 4 | 3 | − |
| C. labiduridarum 3,4 | 2 | − | − |
| C. palmioleophila 3,5 | − | 1 | − |
| C. pseudointermedia 3,5 | − | 1 | − |
| C. quercitrusa | 1 | − | − |
| C. tropicalis | 1 | 3 | − |
| Candida sp. 22,3 | 2 | 1 | − |
| Candida sp. 32,3,5 | − | 2 | − |
| Cryptococcus diffluens 3,5 | − | 1 | − |
| Cr. laurentii 5 | − | 1 | − |
| Debaryomyces hansenii | 5 | 2 | − |
| Kodamaea ohmeri 3,5 | − | 1 | − |
| Lindnera saturnus | 5 | 1 | − |
| Meyerozyma guilliermondii | − | 1 | − |
| Naumavazyma castelli 3,4 | 1 | − | − |
| Scheffersomyces stipitis | 1 | − | + |
| Schwanniomyces polymorphus 3 | 6 | 2 | − |
| Sc. vanrijiae 3 | 2 | 2 | − |
| Spathaspora passalidarum 3,5 | − | 5 | + |
| Spathaspora sp. 12,3 | 1 | 1 | − |
| Spathaspora sp. 32,3,5 | − | 1 | − |
| Trichosporon mycotoxinivorans | 1 | 2 | − |
| Mucajaí Forest Reserve | |||
| A. humicola | 6 | 6 | − |
| Candida amazonensis 2,3 | 3 | 1 | − |
| C. blattae 3,4 | 1 | − | − |
| C. boidinii | 5 | 2 | − |
| C. intermedia | − | 1 | − |
| C. maltosa 3 | 2 | 2 | − |
| C. natalensis 3,5 | − | 1 | − |
| C. quercitrusa | − | 1 | − |
| C. tropicalis | 14 | 10 | − |
| Candida sp. 12,3,5 | − | 2 | − |
| Cr. laurentii 5 | − | 1 | − |
| D. hansenii | 2 | 2 | − |
| L. saturnus | 1 | 1 | − |
| M. guilliermondii | 4 | 1 | − |
| S. stipitis | − | 1 | + |
| Spathaspora sp. 22,3,4 | 1 | − | − |
| T. mycotoxinivorans | − | 3 | − |
Number of samples in which the yeast was isolated.
Novel yeast species.
Occurrence restricted to one isolation site.
Occurrence restricted to YNB-D-xylose medium.
Occurrence restricted to YNB-xylan medium.
The species restricted to the São João da Baliza forest reserve included all Spathaspora passalidarum isolates and four new species (Candida sp. 2, Candida sp. 3, Spathaspora sp. 1 and Spathaspora sp. 3). Candida amazonensis, Candida sp. 1 and Spathaspora sp. 2 were recovered in the Mucajaí forest reserve. Fifteen species were isolated on only one cultivation media, four in YNB-D-xylose medium (C. blattae, C. labiduridarum, Naumavazyma castelli and Spathaspora sp. 2) and 11 species in YNB-xylan medium (C. amphixiae, C. natalensis, C. palmioleophila, C. pseudointermedia, Candida sp. 1, Candida sp. 3, Cryptococcus diffluens, Cr. laurentii, Kodamaea ohmeri, S. passalidarum, and Spathaspora sp. 3). Most known yeast species found in YNB-D-xylose or xylan media have previously been linked to terrestrial environments, such as soil, flowers, fruit, rotting wood, beetle guts, and floricolous insects [42]. Suh et al. [8] reported that C. intermedia, Meyerozyma guilliermondii, Lindnera (Williopsis) saturnus, and S. stipitis were associated with wood-ingesting beetles, and Nguyen et al. [14] observed the same for S. passalidarum. Bhadra et al. [43] isolated Cr. laurentii, D. hansenii and K. ohmeri from tree bark samples. Candida tropicalis and C. maltosa were isolated from rotten fruit by Rao et al. [10]. Schwanniomyces polymorphus and Sc. vanrijiae were also isolated from several samples collected in the São João da Baliza forest reserve. These metabolically versatile species have been isolated from soil, tree exudates, and ant hills [44]. Some species, namely D. hansenii, C. intermedia and M. guilliermondii, were isolated in both culture media and from both collection sites. Two isolates of B. mokoenaii were obtained from the São João da Baliza site. This species is rare and known previously from only a single strain obtained from soil in South Africa [45]. Du Preez et al. [46] reported that B. mokoenaii is thermotolerant and produces an extracellular endo-β-xylanase comparable to that of Aureobasidium pullulans.
Molecular Identification and Phylogenetic Relationships
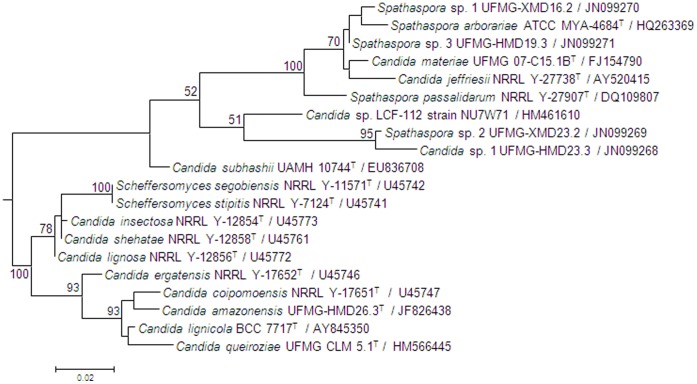
Some yeast isolates showed three or more non-contiguous nucleotide differences in the D1/D2 domains of the LSU rRNA gene when compared with the most closely related known species, indicating that they might represent novel yeast species. According to Kurtzman et al. [42], isolates of the same species typically have zero to two nucleotide differences in the D1/D2 region of the large subunit of the rRNA gene. Based on this concept, seven new yeast species were found in our studies. Five isolates have been described as a new species named Candida amazonensis [47]. The new species belongs to the Scheffersomyces clade and differs by nine nucleotide substitutions and six indels in the D1/D2 region of the LSU rRNA gene from C. coipomoensis, nine nucleotide substitutions and seven indels from C. lignicola and 16 nucleotide substitutions and six indels from C. queiroziae, its closest relatives [47]. All yeast species that are phylogenetically related to C. amazonensis were also isolated from rotting-wood samples or insects associated with this substrate [36], [48], [49]. The yeast isolates identified as Candida sp. 1, Spathaspora sp. 1, Spathaspora sp. 2 and Spathaspora sp. 3 by sequence analysis belong to the Spathaspora clade, which contains D-xylose-fermenting yeasts. Spathaspora sp. 3 (UFMG-HMD-19.3) and Spathaspora sp. 1 (UFMG-XMD-16.2) belong to the same subclade as do S. arborariae, Candida jeffriesii, and Candida materiae. Spathaspora sp. 3 (UFMG-HMD-19.3) differs in the D1/D2 domains by eight substitutions from its least divergent relative, C. materiae. The smallest degree of sequence divergence observed for Spathaspora sp. 1 (UFMG-XMD-16.2) was seven substitutions and four indels from S. arborariae. The other two new species, Spathaspora sp. 2 (UFMG-XMD-23.2) and Candida sp. 1 (UFMG-HMD-23.3), are closely related, forming a separate subclade within the Spathaspora clade. They differ by 76 changes in the combined D1/D2 domains (6 substitutions and 5 indels) and ITS region (19 substitutions and 46 indels) and are well separated phylogenetically from other clade members (Fig. 1). The D1/D2 sequences of both isolates of Spathaspora sp. 1 (UFMG-XMD-16.2 and UFMG-HMD-16.3) were identical. The same was observed for the isolates of Candida sp. 1 (UFMG-HMD-23.3 and UFMG-HMD-25.1). Three isolates were identified as Candida sp. 2 (GenBank accession number is JQ695901). This new species belongs to the Wickerhamomyces clade and it differs by six nucleotide substitutions from C. mycetangii. The later species is associated with tree-boring beetles [36]. Candida sp. 3 (GenBank accession number is JQ695900) belongs to the Lindnera clade and it differs by six nucleotide substitutions from Lindnera (Pichia) japonica, a species associated with tree boring insects [50].
Figure 1. Phylogram of yeast species considered in this study based on the D1/D2 domains of the large subunit ribosomal gene.
The maximum likelihood tree was constructed with the Mega5 program following correction of the distances with the Kimura 2-parameter transformation. A total of 499 nucleotide positions were used in the analysis. Bootstrap values of 50% or greater are shown (100 replicates). Bar 0.02 substitutions per nucleotide position.
Out of the 33 yeast species identified, strains belonging to only two species, S. passalidarum and S. stipitis, showed gas formation from D-xylose in the Durham tube test. Both species had previously been shown to ferment D-xylose [11], [14]. However, although one new species (C. amazonensis) was identified as belonging to the Scheffersomyces clade and four as belonging to the Spathaspora clade, these isolates exhibited no gas production in the Durham tube test, showing that this test alone is insufficient in screening for D-xylose-fermenting yeasts.
Fermentation Assays
D-xylose culture medium
To evaluate D-xylose fermentation and ethanol production, two strains of S. stipitis, six strains of S. passalidarum, five strains of Candida amazonensis and all strains of the new species belonging to the Spathaspora clade were subjected to fermentation assays in D-xylose (50 g/L) culture medium. The results of the fermentation parameters [Yp/s et (g/g), ethanol yield; Yp/s xy (g/g), xylitol yield; Qp (g/L·h), ethanol productivity; η (%), fermentation efficiency; (%), D-xylose consumption (%)] and cells, ethanol and xylitol concentrations (g/L) are summarized in Table 2. These results were calculated according to the fermentation time (time of maximum ethanol production or time of the end of the fermentation experiment) for each yeast strain.
Table 2. Ethanol yield [Yp/s et (g/g)], xylitol yield [Yp/s xy (g/g)], ethanol productivity [Qp (g/L•h)], fermentation efficiency [η (%)], D-xylose consumption (%), cell concentration (g/L), ethanol concentration (g/L) and xylitol concentration (g/L) in D-xylose culture medium assays.
| Yeast species | Yeast strain | D-xylose consumption (%)1 | Cells (g/L) | Yp/s et (g/g)2 | Qp (g/L•h)3 | Η (%)4 | Ethanol concentration(g/L) | Yp/s xy (g/g)5 | Xylitol concentration(g/L) | FermentationTime (h)6 |
| Candida lignosa | CBS 4705 | 76.2 | 7.7 | 0.40 | 0.60 | 79.4 | 14.5 | 0.03 | 1.0 | 24 |
| Scheffersomyces stipitis | NRRL Y-7124 | 89.0 | 10.9 | 0.35 | 0.62 | 69.5 | 15.0 | 0.02 | 1.3 | 24 |
| S. stipitis | UFMG-XMD-15.2 | 84.5 | 12.9 | 0.28 | 0.51 | 55.2 | 12.3 | 0.04 | 1.9 | 24 |
| S. stipitis | UFMG-HMD-32.1 | 98.8 | 14.2 | 0.22 | 0.23 | 44.1 | 11.1 | – | – | 48 |
| C. amazonensis sp. nov. | UFMG-XMD-24.1 | 99.9 | 10.6 | 0.08 | 0.08 | 15.3 | 4.0 | 0.59 | 25.2 | 48 |
| C. amazonensis sp. nov. | UFMG-XMD-26.2 | 99.8 | 10.7 | 0.08 | 0.08 | 14.6 | 4.0 | 0.58 | 25.2 | 48 |
| C. amazonensis sp. nov. | UFMG-HMD-26.3 | 99.9 | 10.7 | 0.07 | 0.08 | 13.4 | 3.7 | 0.57 | 24.8 | 48 |
| C. amazonensis sp. nov. | UFMG-XMD-40.2 | 99.9 | 11.8 | 0.07 | 0.08 | 14.4 | 3.8 | 0.55 | 23.7 | 48 |
| C. amazonensis sp. nov. | UFMG-XMD-40.3 | 99.9 | 11.1 | 0.07 | 0.08 | 14.0 | 3.7 | 0.56 | 23.9 | 48 |
| Spathaspora passalidarum | UFMG-HMD-1.1 | 98.4 | 8.7 | 0.36 | 0.75 | 70.0 | 18.0 | 0.03 | 1.5 | 24 |
| S. passalidarum | UFMG-HMD-1.3 | 98.1 | 9.8 | 0.35 | 0.72 | 68.4 | 17.2 | 0.04 | 2.2 | 24 |
| S.passalidarum | UFMG-HMD-2.1 | 98.4 | 10.7 | 0.31 | 0.62 | 61.0 | 15.0 | 0.02 | 1.1 | 24 |
| S. passalidarum | UFMG-HMD-10.2 | 98.4 | 10.6 | 0.33 | 0.69 | 65.4 | 16.6 | 0.02 | 1.2 | 24 |
| S. passalidarum | UFMG-HMD-14.1 | 98.3 | 10.2 | 0.37 | 0.68 | 71.6 | 16.4 | 0.02 | 1.1 | 24 |
| S. passalidarum | UFMG-HMD-16.2 | 98.3 | 9.9 | 0.33 | 0.64 | 64.2 | 15.3 | 0.02 | 1.0 | 24 |
| Spathaspora sp. 1 | UFMG-XMD-16.2 | 86.8 | 7.4 | 0.33 | 0.27 | 65.4 | 13.1 | 0.21 | 7.8 | 48 |
| Spathaspora sp. 1 | UFMG-HMD-16.3 | 84.0 | 6.3 | 0.27 | 0.22 | 53.8 | 10.7 | 0.17 | 6.7 | 48 |
| Spathaspora sp. 2 | UFMG-XMD-23.2 | 89.8 | 7.4 | 0.26 | 0.21 | 50.8 | 10.2 | 0.19 | 7.0 | 48 |
| Spathaspora sp. 3 | UFMG-HMD-19.3 | 55.2 | 5.6 | 0.13 | 0.07 | 26.2 | 3.3 | 0.16 | 3.9 | 48 |
| Candida sp 1 | UFMG-HMD-23.3 | 60.2 | 9.3 | 0.18 | 0.10 | 35.8 | 4.9 | 0.13 | 3.3 | 48 |
| Candida sp 1 | UFMG-HMD-25.1 | 52.3 | 9.0 | 0.14 | 0.06 | 27.0 | 3.1 | 0.22 | 4.6 | 48 |
D-xylose consumption (%) – percentage of initial D-xylose consumed.
Yp/s et (g/g) – ethanol yield: correlation between ethanol (ΔPethanol) produced and D-xylose (ΔSxylose) consumed.
Qp (g/L•h) – ethanol productivity: ratio of ethanol concentration (g/L) and fermentation time (h).
η (%) – fermentation efficiency: percentage of the maximum theoretical ethanol yield (0.51 g ethanol/g D-xylose).
Yp/s xy (g/g) – xylitol yield: correlation between xylitol (ΔPxylitol) produced and D-xylose (ΔSxylose) consumed.
Time when the maximum ethanol production (g/L) value was attained or time of the end of the fermentation experiment.
Fermentation results revealed that all yeasts tested were able to consume D-xylose, with the consumption rates ranging from 52.3% to practically 100% in 48 h. Da Cunha-Pereira et al. [15] found an efficiency of 84% for D-xylose utilization in 96 h by S. arborariae UFMG-HM 19.1A in synthetic medium containing initial concentrations of 20 g/L xylose, 20 g/L glucose and 10 g/L arabinose. Agbogbo et al. [51] reported 100% D-xylose consumption after 120 h by S. stipitis CBS 6054 in synthetic medium formulated with 60 g/L D-xylose. Agbogbo and Wenger [52] demonstrated a D-xylose consumption over 85% in 48 h by S. stipitis CBS 6054 in synthetic medium with approximately 21 g/L D-xylose and 5.8 g/L glucose. According to du Preez and Prior [53], C. shehatae CBS 2779 consumed 100% xylose in 28 h from medium containing 20 g/L xylose. Among the microorganisms studied in our work, the isolates of C. amazonensis showed the highest D-xylose consumption rates, approaching 100% of consumption (values ranged from 99.8% to 99.9%) in 48 h, whereas S. passalidarum showed the fastest D-xylose consumption rate, 98%, in 24 h. Among the new species, both Spathaspora sp. 1 and Spathaspora sp. 2 consumed up to 80% of D-xylose in 48 h, while Spathaspora sp. 3 and Candida sp. 1 consumed 52.3% to 60.2% of D-xylose, respectively, in 48 h. Strains of S. stipitis achieved similar maximum D-xylose consumption values, approximately 98.8%, in 48 h. These results show, in general, high and fast D-xylose consumption by most of the yeasts studied in this work. Furthermore, we also observed that a specifically high and fast D-xylose consumption behavior can likely be associated with C. amazonensis, S. passalidarum and S. stipitis strains isolated in this work, but not with all species from the genus Spathaspora.
Ethanol production was observed during the fermentation assay, confirming the ability to ferment D-xylose to ethanol by the microorganisms tested. The highest ethanol concentration values were produced by the S. passalidarum strains (a species that exhibited gas production in the Durham tube test), ranging from 15 g/L to 18 g/L in 24 h. Candida amazonensis, Spathaspora sp. 3 and Candida sp. 1 were at the other end of the spectrum, showing the lowest ethanol concentration values, between 3.1 g/L and 4.9 g/L in 48 h. Scheffersomyces stipitis UFMG-XMD-15.2 and UFMG-HMD-32.1, that exhibited gas production in the Durham tube test, produced ethanol with similar concentrations, 12.3 g/L and 11.1 g/L, respectively, but at different fermentation times, 24 h and 48 h, respectively. The variations observed can probably be associated with physiological differences between strains of the same species. Ethanol production of 15 g/L was observed by the yeast S. arborariae UFMG-HM-19.1A in approximately 72 h from 20 g/L xylose, 20 g/L glucose and 10 g/L arabinose [15]. Candida shehatae CBS 2779 reached 13.1 g/L ethanol in 28 h in synthetic medium containing 20 g/L xylose [53]. Scheffersomyces stipitis CBS 6054 was able to produce 24.3 g/L ethanol in 120 h in medium with 60 g/L D-xylose [51]. Agbogbo and Wenger [53] also showed the production of approximately 8.7 g/L ethanol in 48 h by S. stipitis CBS 6054 from 20 g/L xylose. In general, S. passalidarum strains showed the best potential for ethanol production in the conditions tested because they produced the highest ethanol concentrations in a short period of time.
Candida lignosa CBS 4705 (positive control) presented the highest ethanol yield (Yp/s et = 0.4 g/g) and fermentation efficiency (η = 79.4%). S. passalidarum strains showed the highest ethanol productivity (Qp = 0.62 g/L·h to 0.75 g/L·h), with minimum and maximum Yp/s et values between 0.31 g/g and 0.37 g/g and η between 61% and 71.6%. The new species Spathaspora sp. 1 UFMG-XMD-16.2, Spathaspora sp. 1 UFMG-HMD-16.3 and Spathaspora sp. 2 UFMG-XMD-23.2 showed better ethanol fermentation results than C. amazonensis strains, Spathaspora sp. 3, Candida sp. 1 UFMG-HMD-23.3 and Candida sp. 1 UFMG-HMD-25.1 (Table 2). Da Cunha-Pereira et al. [15] found a Yp/s et and Qp equivalent to 0.45 g/g and 0.21 g/L·h, respectively, for S. arborariae UFMG-HM-19.1A. Candida shehatae CBS 2779 and S. stipitis CBS 6054 showed Yp/s et values equal to 0.37 g/g and 0.44 g/g, respectively, and Qp equivalent to 0.47 g/L·h and 0.20 g/L·h, respectively [51], [53]. Overall, S. passalidarum strains showed promising results for ethanol production under the conditions tested, considering their remarkably high ethanol productivity (Qp = 0.62 g/L·h to 0.75 g/L·h) and fermentation efficiency of approximately 70% (η = 61% to 71.6%), in 24 h. However, Yp/s et values found in S. passalidarum strains (0.31 g/g and 0.37 g/g) were lower than those found in S. arborariae UFMG-HM-19.1A and S. stipitis CBS 6054, but it must be taken into consideration the different experimental conditions employed for these yeasts. Spathaspora arborariae UFMG-HM-19.1A produced Yp/s et equal to 0.45 g/g in medium containing 20 g/L xylose, but also 20 g/L glucose and 10 g/L arabinose, and its maximum ethanol production was achieved in approximately 72 h [15], while S. stipitis CBS 6054 showed Yp/s et equal to 0.44 g/g in medium containing 60 g/L xylose and took 120 h to reach its maximum ethanol concentration [51]. In our study, D-xylose (50 g/L) was used as the sole carbon source, and S. passalidarum strains reached maximum ethanol production in 24 h.
A good understanding of ethanol production from xylose requires a knowledge of both the experimental conditions and the enzymes responsible for xylose metabolism. In yeasts, fermentation of xylose to ethanol is strongly dependent on NADH-linked xylose reductase (XR). In Scheffersomyces stipitis, XR preferentially uses NADPH, although it can also use NADH [54]. Recently, it was shown that in S. passalidarum xylose is converted by means of an NADH-preferring XR [16]. These observations are compatible with the higher ethanol yields achieved by strains of S. passalidarum and S. stipitis tested in this work. The impact of XR activities and their co-factor preferences in on ethanol production among these new xylose-fermenting yeasts will be the focus of future work.
Yeast strains that achieved their maximum ethanol concentration in 24 h were observed to undergo a decrease in ethanol concentration after this period (data not shown). This decrease probably results from ethanol assimilation by the yeasts as a consequence of the rapid depletion of sugar from the medium while oxygen remains available. This behavior has previously been reported for yeasts in studies utilizing both synthetic medium and/or lignocellulosic hydrolysates [9], [12], [15], [52], [55].
Although ethanol was the main product obtained in these fermentation assays, by-products such as xylitol and glycerol were also found. Glycerol was present at low concentrations (on average ≤0.05 g/L, maximum value of 1.4 g/L, data not shown), whereas xylitol was produced at higher concentrations. The lowest xylitol values (0 g/L to 2.2 g/L) were obtained for S. stipitis, C. lignosa and S. passalidarum. Ferreira et al. [12] suggested that the formation of low amounts of by-product is important characteristics of the yeast strain, as they allow elevated formation of the main fermentation product, ethanol, to be obtained. The new Spathaspora species that was identified in this study produced xylitol concentrations between 3.3 g/L and 7.8 g/L in 48 h (Yp/s xy between 0.13 g/g and 0.22 g/g) and, together with C. amazonensis, produced lower ethanol concentrations than the other species tested. Another species from Spathaspora clade, S. arborariae UFMG-HMD-19.1A has already been shown to produce xylitol, at 5 g/L in 108 h in synthetic medium [15]. Candida amazonensis was responsible for the highest xylitol production values found in this assay (26.1 g/L to 27.8 g/L in 24 h, Yp/s xi between 0.62 g/g and 0.67 g/g, data not shown). These values were much higher than those found for ethanol production by all yeasts tested.
After 24 h, a decrease in the xylitol concentration occurred, indicating that this product was being used as carbon source, presumably following D-xylose depletion and also due the oxygen availability in the medium. Xylitol is one of the most expensive polyol sweeteners in the world market and has been the subject of specific health claims. It is suitable for diabetics and is recommended for oral health and parenteral nutrition [56]. The biotechnological production of xylitol using microorganisms such as yeasts and fungi is of economic interest and presents some advantages when compared with conventional xylitol production by the chemical reduction of D-xylose, as this latter process is characterized by high costs due to difficulties in separation and purification steps [39], [57], [58]. Thus, C. amazonensis isolates obtained in this work can be studied in the future for direct xylitol production from D-xylose in hemicellulosic hydrolysates. Further studies are needed to establish the best cultivation conditions for this process.
Sugarcane bagasse hemicellulosic hydrolysate
After performing D-xylose fermentation assays, we selected a number of strains for fermentation tests in sugarcane bagasse hydrolysate. We picked the strains or species that presented the highest rates of D-xylose consumption and ethanol yields (Yp/s et), specifically S. passalidarum UFMG-HMD-1.1 and UFMG-HMD-14.1, S. stipitis UFMG-XMD-15.2, Spathaspora sp. 1 UFMG-XMD-16.2 and Spathaspora sp. 2 UFMG-XMD-23.2. Candida amazonensis, Spathaspora sp. 3 and Candida sp. 1 were not selected due to their low ethanol production in D-xylose culture medium.
The fermentation parameter results [Yp/s et (g/g), ethanol yield; Yp/s xy (g/g), xylitol yield; Qp (g/L·h), ethanol productivity; η (%), fermentation efficiency; sugar (D-xylose and glucose) consumption (%)] and cells, ethanol and xylitol concentrations (g/L) in sugarcane bagasse hemicellulosic hydrolysates are summarized in Table 3. These results were also calculated based on the fermentation time (time of maximum ethanol production or time of the end of the fermentation experiment) for each yeast strain.
Table 3. Ethanol yield [Yp/s et (g/g)], xylitol yield [Yp/s xy (g/g)], ethanol productivity [Qp (g/L•h)], fermentation efficiency [η (%)], sugar (D-xylose and glucose) consumption (%), cell concentration (g/L), ethanol concentration (g/L) and xylitol concentration (g/L) in sugarcane bagasse hemicellulosic hydrolysate assays.
| Yeast species | Yeast strains | Sugars consumption (%)1 | Cells (g/L) | Yp/s et (g/g)2 | Qp (g/L•h)3 | Η (%)4 | Ethanol concentration (g/L) | Yp/s xy (g/g)5 | Xylitol concentration (g/L) | Fermentation time (h)6 |
| Candida lignosa | CBS 4705 | 55.3 | 7.8 | 0.16 | 0.05 | 31.6 | 4.6 | – | – | 96 |
| Scheffersomyces stipitis | NRRL Y-7124 | 34.9 | 3.4 | 0.25 | 0.10 | 49.3 | 4.9 | – | – | 48 |
| S. stipitis | UFMG-XMD-15.2 | 80.9 | 12.7 | 0.34 | 0.20 | 65.7 | 14.1 | – | – | 72 |
| Spathaspora passalidarum | UFMG-HMD-1.1 | 84.9 | 13.4 | 0.20 | 0.09 | 40.0 | 8.8 | – | – | 96 |
| S. passalidarum | UFMG-HMD-14.1 | 91.0 | 13.1 | 0.18 | 0.10 | 36.0 | 9.5 | – | – | 96 |
| Spathaspora sp. 1 | UFMG-XMD-16.2 | 72.9 | 7.9 | 0.23 | 0.10 | 46.0 | 9.3 | 0.57 | 18.2 | 96 |
| Spathaspora sp. 2 | UFMG-XMD-23.2 | 68.6 | 6.4 | 0.22 | 0.08 | 42.4 | 7.2 | 0.61 | 17.1 | 96 |
Sugar consumption (%) – percentage of the initial D-xylose and glucose consumed.
Yp/s et (g/g) – ethanol yield: correlation between ethanol (ΔPethanol) produced and D-xylose and glucose (ΔSsugars) consumed.
Yp/s xy (g/g) – xylitol yield: correlation between xylitol (ΔPxylitol) produced and D-xylose (ΔSxylose) consumed.
Qp (g/L·h) – ethanol productivity: ratio of ethanol concentration (g/L) and fermentation time (h).
η (%) – fermentation efficiency: percentage of the maximum theoretical ethanol yield (0.51 g ethanol/g D-xylose and glucose).
Time when the maximum ethanol production (g/L) value was attained or time of the end of the fermentation experiment.
Taking only into account the glucose and D-xylose present in the hydrolysates, S. stipitis UFMG-XMD-15.2 showed the highest sugar consumption, equal to 93.4% in 96 h (data not shown), followed by S. passalidarum strains UFMG-HMD-1.1 (84.9%) and UFMG-HMD-14.1 (91%) in 96 h. Spathaspora sp. 2 UFMG-XMD-23.2 and Spathaspora sp. 1 UFMG-XMD-16.2 presented values of 68.6% and 72.9%, respectively, whereas C. lignosa CBS 4705 (positive control) showed the lowest consumption value, equal to 55.3% in 96 h. Scheffersomyces stipitis NRRL Y-7124 (positive control) consumed 69.8% of glucose and D-xylose (data not shown) in 96 h, similar to the amount consumed by Spathaspora sp. 2. Da Cunha-Pereira et al. [15] observed that S. arborariae UFMG-HM-19.1A consumed glucose and xylose at 100% and 45% in approximately 120 h and 240 h, respectively, in rice hull hydrolysate containing 35 g/L glucose, 13 g/L xylose and 4 g/L arabinose. Scheffersomyces stipitis UFMG-IMH-43.2 consumed, on average, 83.6% of glucose and xylose in 192 h, when cultivated in sugarcane bagasse hydrolysate with an initial concentration of 52.5 g/L xylose [12]. Chandel et al. [59] reported a total sugar consumption of 85.9% after 24 h fermentation by C. shehatae NCIM 3501 in sugarcane bagasse hydrolysate with 20 g/L total sugars. Among the yeasts tested in our assays, glucose and D-xylose were initially metabolized at the same time, but the glucose consumption rate was clearly faster than that of D-xylose. After glucose exhaustion, D-xylose was consumed much faster, and this behavior was observed after 12 h by S. passalidarum strains and S. stipitis UFMG-XMD-15.2, after 24 h by S. stipitis NRRL-Y 7124 and C. lignosa CBS 4705 and after 48 h by Spathaspora sp. 1 and Spathaspora sp. 2 (data not shown). When compared with the results obtained in other studies on hydrolysate fermentation, S. stipitis UFMG-XMD-15.2 and the S. passalidarum strains tested in our work showed excellent D-xylose and glucose consumption results, when the initial D-xylose (50.2 g/L) and glucose (5.3 g/L) concentrations and the time of consumption (96 h) of the majority (84.9% and 93.4%) of these sugars are considered. Unlike D-xylose and glucose, L-arabinose consumption was not observed by the yeasts during the fermentation time studied. The absence of L-arabinose consumption was also observed in fermentations carried out with S. stipitis UFMG-IMH-43.2 in experiments similar to those performed in this work [12].
Ethanol production was observed within 12 h of cultivation by the yeasts tested. Scheffersomyces stipitis NRRL Y-7124 produced a maximum concentration of 4.9 g/L ethanol within 48 h of fermentation. Candida lignosa CBS 4705, S. passalidarum UFMG-HMD-1.1 and UFMG-HMD-14.1 reached maximum ethanol concentrations of 4.6 g/L, 8.8 g/L and 9.5 g/L, respectively, in 96 h. S. stipitis UFMG-XMD-15.2 was responsible for the highest ethanol production, corresponding to 14.1 g/L in 72 h. An ethanol production greater than 15 g/L was observed for S. arborariae UFMG-HM-19.1A after 50 h in rice hull hydrolysate [15]. Candida shehatae NCIM 3501 produced 5.2 g/L ethanol after 24 h in sugarcane bagasse hydrolysate [59]. Spathaspora passalidarum UFMG-HMD-1.1 and UFMG-HMD-14.1 showed ethanol concentration results similar to that reported by S. stipitis UFMG-IMH-43.2, which presented an ethanol production equal to 9.1 g/L, but in 192 h [12]. Still, when the strains tested in this study were compared with S. stipitis UFMG-IMH-43.2, S. stipitis UFMG-XMD-15.2 produced a higher ethanol concentration (14.1 g/L) at a much faster rate (72 h). It must be noted that both studies used sugarcane bagasse hemicellulosic hydrolysate with similar experimental conditions (including the initial concentration of D-xylose, supplementation, agitation and incubation temperature). Considering the initial high D-xylose concentration (50.2 g/L) relative to the glucose concentration (5.3 g/L) and the remarkably high ethanol production by S. stipitis UFMG-XMD-15.2 relative to other species tested here, the hydrolysate fermentation study results suggest that this strain is a promising tool for producing ethanol from D-xylose.
Scheffersomyces stipitis UFMG-XMD-15.2 presented the highest ethanol yield (Yp/s et = 0.34 g/g), ethanol productivity (Qp = 0.2 g.L/h) and fermentation efficiency (η = 65.7%). Spathaspora passalidarum strains UFMG-HMD-1.1 and UFMG-HMD-14.1 showed Yp/s et values between 0.18 g/g and 0.2 g/g, ethanol productivity between 0.09 and 0.1 g/L·h and η between 36% and 40%, whereas Spathaspora sp. 2 and Spathaspora sp. 1 presented ethanol yields between 0.22 g/g and 0.23 g/g, Qp between 0.08 g/L·h and 0.1 g/L·h and η between 42.4% and 46%. Da Cunha-Pereira et al. [15] found a Yp/s et and Qp equivalent to 0.45 g/g and 0.16 g/L·h, respectively, for S. arborariae UFMG-HM-19.1A in a fermentation assay using rice hull hydrolysate. Candida shehatae NCIM 3501 showed Yp/s et equal to 0.3 g/g and Qp equivalent to 0.22 g/L·h in sugarcane bagasse hydrolysate [59], results similar to those found for S. stipitis UFMG-XMD-15.2. Ferreira et al. [12] found Yp/s et and Qp equal to 0.17 g/g and 0.05 g/L·h, respectively, for S. stipitis UFMG-IMH-43.2, showing that ethanol fermentation parameters presented by the strains isolated in our study were better than those observed for S. stipitis UFMG-IMH-43.2. When compared with the ethanol yield of S. arborariae [15], the yeast strains isolated in this study showed lower Yp/s et in hemicellulosic hydrolysate. Spathaspora arborariae UFMG-HM-19.1A produced Yp/s et equal to 0.45 g/g in rice hull hydrolysate containing glucose as the main sugar (35 g/L), with an initial xylose concentration (13 g/L) that was much lower than that employed in our work (50.2 g/L). Despite the higher Yp/s et presented by S. arborariae in rice hull hydrolysates, S. stipitis UFMG-XMD-15.2 showed a Qp (0.2 g/L·h) higher than that observed for S. arborariae (0.16 g/L·h) in our experiments.
Ethanol consumption was observed by S. stipitis NRRL Y-7124 after 48 h and S. stipitis UFMG-XMD-15.2 after 72 h (time of maximum ethanol production). Glycerol was found at low concentrations in S. passalidarum UFMG-HMD-1.1 (0.05 g/L), S. passalidarum UFMG-HMD-14.1 (0.1 g/L), S. stipitis UFMG-XMD-15.2 (0.7 g/L), Spathaspora sp. 1 UFMG-XMD-16.2 (0.2 g/L) and Spathaspora sp. 2 UFMG-XMD-23.2 (0.6 g/L). Except for the new species identified, xylitol production was not observed for the other yeasts studied. Again, as suggested by Ferreira et al. [12], the low production rates of xylitol and glycerol may be important characteristics of the yeast strain that should allow higher production of the main fermentation product, ethanol, by the microorganism, a physiological characteristic desirable in our study.
Spathaspora sp. 1 UFMG-XMD-15.2 and Spathaspora sp. 2 UFMG-XMD-23.2 were the only yeasts that were observed to produce xylitol (18.2 g/L and 17.1 g/L, respectively) in this assay (Table 3). These yeasts produced markedly higher xylitol concentrations in hydrolysate than in D-xylose culture medium. Xylitol yield values that were found for these new species were considerably higher than that found for a previously identified strain of C. tropicalis from the soil (Yp/s xy = 0.45 g/g), tested in a fermentation assay on sugarcane hydrolysate with an initial xylose concentration of 30 g/L [39]. The xylitol production (g/L) and Yp/s xy results found in our work were also higher than those reported by Silva and Roberto [60] for C. gulliermondii FTI 20037. These authors observed a xylitol production of 17 g/L and a Yp/s xy of 0.35 g/g after 120 h for C. gulliermondii FTI 20037 in a rice straw hemicellulosic hydrolysate fermentation assay, with an initial xylose concentration of 90 g/L. The biotechnological production of xylitol from crude hemicellulosic hydrolysates could be a valuable economic alternative to the expensive chemical production of xylitol from D-xylose [39], [61]. Our results for xylitol production by Spathaspora sp. 1 and Spathaspora sp. 2 are promising and warrant future testing of the production of this polyol from xylose fermentation in hemicellulosic hydrolysates.
Our study investigated the ability of yeasts to produce ethanol and/or xylitol while consuming D-xylose in D-xylose culture medium and produce ethanol and/or xylitol while consuming D-xylose and glucose in sugarcane bagasse hemicellulosic hydrolysate. S. stipitis UFMG-XMD-15.2 is of particular interest because it efficiently converted D-xylose to ethanol and grew well in sugarcane bagasse hemicellulosic hydrolysate. Spathaspora passalidarum strains were also observed to grow in hydrolysates and produce ethanol. Candida amazonensis was notable for its potential use in the biotechnological production of xylitol. The newly identified yeast strains Spathaspora sp. 1 and Spathaspora sp. 2 were similarly found to potentially produce xylitol in hemicellulosic hydrolysate. Future studies are needed to test their production of this polyol. As this work is the first to use these yeasts in fermentation assays, further studies are required to optimize cultivation conditions (nutritional dependence, initial substrate concentration, initial cell concentration, pH, temperature and aeration) for the efficient conversion of D-xylose into ethanol or xylitol.
Supporting Information
List of the new yeast species isolated in this work and their respective GenBank deposit numbers.
(DOC)
Acknowledgments
We thank the Empresa Brasileira de Pesquisa Agropecuária (EMBRAPA)-Roraima by their technical support. We also thank Dr. Hélio Tonini for his technical support during the field collections, and the owners, Osvaldo Antônio Sant′ana and José Lopes, of the private lands by the permission to collect the samples in the areas of the private ecological reserves of Mucajaí and São João da Baliza, respectively.
Funding Statement
This work was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq – Brazil, process 560715/2010-2), Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP, process 2008/57926-4), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Procad-NF, process 2280/2008) and the Natural Science and Engineering Research Council of Canada (M.A.L.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pokhrel CP, Yadav RKP, Ohga S (2008) Agricultural waste residues as potential sources of bioethanol. Sci World 6: 19–23. [Google Scholar]
- 2. Kim S, Dale BE (2005) Lyfe cycle assessment of various cropping systems utilized for producing biofuels: bioethanol and biodiesel. Biomass Bioenerg 29: 426–439. [Google Scholar]
- 3. Aristidou A, Penttila M (2000) Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol 11: 187–198. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura N, Yamada R, Katahira S, Tanaka T, Fukuda H, et al. (2008) Effective xylose/cellobiose co-fermentation and ethanol production by xylose-assimilating S. cerevisiae via expression of β-glucosidase on its cell surface. Enzyme MicrobTech 43; 233–236.
- 5. Chandel AK, Singh OV, Chandrasekhar G, Rao LV, Narasu ML (2010) Key-drivers influencing the commercialization of ethanol based biorefineries. J Comm Biotechnol 16: 239–257. [Google Scholar]
- 6. Nigam JN, Ireland RS, Margaritis A, Lachance MA (1985) Isolation and screening of yeasts that ferment D-xylose directly to ethanol. Appl Environ Microbiol 50: 1486–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Toivola A, Yarrow D, van Den Bosch E, van Dijken JP, Scheffers WA (1984) Alcoholic fermentation of D-xylose by yeasts. Appl Environ Microbiol 47: 1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suh SO, Marshall CJ, Hugh JVM, Blackwell M (2003) Wood ingestion by passalid beetles in the presence of xylose-fermenting gut yeasts. Mol Ecol 12: 3137–3145. [DOI] [PubMed] [Google Scholar]
- 9. Cadete RM, Santos RO, Melo MA, Mouro A, Gonçalves DL, et al. (2009) Spathaspora arborariae sp. nov., a D-xylose-fermenting yeast species isolated from rotting wood in Brazil. FEMS Yeast Res 9: 1338–1342. [DOI] [PubMed] [Google Scholar]
- 10. Rao RS, Bhadra B, Shivaji S (2008) Isolation and characterization of ethanol- producing yeasts from fruits and tree barks. Lett Appl Microbiol 47: 19–24. [DOI] [PubMed] [Google Scholar]
- 11. Agbogbo FK, Coward-Kelly G (2008) Cellulosic ethanol production using the naturally occurring xylose-fermenting yeasts, Pichia stipitis . Biotechnol Lett 30: 1515–1524. [DOI] [PubMed] [Google Scholar]
- 12. Ferreira AD, Mussato SI, Cadete RM, Rosa CA, Silva SS (2011) Ethanol production by a new pentose-fermenting yeast strain, Scheffersomyces stipitis UFMG-IMH 43.2, isolated from the Brazilian forest. Yeast 28: 547–554. [DOI] [PubMed] [Google Scholar]
- 13. Wohlbach DJ, Kuo A, Sato TK, Potts KM, Salamov AA, et al. (2011) Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc Natl Acad Sci U S A 108: 13212–13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen NH, Suha SO, Marshall CJ, Blackwell M (2006) Morphological and ecological similarities: wood-boring beetles associated with novel xylose-fermenting yeasts, Spathaspora passalidarum gen. sp. nov. and Candida jeffriesii sp. nov. Mycol Res 110: 1232–1241. [DOI] [PubMed] [Google Scholar]
- 15. da Cunha-Pereira F, Hickert LR, Sehnem NT, de Souza-Cruz PB, Rosa CA, et al. (2011) Conversion of sugars present in rice hull hydrolysates into ethanol by Spathaspora arborariae, Saccharomyces cerevisiae, and their co-fermentations. Bioresource Tech 102: 4218–4225. [DOI] [PubMed] [Google Scholar]
- 16. Hou X (2012) Anaerobic xylose fermentation by Spathaspora passalidarum . Appl Microbiol Biotechnol 94: 205–214. [DOI] [PubMed] [Google Scholar]
- 17. Hahn-Hägerdal B, Pamment N (2004) Microbial pentose metabolism. Appl Biochem Biotechnol 113: 1207–1209. [DOI] [PubMed] [Google Scholar]
- 18. Chandel AK, Chandrasekhar G, Radhika K, Ravinder R, Ravindra P (2011) Bioconversion of pentose sugars into ethanol: A review and future directions. Biotechnol Mol Biol Rev 6: 08–20. [Google Scholar]
- 19. Jeffries TW, Kurtzman CP (1994) Strain selection, taxonomy, and genetics of xylose-fermenting yeasts. Enzyme Microb Technol 16: 922–932. [Google Scholar]
- 20. Jeffries TW (1985) Emerging technology for fermenting D-xylose. Trends Biotechnol 3: 208–212. [Google Scholar]
- 21. van Maris AJA, Abbott DA, Bellissimi E, van Den Brink J, Kuyper M, et al. (2006) Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie van Leeuwenhoek 90: 391–418. [DOI] [PubMed] [Google Scholar]
- 22. Salati E, Vose PB (1984) Amazon basin: a system in equilibrium. Science 225: 129–138. [DOI] [PubMed] [Google Scholar]
- 23. Peres CA, Gardner TA, Barlow J, Zuanon J, Michalski F, et al. (2010) Biodiversity conservation in human-modified Amazonian forest landscapes. Biol Conserv 143: 2314–2327. [Google Scholar]
- 24. Morais PB, Martins MB, Klaczko LB, Mendonça-Hagler LC, Hagler AN (1995) Yeast succession in the Amazon fruit Parahancornia amapa as resource partitioning among Drosophila spp. Appl Environ Microbiol 61: 4251–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morais PB, Rosa CA, Meyer SA, Mendonça-Hagler LC, Hagler AN (1995) Candida amapae, a new amino acid-requiring yeast from the Amazonian fruit Parahancornia amapa . J Ind Microbiol Biotechnol 14: 531–535. [Google Scholar]
- 26. Vital MJS, Abranches J, Hagler AN, Mendonça-Hagler LC (2002) Mycocinogenic yeasts isolated from Amazon soils of the Maracá Ecological Station, Roraima-Brazil Braz J Microbiol. 33: 230–236. [Google Scholar]
- 27.Pires JM (1972) Estudos dos principais tipos de vegetação do estuário amazônico. Tese de Doutorado. Piracicaba, Brazil: ESALQ, USP. 183 p.
- 28.Kurtzman CP, Fell JW, Boekhout T, Robert V (2011) Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts: a taxonomic study. Fifth edition. Elsevier. 87–110.
- 29. de Barros Lopes M, Soden A, Henschke PA Langridge P (1996) PCR differentiation of commercial yeasts strains using intron splice site primers. Appl Environ Microbiol 62: 4514–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rosa CA, Lachance MA, Teixeira LCRS, Pimenta RS, Morais PB (2007) Metschnikowia cerradonensis sp. nov., a yeast species isolated from ephemeral flowers and their nitidulid beetles in Brazil. Int J Syst Evol Microbiol 57: 161–165. [DOI] [PubMed] [Google Scholar]
- 31. Lachance MA, Bowles JM, Starmer WT, Barker JSF (1999) Kodamaea kakaduensis and Candida tolerans, two new yeast species from Australian Hibiscus flowers. Can J Microbiol 45: 172–177. [DOI] [PubMed] [Google Scholar]
- 32. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carvalho W, Silva SS, Converti A, Vitolo M, Felipe MGA, et al. (2002) Use of immobilized Candida yeast cells for xylitol production from sugarcane bagasse hydrolysate: cell immobilization conditions. Appl Biochem Biotechnol 98–100: 489–496. [DOI] [PubMed] [Google Scholar]
- 34. Alves LA, Felipe MGA, Silva JBAE, Silva SS, Prata AMR (1998) Pretreatment of sugarcane bagasse hemicelluloses hydrolysate for xylitol production by Candida guilliermondii . Appl Biochem Biotechnol 70–72: 89–98. [Google Scholar]
- 35.Schmidell W, Lima UDA, Aquarone E, Borzani W (2001) Modeladem matemática e simulação de processos fermentativos. Biotecnologia Industrial. São Paulo: Edgard Blücher, 123–178.
- 36.Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP (2011) Candida Berkhout (1923). In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts: a taxonomic study. Fifth edition. Elsevier. 987–1278.
- 37. Jeffries TW (1981) Conversion of xylose to ethanol under aerobic conditions by Candida tropicalis . Biotechnol Lett 3: 213–218. [Google Scholar]
- 38. Kim TB, Oh DK (2003) Xylitol production by Candida tropicalis in a chemically defined medium. Biotechnol Lett 25: 2085–2088. [DOI] [PubMed] [Google Scholar]
- 39. Rao RS, Jyothi ChP, Prakasham RS, Sarma PN, Rao LV (2006) Xylitol production from corn fiber and sugarcane hydrolysates by Candida tropicalis . Bioresource Technol 97: 1974–1978. [DOI] [PubMed] [Google Scholar]
- 40. Sánchez S, Bravo V, García JF, Cruz N, Cuevas M (2008) Fermentation of D-glucose and D-xylose mixtures by Candida tropicalis NBRC 0618 for xylitol production. World J Microbiol Biotechnol 24: 709–716. [Google Scholar]
- 41. Liu YR, Huang LY, Young SS, Chang CF, Lee CF (2011) Asterotremella meifongana sp. nov. and Asterotremella nantouana sp. nov., two anamorphic basidiomycetous yeasts isolated from soil and mushrooms. Antonie van Leeuwenhoek 99: 643–650. [DOI] [PubMed] [Google Scholar]
- 42.Kurtzman CP, Fell JW, Boekhout T (2011) The yeasts: a taxonomic study. Fifth edition. Elsevier. 2080 p.
- 43. Bhadra B, Rao RS, Singh PK, Sarkar PK, Shivaji S (2008) Yeasts and yeast-like fungi associated with tree bark: diversity and identification of yeasts producing extracellular endoxylanases. Curr Microbiol 56: 489–494. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki M, Kurtzman CP (2011) Schwanniomyces Klocker emend. M. Suzuki & Kurtzman (2010). In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts: a taxonomic study. Fifth edition. Elsevier. 785–794.
- 45.Smith MTh, de Hoog GS, Statzell-Tallman A, Kurtzman CP (2011) Blastobotrys von Klopotek (1967). In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts: a taxonomic study. Fifth edition. Elsevier. 959–977.
- 46. du Preez J, de Goede E, Myburgh J (2009) Blastobotrys mokoenaii: a thermotolerant yeast that produces extracellular endo-β-xylanase. New Biotechnology. Barcelona, Spain, 13–16 September: Abstracts of the 14th European Congress on Biotechnology 25: S52. [Google Scholar]
- 47. Cadete RM, Melo MA, Lopes MR, Pereira GMD, Zilli JE, et al. (2012) Candida amazonensis sp. nov., an ascomycetous yeast isolated from rotting wood in Amazonian Forest, Brazil. Int J Syst Evol Microbiol 62: 1438–1440. [DOI] [PubMed] [Google Scholar]
- 48. Jindamorakot S, Limtong S, Yongmanitchai W, Tuntirungkij M, Potacharoen W, et al. (2007) Two new anamorphic yeasts, Candida thailandica sp. nov., and Candida lignicola sp. nov., isolated from insect frass in Thailand. FEMS Yeast Res 7: 1409–1414. [DOI] [PubMed] [Google Scholar]
- 49. Santos RO, Cadete RM, Badotti F, Mouro A, Wallheim DO, et al. (2011) Candida queiroziae sp. nov., a cellobiose-fermeting yeast species isolated from rotting wood in Atlantic Rain Forest. Antonie van Leeuwenhoek 99: 635–642. [DOI] [PubMed] [Google Scholar]
- 50.Kurtzman CP (2011) Lindnera Kurtzman, Robnett & Basehoar-Powers (2008). In: Kurtzman CP, Fell JW, Boekhout T, editors. The yeasts: a taxonomic study. Fifth edition. Elsevier. 521–543.
- 51. Agbogbo FK, Coward-Kelly G, Torry-Smith M, Wenger KS (2006) Fermentation of glucose/xylose mixtures using Pichia stipitis . Process Biochem 41: 2333–2336. [Google Scholar]
- 52. Agbogbo FK, Wenger KS (2007) Production of ethanol from corn stover hemicellulose hydrolysate using Pichia stipitis . J Ind Microbiol Biotechnol 34: 723–727. [DOI] [PubMed] [Google Scholar]
- 53. du Preez JC, Bosch M, Prior BA (1986) The fermentation of hexoses and pentoses sugars by Candida shehatae and Pichia stipitis . Appl Microbiol Biotechnol 23: 228–233. [Google Scholar]
- 54. Bruinenberg PM, de But PHM, van Dijken JP, Scheffers WA (1984) NADH-linked aldose reductase: the key to anaerobic alcoholic fermentation of xylose by yeasts. Appl Microbiol Biotechnol 19: 256–260. [Google Scholar]
- 55. Fonseca C, Spencer-Martins I, Hahn-Hägerdal B (2007) L-Arabinose metabolism in Candida arabinofermentans PYCC 5603T and Pichia guilliermondii PYCC 3012: influence of sugar and oxygen on product formation. Appl Microbiol Biotechnol 75: 303–310. [DOI] [PubMed] [Google Scholar]
- 56. Saha BC (2003) Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30: 279–291. [DOI] [PubMed] [Google Scholar]
- 57. Nigam I, Singh D (1995) Processes for fermentative production of xylitol – a sugar substitute. Process Biochem 30: 117–124. [Google Scholar]
- 58. Silva SS, Felipe MGA, Mancilha IM (1998) Factors that affect the Biosynthesis of xylitol by xylose-fermenting yeasts. Appl Biochem Biotechnol 70–72: 331–339. [DOI] [PubMed] [Google Scholar]
- 59. Chandel AK, Kapoor RK, Singh A, Kuhad RC (2007) Detoxification of sugarcane bagasse hydrolysate improves ethanol production by Candida shehatae NCIM 3501. Bioresource Technol 98: 1947–1950. [DOI] [PubMed] [Google Scholar]
- 60. Silva CJSM, Roberto IC (2001) Improvement of xylitol production by Candida guilliermondii FTI 20037 previously adapted to rice straw hemicellulosic hydrolysate. Lett Appl Microbiol 32: 248–252. [DOI] [PubMed] [Google Scholar]
- 61. Branco RF, dos Santos JC, Silva SS (2011) A novel use for sugarcane bagasse hemicellulosic fraction: xylitol enzymatic production. Biomass Bioenerg 35: 3241–3246. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the new yeast species isolated in this work and their respective GenBank deposit numbers.
(DOC)