NAC acts as a modulator of SRP function. It can bind to signal sequences directly. SRP initially displaces NAC from RNCs; however, when the signal sequence emerges, trimeric NAC·RNC·SRP complexes form. Upon docking NAC·RNC·SRP complexes to the ER, NAC remains bound, allowing NAC to shield cytosolically exposed nascent chain domains.
Abstract
Nascent polypeptide-associated complex (NAC) was initially found to bind to any segment of the nascent chain except signal sequences. In this way, NAC is believed to prevent mistargeting due to binding of signal recognition particle (SRP) to signalless ribosome nascent chain complexes (RNCs). Here we revisit the interplay between NAC and SRP. NAC does not affect SRP function with respect to signalless RNCs; however, NAC does affect SRP function with respect to RNCs targeted to the endoplasmic reticulum (ER). First, early recruitment of SRP to RNCs containing a signal sequence within the ribosomal tunnel is NAC dependent. Second, NAC is able to directly and tightly bind to nascent signal sequences. Third, SRP initially displaces NAC from RNCs; however, when the signal sequence emerges further, trimeric NAC·RNC·SRP complexes form. Fourth, upon docking to the ER membrane NAC remains bound to RNCs, allowing NAC to shield cytosolically exposed nascent chain domains not only before but also during cotranslational translocation. The combined data indicate a functional interplay between NAC and SRP on ER-targeted RNCs, which is based on the ability of the two complexes to bind simultaneously to distinct segments of a single nascent chain.
INTRODUCTION
Nascent polypeptide-associated complex (NAC) is a heterodimeric complex consisting of α-NAC and β-NAC, which in yeast are termed Egd2 and Egd1, respectively. NAC, which is confined to eukaryotic cells, associates with cytosolic ribosomes close to the exit of the polypeptide tunnel (Wiedmann et al., 1994; Wegrzyn et al., 2006; Pech et al., 2010). Yeast NAC is expressed at concentrations approximately equimolar to ribosomes, and the bulk of it is bound to ribosomes at steady state (Raue et al., 2007). NAC interacts with nascent polypeptides emerging from the ribosomal tunnel (Wiedmann et al., 1994; Gautschi et al., 2003; Raue et al., 2007; Berndt et al., 2009). Based on these fundamental findings and additional experimental observations, three major functions of NAC have been put forward. First, NAC is believed to be involved in cotranslational protein folding (Bukau et al., 2000; Hartl and Hayer-Hartl, 2002; Rospert et al., 2002; Wegrzyn and Deuerling, 2005). Of interest, recent evidence revealed that NAC has a specific role in de novo folding of ribosomal proteins (Koplin et al., 2010). Second, in vitro (Fünfschilling and Rospert, 1999) and in vivo (George et al., 1998, 2002; Yogev et al., 2007) evidence suggests that NAC is involved in cotranslational delivery of mitochondrial precursor proteins. Third, as detailed later, NAC is believed to act as a negative regulator of the signal recognition particle (SRP). Overall, the role of NAC in the aforementioned processes is only poorly defined. A long-standing debate persists about the role of NAC in SRP-dependent targeting of proteins to the endoplasmic reticulum (ER).
SRP cotranslationally targets membrane and secretory proteins to the translocon in the ER membrane (Shan and Walter, 2005; Cross et al., 2009; Grudnik et al., 2009; Saraogi and Shan, 2011). To that end, SRP interacts with hydrophobic signal sequences (Martoglio and Dobberstein, 1998) and directs ribosome nascent chain complexes (RNCs) to the translocon. In yeast, SRP prefers highly hydrophobic signal sequences such as the so-called signal anchor (SA) sequences of transmembrane proteins, which remain uncleaved and serve as membrane anchors. The best-studied example is the vacuolar type II membrane protein dipeptidyl aminopeptidase B (Dap2; Roberts et al., 1989; Ng et al., 1996; Cheng and Gilmore, 2006). ER-targeted proteins that possess signal sequences that fall below a threshold hydrophobicity are targeted in a posttranslational, SRP-independent manner (Ng et al., 1996; Hegde and Bernstein, 2006).Owing to the hydrophobic interaction between the Srp54 subunit and hydrophobic signal sequences, SRP binds with high affinity and in a salt-resistant manner to substrate RNCs (Walter et al., 1981; Powers and Walter, 1996; Flanagan et al., 2003; Berndt et al., 2009). SRP binds with low affinity also to nonsubstrate RNCs and even empty ribosomes (Walter et al., 1981; Powers et al., 1996; Flanagan et al., 2003; Berndt et al., 2009). Contact with the ribosome is mainly established via ribosomal proteins Rpl25/Rpl35 at the tunnel exit. Additional contacts involve rRNA segments at the tunnel exit close to Rpl17, which is localized on the side opposite to Rpl25/Rpl35 (Pool et al., 2002; Halic et al., 2004). SRP recognizes a conformational difference between translating and nontranslating ribosomes, even if ribosomes are involved in the translation of nonsubstrate proteins (Walter et al., 1981; Powers and Walter, 1996; Flanagan et al., 2003). High-affinity binding of SRP is established when the Srp54 subunit binds to a signal sequence (Walter et al., 1981; Powers and Walter, 1996; Flanagan et al., 2003). Recently it was observed that SRP can distinguish between RNCs that contain a hydrophobic SA sequence inside the ribosomal tunnel even before direct contact between SRP and the SA is established (Berndt et al., 2009; Mariappan et al., 2010). This leads to the early recruitment of SRP to RNCs, translating substrates targeted for cotranslational translocation.
Different models suggest a role of NAC in negatively modulating SRP function. The models have in common that NAC is believed to protect signalless RNCs from inappropriate interactions. One model, termed the NAC-translocon model hereafter, suggests that NAC prevents the binding of empty ribosomes or signalless RNCs to the translocon (Wiedmann et al., 1994; Lauring et al., 1995a, b; Möller et al., 1998). Follow-up studies from independent groups, however, have questioned the NAC-translocon model (Neuhof et al., 1998; Raden and Gilmore, 1998). The NAC-SRP model is based on the observation that NAC binds to nascent chains unspecifically, with the exception of nascent chains containing signal sequences (Wiedmann et al., 1994). The NAC-SRP model predicts that binding of NAC to a nascent chain prevents binding of SRP (Wiedmann et al., 1994; Wiedmann and Prehn, 1999; Lauring et al., 1995a, b; Reimann et al., 1999). A modified NAC-SRP model was suggested when NAC was found to prevent the salt-sensitive, but not the salt-resistant, binding of SRP to RNCs. This model predicts that NAC and SRP possess at least partly overlapping ribosomal binding sites and that SRP replaces NAC as soon as a signal sequence emerges out of the ribosomal tunnel (Powers and Walter, 1996). Because the translocon as well as SRP interact with the ribosome via Rpl25 (Halic et al., 2006b; Becker et al., 2009), the NAC-translocon and the modified NAC-SRP models received support when it was reported that NAC also interacts with Rpl25 (Wegrzyn et al., 2006). Recently, however, another study identified Rpl31 as the major ribosomal binding site of NAC (Pech et al., 2010). Because NAC binds to ribosomes via a short segment in its N-terminus (Franke et al., 2001; Pech et al., 2010) and because Rpl25 and Rpl31 are not adjacent to each other (Supplemental Figure S1), it seems unlikely that NAC interacts simultaneously with both ribosomal proteins. In light of the currently discussed models, it came as a surprise that a recent in vivo study revealed that yeast NAC was bound to RNCs carrying SRP substrates (Del Alamo et al., 2011). Here we revisit the interplay between NAC and SRP, using a homologous in vitro system in which the ratio of the components closely resembles those of a living cell. The results sort out some of the long-standing discrepancies with respect to NAC action and reveal a coordinated and dynamic interplay between NAC and SRP. NAC unexpectedly assists rather than restrains SRP function by modulating the early steps of the SRP cycle. The ability to bind to any nascent chain segment not covered by SRP allows NAC not only to shield hydrophobic signal sequences if SRP binding is delayed, but also to shield cytosolically exposed nascent chain domains during cotranslational translocation.
RESULTS
NAC is not required to prevent binding of SRP to RNCs lacking a signal sequence
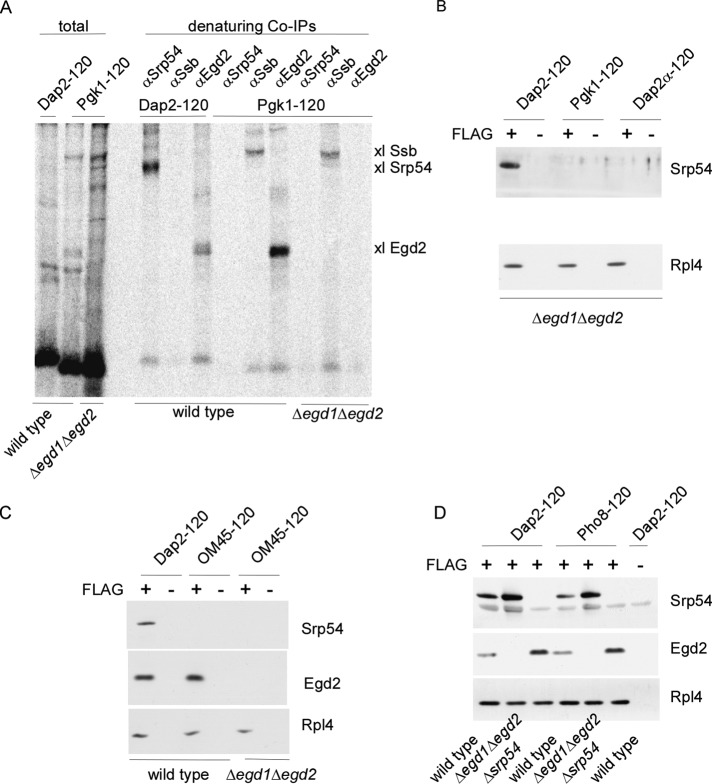
We explored the interplay between NAC and SRP in a homologous in vitro system. To that end, we generated RNCs in translation extracts derived from wild type or a set of yeast mutant strains (see subsequent discussion). In wild-type translation extracts the concentration of free NAC was ∼10-fold higher than the concentration of free SRP (see Materials and Methods). In this system, nascent Pgk1 or a mutant version of Dap2 termed Dap2α (Supplemental Figure S2), both of which lack hydrophobic SA segments, are in close proximity to NAC and the Hsp70 homologue Ssb. Nascent Dap2, which contains a hydrophobic SA segment, is in close proximity to NAC and SRP (Raue et al., 2007; Berndt et al., 2009). Chemical cross-linking experiments (Supplemental Figure S3A) confirmed these earlier observations: Pgk1-120 was in close proximity to Ssb and the Egd2 subunit of NAC, whereas Dap2-120 was in close proximity to Srp54 and Egd2 (Figure 1A). When Pgk1-120 RNCs were generated in a Δegd1Δegd2 translation extract, that is, in the absence of NAC, still no cross-link between nascent Pgk1-120 and SRP was observed (Figure 1A). We next tested the possibility that SRP, although not in direct contact with signalless nascent chains, was still bound to signalless RNCs in the absence of NAC. To that end, we performed FLAG–nascent chain pull downs (Supplemental Figure S3B) in a translation extract derived from a Δegd1Δegd2 strain. SRP was not detected on NAC-free Pgk1-RNCs or Dap2α-RNCs (Figure 1B). In addition, SRP was not recruited to NAC-free RNCs, exposing the transmembrane domain of the mitochondrial outer membrane protein OM45 (Figure 1C), which displays similar hydrophobicity compared with the SA segments of the ER-targeted protein Dap2 or Pho8 (Supplemental Figure S2; Ng et al., 1996; Waizenegger et al., 2003). Thus, in a complex homologous system the affinity of SRP for different types of RNCs and nascent chains lacking ER-targeting signals was below the detection limit, even in the absence of NAC.
FIGURE 1:
SRP does not bind to signalless RNCs in the absence of NAC. Chemical cross-linking with BS3 was performed with untagged, 35S-labeled nascent chains. FLAG–nascent chain pull downs were performed with RNCs carrying FLAG-tagged (FLAG +) or untagged (FLAG –) nascent chains generated in the absence of35S and were analyzed for the relative amount of RNCs (Rpl4), SRP (Srp54), and NAC (Egd2) via immunoblotting. To allow for a direct comparison of RNC occupation with NAC or SRP, similar amounts of RNCs (Rpl4) were loaded to a single gel. In each experiment Dap2-120-RNCs served as a control for SRP and NAC binding. For details on the nascent chains and methods see Supplemental Figures S2 and S3. (A) Chemical cross-linking of RNCs carrying 120-residue Dap2 or Pgk1 generated in a wild-type or Δegd1Δegd2 translation extract. Aliquots of the total after BS3 cross-linking were applied to immunoprecipitation reactions under denaturing conditions using antibodies as indicated. Samples were run on Tris-tricine gels and were analyzed via autoradiography. The total represents 5% of the amount added to each immunoprecipitation reaction. The size of the cross-links (xl) between the nascent chains and Srp54, Ssb, or Egd2, respectively, is indicated at the right. (B) FLAG–nascent chain pull down of RNCs carrying 120-residue Dap2, Pgk1, or Dap2α generated in a Δegd1Δegd2 translation extract. (C) FLAG–nascent chain pull down of RNCs carrying 120-residue Dap2 or OM45 generated in a wild-type or Δegd1Δegd2 translation extract as indicated. (D) FLAG–nascent chain pull down of RNCs carrying 120-residue Dap2 or Pho8 generated in a wild-type, Δegd1Δegd2, or Δsrp54 translation extract.
NAC binds to RNCs, exposing a signal sequence, and competes with SRP
Dap2 and Pho8 of 120 residues both expose a SA segment far outside of the ribosomal tunnel (Supplemental Figure S2; Ng et al., 1996). When RNCs were generated in a wild-type translation extract, both nascent chains recruited NAC, as well as SRP (Figure 1D). When RNCs were generated in a Δsrp54 translation extract, occupation of Dap2- or Pho8-RNCs with NAC was significantly enhanced (Figure 1D). This is consistent with a model in which SRP displaces NAC on RNCs when a SA segment becomes accessible (also see subsequent discussion). When RNCs were generated in a Δegd1Δegd2 translation extract, occupation of Dap2- or Pho8-RNCs with SRP was significantly enhanced (Figure 1D). This observation was unexpected because it suggested that in the wild-type extract NAC partly prevented the interaction of SRP with RNCs, exposing signal sequences.
The effect was analyzed in more detail via quantitative FLAG–nascent chain pull-down experiments (Supplemental Figure S3B; Raue et al., 2007). When translation was performed in a wild-type extract, ∼39% of Dap2-120-RNCs were occupied with NAC and ∼26% of Dap2-120-RNCs were occupied with SRP (Figure 2A). When SRP was absent, NAC binding was enhanced ∼2.3-fold. When NAC was absent, SRP binding was enhanced about threefold (Figure 2, A and B). Taking into account that the quantification procedure involves elements of uncertainty (discussed in Raue et al., 2007), we see that the data are compatible with a model in which the bulk of Dap2-120-RNCs was occupied by NAC in the absence of SRP and vice versa. We next tested whether NAC was able to bind to RNCs, exposing exclusively the SA segment. To that end, we constructed Dap2Δ, which lacks the segment located N-terminal of the SA. Nascent Dap2Δ of 62 residues exposes only the SA segment outside of the ribosomal tunnel (Supplemental Figure S2). In a wild-type extract Dap2Δ-62-RNCs were occupied with SRP and NAC (Figure 2C). When RNCs were generated in a Δsrp54 translation extract, NAC binding was increased ∼2.5-fold, suggesting that the bulk of Dap2Δ-62-RNCs were now occupied by NAC (Figure 2, C and D).
FIGURE 2:
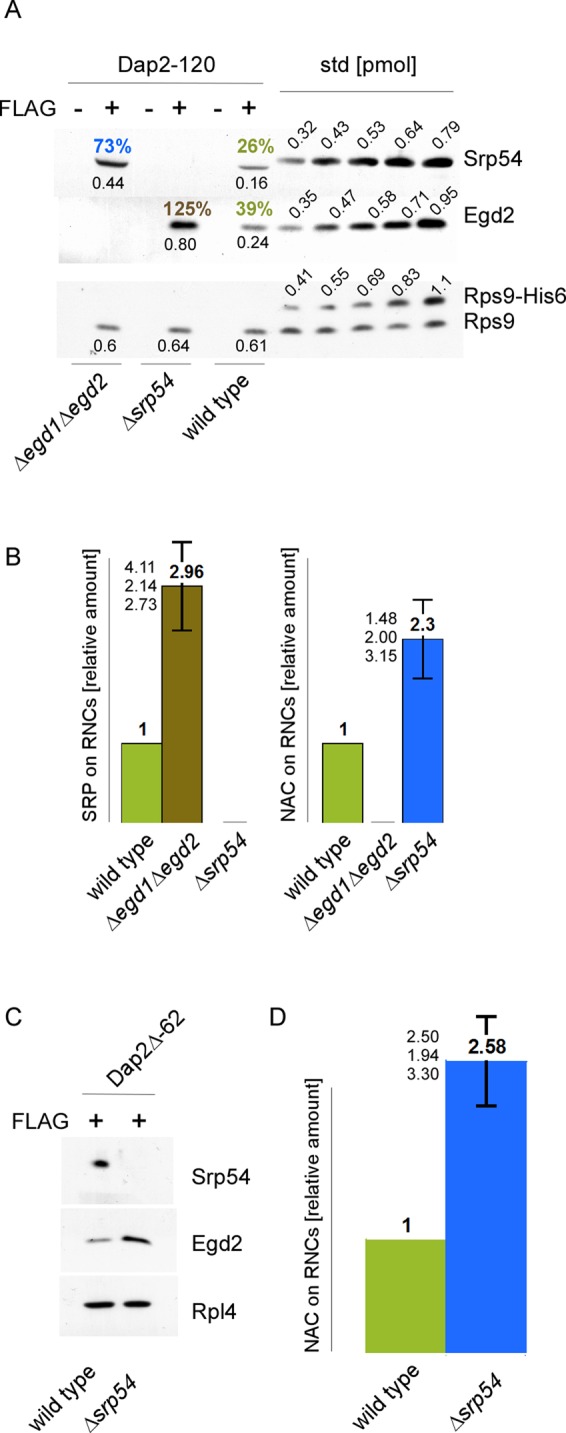
NAC and SRP compete for RNCs, exposing SA segments. FLAG–nascent chain pull downs were performed as described in Figure 1 with RNCs carrying FLAG-tagged (FLAG +) or untagged (FLAG –) nascent chains. (A) FLAG–nascent chain pull downs of RNCs carrying 120-residue Dap2 generated in wild-type, Δsrp54, or Δegd1Δegd2 translation extract. Increasing amounts of purified Rps9a-His6 (Rps9-His6), His6-Srp54 (Srp54), and NAC (Egd2) were used as standard. The amount of standard proteins loaded is given in picomoles above the protein bands. Black numbers below the protein bands indicate the amount of NAC, SRP, and RNCs in picomoles as determined based on calibration curves derived from the standard proteins. Colored numbers above the protein bands indicate the calculated percentage of RNCs occupied by NAC or SRP, respectively. (B) Relative occupation of RNCs exposing the SA segment far outside of the ribosomal tunnel. Experiments were performed as in A. Occupation of Dap2-120-RNCs generated in a wild-type translation extract was set to 1. Occupation of Dap2-120-RNCs generated in Δsrp54 or Δegd1Δegd2 translation extract is given relative to the occupation of wild-type RNCs (Materials and Methods). Error bars indicate the SE of three independent experiments; the actual individual results are indicated to the left of the error bar. (C) FLAG–nascent chain pull downs of RNCs carrying 62-residue Dap2Δ (Supplemental Figure S2) generated in wild-type or Δsrp54 translation extract. (D) Relative occupation of Dap2Δ-62-RNCs exposing the SA segment adjacent to the ribosomal tunnel. Analysis was performed as described in B. Error bars indicate the SE of three independent experiments; the actual individual results are indicated to the left of the error bar.
NAC binds to ribosomes via Rpl31
Chemical cross-linking experiments in total yeast extracts were performed to determine whether NAC interacted with Rpl25 and/or with Rpl31 (see Introduction and Supplemental Figure S1). Using an antibody specific for Rpl25, we detected a variety of cross-link products (Figure 3A). Rpl25 cross-links of higher molecular mass were significantly reduced after high-salt treatment, indicating that these cross-links represented proteins associated with Rpl25 in a salt-sensitive manner. Three prominent Rpl25 cross-links of lower molecular mass were salt resistant and thus most likely represented ribosomal proteins in close proximity to Rpl25 (Figure 3A). The cross-linking pattern detected with the Rpl25 antibody remained unaltered when the experiment was performed in a Δegd1Δegd2 extract (Figure 3A), indicating that Rpl25 did not form efficient cross-links with Egd1 or Egd2. Using an antibody specific for Rpl31, we also detected a variety of cross-link products (Figure 3B). The most prominent cross-link product, of ∼70 kDa, was previously shown to contain Zuo1 (Peisker et al., 2008). In the lower–molecular mass range three Rpl31 cross-links of ∼35 kDa were detected. One of the 35-kD cross-link products was absent when cross-linking was performed in an extract derived from Δegd1Δegd2 (Figure 3B). In the wild-type extract this 35-kD cross-link was also detected using an Egd1 antibody (Figure 3C). To confirm that the 35-kD cross-link was between Rpl31 and Egd1, a C-terminally histidine (His)-tagged version of Rpl31 (Rpl31-His8) was expressed in a Δrpl31aΔrpl31b strain, termed the His-Rpl31 strain. As expected, the cross-link between Rpl31-His8 and Egd1 was shifted to higher molecular mass (Figure 3C). The molecular mass shift was also detected with the antibody directed against Rpl31 (Figure 3D).
FIGURE 3:
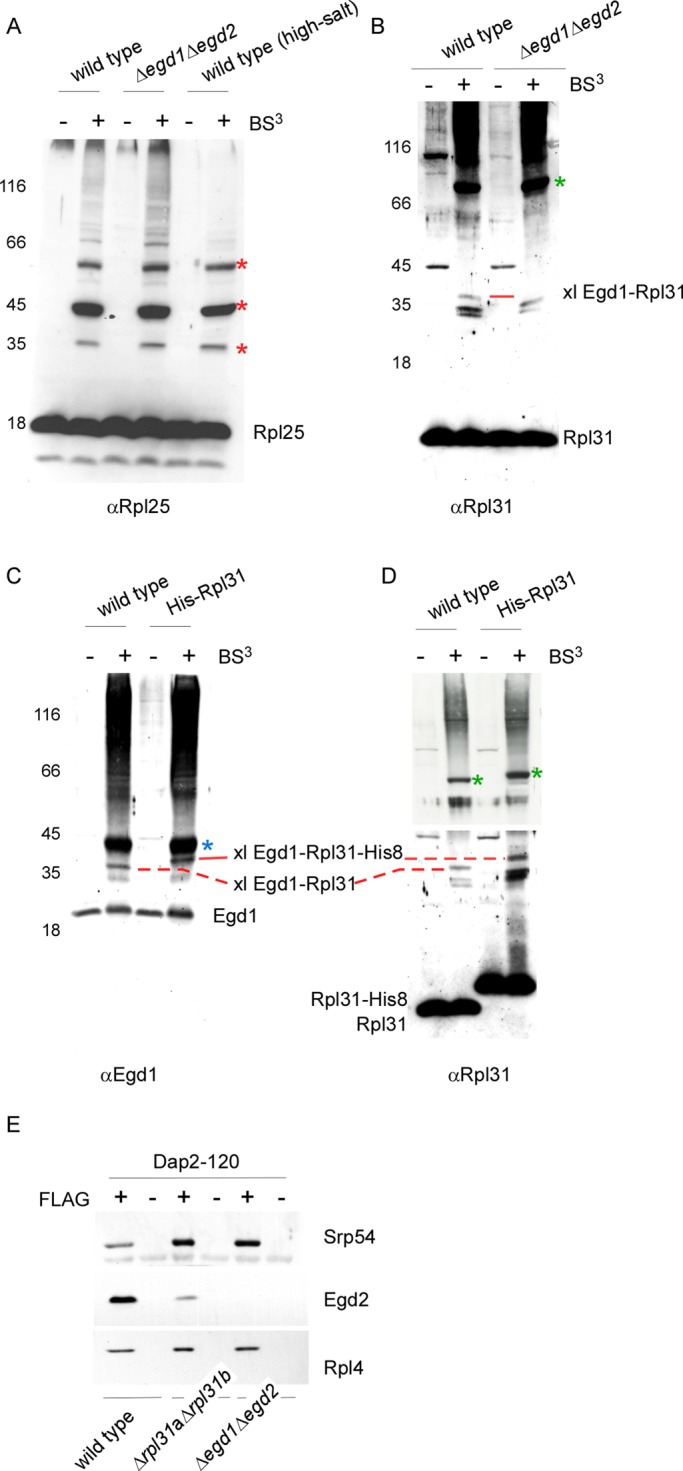
Egd1 forms a cross-link with Rpl31 but does not from a cross-link with Rpl25. Cross-linking was performed using ribosomes isolated under low-salt conditions or, if indicated, under high-salt conditions. Ribosome preparations generated from wild type, the Δegd1Δegd2, or the His-Rpl31 strain were incubated in either the presence (+) or the absence (–) of BS3. The molecular masses of the proteins of interest are Rpl25 (16 kDa), Rpl31 (13 kDa), Egd1 (17 kDa), and Egd2 (19 kDa). FLAG–nascent chain pull downs were performed as described in Figure 1. (A) Aliquots of a cross-linking reaction were analyzed using an antibody directed against Rpl25 (αRpl25). Cross-link products observed after high-salt treatment (labeled with red asterisks) are most likely to be unidentified ribosomal neighbors of Rpl25. (B) Aliquots were analyzed with an antibody directed against Rpl31 (αRpl31). The prominent cross-link between Rpl31 and Zuo1 (Peisker et al., 2008) is labeled with a green asterisk. The cross-link between Egd1 and Rpl31 (xl Egd1-Rpl31) is labeled with a red bar. (C) Aliquots were analyzed with an antibody recognizing the Egd1 subunit of NAC (αEgd1). The prominent cross-link between Egd1 and Egd2 is labeled with a blue asterisk. The cross-link between wild-type Rpl31 and Egd1 (xl Egd1-Rpl31) and the size-shifted cross-link between Rpl31-His8 and Egd1 (xl Egd1-Rpl31-His8) are indicated with red bars. (D) Aliquots as shown in C were analyzed using antibodies directed against Rpl31 (αRpl31). The cross-link between wild-type Rpl31 and Egd1 (xl Egd1-Rpl31) and the size-shifted cross-link between Rpl31-His8 and Egd1 (xl Egd1-Rpl31-His8), which are detected with αRpl31, comigrated with the cross-links recognized by αEgd1 shown in C. The cross-link between Rpl31 and Zuo1 and the size-shifted cross-link between Rpl31-His8 and Zuo1 are labeled with green asterisks. Because the strong cross-link product between Zuo1 and Rpl31 is otherwise overexposed, a shorter exposure of the same immunoblot is shown for the upper part. (E) FLAG–nascent chain pull downs of RNCs carrying 120-residue Dap2 generated in wild-type, Δrpl31aΔrpl31b, or Δegd1Δegd2 translation extract were performed as described in Figure 1.
In contrast to Rpl25, Rpl31 is not essential (Peisker et al., 2008). This allowed us to determine NAC binding to RNCs lacking Rpl31. Consistent with the cross-linking experiments shown in Figure 3 and with the work of Beatrix and coworkers (Pech et al., 2010), only a residual amount of NAC was bound to Dap2-120-RNCs lacking Rpl31 (Figure 3E). Of importance, at the same time significantly more SRP was recruited to Dap2-120-RNCs lacking Rpl31 (Figure 3E). Thus SRP binding was enhanced not only in the absence of NAC (Figures 1D and 2, A and B), but also in the absence of Rpl31, which resulted in a depletion of RNCs from NAC (Figure 3E). Moreover, the data revealed that Rpl31 was not required for SRP binding, indicating that the major binding sites of NAC and SRP were distinct.
NAC and SRP can bind simultaneously to a single RNC
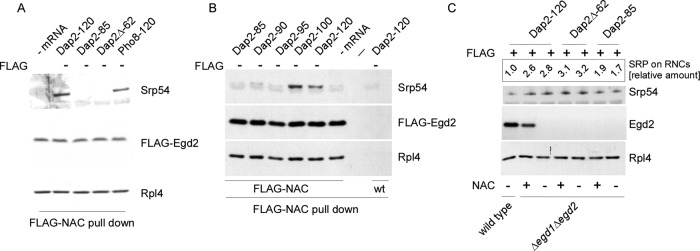
To test whether NAC and SRP were able to bind simultaneously to a single RNC, we used two independent approaches. A yeast strain in which Egd2 was replaced by FLAG-Egd2 was used to isolate specifically those RNCs that were occupied by NAC (termed NAC·RNCs hereafter; see Supplemental Figures S3C and S4A). In this setup, nontranslating ribosomes were coisolated with NAC·RNCs because NAC efficiently forms NAC·ribosome complexes (Raue et al., 2007). Of importance, the bulk of RNCs generated in the in vitro translation system carried nascent chains of uniform length, indicating that disomes or polysomes were absent or represented only minor species (Supplemental Figure S4, B and C, and see Materials and Methods). Four different nascent chains were tested for their ability to recruit SRP to NAC·RNCs: Dap2-120 and Pho8-120, exposing the SA segment far outside of the ribosomal tunnel, and Dap2-85 and Dap2Δ-62, exposing the SA segment in close proximity to the tunnel exit (Supplemental Figure S2B). SRP was bound to NAC·RNCs carrying Dap2-120 or Pho8-120; however, it was not bound to NAC·RNCs carrying Dap2-85 or Dap2Δ-62 (Figure 4A). When the length of nascent Dap2 was successively increased from 85 to 120-residues, SRP was bound to NAC·RNCs starting at a nascent chain length of 100 residues (Figure 4B). Thus SRP and NAC were able to bind to a single RNC only if the SA segment had departed significantly from the tunnel exit.
FIGURE 4:
NAC and SRP can bind to the same RNC molecule if the SA segment is positioned far outside of the ribosomal tunnel. (A) RNCs carrying untagged (FLAG –) nascent chains were generated in a FLAG-Egd2 translation extract. A mock translation reaction lacking mRNA (– mRNA) was performed in the FLAG-Egd2 translation extract as a control. FLAG-NAC pull downs (Supplemental Figure S3) were then performed to isolate ribosomes and RNCs bound to FLAG-NAC. The isolated material was analyzed via immunoblots using αSrp54, αEgd2 to detect FLAG-NAC (Egd2-FLAG), and αRpl4 to detect ribosomes and RNCs. (B) As in A, using FLAG-NAC RNCs carrying nascent Dap2 of increasing length as indicated (Supplemental Figure S2). A translation reaction lacking mRNA (– mRNA) and wild-type RNCs carrying Dap2-120 were used as controls. (C) RNCs carrying FLAG-tagged (FLAG +) or untagged (FLAG –) nascent chains were generated in a translation extract derived from wild type or the Δegd1Δegd2 strain. If indicated, after cycloheximide addition, purified NAC (NAC +) was added to a final concentration of 1 μM and was allowed to bind to ribosomes and RNCs for 10 min at 20°C. Subsequently, FLAG–nascent chain pull downs (Supplemental Figure S3) were performed as described in Figure 1. Relative occupation of RNCs with SRP (SRP on RNCs) was calculated as described in Materials and Methods. The occupation of Dap2-120-RNCs generated in a wild-type translation extract was set to 1.
In a second approach, we used the observation that the bulk of RNCs exposing a SA segment was occupied by SRP if translations were performed in a NAC-free environment (Figures 2, A and B, and 3E). On the basis of this observation, we generated SRP·RNCs carrying nascent Dap2-85, Dap2Δ-62, or Dap2-120 in a Δegd1Δegd2 extract. Translation was then inhibited by the addition of cycloheximide, and finally purified NAC (Supplemental Figure S5A) was added. When RNCs were subsequently isolated, NAC did not bind to Dap2-85-RNCs or Dap2Δ-62-RNCs; however, NAC did bind to Dap2-120-RNCs (Figure 4C). The result is compatible with a model in which SRP prevents the binding of NAC to RNCs, exposing the SA segment close to the tunnel exit, but does not prevent binding of NAC if the SA has departed from the tunnel exit.
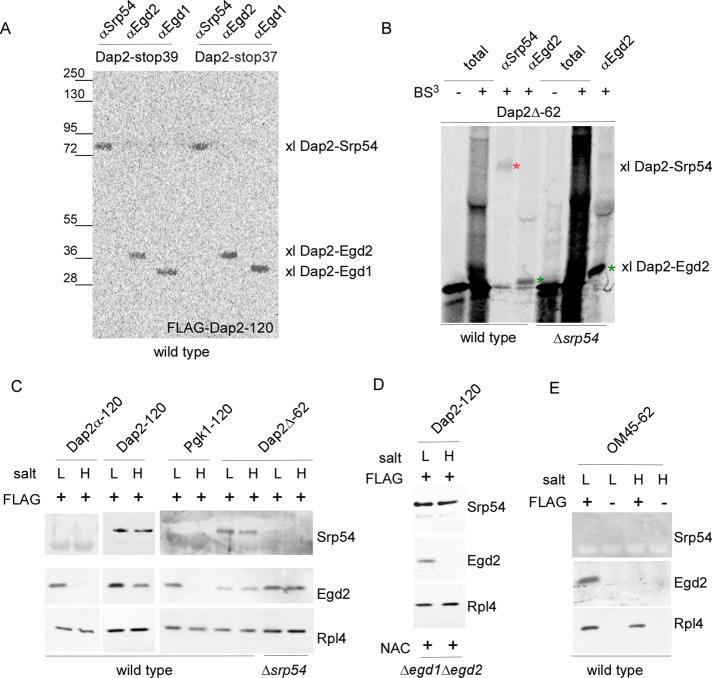
NAC interacts directly and in a salt-resistant manner with nascent SA segments
Direct contact of NAC with signal sequences was tested via two different approaches: site-specific cross-linking and chemical cross-linking (Supplemental Figure S3). When a photo probe was incorporated into the SA segment of Dap2-120 (Supplemental Figure S2), both subunits of NAC (Egd1 and Egd2) formed a cross-link (Figure 5A). Chemical cross-linking was performed by using the Dap2Δ-62 nascent chain, which exposes only the SA segment outside of the ribosomal tunnel (Supplemental Figure S2B). In a wild-type extract Srp54, as well as Egd2, formed a cross-link with Dap2Δ-62; however, both cross-links were relatively weak (Figure 5B). Because NAC and SRP can bind to Dap2Δ-62-RNCs efficiently (Figure 2C), this is likely due to improper positioning of the only two exposed amino groups within nascent Dap2Δ-62 (Supplemental Figure S2A). Of importance, the cross-link between Dap2Δ-62 and Egd2 was significantly enhanced when SRP was absent (Figure 5B).
FIGURE 5:
NAC binds to the SA segment directly and in a salt-resistant manner. (A) Site-specific cross-linking to 35S-labeled nascent chains (Supplemental Figure S3D). The photo probe was positioned within the SA segment at residue 39 (Dap2-stop39) or 37 (Dap2-stop37) of Dap2-120 (Supplemental Figure S2). Cross-link products were isolated via immunoprecipitation under denaturing conditions using αSrp54, αEgd2, or αEgd1 as indicated and were subsequently visualized via autoradiography. xl Dap2-Srp54, cross-link product between the nascent chain and Srp54; xl Dap2-Egd2, cross-link product between the nascent chain and Egd2; and xl Dap2-Egd1, cross-link product between the nascent chain and Egd1. (B) Chemical cross-linking (Supplemental Figure S3) of RNCs carrying radiolabeled Dap2Δ-62. RNCs were generated in either a wild-type or a Δsrp54 translation extract as indicated. Analysis of the cross-link products was as described in A using αSrp54 and αEgd2. The total represents 33% of the amount added to immunoprecipitation reactions. Samples of cross-link experiments were analyzed via autoradiography. The cross-link between Dap2Δ-62 and Srp54 is labeled with a red asterisk; the cross-link between Dap2Δ-62 and Egd2 is labeled with a green asterisk. (C) RNCs carrying FLAG-tagged (FLAG +) nascent chains as indicated were generated in a wild-type or Δsrp54 translation extract. FLAG–nascent chain pull downs were performed with RNCs isolated under low-salt (L) or high-salt (H) conditions (see Materials and Methods). The isolated material was then analyzed for the presence of SRP (Srp54), NAC (Egd2), and RNCs (Rpl4) via immunoblotting. (D) Dap2-120-RNCs were generated in a translation extract derived from the Δegd1Δegd2 strain. After inhibition of translation, purified NAC was added as described in Figure 4. Subsequently, RNCs were isolated under low-salt (L) or high-salt (H) conditions, and the isolated material was analyzed as in C. (E) RNCs carrying FLAG-tagged (FLAG +) or untagged (FLAG –) 62-residue nascent OM45 (Supplemental Figure S2) were generated in wild-type translation extract. FLAG–nascent chain pull downs were preformed with low-salt-treated (L) or high-salt-treated (H) RNCs and were analyzed as described.
To determine whether the cross-links between NAC and the SA segment were merely due to close proximity or instead reflected a functional interaction, we tested the salt resistance of NAC binding. NAC was efficiently released from Dap2α-120-RNCs or Pgk1-120-RNCs; however, NAC was only partly released from Dap2-120-RNCs by high-salt treatment (Figure 5C). This suggested the presence of two distinct pools of NAC·RNCs carrying 120-residue Dap2—one in which NAC was bound to a hydrophilic stretch of Dap2 in a salt-sensitive manner, and one in which NAC was bound to the SA segment of Dap2 in a salt-resistant manner. To test for this possibility, we generated Dap2-120-RNCs predominantly occupied by SRP in a NAC-free translation extract derived from the Δegd1Δegd2 strain. Purified NAC was then added, and trimeric NAC·RNC·SRP complexes were allowed to form (see Figure 4C). In this case, NAC binding remained fully salt sensitive, as expected if in the preformed RNC·SRP complexes SRP was bound to the SA segment, whereas NAC, which entered the complex later, was bound to a hydrophilic stretch of the nascent chain more adjacent to the tunnel exit (Figure 5D). In support of the chemical cross-linking data (Figure 5B), NAC bound to Dap2Δ-62-RNCs was fully salt resistant, suggesting that in this complex the bulk of NAC was in contact with the SA segment (Figure 5C). Because NAC and SRP binding to Dap2Δ-62-RNCs were mutually exclusive (Figure 4A), salt resistance of NAC in this case was independent of the presence or absence of SRP (Figure 5C). Of importance, NAC did not unspecifically bind to hydrophobic nascent chain segments in a salt-resistant manner. For example, NAC was effectively released from OM45-62-RNCs upon high-salt treatment (Figure 5E). The combined data indicate that NAC was able to establish salt-resistant interactions with a nascent SA segment, which was not bound to SRP. When SRP was bound to the SA segment, NAC was able to establish salt-sensitive interactions with segments of the nascent chain located C-terminally of the SA.
NAC is not released upon docking of NAC·RNC·SRP to the ER membrane
We next tested whether NAC affected targeting of RNCs to the ER membrane. To that end, we generated RNCs in a Δegd1Δegd2 translation extract. NAC was added after inhibition of translation, and RNCs were subsequently allowed to bind to translocation-competent microsomes (Supplemental Figures S3E and S6A). Pgk1-120-RNCs generated in the absence of NAC were not targeted to the ER membrane whether or not NAC was allowed to bind after inhibition of translation (Figure 6A). Thus NAC was not required to prevent binding of a signalless RNC to the ER membrane. When RNCs carrying nascent Dap2-120 were generated in the absence of NAC, the resulting RNC·SRP complexes were efficiently targeted to the ER membrane. Targeting efficiency of Dap2-120-RNCs was not affected when NAC was added after translation to allow the formation of NAC·RNC·SRP complexes (Figure 6A; also see Figure 4C). In this experiment NAC was bound to nascent chain segments distal of the SA segment (Figure 5D). The fate of NAC and SRP upon docking of RNCs to the ER membrane was analyzed via immunoblotting. Note that microsomes prepared from the Δegd1Δegd2 strain were not only free of NAC, but also were essentially free of ribosomes and SRP (Supplemental Figure S6B). After incubation of RNC·SRP or NAC·RNC·SRP complexes with microsomes, ribosomes, as well as SRP, were detected in the reisolated membrane fraction (Figure 6B). When present, NAC was also bound to the reisolated membranes (Figure 6B). As a control, ribosomes, SRP, and NAC were not detected in reisolated membranes after incubation with Pgk1-120-RNCs (Figure 6B). The data indicate that NAC was not released from NAC·RNC·SRP complexes upon initial binding to the ER membrane.
FIGURE 6:
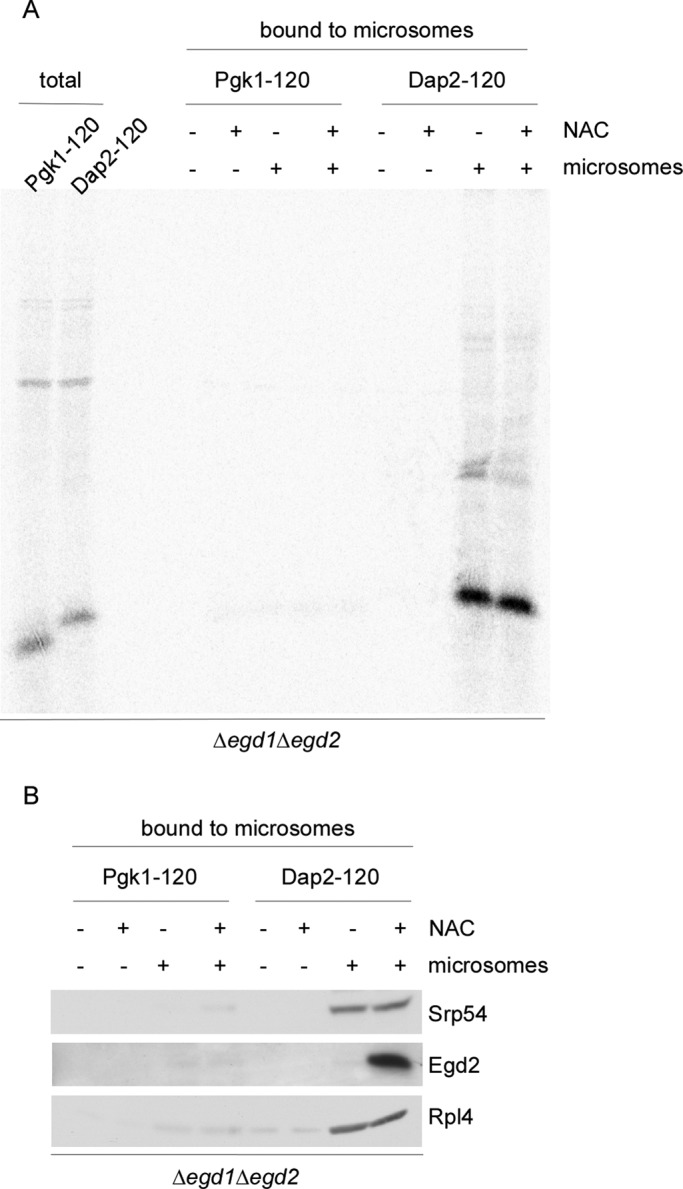
NAC·RNC·SRP complexes are targeted to ER membranes. RNCs were generated in Δegd1Δegd2 translation extract. Wild-type (+ NAC) or Δegd1Δegd2 (– NAC) extract was added after inhibition of translation). (A) 35S-Labeled RNCs complemented with either wild-type translation extract (+ NAC) or ∆egd1∆egd2 translation extract (– NAC) were allowed to bind to high-salt-washed microsomal membranes (+ microsomes). After reisolation, microsomal membranes and bound RNCs were run on Tris-tricine gels and were analyzed via autoradiography (Supplemental Figure S3E). The total represents 10% of Pgk1-120 or Dap2-120-RNCs added to high-salt-washed microsomes. (B) RNCs generated as in A but in the absence of 35S were allowed to bind to microsomal membranes. After reisolation of microsomal membranes, aliquots were analyzed via immunoblotting using the indicated antibodies.
SRP replaces NAC when a SA segment is exposed outside of the ribosomal tunnel
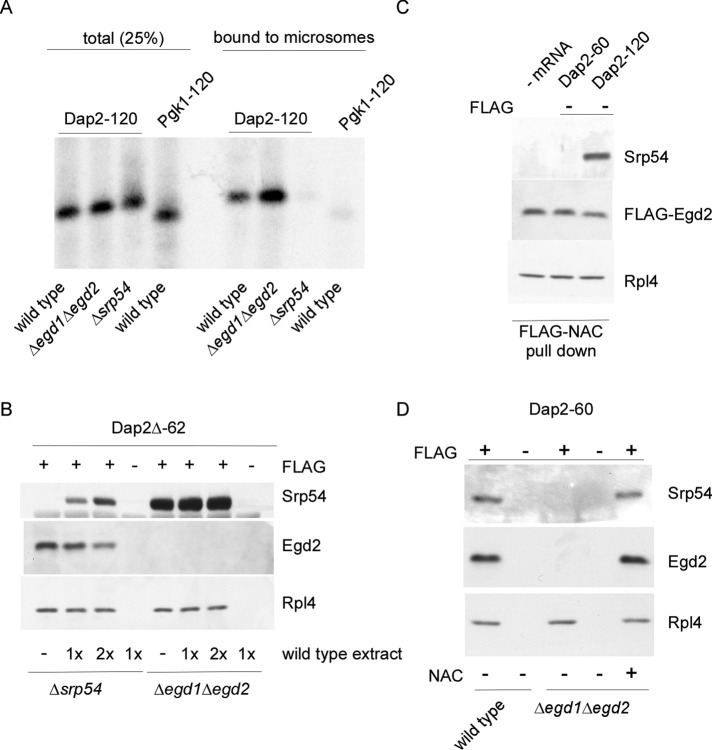
We next compared targeting of RNCs exposing a SA segment bound to either SRP or NAC. To that end, we generated radiolabeled Dap2-120-RNCs in Δsrp54 (SA segment bound to NAC) or Δegd1Δegd2 (SA segment bound to SRP) translation extract. As expected, Dap2-120-RNCs were efficiently targeted to microsomal membranes when bound to SRP. Dap2-120-RNCs were not targeted to microsomal membranes when bound to NAC (Figure 7A). The result confirms that SRP is essential for efficient cotranslational targeting of Dap2 (Ng et al., 1996). Consistent with the enhanced occupation of Dap2-120-RNCs with SRP in the absence of NAC (Figure 2, A and B), the binding efficiency of Dap2-120-RNCs to microsomes was enhanced when NAC was absent (Figure 7A).
FIGURE 7:
NAC is required for the recruitment of SRP when the SA segment is localized inside the ribosomal tunnel. (A) 35S-Labeled RNCs generated in wild-type, Δegd1Δegd2, or Δsrp54 translation extract were allowed to bind to high-salt-washed microsomal membranes. After reisolation, microsomal membranes and bound RNCs were run on Tris-tricine gels and were analyzed via autoradiography. The total represents 25% of Pgk1-120 or Dap2-120-RNCs added to high-salt-washed microsomes. (B) FLAG–nascent chain pull downs with RNCs carrying Dap2∆-62 (Supplemental Figure S2). RNCs were generated in Δegd1Δegd2 or ∆srp54 translation extract to which wild-type extract containing NAC and SRP was added after inhibition of translation. The amount of wild-type extract corresponded to 1× or 2× the volume of the Δegd1Δegd2 or a ∆srp54 translation extract used in the translation reaction. FLAG–nascent chain pull downs (Supplemental Figure S3B) were then performed and were analyzed as described in Figure 1. (C) RNCs carrying untagged Dap2-60 or Dap2-120 were generated in a FLAG-Egd2 translation extract. Subsequently FLAG-NAC pull downs (Supplemental Figure S3C) were performed and were analyzed as in Figure 4. (D) RNCs carrying FLAG-tagged (FLAG +) or untagged (FLAG –) Dap2-60 were generated in wild-type or Δegd1Δegd2 translation extract. As indicated, the Δegd1Δegd2 translation extract was complemented with a final concentration of 1 μM purified NAC (+ NAC) before the translation reaction.
The foregoing data indicated that efficient targeting of RNCs to the ER membrane required the release of NAC from the SA segment. To test whether NAC was irreversibly replaced by SRP, we produced Dap2Δ-62-RNCs carrying either NAC (generated in Δsrp54 extract) or SRP (generated in Δegd1Δegd2 extract; Figure 7B). After inhibition of translation, reactions were complemented with NAC or SRP as indicated. In this setup, NAC was unable to bind to Dap2Δ-62-RNCs generated in Δegd1Δegd2 extract, whereas SRP was able to bind to Dap2Δ-62-RNCs generated in Δsrp54 extract (Figure 7B). Binding of SRP was accompanied by release of NAC, confirming that Dap2Δ-62-RNCs were unable to bind NAC and SRP simultaneously (Figure 7B). Thus, even though NAC was directly and salt-resistantly bound to the SA segment, SRP was able to get access to and replace NAC. This process was unidirectional because NAC was unable to dispose SRP.
NAC is required for the early recruitment of SRP to RNCs
The SA segment of 60-residue Dap2 is localized inside the ribosomal tunnel (Supplemental Figure S2) and therefore cannot directly interact with SRP (Berndt et al., 2009). However, the affinity of SRP to Dap2-60-RNCs compared with RNCs that do not contain a SA segment inside the tunnel is significantly increased (Berndt et al., 2009). We now tested whether NAC and SRP were able to simultaneously bind to Dap2-60-RNCs via FLAG-NAC pull downs (Supplemental Figure S3C). In contrast to Dap2-120-RNCs, Dap2-60-RNCs did not form NAC·RNC·SRP complexes (Figure 7C). Thus, even though there was no competition for direct binding to the SA segment, simply because the SA was not accessible, NAC and SRP excluded each other from Dap2-60-RNCs. Binding of SRP to Dap2-60-RNCs was next tested in the absence of NAC. Unexpectedly, Dap2-60-RNCs generated in a Δegd1Δegd2 translation extract failed to recruit SRP (Figure 7D). The effect was indeed due to the absence of NAC, because purified NAC added to the Δegd1Δegd2 extract before the translation reaction fully restored SRP binding to Dap2-60-RNCs (Figure 7D). Thus NAC and SRP did not simultaneously bind to RNCs carrying 60-residue Dap2. However, SRP was recruited to RNCs containing the SA segment inside the ribosomal tunnel only if NAC was present.
DISCUSSION
Most studies addressing the interplay between NAC and SRP used heterologous combinations of wheat germ RNCs, canine SRP, and bovine NAC (Wiedmann et al., 1994; Powers and Walter, 1996; Flanagan et al., 2003). Affinities between SRP, NAC, and ribosomes or RNCs were determined using high-salt-washed RNCs, purified SRP, and purified NAC (Wiedmann et al., 1994; Powers and Walter, 1996; Flanagan et al., 2003). In this setup, the absence of NAC caused faulty interaction of SRP with ribosomes and RNCs carrying a variety of nonsubstrate nascent chains (see Introduction, NAC-SRP model). In contrast, the affinity of SRP for NAC-free, signalless RNCs tested in the course of this study was below the detection limit. A simple explanation for the apparent discrepancy is that the affinity between yeast RNCs and SRP differs from that of the higher eukaryotic components. In addition, SRP binding is likely affected not only by NAC, but also by other ribosome-bound protein biogenesis factors (RPBs), which can be removed by high-salt treatment (Rospert et al., 2005; Raue et al., 2007). Of note, the binding sites for RPBs close to the tunnel exit partly overlap. For example, NAC and the ribosome-associated complex bind close to Rpl31, and SRP and the N-acetyltransferase NatA bind close to Rpl25 (Pool et al., 2002; Peisker et al., 2008; Polevoda et al., 2008; Pech et al., 2010), and here we show that NAC and SRP affect each other's binding. Of interest, recent in vivo data also revealed that NAC does not in general prevent SRP binding to RNCs carrying cytosolic proteins. Instead, only a specific subset of highly abundant RNCs was affected (Del Alamo et al., 2011). What exactly makes a signalless RNC a substrate for SRP in the absence of NAC is not understood. An intriguing possibility is that the recently discovered effect of N-terminal nascent chain processing on ER targeting might be connected also to NAC's function (Forte et al., 2011). Of note, N-terminal acetyltransferases (Van Damme et al., 2011), as well as methionine aminopeptidases (Giglione et al., 2009), bind close to the tunnel exit of their substrate RNCs.
Signalless RNCs were not targeted to ER membranes in the absence of NAC in the yeast in vitro system (see Introduction, NAC-translocon model). This observation is consistent with previous data (Neuhof et al., 1998; Raden and Gilmore, 1998). However, in the absence of SRP, NAC prevented targeting of RNCs, exposing a signal sequence. Consistently, NAC was found to interact with a subset of RNCs exposing signal sequences in vivo (Del Alamo et al., 2011). Our data reveal that NAC is able to affect targeting to the translocon because NAC can directly interact with signal sequences.
To understand the interplay between NAC and SRP, it is important to know how the two complexes bind to RNCs relative to each other in time and space. Although it is widely agreed that Rpl25/Rpl35 provides the major binding site for SRP (Pool et al., 2002; Halic et al., 2004), there are contradictory reports about the ribosomal binding site for NAC (Wegrzyn et al., 2006; Pech et al., 2010). We revisited the question and found, in agreement with the work of Beatrix and coworkers (Pech et al., 2010), that Rpl31 contacts the Egd1 subunit of NAC and is required for efficient ribosome association. Distinct ribosomal binding sites for NAC and SRP are consistent with the formation of NAC·RNC·SRP complexes. However, NAC and SRP affect each other with respect to ribosome binding (Figure 8). This is likely due to the fact that NAC and SRP not only contact ribosomes via Rpl31 or Rpl25/Rpl35, respectively, but also make additional contacts, resulting in sterical constraints (Pool et al., 2002; Halic et al., 2004; Pech et al., 2010). Only when NAC got hold of a nascent chain segment C-terminal of a SA did the affinity of NAC for RNC·SRP complexes become sufficiently high to allow for the formation of trimeric NAC·RNC·SRP complexes. In this model the nascent chain regulates the interaction of NAC with RNC·SRP complexes: if SRP binding and subsequent translocation to the ER occur in a timely manner, NAC will not rebind to RNCs. However, if translation continues because SRP binding or the subsequent targeting process is delayed, NAC will rebind (Figure 8A).
FIGURE 8:
Model of the interactions between NAC and SRP on ribosomes involved in the translation of a protein destined for cotranslational translocation to the ER. (A) Sequence of events in the presence of NAC and SRP. 1) NAC binds to empty ribosomes and 2) remains bound when translation initiates. 3) NAC·RNC complexes remain stable when a SA segment emerges inside the ribosomal tunnel. The SA segment may induce conformational changes in the SRP binding site. 4) SRP is recruited, and NAC is concomitantly released. At this stage trimeric complexes are unstable. This is likely due to sterical hindrance between domains of NAC and SRP, which are not directly involved in ribosome binding. 5, 6) When the SA segment emerges SRP binds to it directly and in a salt-resistant manner. At this stage NAC and SRP cannot bind simultaneously to an RNC. 7, 8) On further elongation of the nascent chain, segments C-terminal of the SA exit the tunnel. Now NAC can rebind, and trimeric NAC·RNC·SRP complexes form, which can interact with the translocation machinery in the ER membrane. (B) Sequence of events when SRP is absent or SRP binding is delayed. 1–3) as in A. 4,5) Because SRP is absent, NAC·RNC complexes remain stable, and NAC establishes salt-resistant interactions with the nascent SA segment. If added to the system at stage 4, SRP displaces NAC. This order of events allows NAC to shield a SA segment until SRP is available. (C) Sequence of events when NAC is absent. 1, 2) SRP binds poorly to nontranslating ribosomes or signalless RNCs. 3) SRP is not recruited to RNCs containing a SA segment inside the ribosomal tunnel. 4, 5) As soon as the SA segment emerges outside of the tunnel, SRP binds to the RNC and the SA segment in a salt-resistant manner. If added to the system at stage 4, NAC is unable to displace SRP. For details see Results and Discussion. The ribosome is shown in gray, NAC is shown in blue, and SRP is shown in green. The nascent chain is depicted in light blue, and the SA segment is indicated in black. The red arrow points to the SRP-binding site. The major ribosomal binding sites of SRP (Rpl25) and NAC (Rpl31) are indicated.
The existence of NAC·RNC·SRP complexes was previously suggested. It remained unclear, however, whether NAC and SRP were bound to a single ribosome or to different ribosomes contained in the same polysome (Del Alamo et al., 2011). With respect to the time window required for rebinding of NAC, it is of interest to recall that the rate of Dap2 translocation is slow relative to the rate of Dap2 synthesis. As a consequence, membrane integration of Dap2 is completed only at a nascent chain length of ∼200 amino acids (Cheng and Gilmore, 2006). We find that NAC is able to rebind to RNCs carrying Dap2 of 100 amino acids, providing a time window sufficient for the formation of NAC·RNC·SRP complexes in vivo.
The previously unrecognized complexity of the interplay between NAC and SRP is due to at least three previously undiscovered, mechanistically important properties of NAC.
First, NAC interacted with any nascent chain explicitly, including signal sequences, such as SA segments. If not replaced by SRP, NAC remained bound to a SA segment of a growing nascent chain. It is likely that in this situation the SA segment was locked at the so-called signal sequence binding site close to Rpl25/Rpl35, which upon elongation of the nascent chain results in a loop structure (Ullers et al., 2003; Halic et al., 2006a; Figure 8B).
Second, when NAC was in contact with a nascent SA segment, the interaction with the RNC was salt resistant. Because salt resistance was not observed with any other nascent chain, this suggests a specific mode of interaction between NAC and bona fide SRP substrates. Although structural information about the NAC subunits and domains has been emerging (Spreter et al., 2005; Liu et al., 2010; Wang et al., 2010), it has remained enigmatic how exactly NAC interacts with nascent chains. Our finding that both NAC subunits can directly contact the SA segment suggests that NAC binds via a rather extended surface spanning both subunits.
Third, NAC was required for the early recruitment of SRP to RNCs. In a previous study we found that formation of the RNC·SRP complex is induced early, when the SA segment is still inside the tunnel (Berndt et al., 2009). A similar finding was recently made with respect to the targeting of tail-anchored proteins, which recruit the Bat3 complex from within the ribosomal tunnel (Mariappan et al., 2010; Saraogi and Shan, 2011). We now show that NAC is required specifically for the early recruitment of SRP to RNCs (Figure 8C), whereas the later, high-affinity recruitment of SRP to RNCs exposing signal sequences was unaffected by NAC. This suggests that NAC is involved in transmitting information about the nature of the emerging nascent chain from within the tunnel to the ribosomal SRP-binding site. The exact mechanism for this effect is unknown. It is possible that NAC alters the conformational dynamics of translating ribosomes (Petrov et al., 2011). This may enhance the low-affinity binding of SRP to ribosomes that contain a hydrophobic signal sequence inside the ribosomal tunnel.
Based on these properties, NAC may affect not only the initial steps of cotranslational targeting, but also the subsequent translocation process. Transmembrane segments inside the exit tunnel alter the environment of Rpl17 such that the ribosomal protein now contacts a component of the translocon (Pool, 2009). Because Egd2 contacts Rpl17 (Pech et al., 2010), NAC might contribute to the nascent chain–induced changes of the Rpl17 environment. Of interest, it was recently shown that the two homologous translocons of yeast, Sec61 and Ssh1, differ in their preference for translocation substrates (Spiller and Stirling, 2011). It is possible that NAC·RNC·SRP complexes have a preference for one or the other type of translocon. Our findings revealed that NAC was not released from NAC·RNC·SRP complexes upon binding to the ER translocation machinery. This is consistent with the fact that RNCs bind to the translocon via the ribosomal protein Rpl25 (Becker et al., 2009), whereas NAC binds to RNCs via Rpl31 (Pech et al., 2010; and this study). This might be important because cytosolic exposure of transmembrane domains and lumenal domains negatively affects the fidelity of membrane protein integration if cells do not prevent unwanted interactions with components in the cytosol (Cheng and Gilmore, 2006). Such a role is also supported by the fact that NAC associates with RNCs carrying nascent SRP substrates in vivo (Del Alamo et al., 2011). NAC would be ideally suited to participate in the stabilization of translocation intermediates of transmembrane proteins on the cytosolic side of the ER membrane.
MATERIALS AND METHODS
Plasmids
Plasmids encoding Dap2 (pSPUTK-Dap2), Dap2α (pSPUTK-Dap2α), Pgk1 (pSPUTK-Pgk1), and the FLAG-tagged versions pSPUTK-FLAG-Dap2, pSPUTK-FLAG-Dap2α, and pSPUTK-FLAG-Pgk1, which contain the DYKDDDDK peptide behind the initiator methionine, are based on pSPUTK (Stratagene, Santa Clara, CA) and have been described (Raue et al., 2007; Berndt et al., 2009). pSPUTK-Dap2∆ is a version of Dap2 in which the N-terminal 23 amino acids were deleted. Replacement of the N-terminal 23 amino acids with a FLAG tag yielded plasmid pSPUTK-FLAG-Dap2∆. The gene encoding Pho8 (yeast repressible alkaline phosphatase) was amplified from genomic DNA and was cloned into EcoRI/PstI site of pSP65 (Promega, Madison, WI). A FLAG-tagged version of Pho8 was constructed by placing a sequence encoding DYKDDDDK behind the ATG start codon of Pho8. The resulting plasmids are pSP65-Pho8 and pSP65-FLAG-Pho8. The gene encoding OM45 (yeast mitochondrial outer membrane protein OM45) was amplified from genomic DNA and was cloned into PstI/BamHI site of pSP64 (Promega). A FLAG-tagged version of OM45 was constructed by placing a sequence encoding DYKDDDDK behind the ATG start codon of OM45. The resulting plasmids are pSP64-OM45 and pSP64-FLAG-OM45. For photo–cross-linking experiments, CTG coding for Leu at position 37 or TGG coding for Trp at position 39 of Dap2 was replaced with the TAG amber stop codon via PCR. The fragment containing the amber mutation was exchanged for the wild-type fragment using the BsrGI and BstZ17I sites in pSPUTK-FLAG-Dap2. The resulting plasmids are termed pSPUTK-FLAG-Dap2-stop37 and pSPUTK-FLAG-Dap2-stop39. For the generation of the FLAG-tagged version of Egd2 the FLAG tag was fused to the N-terminus of Egd2 by inserting a sequence encoding for the DYKDDDDK peptide behind the ATG start codon of EGD2 plus 400 base pairs upstream and downstream of the orf. FLAG-Egd2 was cloned into the pYCPlac111 low-copy plasmid (LEU2 marker; Gietz and Sugino, 1988); the resulting plasmid is termed pYCPlac111-FLAG-Egd2. A C-terminal His tag was fused to Rpl31a by inserting a sequence encoding for eight consecutive histidine residues in front of the TAA stop codon of RPL31a plus 600 base pairs upstream and 300 base pairs downstream of the orf. Rpl31-His8 was cloned into the pYCPlac33 low-copy plasmid (URA3 marker; Gietz and Sugino, 1988); the resulting plasmid is termed pYCPlac33-Rpl31-His8.
Yeast strains
MH272-3f a/α (ura3/ura3, leu2/leu2, his3/his3, trp1/trp1, ade2/ade2; Gautschi et al., 2002) was the parental strain for the yeast mutant strains used in this study. The ∆rpl31a∆rpl31b strain carries the rpl31a::TRP1 and rpl31b::ADE2 mutation and was previously described (Peisker et al., 2008). The Δegd1Δegd2 strain (YRG16) carries the egd1::URA3 and egd2::ADE2 mutation (George et al., 2002). The Δsrp54 strain was generated by replacing the SRP54 orf with the kanMX module according to Wach et al. (1994) in the diploid parental strain. A haploid srp54::kanMX strain was isolated after sporulation of the diploid, followed by dissection and tetrad analysis. Only few of the spores containing the srp54::kanMX disruption were recovered after dissection. The expression level of SRP in the Δegd1Δegd2 strain was similar to the wild-type expression level. Conversely, in the Δsrp54 strain NAC was expressed at normal levels (Supplemental Figure S5B). Δegd1Δegd2 expressing FLAG-Egd2 and Egd1 from plasmids pYCPlac111-FLAG-Egd2 and pYCPlac22-Egd1 was termed the FLAG-Egd2 strain. ∆rpl31a∆rpl31b expressing the pYCPlac33-Rpl31-His8 was termed the His-Rpl31 strain. NAC was purified from the protease-deficient C13-ABYS-86 strain (Heinemeyer et al., 1991; Fünfschilling and Rospert, 1999).
Generation of RNCs
Transcription was performed as described using SP6 polymerase (Garcia et al., 1991). DNA templates of various lengths were generated by PCR using the pSPUTK or pSP65/pSP64 plasmids containing tagged or untagged genes as a template (Raue et al., 2007). Nascent chains resulting from translation of the various transcripts are shown in Supplemental Figure S2. The length of the nascent chains is indicated as the number that follows the hyphen (e.g., Dap2-120 is a nascent chain that consists of the N-terminal 120 amino acids of Dap2). The FLAG tag was not included in calculating the nascent chain length. All transcripts used to generate RNCs lacked terminal stop codons. As a result, translation products remained bound to ribosomes as peptidyl-tRNAs generating stable RNCs. When translation was performed in the presence of 35S-methionine (PerkinElmer, Waltham, MA), nascent chains of uniform length were detected in autoradiographs (Supplemental Figure S4, A–C) This indicates that in vitro–generated RNCs predominantly consisted of monosomes (Hüttenhofer and Noller, 1994). Yeast translation extract was prepared as previously described (Garcia et al., 1991) from wild type (MH272-3f), strains ∆srp54, Δegd1Δegd2, ∆rpl31a∆rpl31b, or FLAG-Egd2. Translation reactions for site-specific and chemical cross-linking were performed in the presence of 35S-methionine, and translation reactions for FLAG–nascent chain pull downs and FLAG-NAC pull downs were performed in the absence of labeled methionine as described (Garcia et al., 1991; Fünfschilling and Rospert, 1999; Berndt et al., 2009). Where indicated, purified NAC (Supplemental Figure S5A) was added to the translation reactions at a final concentration of 1 μM. As indicated in the figure legends, NAC was added either before the addition of mRNA or after translation was terminated by addition of cycloheximide. Wild-type translation extract was used to replenish Δegd1Δegd2 or a ∆srp54 reactions with NAC or SRP side by side. The volume of wild-type extract was adjusted with respect to the volume of Δegd1Δegd2 or a ∆srp54 extract used in the translation reaction. After addition of wild-type extract, samples were incubated at 20°C for 10 min, and RNCs were isolated via centrifugation at 250,000 × g. Ribosomal pellets were resuspended in 200 μl of immunoprecipitation buffer, and FLAG–nascent chain pull downs were performed.
Estimation of the concentration of ribosomes, RNCs, NAC, and SRP in translation reactions
The concentration of the components of interest can be estimated as follows: 1) a haploid yeast cell has a volume of ∼70 fl (Sherman, 1991); 2) a haploid yeast cell contains 300,000 ribosomes, 400,000 molecules of NAC, and 8000 molecules of SRP (Raue et al., 2007); 3) due to generation of translation extract, cytosolic proteins are diluted ∼10-fold compared with the cytosol of living cells; and 4) a fraction of ∼1.5% of total ribosomes in a translation extract forms RNCs with the nascent chain encoded by the mRNA added to the reaction (Raue et al., 2007). Based on these assumptions, a wild-type translation reaction contains ∼750 nM empty ribosomes, 11.5 nM RNCs, 20 nM SRP, and 1000 nM NAC. Because NAC binds to empty ribosomes, whereas SRP is mainly free in solution (Raue et al., 2007), the pool of unbound SRP is ∼20 nM, whereas that of unbound NAC is ∼250 nM.
FLAG–nascent chain pull-down and FLAG-NAC pull-down reactions
FLAG–nascent chain or FLAG-NAC pull-down reactions were performed as outlined in Supplemental Figure S3. The pull-down experiments were analyzed via immunoblotting;35S-methionine was omitted from the reactions. For a typical experiment, 80-μl translation reactions were performed at 20°C for 50 min and were terminated by the addition of cycloheximide to a final concentration of 200 μg/ml. Translation reactions were then added to 40 μl of prewashed ANTI-FLAG M2 affinity gel (α-FLAG-beads; Sigma-Aldrich, St. Louis, MO) resuspended in 500 μl of immunoprecipitation buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]–KOH, pH 7.4, 150 mM KAc, 2 mM MgAc2, 50 μg/ml trypsin inhibitor, 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor mix [1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin, 0.25 μg/ml elastinal, 5 μg/ml pepstatin A]), and the reactions were incubated for 1 h at 4°C on a shaker. Beads were separated from the supernatant by centrifugation and were washed twice with 500 μl of ice-cold immunoprecipitation buffer. NAC and SRP were stably bound and were not lost during the washes under these conditions (Raue et al., 2007). For experiments in which RNCs treated with either low salt or high salt were compared, 80-μl translation reactions were loaded onto either a 120 μl of low-salt sucrose cushion (25% sucrose, 20 mM HEPES-KOH, pH 7.4, 2 mM MgAc2, 120 mM KAc, 2 mM dithiothreitol [DTT], 1 mM PMSF, protease inhibitor mix) or high-salt sucrose cushion (25% sucrose, 20 mM HEPES-KOH, pH 7.4, 2 mM MgAc2, 800 mM KAc, 2 mM DTT, 1 mM PMSF, protease inhibitor mix). After centrifugation for 22 min at 400,000 × g at 4°C, ribosomal pellets were resuspended in immunoprecipitation buffer and were applied to FLAG-NAC pull-down reactions. Proteins bound to α-FLAG beads were released by incubation in SDS–PAGE sample buffer for 10 min at 95°C, and aliquots were run on 10% Tris-tricine gels (Schägger and von Jagow, 1987), transferred to nitrocellulose membrane, and analyzed by immunoblotting. Nontagged versions of nascent chains were analyzed in parallel with the FLAG-tagged nascent chains to determine background binding to the α-FLAG beads. The size of Srp54 (59.6 kDa), Egd2 (18.7 kDa), and Rpl4 (39.1 kDa) allowed for the analysis of SRP, NAC, and ribosomes on a single immunoblot. Similar amounts of RNCs (adjusted via the intensity of the Rpl4 band) were loaded to allow for a direct comparison of bound NAC (Egd2) and SRP (Srp54). His6-Rps9 (22.4 kDa) was used when RNCs were quantified using purified standard proteins. Purification of standard proteins (His6-Srp54 for the quantification of SRP, His6-Rps9a for the quantification of ribosomes) was done as described (Raue et al., 2007). Calibration curves for each standard protein in the linear range were used to quantify the protein content (pmol) in the pull-down reactions. Resulting values for Rps9a were divided by the factor 0.7 (Raue et al., 2007). Occupation of ribosomes with NAC or SRP in percent was calculated as described (Raue et al., 2007). Relative changes in the occupation of RNCs (relative occupation) derived from wild-type or mutant yeast strains were determined as follows. Similar amounts of isolated RNCs (Rpl4) from pull-down reactions were analyzed on a single immunoblot. The intensities of the bands corresponding to Rpl4/Rps9, Srp54, or Egd2 were determined densitometrically. Band intensities Srp54 (SRP) or Egd2 (NAC) were normalized to the band intensities of Rpl4 or Rps9 (ribosomes), respectively. Resulting values (NAC per ribosome or SRP per ribosome) obtained from pull-down reactions performed in wild-type extracts were set to 1. Values obtained from pull-down reactions performed in Δsrp54 or Δegd1Δegd2 extracts are given relative to wild type. Statistical analysis was performed with KaleidaGraph 4.0 software.
Chemical cross-linking and site-specific cross-linking
To identify cross-link partners of nascent chains, we generated RNCs in the presence of 35S-methionine. Chemical cross-linking and immunoprecipitations under denaturing conditions were performed as previously described (Gautschi et al., 2002) using the homobifunctional cross-linker bis-(sulfosuccinimidyl)-suberate (BS3; spacer length, 11.4 Å; Pierce, Thermo Fisher Scientific, Rockford, IL). Site-specific cross-linking in the yeast system was performed as described (Berndt et al., 2009). The photoactivatable probe was placed at position 37 or 39 of FLAG-Dap2 (Supplemental Figures S2 and S3) by adding εANB-Lys-tRNAamb (Trna Probes, College Station, TX) to translation reactions programmed with mRNA derived from pSPUTK-FLAG-Dap2-stop37 or pSPUTK-FLAG-Dap2-stop39. A standard 80-μl photo–cross-linking reaction contained 48 pmol of εANB-Lys-tRNAamb. Translations were performed in the dark for 40 min at 20°C. Samples were then photolyzed in an ice/water mixture for 10 min using a 500-W mercury arc lamp. RNCs were immunoprecipitated under denaturing conditions using protein A–Sepharose precoated with antibodies directed against Srp54, Egd1, or Egd2.
Chemical cross-linking of ribosomal proteins to the subunits NAC was performed with BS3 in glass bead extracts. To that end, yeast strains were grown to an OD600 of 0.8–1.0, and then cycloheximide was added to a final concentration of 100 μg/ml. Cells were harvested by centrifugation, and glass bead disruption was performed in lysis buffer (20 mM HEPES-KOH, pH 7.4, 150 mM KAc, 2 mM MgAc2, 0.5 mM DTT, 0.5 mM PMSF, and protease inhibitor mix [1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin, 0.25 μg/ml elastinal, 5 μg/ml pepstatin A]) as described (Ashe et al., 2000). Cell debris and unbroken cells were removed by centrifugation at 10.000 × g for 20 min at 4°C. We loaded 90 μl of the resulting supernatant onto a 120-μl low-salt sucrose cushion or high-salt sucrose cushion (see previous discussion). After centrifugation for 22 min at 400,000 × g at 4°C, ribosomal pellets were resuspended in 300 μl of lysis buffer and incubated in the presence or absence of 400 μM BS3 for 20 min on ice. Cross-linking reactions were quenched by adding Tris-HCl, pH 7.5, to a final concentration of 50 mM. Proteins were precipitated by adding trichloric acid (TCA) to a final concentration of 5% and were subsequently analyzed via immunoblotting.
Preparation of microsomal membranes
A 6-l culture of the ∆egd1∆egd2 strain was grown on yeast extract/peptone/dextrose (YPD) at 30°C, and cells were harvested at an OD600 of 0.8–1.0. Crude cell extracts were prepared as described (Fünfschilling and Rospert, 1999). In brief, cells (25 g wet weight) were converted to spheroplasts by Zymolyase treatment and were lysed in 15 ml of lysis buffer (20 mM HEPES-KOH, pH 6.8; 50 mM KAc, 100 mM sorbitol, 2 mM EDTA, 1 mM PMSF, 1× protease inhibitor mix) using an all-glass Dounce homogenizer. The crude extract was centrifuged at 27,000 × g for 10 min, and the resulting pellet was resuspended in 1 ml of lysis buffer and carefully loaded onto a two step-gradient (5 ml of 1.2 M sucrose/5 ml of 1.5 M sucrose in lysis buffer). After centrifugation in a swing-out rotor at 100,000 × g for 1 h, microsomes were collected at the 1.2/1.5 M sucrose interface. The microsomal fraction was diluted 10-fold into high-salt buffer (20 mM HEPES-KOH, pH 6.8, 600 mM KAc, 100 mM sorbitol, 2 mM EDTA, 1 mM PMSF, 1× protease inhibitor). High-salt-treated microsomal membranes were collected via centrifugation at 27,000 × g for 10 min at 4°C and were resuspended in 1 ml of 20 mM HEPES-KOH, pH 7.4, 120 mM KAc, 240 mM sorbitol, and 2 mM MgAc2, and aliquots were stored at −80°C. High-salt-treated microsomal membranes contained the ER marker protein Kar2 but did not contain detectable amounts of SRP (Srp54), ribosomes (Rps9 and Rpl4), or the cytosolic protein Sse1 (Supplemental Figure S6B). The recovery of the microsomal marker Kar2 was ∼14% (Supplemental Figure S6B). To test for translocation competence, we used the ER-targeted yeast prepro-α-factor (ppα-factor; Raue et al., 2007). To that end, we added 4 μl of high-salt-washed microsomal membranes to a 20-μl translation reaction programmed with ppα-factor mRNA in the presence of 35S-methionine. Analysis via autoradiography revealed glycosylation of ppα-factor at three sites in the presence of microsomal membranes (Supplemental Figure S6A) as previously described (Waters et al., 1988).
ER-targeting assay
To test for the effect of NAC on the binding of RNCs to ER membranes, we performed in vitro translations in a volume of 80 μl using ∆egd1∆egd2 translation extract. As indicated in the figure legends, RNCs were generated in either the presence or the absence of 35S-methionine. In either case, cycloheximide was added to a final concentration of 200 μg/ml after a 50-min translation period at 20°C. Subsequently, 40 μl of wild-type translation extract or ∆egd1∆egd2 translation extract (lacking NAC, as a control) was added, and samples were incubated for 5 min at 20°C to allow for the binding of NAC to RNCs. After that, 40 μl of high-salt-washed microsomal membranes was added, and the reactions were incubated for 10 min at 20°C. Microsomal membranes and bound RNCs were collected via centrifugation at 30,000 × g for 10 min and were resuspended in 200 μl of 20 mM HEPES-KOH, pH 7.4, 120 mM KAc, 2 mM MgAc2, and 1 mM PMSF. Proteins were precipitated with 5% TCA and were separated on 10% Tris-tricine gels. Analysis was via either autoradiography (35S-labeled nascent chains) or immunoblotting using the antibodies indicated in the figure legends.
Purification of NAC from yeast
For the purification of native, untagged NAC, 12-l YPD cultures of C13-ABYS-86 were harvested at an OD600 of 0.8–1.0. Preparation of cell lysate and preparation of ribosomes from the cytosolic fraction of the cell lysate were performed as described (Fünfschilling and Rospert, 1999). NAC was purified from the material released from ribosomes by a high-salt wash. To that end, the ribosomal pellet was resuspended in buffer A (20 mM HEPES-KOH, pH 7.4, 5 mM MgAc2, 2 mM DTT, 0.5 mM PMSF, 120 mM KAc). The potassium concentration was adjusted to 800 mM with buffer B (20 mM HEPES-KOH, pH 7.4, 2 M KAc, 5 mM MgAc2, 2 mM DTT, 0.5 mM PMSF) to release the proteins bound in a salt-sensitive manner. After centrifugation at 200,000 × g in a 70.1 Ti rotor at 4°C, the clear supernatant was collected and was diluted with buffer C (20 mM HEPES-KOH, pH 7.4, 5 mM MgAc2, 2 mM DTT, 0.5 mM PMSF) to a potassium concentration of 200 mM. The material was loaded onto a Resource Q column (6 ml), and the column was washed with five volumes of HEPES-KOH, pH 7.4, and 200 mM KAc. NAC was then eluted with 30 ml of 200–600 mM KAc gradient in HEPES-KOH, pH 7.4. Fractions containing NAC were combined and loaded onto a Mono Q column (8 ml). The column was washed with five volumes of HEPES-KOH, pH 7.4 and 500 mM KAc, and NAC was eluted with 64 ml of 500–700 mM KAc gradient. Fractions containing NAC were combined and were diluted 1:5 with buffer D (20 mM HEPES-KOH, pH 7.4, 50 mM KAc, 5 mM MgAc2, 2 mM DTT, 2 mM PMSF, 1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin, 0.25 μg/ml elastinal, 5 μg/ml pepstatin A). The material was concentrated approximately fivefold in a Millipore concentrator (15 ml; cutoff size, 10 kDa), diluted for a second time fivefold with 1:5 with buffer D, and concentrated to the original volume. The material was then loaded onto a Mono S column (1 ml). The column was washed with two column volumes of 60 mM KAc in HEPES-KOH, pH 7.0, and NAC was eluted with a gradient from 60 to 800 mM KAc in HEPES-KOH, pH 7.0. Fractions containing purified NAC (Supplemental Figure S5A) were pooled and were dialyzed against 20 mM HEPES-KOH, pH 7.4, and 100 mM KAc. Aliquots of purified NAC (0.35 mg/ml) were frozen in liquid nitrogen and were stored at −80°C.
Miscellaneous
Yeast strains were grown to log phase on 1% yeast extract/2% peptone/2% dextrose. Immunoblots were developed using enhanced chemiluminescence with horseradish peroxidase–conjugated goat anti–rabbit immunoglobulin G (Pierce) as the secondary antibody, as described in (Raue et al., 2007). Densitometric analysis was performed using the AIDA ImageAnalyzer (Raytest, Straubenhardt, Germany). The concentration of purified NAC, Rps9a-His6, and Srp54-His6 was determined via Bradford (Bio-Rad, Hercules, CA) and bicinchoninic acid (Sigma-Aldrich) assays, using bovine serum albumin as a standard. Polyclonal antibodies were raised in rabbits and are described in Raue et al. (2007).
Supplementary Material
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft Grant SFB 746, Forschergruppe 967, and the Excellence Initiative of the German Federal and State Governments (EXC 294; to S.R.).
Abbreviations used:
- Dap2
dipeptidyl aminopeptidase B
- NAC
nascent polypeptide-associated complex
- RNCs
ribosome-nascent chain complexes
- RPBs
ribosome-bound protein biogenesis factors
- SA
signal anchor
- SRP
signal recognition particle
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-02-0112) on June 27, 2012.
REFERENCES
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci USA. 2009;106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Deuerling E, Pfund C, Craig EA. Getting newly synthesized proteins into shape. Cell. 2000;101:119–122. doi: 10.1016/S0092-8674(00)80806-5. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Gilmore R. Slow translocon gating causes cytosolic exposure of transmembrane and lumenal domains during membrane protein integration. Nat Struct Mol Biol. 2006;13:930–936. doi: 10.1038/nsmb1146. [DOI] [PubMed] [Google Scholar]
- Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- Del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JJ, Chen JC, Miao Y, Shao Y, Lin J, Bock PE, Johnson AE. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J Biol Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- Forte GM, Pool MR, Stirling CJ. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 2011;9:e1001073. doi: 10.1371/journal.pbio.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke J, Reimann B, Hartmann E, Kohlerl M, Wiedmann B. Evidence for a nuclear passage of nascent polypeptide-associated complex subunits in yeast. J Cell Sci. 2001;114:2641–2648. doi: 10.1242/jcs.114.14.2641. [DOI] [PubMed] [Google Scholar]
- Fünfschilling U, Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol Biol Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PD, Hansen W, Walter P. In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:675–682. doi: 10.1016/0076-6879(91)94049-i. [DOI] [PubMed] [Google Scholar]
- Gautschi M, Just S, Mun A, Ross S, Rücknagel P, Dubaquié Y, Ehrenhofer-Murray A, Rospert S. The yeast Nα-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol Cell Biol. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Mun A, Ross S, Rospert S. A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci USA. 2002;99:4209–4214. doi: 10.1073/pnas.062048599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R, Beddoe T, Landl K, Lithgow T. The yeast nascent polypeptide-associated complex initiates protein targeting to mitochondria in vivo. Proc Natl Acad Sci USA. 1998;95:2296–2301. doi: 10.1073/pnas.95.5.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George R, Walsh P, Beddoe T, Lithgow T. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 2002;516:213–216. doi: 10.1016/s0014-5793(02)02528-0. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Giglione C, Fieulaine S, Meinnel T. Cotranslational processing mechanisms: towards a dynamic 3D model. Trends Biochem Sci. 2009;34:417–426. doi: 10.1016/j.tibs.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Grudnik P, Bange G, Sinning I. Protein targeting by the signal recognition particle. Biol Chem. 2009;390:775–782. doi: 10.1515/BC.2009.102. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006a;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006b;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Noller HF. Footprinting mRNA-ribosome complexes with chemical probes. EMBO J. 1994;13:3892–3901. doi: 10.1002/j.1460-2075.1994.tb06700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol. 2010;189:57–68. doi: 10.1083/jcb.200910074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring B, Kreibich G, Weidmann M. The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex. Proc Natl Acad Sci USA. 1995a;92:9435–9439. doi: 10.1073/pnas.92.21.9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring B, Sakai H, Kreibich G, Wiedmann M. Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc Natl Acad Sci USA. 1995b;92:5411–5415. doi: 10.1073/pnas.92.12.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu Y, Li X, Niu L, Teng M. The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit. Biochemistry. 2010;49:2890–2896. doi: 10.1021/bi902050p. [DOI] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- Möller I, Beatrix B, Kreibich G, Sakai H, Lauring B, Wiedmann M. Unregulated exposure of the ribosomal M-site caused by NAC depletion results in delivery of non-secretory polypeptides to the Sec61 complex. FEBS Lett. 1998;441:1–5. doi: 10.1016/s0014-5793(98)01440-9. [DOI] [PubMed] [Google Scholar]
- Neuhof A, Rolls MM, Jungnickel B, Kalies KU, Rapoport TA. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol Biol Cell. 1998;9:103–115. doi: 10.1091/mbc.9.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M, Spreter T, Beckmann R, Beatrix B. Dual binding mode of the nascent polypeptide-associated complex reveals a novel universal adapter site on the ribosome. J Biol Chem. 2010;285:19679–19687. doi: 10.1074/jbc.M109.092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisker K, Braun D, Wölfle T, Hentschel J, Fünfschilling U, Fischer G, Sickmann A, Rospert S. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol Biol Cell. 2008;19:5279–5288. doi: 10.1091/mbc.E08-06-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov A, Kornberg G, O'Leary S, Tsai A, Uemura S, Puglisi JD. Dynamics of the translational machinery. Curr Opin Struct Biol. 2011;21:137–145. doi: 10.1016/j.sbi.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Brown S, Cardillo TS, Rigby S, Sherman F. Yeast N(alpha)-terminal acetyltransferases are associated with ribosomes. J Cell Biochem. 2008;103:492–508. doi: 10.1002/jcb.21418. [DOI] [PubMed] [Google Scholar]
- Pool MR. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J Cell Biol. 2009;185:889–902. doi: 10.1083/jcb.200807066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr Biol. 1996;6:331–338. doi: 10.1016/s0960-9822(02)00484-0. [DOI] [PubMed] [Google Scholar]
- Raden D, Gilmore R. Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol Biol Cell. 1998;9:117–130. doi: 10.1091/mbc.9.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J Biol Chem. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- Reimann B, Bradsher J, Franke J, Hartmann E, Wiedmann M, Prehn S, Wiedmann B. Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast. 1999;15:397–407. doi: 10.1002/(SICI)1097-0061(19990330)15:5<397::AID-YEA384>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Pohlig G, Rothman JH, Stevens TH. Structure, biosynthesis, and localization of dipeptidyl aminopeptidase B, an integral membrane glycoprotein of the yeast vacuole. J Cell Biol. 1989;108:1363–1373. doi: 10.1083/jcb.108.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospert S, Dubaquié Y, Gautschi M. Nascent-polypeptide-associated complex. Cell Mol Life Sci. 2002;59:1632–1639. doi: 10.1007/PL00012490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rospert S, Gautschi M, Rakwalska M, Raue U. Ribosome-bound proteins acting on newly synthesized polypeptide chains. In: Buchner J, Kiefhaber T, editors. Protein Folding Handbook. II.1. Weinheim, Germany: Wiley-VCH; 20052005. pp. 429–458. [Google Scholar]
- Saraogi I, Shan SO. Molecular mechanism of co-translational protein targeting by the signal recognition particle. Traffic. 2011;12:535–542. doi: 10.1111/j.1600-0854.2011.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shan SO, Walter P. Co-translational protein targeting by the signal recognition particle. FEBS Lett. 2005;579:921–926. doi: 10.1016/j.febslet.2004.11.049. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Spiller MP, Stirling CJ. Preferential targeting of a signal recognition particle-dependent precursor to the Ssh1p translocon in yeast. J Biol Chem. 2011;286:21953–21960. doi: 10.1074/jbc.M111.219568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreter T, Pech M, Beatrix B. The crystal structure of archaeal nascent polypeptide-associated complex (NAC) reveals a unique fold and the presence of a ubiquitin-associated domain. J Biol Chem. 2005;280:15849–15854. doi: 10.1074/jbc.M500160200. [DOI] [PubMed] [Google Scholar]
- Ullers RS, Houben EN, Raine A, ten Hagen-Jongman CM, Ehrenberg M, Brunner J, Oudega B, Harms N, Luirink J. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J Cell Biol. 2003;161:679–684. doi: 10.1083/jcb.200302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Arnesen T, Gevaert K. Protein alpha-N-acetylation studied by N-terminomics. FEBS J. 2011;278:3822–3834. doi: 10.1111/j.1742-4658.2011.08230.x. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Waizenegger T, Stan T, Neupert W, Rapaport D. Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics. J Biol Chem. 2003;278:42064–42071. doi: 10.1074/jbc.M305736200. [DOI] [PubMed] [Google Scholar]
- Walter P, Ibrahimi I, Blobel G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang W, Zhang XC, Li X, Rao Z. Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its alphaNAC subunit. Protein Cell. 2010;1:406–416. doi: 10.1007/s13238-010-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MG, Evans EA, Blobel G. Prepro-alpha-factor has a cleavable signal sequence. J Biol Chem. 1988;263:6209–6214. [PubMed] [Google Scholar]
- Wegrzyn RD, Deuerling E. Molecular guardians for newborn proteins: ribosome-associated chaperones and their role in protein folding. Cell Mol Life Sci. 2005;62:2727–2738. doi: 10.1007/s00018-005-5292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn RD, Hofmann D, Merz F, Nikolay R, Rauch T, Graf C, Deuerling E. A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J Biol Chem. 2006;281:2847–2857. doi: 10.1074/jbc.M511420200. [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Prehn S. The nascent polypeptide-associated complex (NAC) of yeast functions in the targeting process of ribosomes to the ER membrane. FEBS Lett. 1999;458:51–54. doi: 10.1016/s0014-5793(99)01118-7. [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M. A protein complex required for signal-sequence-specific sorting and translocation. Nature. 1994;370:434–440. doi: 10.1038/370434a0. [DOI] [PubMed] [Google Scholar]
- Yogev O, Karniely S, Pines O. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J Biol Chem. 2007;282:29222–29229. doi: 10.1074/jbc.M704201200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.