The deposition of misfolded proteins in cytoplasmic protein bodies requires the concerted action of stress-inducible protein-sorting factors and molecular chaperones. Protein sequestration during acute stress is a cellular strategy that adjusts the flux of misfolded proteins to the capacities of the protein quality control system.
Abstract
Acute stress causes a rapid redistribution of protein quality control components and aggregation-prone proteins to diverse subcellular compartments. How these remarkable changes come about is not well understood. Using a phenotypic reporter for a synthetic yeast prion, we identified two protein-sorting factors of the Hook family, termed Btn2 and Cur1, as key regulators of spatial protein quality control in Saccharomyces cerevisiae. Btn2 and Cur1 are undetectable under normal growth conditions but accumulate in stressed cells due to increased gene expression and reduced proteasomal turnover. Newly synthesized Btn2 can associate with the small heat shock protein Hsp42 to promote the sorting of misfolded proteins to a peripheral protein deposition site. Alternatively, Btn2 can bind to the chaperone Sis1 to facilitate the targeting of misfolded proteins to a juxtanuclear compartment. Protein redistribution by Btn2 is accompanied by a gradual depletion of Sis1 from the cytosol, which is mediated by the sorting factor Cur1. On the basis of these findings, we propose a dynamic model that explains the subcellular distribution of misfolded proteins as a function of the cytosolic concentrations of molecular chaperones and protein-sorting factors. Our model suggests that protein aggregation is not a haphazard process but rather an orchestrated cellular response that adjusts the flux of misfolded proteins to the capacities of the protein quality control system.
INTRODUCTION
Due to the unstable nature of protein conformations, cells have to manage an unremitting burden of misfolded and aggregation-prone proteins. Proteins with aberrant folds can disrupt cellular homeostasis and cause pathological changes and aging. Because the consequences of protein misfolding can be severe, eukaryotic cells have evolved an elaborate system for protein quality control (PQC). Three different strategies come into play: molecular chaperones that recognize and refold nonnative proteins, energy-dependent proteases that remove misfolded proteins by degradation, and autophagy—the selective uptake and degradation of aberrant proteins in membrane-enclosed compartments (Hartl and Hayer-Hartl, 2009; Buchberger et al., 2010; Tyedmers et al., 2010).
To maintain proteome functionality throughout the lifetime of an organism, protein conformations are subject to constant surveillance by molecular chaperones. Several different classes of structurally unrelated chaperones exist in eukaryotic cells. Members of these protein families are known as heat shock proteins (HSPs), as they are upregulated under conditions of stress in which the concentrations of aggregation-prone proteins can increase substantially. Molecular chaperones are typically classified according to their molecular weight (HSP40, HSP60, HSP70, HSP90, HSP100, and the small HSPs). Together they form a cooperative network that is involved in a multitude of cellular functions, including de novo folding, refolding of damaged proteins, assembly and disassembly of oligomers, and assistance of protein trafficking and degradation.
The study of yeast prions has provided important insight into the interplay between molecular chaperones and aggregation-prone proteins. Yeast prions can exist in two structural and functional states, at least one of which is a self-sustaining prion form (Ross et al., 2005; Shorter and Lindquist, 2005; Chernoff, 2007). The prion state is formed by an unusually stable β sheet–rich aggregate or amyloid fiber. Prion fibers are inherited through a process of repeated fiber growth and chaperone-mediated fiber division. The molecular machinery that drives this prion replication cycle involves the protein disaggregase Hsp104 (Chernoff et al., 1995). Hsp104 promotes the fragmentation of prion fibers and generates heritable seeds that are disseminated between mother and daughter during cell divisions (Paushkin et al., 1996; Borchsenius et al., 2001; Wegrzyn et al., 2001; Satpute-Krishnan et al., 2007).
In recent years, important progress has been made that greatly enhances our understanding of chaperone action on amyloids in the cellular milieu. The prion forms of the yeast prion proteins Sup35 and Rnq1, for example, make extensive physical contact with the Hsp70 Ssa1 (Allen et al., 2005; Bagriantsev et al., 2008) and contain significant amounts of the Hsp40 chaperone Sis1 (Sondheimer et al., 2001; Lopez et al., 2003; Bagriantsev et al., 2008). Moreover, evidence is now accumulating that Sis1 has an essential function in prion replication. Depletion of Sis1 caused an initial increase in prion polymer size and a subsequent loss of the prion state in dividing yeast cell populations (Aron et al., 2007; Higurashi et al., 2008; Tipton et al., 2008). Weissman and colleagues provided a compelling explanation for the critical role of Sis1 (Tipton et al., 2008). They found that Sis1 is required for substrate recognition and subsequent translocation of the substrate through the central pore of Hsp104.
Prions and other misfolded proteins are not randomly distributed in cells but are directed to specific sites. Mechanisms of spatial PQC have been identified in organisms as diverse as bacteria and humans (Tyedmers et al., 2010). Two distinct stress-inducible compartments for misfolded proteins were recently described in the cytoplasm of yeast and mammalian cells (Kaganovich et al., 2008). The two compartments were termed the insoluble protein deposit (IPOD) and the juxtanuclear quality control (JUNQ). The IPOD is localized in the peripheral part of yeast cells and contains terminally misfolded proteins such as amyloids and prions. The JUNQ resides in proximity of the nucleus and contains soluble proteins that rapidly exchange with the surrounding cytosol. Experimental manipulations that changed the ubiquitylation status of misfolded proteins altered their partitioning between IPOD and JUNQ (Kaganovich et al., 2008). A recent study identified the small heat shock protein Hsp42 as an important aggregate-sorting factor that mediates the retention of misfolded proteins in a peripheral location of yeast cells (Specht et al., 2011). Despite these advances, however, the molecular details of spatial PQC have remained largely unknown.
Using a phenotypic reporter for prion aggregation, we identified two proteins of the Hook family, termed Bnt2 and Cur1, as central regulators of spatial PQC in yeast. Btn2 and Cur1 physically and functionally interact with chaperones to promote the sorting and deposition of misfolded proteins into cytosolic compartments. On the basis of our findings, we propose a dynamic model that describes the spatiotemporal organization of misfolded proteins during mild heat stress as a result of the concerted action of protein-sorting factors and molecular chaperones.
RESULTS
Stress-inducible Btn2 and Cur1 interfere with prion inheritance
Molecular chaperones act on misfolded proteins and relieve the cell from proteotoxic stress, but at the same time they are required for the inheritance of yeast prions. To gain insight into the functional interactions between chaperones and prions, we investigated a set of candidate yeast prions that was recently identified in a systematic prion survey (Alberti et al., 2009). Here we will focus on the prion domain (PrD) of one of these proteins, Nrp1, as it turned out to be particularly sensitive to mild heat stress when fused to a reporter (see next paragraph). To track the aggregation state of Nrp1PrD, we generated a chimera between Nrp1PrD and the C terminus of Sup35 (Alberti et al., 2009). Conformational conversion of Sup35 to the prion state leads to reduced translation termination activity, which causes ribosomes to read through stop codons at an increased frequency. In yeast strains carrying a premature stop codon in the ADE1 gene, readthrough results in a white colony color phenotype in the presence of the prion (Figure 1A). In the absence of the prion, the colony color is red. Similar to other yeast prions, the prion state of the synthetic Nrp1PrD-Sup35C construct (termed [NRP1C+]) required the continuous activity of Hsp104 and was independent of the prion-inducing factor Rnq1 once it was formed (Supplemental Figure S1A).
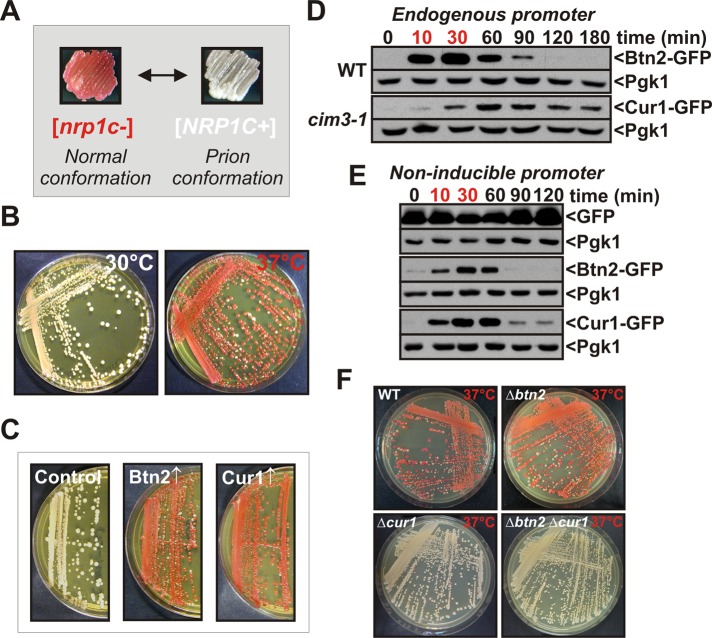
FIGURE 1:
Stress-inducible Btn2 and Cur1 interfere with prion inheritance. (A) A prion reporter based on the translation termination activity of the C-terminal domain of Sup35 was used to monitor [NRP1C+] in yeast (see the text for details). (B) [NRP1C+] yeast cells were incubated on YPD plates at 30°C or 37°C for 3 d. (C) Galactose-regulatable expression plasmids coding for BTN2 or CUR1 were introduced into [NRP1C+] yeast. The transformants were transferred onto galactose-containing plates and incubated for 3 d at 30°C. Subsequently, the strains were transferred onto fresh YPD plates to allow for colony color development. (D) Chromosomal BTN2 was tagged with GFP in the BY4741 strain background. CUR1 was modified with GFP in a strain carrying a temperature-sensitive mutation in the proteasome subunit CIM3. The strains were grown at 25°C, shifted to 39°C for 30 min (time points in red), and then again exposed to 25°C. Cell lysates were prepared at the indicated time points and analyzed by immunoblotting with an anti-GFP antibody. The extended presence of Cur1 is probably an artifact of the reduced proteasomal activity of the cim3-1 strain. (E) Cells were treated as in D except that Btn2-GFP and Cur1-GFP were expressed from a low-copy plasmid that carried a GPD promoter. (F) [NRP1C+] yeast carrying deletions of BTN2 and/or CUR1 were incubated at 37°C for 3 d.
All known yeast prions are dependent on Hsp104, but other chaperones can play important functional roles as well. The expression of chaperones is subject to dramatic changes during stress. We therefore investigated whether [NRP1C+] is affected by heat stress. Surprisingly, when we grew the [NRP1C+] strain at 37°C, we found that the prion state was almost completely lost (Figure 1B). To test whether heat shock protein induction was involved in prion destabilization, we overexpressed a comprehensive set of chaperones, cochaperones, and other heat shock proteins that had been implicated as prion modifiers in [NRP1C+] yeast. The propagation of [NRP1C+] was relatively stable in all but two strains (Figure 1C and Supplemental Figure S1B). These strains contained plasmids for the expression of Btn2 or Cur1, respectively.
The transcripts of BTN2 and CUR1 are highly induced by heat stress (Gasch et al., 2000). To determine whether the protein levels of Btn2 and Cur1 are also upregulated at higher temperatures, we introduced a green fluorescent protein (GFP) tag into the chromosomal loci. Btn2 was barely detectable in cells growing at 25°C. However, after growth at 39°C for 1 hr, Btn2 expression was strongly induced (Supplemental Figure S1C). Surprisingly, we were unable to detect Cur1-GFP in cells growing at either 25 or 39°C (data not shown). We noticed, however, that the steady-state levels of overexpressed Cur1 and Btn2 were significantly elevated in cells with impaired proteasomes (Supplemental Figure S1D). We therefore performed a heat shock experiment with a strain that carried a temperature-sensitive mutation in a proteasome subunit (cim3-1). In this strain background, Cur1 expression was detectable at both 25 and 39°C, with cells grown at 39°C showing a marked increase of expression over cells grown at 25°C (Supplemental Figure S1E).
To analyze the temporal changes in Btn2 and Cur1 expression during acute thermal stress, we exposed cells expressing Btn2-GFP or Cur1-GFP from the endogenous locus to a brief heat shock at 39°C. Both Btn2 and Cur1 were induced after a 10-min exposure to heat, and the protein levels rapidly declined when the stress subsided (Figure 1D). These changes were likely due to changes in gene expression. However, as Btn2 and Cur1 are rapidly turned over by the proteasome and the activity of the proteasome could be altered by stress, we tested whether the protein levels also increase when Btn2 and Cur1 are expressed from a noninducible promoter. Indeed, Btn2 and Cur1 were stabilized in stressed cells that expressed these proteins from the constitutive GPD promoter (Figure 1E). Moreover, when the temperature dropped back to 25°C, the protein concentrations rapidly returned to prestress levels. Thus changes in gene expression and a transient decrease in proteasomal activity jointly restrict Btn2 and Cur1 expression to periods of acute stress.
To determine whether induction of BTN2 and CUR1 was causally responsible for heat-induced prion loss, we generated [NRP1C+] strains that carried a single deletion of BTN2 or CUR1 or a double deletion of both genes. Remarkably, the two strains that lacked a functional copy of CUR1 were now able to stably propagate [NRP1C+] at 37°C (Figure 1F). The BTN2-deficient strain, however, was still vulnerable to heat-induced prion loss. Thus induction of CUR1 by heat was sufficient to interfere with prion propagation. BTN2 induction, however, did not cause prion loss under the same conditions, despite highly increased protein levels. This is due to important mechanistic differences that will be discussed later.
Similar to [NRP1C+], the yeast prion [PSI+] is vulnerable to mild heat stress. The cause of heat-induced [PSI+] loss was determined to be an imbalance in the gene expression of molecular chaperones (Newnam et al., 2011). However, no differences in the steady-state levels of Hsp104, Sis1, or Ssa1/2 were detected in the absence of BTN2 and/or CUR1 (Supplemental Figure S1F). Thus [NRP1C+] loss during acute heat stress is not caused by global alterations in chaperone expression, but is likely due to direct effects on prion aggregates or prion-regulating factors.
Btn2 and Cur1 physically and functionally interact with Sis1 to modify prion inheritance
Btn2 and Cur1 had previously been identified as factors that induce the loss of the yeast prion [URE3] when overexpressed (Kryndushkin et al., 2008). The physiological relevance of this observation, as well as the molecular mechanism behind prion loss, however, remained undetermined. Because no direct interaction between prion aggregates and Btn2 or Cur1 was detected, we hypothesized that Btn2 and Cur1 could act as inhibitors of a factor that is required for prion propagation. To investigate this possibility, we overexpressed a collection of heat shock proteins (see preceding section) in [NRP1C+] yeast at 37°C. Remarkably, we observed a weak stabilization of the prion state in Hsp104-expressing cells and a substantial stabilization in the strain that expressed Sis1 (Figure 2A and Supplemental Figure S2A). To test whether Sis1 also affects the prion loss that is induced by overexpression of Btn2 and Cur1, we next generated a [NRP1C+] strain that constitutively expressed Sis1 at higher levels. In this strain, the prion state was stable in the presence of increased levels of Btn2 or Cur1 (Supplemental Figure S2B). Thus a higher cellular Sis1 concentration can counteract the prion-inhibiting activities of Btn2 and Cur1.
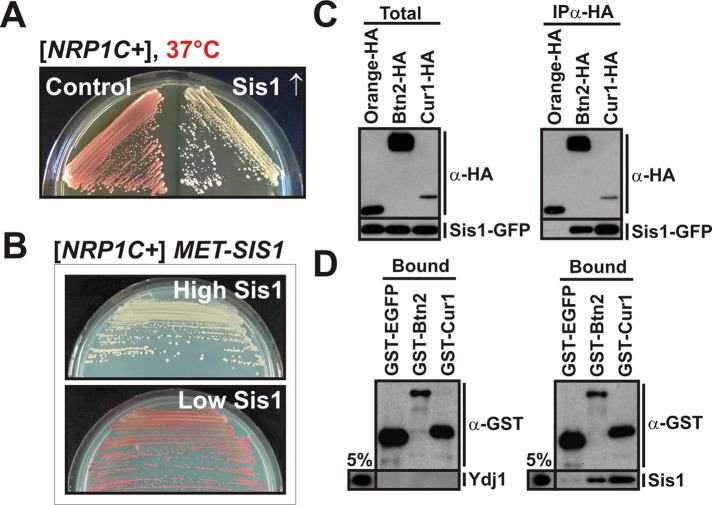
FIGURE 2:
Btn2 and Cur1 functionally and physically interact with Sis1 to modify prion inheritance. (A) [NRP1C+] cells were transformed with a control plasmid or a low-copy expression plasmid coding for SIS1 and incubated at 37°C for 3 d. (B) Endogenous SIS1 was deleted in [NRP1C+] yeast, and the deletion was covered with a plasmid carrying SIS1 behind a methionine-regulatable promoter. The cells were grown in the presence (low Sis1) or absence (high Sis1) of methionine. (C) BY4741 yeast cells carrying a GFP-tagged chromosomal copy of SIS1 were transformed with low-copy expression plasmid for HA-tagged Orange (control), BTN2, or CUR1. HA-tagged proteins were immunoprecipitated from cell lysates with a HA-specific antibody. (D) Protein-binding assay with bacterially purified GST-EGFP (control), GST-Btn2, GST-Cur1, His6-Ydj1 (control), or His6-Sis1. Proteins were detected by immunoblotting with an anti-GST or anti-His antibody. The pull-down efficiency was ∼20% for GST-tagged proteins. Five percent of the input is shown for comparison.
Sis1 is required for the propagation of several yeast prions (Aron et al., 2007; Higurashi et al., 2008; Tipton et al., 2008). We therefore investigated whether Sis1 is also necessary for the maintenance of [NRP1C+]. To do this, we generated a strain in which cellular Sis1 levels could be manipulated by changing the concentration of methionine in the medium. When this strain was grown in the presence of methionine (low expression level of Sis1), the prion state was lost (Figure 2B). A lower cellular Sis1 concentration also led to a strong reduction in the number of cells with visible Nrp1PrD aggregates (Supplemental Figure S2C). Thus, to determine whether Sis1 is required for prion fragmentation, we investigated the size distribution of [NRP1C+] polymers in cell lysates using gel filtration. In the control cell lysate, Nrp1PrD-Sup35C was partitioned into two broad peaks, one of which contained polymeric prion aggregates (Supplemental Figure S2D). In cells that expressed Sis1 at higher levels, the prion polymers were significantly smaller. As shown in Supplemental Figure S2E, Nrp1PrD aggregates also colocalized with Sis1. In addition, Sis1 was specifically associated with immunoprecipitated Nrp1PrD (Supplemental Figure S2F). Collectively these data indicate that propagation of [NRP1C+] not only is dependent on Hsp104 but also requires Sis1.
To investigate whether Btn2 and Cur1 affect [NRP1C+] inheritance through changes in prion fragmentation, we separated the lysate of prion-containing yeast by semidenaturing detergent–agarose gel electrophoresis (Alberti et al., 2010). As shown in Supplemental Figure S2G, the amount of small- to medium-sized polymers was significantly increased in cells deleted for BTN2 and/or CUR1. A similar effect was observed in cells that overproduced Sis1 (Supplemental Figure S2G), suggesting that Btn2 and Cur1 modify the activity or accessibility of Sis1. Therefore, to determine whether Btn2 and/or Cur1 physically interact with Sis1, we performed a coimmunoprecipitation experiment with yeast cell lysate. Sis1 was easily detectable in Btn2 and Cur1 immunocomplexes but not in the immunocomplex of a control protein (Figure 2C). The interaction with Sis1 could be a direct one, or other proteins present in the cell lysate could mediate it. To differentiate between these two possibilities, we performed a binding assay with proteins purified from bacteria. As shown in Figure 2D, Sis1 was specifically retained on beads containing glutathione S-transferase (GST)–tagged Btn2 or Cur1. The functionally related Hsp40 Ydj1, however, did not associate with Btn2 or Cur1. Thus Btn2 and Cur1 directly and specifically bind to Sis1 with direct consequences for its function.
Btn2 and Cur1 promote sorting of Sis1 to the nucleus and to stress-inducible cytosolic compartments
Harnessing the phenotypic properties of a synthetic yeast prion, we were able to identify stress-inducible Btn2 and Cur1 as interactors and functional modifiers of Sis1. To gain further insight into the function of Sis1 during stress, we explored the behavior of Sis1-GFP in yeast that were subjected to a temperature shift from 25 to 37°C. After 1 h at 37°C, there was a significant reduction in the amount of diffuse cytosolic Sis1 (Figure 3A). This change was accompanied by an accumulation of Sis1 in the nucleus and the coalescence of Sis1 into several peripheral and juxtanuclear foci (arrows in Figure 3A). Thus the spatial distribution of Sis1 is subject to considerable changes during heat stress.
FIGURE 3:
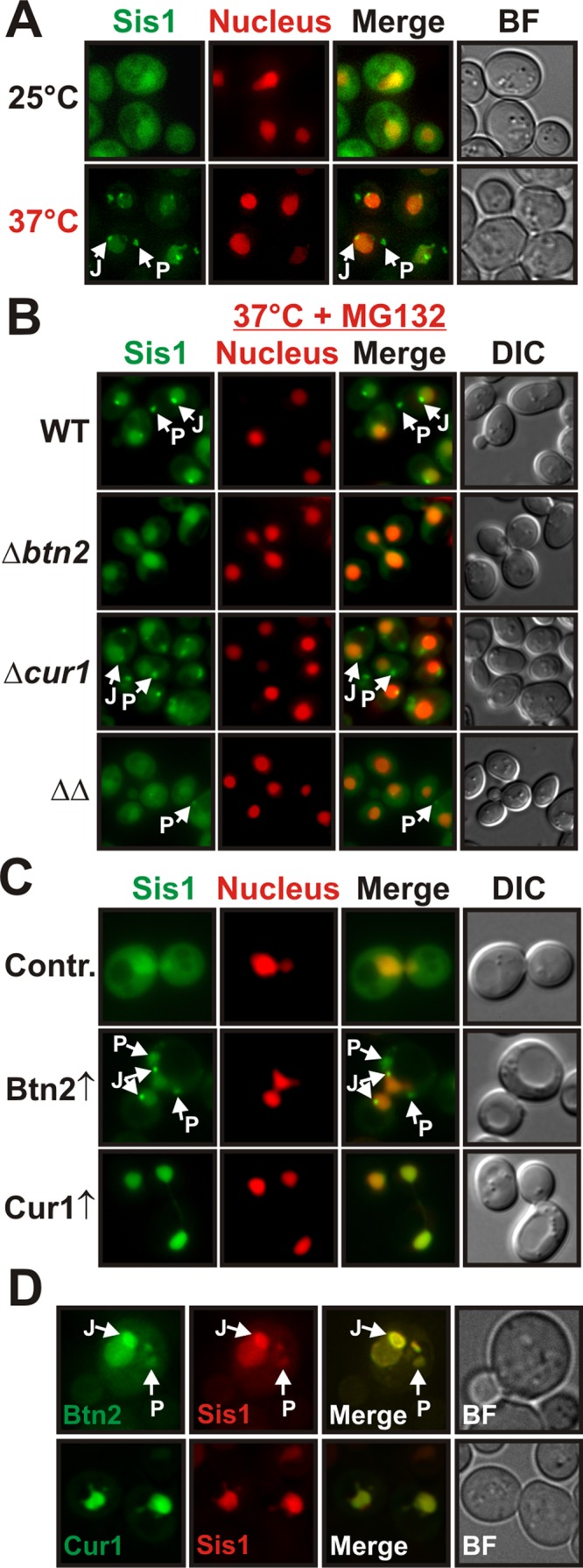
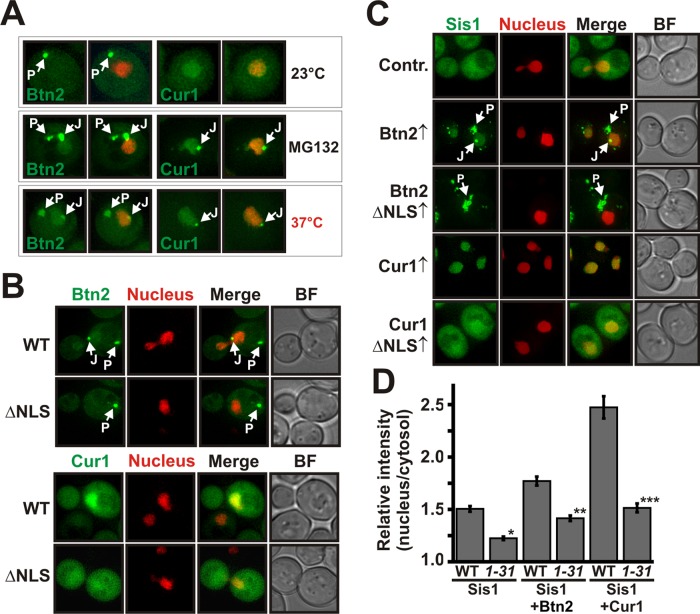
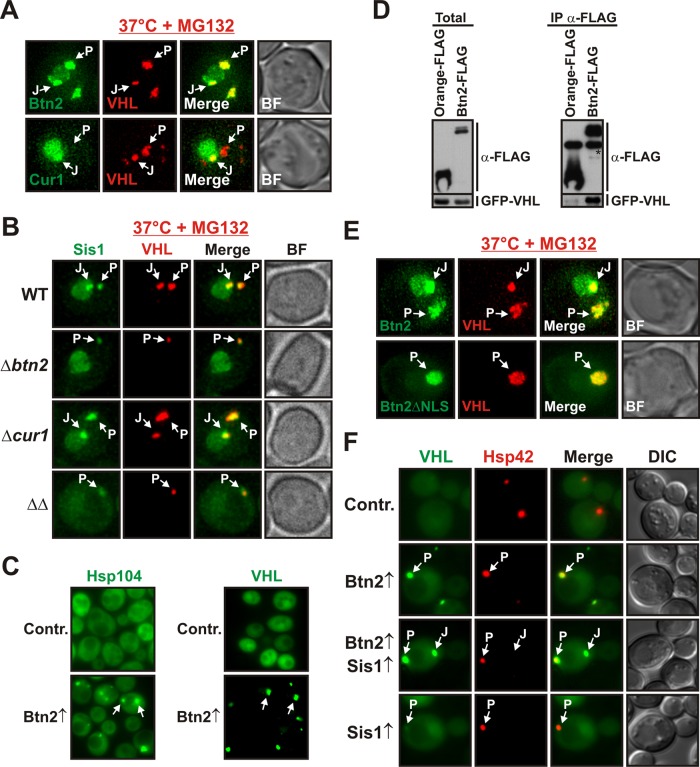
Btn2 and Cur1 promote the sorting of Sis1 to the nucleus and to stress-inducible cytosolic compartments. (A) BY4741 yeast cells carrying a GFP-tagged chromosomal copy of SIS1 were grown at 25 or 37°C and subjected to fluorescence microscopy. J and P denote juxtanuclear and peripheral compartments, respectively. See Supplemental Information for details on image interpretation, as well as on control experiments. (B) Wild-type, Δbtn2, Δcur1, or Δbtn2 Δcur1 BY4741 cells carrying a GFP-tagged chromosomal copy of SIS1 were grown at 37°C in the presence of MG132 (MG132 was used because compartment formation was more pronounced) and were subjected to fluorescence microscopy. (C) Low-copy expression plasmids for BTN2 and CUR1 were introduced into a BY4741 strain expressing Sis1-GFP and an mCherry-tagged nuclear marker. Fluorescence microscopy was performed at 25°C. (D) Plasmid-expressed Btn2-GFP or Cur1-GFP was coexpressed with Sis1-mCherry in BY4741 yeast for colocalization analysis at 25°C.
Btn2 and Cur1 are homologues of the Hook family of proteins, which function as cytoskeleton-associated transport factors mediating the distribution of organelles in mammalian cells (Kama et al., 2007; Kryndushkin et al., 2008). This suggested that Btn2 and Cur1 could be sorting factors for Sis1. To investigate this possibility, we analyzed the localization patterns of GFP-tagged endogenous Sis1 in stressed yeast that carried deletions of BTN2 and/or CUR1. Indeed, Btn2-deleted cells showed a strong reduction in Sis1-positive foci when compared with wild-type cells (Figure 3B and Supplemental Movie S1). Cells lacking Cur1 exhibited a diminished nuclear Sis1 signal and a concomitant increase in the amount of diffuse cytosolic Sis1 (Figure 3B and Supplemental Movie S1). The nuclear Sis1 signal was even more strongly reduced in cells that were eliminated for both Btn2 and Cur1 (Figure 3B and Supplemental Movie S1). Thus Btn2 and Cur1 are required for the redistribution of Sis1 during acute stress.
To determine whether an increase in cellular Btn2 and Cur1 levels is sufficient to induce changes in Sis1 localization, we overexpressed Btn2 or Cur1 in unstressed yeast that carried a GFP-tagged chromosomal copy of SIS1. Sis1 was only slightly enriched in the nucleus in control cells growing at 25°C. In Btn2-expressing cells, however, the nuclear:cytosolic ratio of Sis1 was increased by a factor of two (Figure 3C and Supplemental Figure S3A). In cells that expressed Cur1, Sis1 was even more strongly enriched in the nucleus and barely detectable in the cytosol (Figure 3C and Supplemental Figure S3A). In addition, we frequently observed that Sis1 accumulated in peripheral and juxtanuclear foci in Btn2-expressing cells (Figure 3C and Supplemental Figure S3B). To determine whether these changes were due to an interaction with Sis1 in the cellular environment, we subjected yeast cells expressing Sis1-mCherry and GFP-tagged Btn2 or Cur1 to colocalization analysis. The signals in the GFP and mCherry channel showed extensive overlap (Figure 3D). Given that Sis1 interacts with Btn2 and Cur1 in vitro (see Figure 2D), we conclude that Btn2 and Cur1 are sorting factors for Sis1.
The structure of Sis1 has been studied extensively (Yan and Craig, 1999; Sha et al., 2000; Lee et al., 2002). Sis1 contains an N-terminal J domain that regulates the ATPase activity of Hsp70s, a C-terminal substrate-binding domain, and a dimerization motif at the extreme C-terminus (Supplemental Figure S3C). To investigate whether these domains are required for Sis1 sorting, we coexpressed Btn2 or Cur1 with several GFP-tagged mutant variants of Sis1. Surprisingly, we found that Sis1 redistribution was not dependent on a functional substrate-binding site or J domain but was impaired in a mutant that lacked the dimerization motif (Supplemental Figure S3, C–E). Thus Btn2 and Cur1 are not client proteins of Sis1, but instead have an important sorting function that requires Sis1 dimerization.
Nuclear targeting of Sis1 is dependent on nuclear localization sequences in Btn2 and Cur1 and requires the α-importin Srp1
In cells that expressed Sis1 together with Btn2 or Cur1, we noticed that Cur1 and to a lesser extent Btn2 were enriched in the nucleus (see Figure 3D). Nuclear localization of Cur1 had been described previously (Kryndushkin et al., 2008), but nuclear enrichment of Btn2 has not been reported, despite the fact that a number of studies investigated the localization of Btn2 (Chattopadhyay and Pearce, 2002; Kama et al., 2007; Kryndushkin et al., 2008; Kanneganti et al., 2011). Because Btn2 and Cur1 are strongly stabilized during stress or proteasomal impairment, we hypothesized that this discrepancy could be due to the fact that earlier studies were performed under normal growth conditions. To investigate this possibility, we grew yeast cells expressing Btn2-GFP or Cur1-GFP at 23°C (control) or 37°C or in the presence of the proteasome inhibitor MG132. As can be seen in Figure 4A, yeast cells that were exposed to MG132 or increased temperatures showed a significant enrichment of Btn2 in the nucleus. However, in cells that grew under control conditions, nuclear enrichment was barely detectable. Of importance, Btn2 accumulation in the nucleus was accompanied by the formation of juxtanuclear foci that contained high amounts of Btn2 (Figure 4A). Cur1 was also enriched in the nucleus and likewise accumulated in juxtanuclear sites. Together, these data indicate that acute stress conditions lead to accumulation of Btn2 and Cur1 in the nucleus and in juxtanuclear foci due to decreased proteasomal activity.
FIGURE 4:
Nuclear targeting of Sis1 is dependent on nuclear localization sequences in Btn2 and Cur1 and requires the α-importin Srp1. (A) W303 yeast expressing GFP-tagged Btn2 or Cur1 from a low-copy plasmid were grown at 23 or 37°C or in the presence of the proteasome inhibitor MG132. Left, the GFP channel. Right, an overlay with an mCherry-tagged nuclear marker. (B) W303 yeast cells expressing GFP-tagged Btn2, Cur1, Btn2ΔNLS, or Cur1ΔNLS were subjected to fluorescence microscopy at 30°C. (C) BY4741 yeast cells carrying a GFP-tagged chromosomal copy of SIS1 were transformed with low-copy plasmids for Btn2, Cur1, Btn2ΔNLS, or Cur1ΔNLS. The resulting transformants were grown at 25°C and subjected to fluorescence microscopy. (D) Wild-type yeast (WT) or yeast carrying a temperature-sensitive mutation in SRP1 (srp1-31) were cotransformed with expression plasmids for Sis1-GFP and Orange (control), Btn2, or Cur1. The cells were subjected to fluorescence microscopy after a shift to the nonpermissive temperature for 1 h. The relative nuclear:cytosolic GFP pixel intensity of the strains was determined. *p = 2.7 × 10−8; **p = 0.0027; ***p = 2.9 × 10−8.
We previously observed that nuclear accumulation of Sis1 was strongly dependent on the presence of Btn2 and Cur1 (see Figure 3, B and C). This finding suggested that Btn2 and Cur1 could be nuclear targeting factors for Sis1. To collect evidence for such a function, we investigated the amino acid sequences of Btn2, Cur1, and Sis1 for the presence of nuclear localization sequences (NLSs). Indeed, we identified a classic NLS in the N-terminal region of Btn2 and Cur1 (see Supplemental Information for details on NLS prediction). We did not, however, find an NLS in Sis1. To determine whether the identified sequence motifs are genuine nuclear targeting signals, we generated mutant versions of Btn2 and Cur1 without NLS motifs. Wild-type and mutant proteins were expressed as GFP fusions and analyzed by fluorescence microscopy. As shown in Figure 4B and Supplemental Figure S4A, the wild-type proteins were enriched in the nucleus, whereas the NLS-deleted versions of Btn2 and Cur1 were equally distributed between cytosol and nucleus. Thus the NLS motifs are functional nuclear targeting signals. Importantly, we also observed that Btn2ΔNLS no longer accumulated in juxtanuclear sites (Figure 4B). This implies that the NLS of Btn2 is required not only for nuclear import, but also for targeting to a juxtanuclear compartment.
To investigate whether Sis1 accumulation in the nucleus requires functional NLS motifs, we overexpressed wild-type or mutant Btn2 or Cur1 in yeast cells that expressed Sis1-GFP from the chromosomal locus. As shown in Figure 4C, Sis1 was no longer enriched in the nucleus in cells that expressed mutant Btn2 or Cur1. In addition, mutant Btn2 did not promote the recruitment of Sis1 to juxtanuclear sites. Importantly, these effects were not due to an altered interaction with Sis1, as the mutant versions of Btn2 and Cur1 were still able to associate with coimmunoprecipitated Sis1 (Supplemental Figure S4B). Moreover, deletion of the NLS motif did not decrease the steady-state levels of Btn2 and Cur1 but instead led to a strong stabilization (Supplemental Figure S4C). Thus sorting of Sis1 to the nucleus is dependent on nuclear localization sequences in Btn2 and Cur1.
The α-importin Srp1 promotes nuclear targeting of NLS-containing proteins (Tabb et al., 2000). To determine whether Btn2, Cur1, and Sis1 are transported to the nucleus in an Srp1-dependent manner, we monitored nuclear accumulation of Sis1 in cells carrying a temperature-sensitive mutation in SRP1 (srp1-31). When srp1-31 cells were exposed to the nonpermissive temperature, nuclear import of Sis1 was impaired in control cells as well as in cells that produced Btn2 or Cur1 (Figure 4D and Supplemental Figure S4D). Srp1 also physically interacted with Btn2 and Cur1, as shown by coimmunoprecipitation (Supplemental Figure S4E) and an in vitro binding assay with purified Srp1 and wild-type or NLS-deleted Btn2 or Cur1 (Supplemental Figure S4F). Thus we conclude that nuclear targeting of Btn2, Cur1, and Sis1 requires NLS motifs in Btn2 and Cur1 and association with the nuclear import factor Srp1.
Complex formation of Btn2 or Cur1 with Sis1 is required for targeting to the nucleus
Our previous data indicated that Btn2 and Cur1 promote the accumulation of Sis1 in the nucleus (see Figure 4C). However, it was unclear whether Btn2 and Cur1 can enter the nucleus unaccompanied or need to associate with Sis1 to be imported into the nucleus. To differentiate between these two possibilities, we overproduced Sis1 in cells that expressed GFP-tagged Btn2 or Cur1. As shown in Figure 5, A and B, the amount of nuclear Btn2 and Cur1 was strongly increased relative to control cells that expressed Sis1 at endogenous levels. Thus higher levels of Sis1 cause an enrichment of Btn2 and Cur1 in the nucleus. Interestingly, we not only observed an accumulation of Btn2 in the nucleus, but we also noticed a strong increase in the frequency of cells with Btn2-containing juxtanuclear foci (Figure 5C). This suggested that Sis1 affects the partitioning of Btn2 between the peripheral and juxtanuclear site.
FIGURE 5:
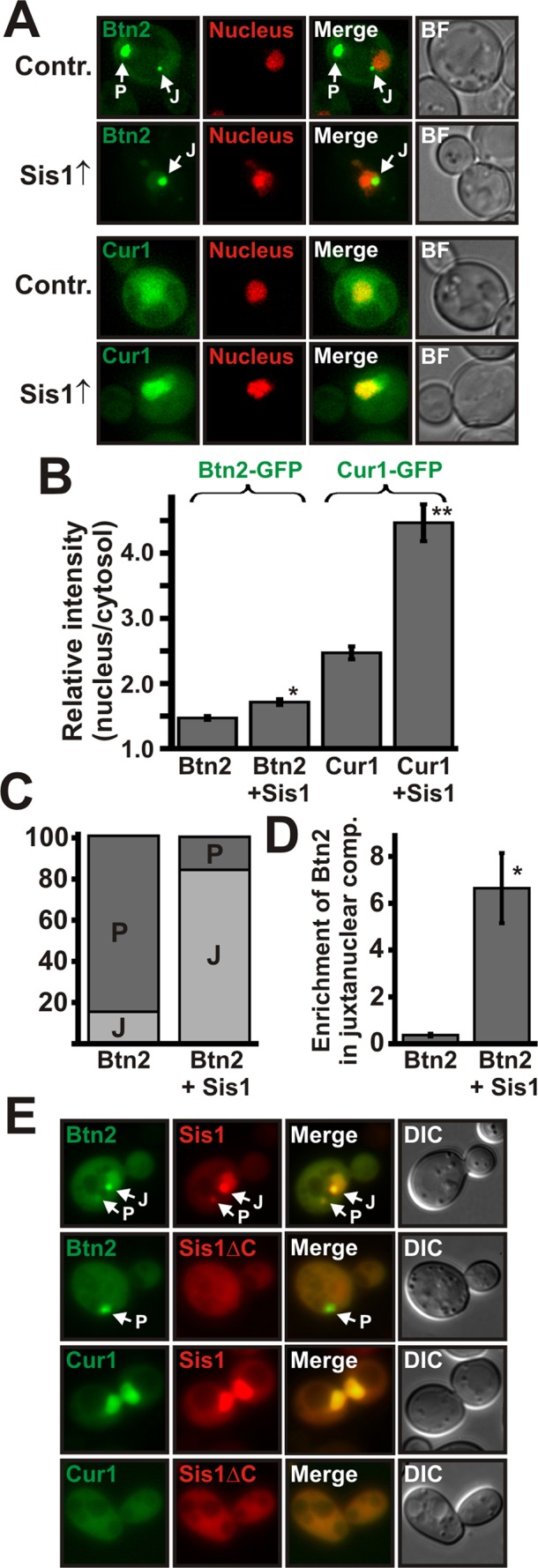
Complex formation between Sis1 and Btn2 or Sis1 and Cur1 is required for targeting to the nucleus. (A) Low-copy expression plasmids for GFP-tagged Btn2 and Cur1 were introduced into a BY4741 strain that contained a control plasmid or a low-copy expression plasmid for Sis1. Fluorescence microscopy was performed at 25°C. (B) Quantification of the relative nuclear:cytosolic GFP pixel intensity of the strains shown in A. *p = 3.3 × 10−5; **p = 2.6 × 10−10. (C) Quantification of the fraction of cells containing Btn2-positive juxtanuclear (J) and/or peripheral (P) foci. On the basis of the distribution of Bnt2-GFP, we arbitrarily divided cells into the two categories J and P (see Materials and Methods for details). (D) Juxtanuclear and peripheral signals were quantified (total integrated pixel intensity) in 30 cells that simultaneously contained one juxtanuclear and one peripheral compartment; p = 0.000094. (E) Chromosomal SIS1 was deleted in BY4741 yeast, and the deletion was covered with expression plasmids for mCherry-tagged SIS1 or SISΔC. Expression plasmids for GFP-tagged BTN2 and CUR1 were introduced, and the cells were observed by fluorescence microscopy.
To investigate this possibility, we analyzed yeast cells with a single juxtanuclear and a single peripheral focus and compared the amount of juxtanuclear Btn2 in control cells and cells that overexpressed Sis1. Remarkably, in the presence of a high Sis1 concentration, the relative amount of Btn2 in juxtanuclear sites was strongly increased (Figure 5D). Thus Sis1 can reroute Btn2 from a peripheral to a juxtanuclear location. To investigate whether these alterations require a functional NLS, we overexpressed Sis1 in yeast cells that coexpressed GFP-tagged Btn2ΔNLS or Cur1ΔNLS. As shown in Supplemental Figure S5A, additional Sis1 was not able to overcome the nuclear targeting defect of mutant Btn2 and Cur1. Collectively these data argue for the interpretation that the NLS of Btn2 and Cur1 only become functional after a complex with Sis1 has been formed.
To unequivocally demonstrate that complex formation with Sis1 is necessary for nuclear targeting of Btn2 and Cur1, we analyzed the localization of Btn2 and Cur1 in cells in which we had replaced endogenous Sis1 with a variant that is unable to associate with Btn2 and Cur1 (Sis1ΔC). In these cells, Sis1, Btn2, and Cur1 no longer accumulated in the nucleus (Figure 5E and Supplemental Figure S5B). In addition, a Btn2-positive juxtanuclear signal was no longer detectable. A Btn2-containing peripheral signal was still present, but it did not colocalize with Sis1ΔC (Figure 5E). Thus complex formation of Btn2 or Cur1 with Sis1 is required for nuclear transport and sorting to a juxtanuclear site. Sis1 is not, however, necessary for targeting Btn2 to a peripheral compartment.
Sis1 localizes to stress-inducible compartments that contain misfolded proteins and molecular chaperones
In stressed yeast cells Sis1 coalesced into foci that were reminiscent of previously reported spatial PQC compartments (Kaganovich et al., 2008; Specht et al., 2011). Therefore as a next step we investigated colocalization of Sis1 with the two misfolding-prone marker proteins VHL and Ubc9ts. As reported previously (Kaganovich et al., 2008; Specht et al., 2011), VHL and Ubc9ts formed aggregates that partitioned to peripheral and juxtanuclear compartments in cells that were exposed to elevated temperatures and MG132. The Sis1 signal showed substantial overlap with both aggregated VHL and Ubc9ts (Figure 6A). Thus, during acute heat stress Sis1 accumulates in cytosolic deposition sites that contain aggregation-prone proteins.
FIGURE 6:
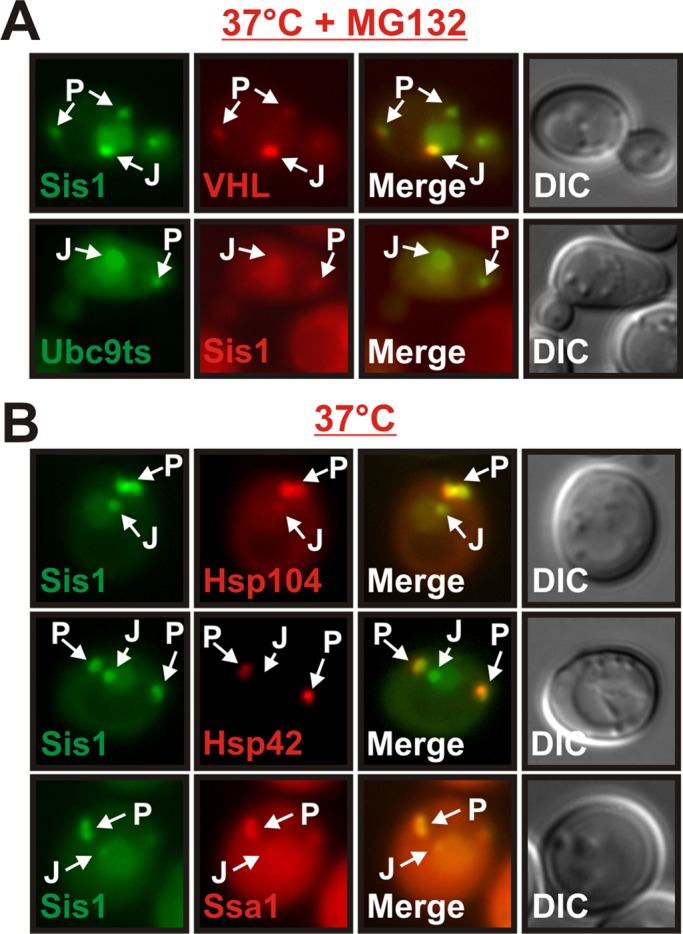
Sis1 localizes to stress-inducible compartments that contain misfolded proteins and molecular chaperones. (A) Fluorescence microscopy of BY4741 yeast cells expressing Sis1-GFP and mCherry-VHL or GFP-Ubc9ts and Sis1-mCherry. The cells were incubated at 37°C for 1 h in the presence of MG132. (B) BY4741 yeast cells carrying a GFP-tagged chromosomal copy of SIS1 and an mCherry-tagged chromosomal copy of HSP104, HSP42, or SSA1 were observed by fluorescence microscopy after growth at 37°C for 1 h.
The protein disaggregase Hsp104 localizes to juxtanuclear and peripheral compartments that contain VHL (Kaganovich et al., 2008; Specht et al., 2011). In addition, a recent study reported that Hsp42 is enriched in the peripheral compartment (Specht et al., 2011). Thus, to investigate whether Hsp42 and Hsp104 are recruited to Sis1-containing cytosolic compartments, we performed a colocalization analysis with GFP-tagged Sis1 and mCherry-tagged Hsp104 or Hsp42 at 37°C. Peripheral Sis1 foci indeed exhibited a very strong enrichment of Hsp104 and Hsp42 (Figure 6B). Juxtanuclear foci, however, contained only low amounts of Hsp104 and were devoid of Hsp42. In addition, we found that juxtanuclear and peripheral sites contained Ssa1, an Hsp70 whose ATPase activity is regulated by Sis1 (Figure 6B). Therefore, in addition to misfolded proteins, Sis1-positive compartments contain a variety of chaperones. This indicates that Sis1 is sorted to stress-inducible compartments that function as deposition sites for aggregation-prone proteins.
Localization of Btn2 to a peripheral compartment is dependent on Hsp42
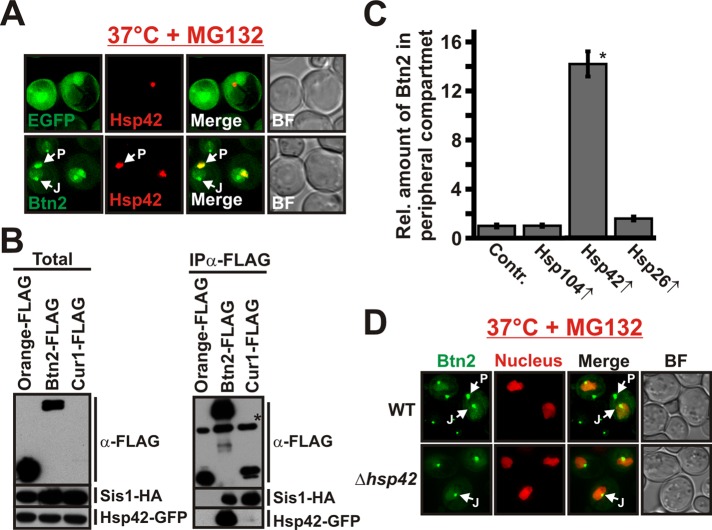
A property that distinguished Btn2 from Cur1 was that Btn2 strongly localized to a peripheral protein deposition site (see Figure 4A). Given the central role of Hsp42 in controlling the formation of this compartment (Specht et al., 2011), we hypothesized that Btn2 could be sorted to the peripheral site in an Hsp42-dependent manner. To investigate this idea, we examined the localization of Btn2-GFP in cells that expressed mCherry-tagged Hsp42 from the endogenous locus. As shown in Figure 7A, stressed yeast cells exhibited peripheral foci that contained large amounts of both proteins. Hsp42 was also strongly enriched in coimmunocomplexes of Btn2 isolated from yeast cell lysate (Figure 7B), whereas Hsp42 was undetectable in coimmunocomplexes of Cur1. Furthermore, we found that Btn2 was concentrated in the peripheral compartment in cells that overexpressed Hsp42 but showed a normal distribution in cells that overexpressed Hsp26 or Hsp104 (Figure 7C and Supplemental Figure S6A). In contrast, the amount of Cur1 that colocalized with overexpressed Hsp42 was very low (mutant Cur1 was used in this experiment to increase cytosolic protein levels; Supplemental Figure S6B). However, to unequivocally demonstrate that Hsp42 is required for enrichment of Btn2 in the peripheral compartment, we expressed GFP-tagged Btn2 in cells that lacked functional Hsp42. In these cells, Btn2 was still accumulating in the nucleus and in a juxtanuclear compartment but was no longer recruited to peripheral sites (Figure 7D). Thus association of Btn2 with Hsp42 is required for targeting to a peripheral compartment.
FIGURE 7:
Btn2 localization to a peripheral compartment is dependent on Hsp42. (A) BY4741 yeast carrying an mCherry-tagged chromosomal copy of HSP42 were transformed with low-copy expression plasmids for GFP (control) or GFP-tagged Btn2. (B) BY4741 yeast carrying a GFP-tagged chromosomal copy of HSP42 were transformed with low-copy expression plasmids for HA-tagged Sis1 and FLAG-tagged Orange (control), Btn2, or Cur1. FLAG-tagged proteins were immunoprecipitated with a specific antibody. The anti-FLAG immunoblot on the top right received only 1/10 of the control (Orange-FLAG) sample. Due to its low expression level, Cur1 was only detected in the total after longer exposure times (data not shown). The asterisk marks the heavy chain of the antibody that was used for immunoprecipitation. (C) The amount of Btn2-GFP in the peripheral compartment was quantified (total integrated pixel intensity) in BY4741 cells that expressed Hsp26, Hsp104, or Hsp42 from a low-copy plasmid; *p = 9.4 × 10−23. (D) Wild-type or Hsp42-deficient BY4741 yeast were transformed with a low-copy expression plasmid for GFP-tagged Btn2. The cells were incubated at 37°C for 1 h in the presence of MG132.
Btn2 promotes the sorting of misfolded proteins to cytosolic protein deposition sites
Our data so far suggested that Btn2 and Cur1 could be sorting factors for misfolded proteins. To provide evidence for this hypothesis, we first performed a colocalization analysis with the aggregation-prone protein VHL. As shown in Figure 8A, Btn2 was strongly enriched in juxtanuclear and peripheral sites that contained aggregated VHL. Cur1 also colocalized with VHL in the juxtanuclear compartment, but it did not overlap with VHL aggregates in the periphery (Figure 8A). To further investigate the role of Btn2 and Cur1 in aggregate sorting, we performed a time-resolved analysis of mCherry-VHL aggregation in cells that were eliminated for BTN2 and/or CUR1. In wild-type cells, VHL accumulated in juxtanuclear and peripheral foci that colocalized with GFP-tagged Sis1 (Figure 8B and Supplemental Movie S2). In Btn2-deficient cells, however, VHL was no longer sorted to juxtanuclear foci, and the size of peripheral VHL foci was strongly reduced (Figure 8B and Supplemental Movie S2). Surprisingly, sorting to both cytosolic sites was unimpaired in cells that lacked Cur1 (Figure 8B and Supplemental Movie S2). Thus, while elimination of Btn2 strongly interfered with aggregate sorting, deletion of Cur1 did not have such an effect. Therefore, as we were unable to detect a direct role for Cur1 in localizing VHL, we focused our efforts on elucidating an aggregate sorting function for Btn2.
FIGURE 8:
Btn2 promotes the sorting of misfolded proteins to cytosolic protein deposition sites. (A) Low-copy expression plasmids for Btn2-GFP or Cur1-GFP were introduced into a W303 strain that expressed mCherry-VHL. Fluorescence microscopy was performed at 37°C in the presence of MG132. (B) Wild-type, Δbtn2, Δcur1, or Δbtn2 Δcur1 BY4741 cells carrying a GFP-tagged chromosomal copy of SIS1 were transformed with a plasmid for expression of mCherry-VHL. Cells were treated as in A. (C) Low-copy expression plasmids coding for Btn2 were introduced into a BY4741 [pin-] strain expressing Hsp104-GFP from its chromosomal locus (left) or a BY4741 strain that contained an expression plasmid for GFP-VHL (right). The cells were processed for fluorescence microscopy at 25°C. (D) BY4741 yeast cells were transformed with low-copy expression plasmids for GFP-VHL and FLAG-tagged Orange (control) or Btn2. FLAG-tagged proteins were immunoprecipitated with a specific antibody. The asterisk denotes the heavy chain of the antibody that was used for immunoprecipitation. (E) Low-copy expression plasmids for Btn2-GFP or Btn2ΔNLS-GFP were introduced into W303 cells that expressed mCherry-VHL. Fluorescence microscopy was performed at 37°C in the presence of MG132. (F) Low-copy expression plasmids for Btn2 and Cur1 were introduced into a W303 strain that expressed GFP-VHL from a plasmid and Hsp42-mCherry from the endogenous locus. Strains that overexpressed Sis1 contained an additional copy of SIS1 that was integrated into the genome. Fluorescence microscopy was performed at 25°C.
To investigate the role of Btn2 in aggregate sorting, we overexpressed Btn2 in unstressed cells that expressed Hsp104-GFP as an in situ marker for aggregated proteins. Btn2 expression induced the formation of Hsp104-positive foci, whereas Hsp104 was diffusely distributed in control cells (Figure 8C, left). Likewise, in unstressed cells that coexpressed VHL and Btn2, VHL was no longer diffusely localized as in control cells but instead coalesced into foci (Figure 8C, right). Moreover, we found that VHL was specifically associated with Btn2 coimmunocomplexes that were isolated from yeast cell lysate (Figure 8D). Together, these data indicate that Btn2 actively promotes the sorting of misfolded proteins to protein deposition sites.
A previous study proposed that sorting of VHL to peripheral sites is dependent on Hsp42 (Specht et al., 2011). In agreement with this study, we found that the incidence of cytosolic deposition sites was strongly reduced in the absence of Hsp42 (Supplemental Figure S7A). The remaining fluorescent foci typically assumed a juxtanuclear position (Supplemental Figure S7B). Thus aggregate sorting to the peripheral compartment is dependent on Hsp42. To rule out the possibility that Btn2 causes VHL aggregation through induction of HSP42, we analyzed the steady-state levels of Hsp42 in control cells and cells that overexpressed Btn2. However, as can be seen in Supplemental Figure S7C, additional Btn2 did not increase the expression of Hsp42. Thus, to investigate whether Btn2 directly promotes aggregate sorting to the peripheral site, we overexpressed NLS-deleted Btn2 in stressed yeast cells that simultaneously expressed mCherry-VHL. Remarkably, Btn2ΔNLS redirected aggregated VHL away from a juxtanuclear to a peripheral site (Figure 8E). Together with the finding that peripheral VHL aggregates have a much smaller size in Btn2-deficient cells (Figure 8B and Supplemental Movie S2), this indicates that Hsp42 and Btn2 cooperate to sort misfolded proteins to a peripheral deposition site.
Btn2-deficient cells showed defects in sorting to both the peripheral and the juxtanuclear compartment. This also suggested an active role for Btn2 in the sorting of proteins to the juxtanuclear site. To validate this hypothesis, we made use of the fact that additional Sis1 can divert Btn2 to the nucleus. We coexpressed Sis1 with Btn2 in cells that simultaneously produced VHL. In the strain that expressed both Btn2 and Sis1, we observed one additional VHL focus per cell that did not colocalize with Hsp42 (Figure 8F). Thus Btn2 can redirect misfolded proteins to the juxtanuclear compartment in a Sis1-dependent manner. This is further substantiated by the finding that Sis1 is specifically associated with VHL in yeast cell lysates (Supplemental Figure S7D). Thus we conclude that Btn2 forms a complex with Sis1 to shuttle misfolded proteins to the juxtanuclear compartment.
Btn2 and Cur1 influence prion propagation indirectly through changes in the availability of Sis1
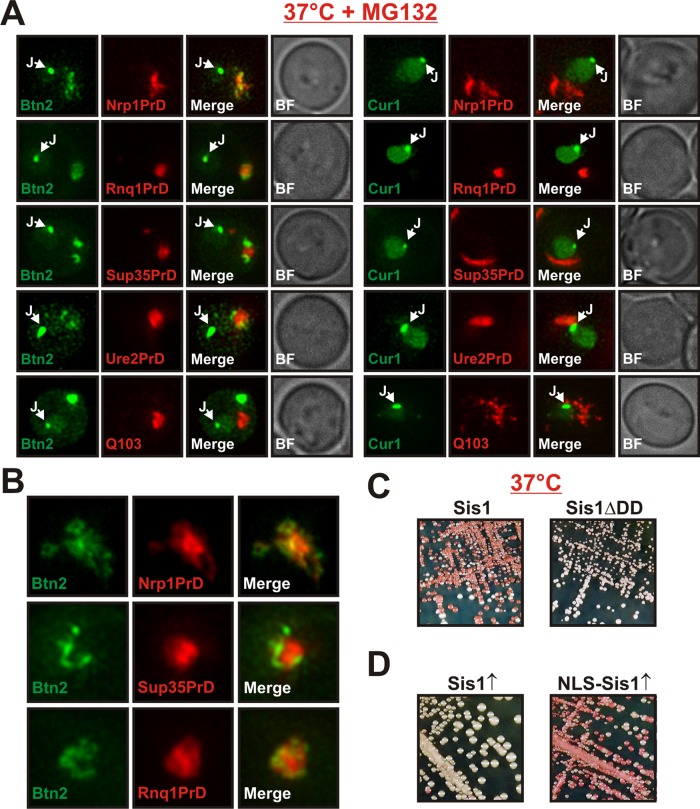
Btn2 and Cur1 were initially identified as modifiers of prion propagation (Kryndushkin et al., 2008). This opened up the possibility that Btn2 and Cur1 could target prion aggregates to cytosolic protein deposition sites. To investigate this idea, we compared the subcellular distribution of different amyloidogenic proteins in cells that coexpressed Btn2 or Cur1 using improved imaging technology (see Materials and Methods for details). The amyloidogenic proteins formed large aggregates in the cytosol of yeast cells (Figure 9A). However, these aggregates showed no overlap with Btn2 or Cur1 at the juxtanuclear site and only limited colocalization in the periphery (Figure 9, A and B). This is in agreement with a previous study, which found that Hsp42-containing compartments are spatially separated from aggregates of the prion protein Rnq1 (Specht et al., 2011). Thus yeast cells have at least two functionally distinct peripheral compartments—one for amorphous aggregates and one for terminally misfolded proteins such as prions.
FIGURE 9:
Btn2 and Cur1 influence prion propagation indirectly through changes in the availability of Sis1. (A) Low-copy expression plasmids for the expression of mCherry-tagged Nrp1PrD, Rnq1PrD, Sup35PrD, Ure2PrD, or glutamine-expanded huntingtin exon 1 (Q103) were introduced into BY4741 yeast cells that expressed GFP-tagged Btn2 or Cur1 from a plasmid. Fluorescence microscopy was performed at 25°C. (B) Magnification of peripheral aggregates in cells that coexpressed Btn2-GFP and mCherry-tagged Nrp1PrD, Sup35PrD, or Rnq1PrD. (C) Wild-type [NRP1C+] cells or cells in which chromosomal Sis1 was replaced with Sis1ΔDD were incubated at 37°C for 3 d. (D) [NRP1C+] cells were transformed with galactose-regulatable expression plasmids for Sis1 or NLS-Sis1. The transformants were streaked onto galactose plates, incubated for 3 d, transferred onto YPD plates for color development, and photographed.
The limited spatial overlap of Btn2 and Cur1 with amyloidogenic proteins and the absence of these proteins in juxtanuclear sites suggested that Btn2 and Cur1 do not directly influence the propagation of prion aggregates, as previously suggested (Kryndushkin et al., 2008). Instead, these findings hinted at the possibility that prion loss could be due to changes in the availability of Sis1. To investigate this idea, we generated a [NRP1C+] strain in which we replaced endogenous Sis1 with a mutant version that lacked the dimerization domain. The mutant strain was still able to propagate [NRP1C+] (Supplemental Figure S8A), suggesting that the dimerization motif is not required for prion maintenance. However, when we incubated mutant and wild-type [NRP1C+] strains at 37°C, the prion state was lost only in the wild-type background (Figure 9C). Thus yeast cells that contain a Sis1 variant that cannot interact with Btn2 or Cur1 are immune to stress-induced prion loss. Given the central role of Sis1 for the propagation of yeast prions (Higurashi et al., 2008), this argues for the possibility that Btn2 and Cur1 have more general effects on prion propagation than previously anticipated (Kryndushkin et al., 2008).
To investigate whether relocalization of Sis1 to the nucleus is sufficient to induce prion loss, we engineered a version of Sis1 that exclusively directs it to the nucleus. This version (termed NLS-Sis1) contained the well-characterized NLS of the viral SV40 protein. Expression of GFP-tagged NLS-Sis1 in yeast cells validated that it predominantly localized to the nucleus (Supplemental Figure S8B). Because functional Sis1 forms a homodimer (Sha et al., 2000), we hypothesized that overexpression of NLS-Sis1 could redirect endogenous Sis1. Indeed, in cells expressing NLS-Sis1, we observed accumulation of endogenous Sis1 in the nucleus to a degree that was comparable to that of stressed yeast cells (Supplemental Figure S8C). Remarkably, we found that NLS-Sis1 expression alone was able to destabilize [NRP1C+], whereas expression of wild-type Sis1 was not (Figure 9D). In agreement with this result, we also observed that the NLS-deleted versions of Btn2 and Cur1 had a strongly reduced ability to promote prion loss (Supplemental Figure S8D), despite increased expression levels (see, e.g., Supplemental Figure S4C). Thus stress-induced relocalization of Sis1 to the nucleus is sufficient to cause the loss of [NRP1C+] in dividing yeast cells.
Collectively these data indicate that sorting to the correct destination compartment is strongly dependent on the structure of a misfolded protein. They also imply that yeast cells contain several functionally distinct protein deposition sites that compete for PQC components.
Cur1 regulates the partitioning of substrate proteins between juxtanuclear and peripheral compartments
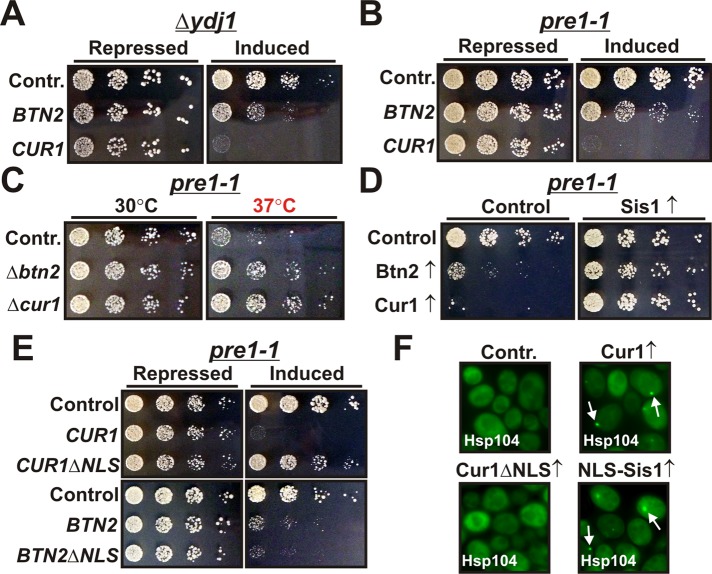
Our findings so far identified Btn2 as an important sorting factor for soluble misfolded proteins. Surprisingly, this function was unique to Btn2 and not shared by the paralogue Cur1 (see Figure 8). Therefore, to elucidate the role of Cur1, we compared the functional effects of Btn2 and Cur1 in yeast cells that contained mutations in PQC components. First, we investigated cells that were deficient for Ydj1, the other major Hsp40 in the yeast cytosol. Importantly, deletion of the nonessential YDJ1 is associated with a growth defect that can partially be restored by excess Sis1 (Caplan and Douglas, 1991). As can be seen in Figure 10A, expression of both Btn2 and Cur1 strongly aggravated the slow-growth phenotype of YDJ1-deficient cells. However, the growth-inhibiting effect of Cur1 was much more pronounced than that of Btn2, despite lower expression levels (see, e.g., Figure 2C). Growth was also negatively affected in wild-type cells but to a much lesser degree (Supplemental Figure S9A).
FIGURE 10:
Cur1 regulates the partitioning of substrate proteins between the juxtanuclear and peripheral compartments. (A) BY4741 yeast cells carrying a chromosomal deletion of YDJ1 were transformed with galactose-regulatable expression plasmids for BTN2 or CUR1. Cells were grown overnight in glucose-containing media. Fivefold serial dilutions were prepared and spotted onto either glucose-containing (repressing) or galactose-containing (inducing) plates. (B) Yeast cells carrying a temperature-dependent mutation in PRE1 (pre1-1) were transformed with galactose-regulatable expression plasmids for BTN2 and CUR1. Cells were treated as in A. (C) Deletions of BTN2 or CUR1 were introduced into the pre1-1 strain and tested for growth on rich medium at the indicated temperatures. (D) Pre1-1 yeast cells were transformed with a plasmid for constitutive Sis1 expression and galactose-regulatable expression plasmids for Btn2 and Cur1. The cells were spotted onto either glucose- or galactose-containing plates. The plates were incubated at 30°C. (E) Cells were treated as in D using plasmids for the indicated proteins. (F) BY4741 yeast expressing Hsp104-GFP from the endogenous locus were transformed with low-copy expression plasmids for Cur1, Cur1ΔNLS, or NLS-Sis1. The transformants were grown at 25°C and analyzed by fluorescence microscopy.
We next studied the functional effects of Btn2 and Cur1 in yeast cells with defects in the proteasome. For this purpose, we used a mutant that carried a temperature-sensitive mutation in the proteasome subunit PRE1. Of note, this mutant had previously been reported to show a functional interaction with SIS1 (Ohba, 1997). Similar to our previous findings with YDJ1-deficient cells, Cur1-expressing cells exhibited a stronger growth defect than cells that expressed Btn2 (Figure 10B). Importantly, stressed pre1-1 cells that were eliminated for Btn2 or Cur1 displayed the opposite phenotype, namely improved growth (Figure 10C; we did not test the effects of Btn2 and Cur1 deletion in the Δydj1 background because the strain was unable to grow at temperatures >30°C). Collectively these findings suggested that Cur1 and to a lesser extent Btn2 could negatively affect the function of Sis1.
To demonstrate that the observed growth effects were indeed caused by changes in the activity or availability of Sis1, we performed growth assays with cells that expressed a higher amount of Sis1. In these cells, the phenotype of Cur1-expressing cells was strongly improved (Figure 10D and Supplemental Figure S9B). The growth of Btn2-expressing cells was also alleviated but to a lesser degree (Figure 10D and Supplemental Figure S9B). To further investigate whether the growth-inhibiting effects of Btn2 and Cur1 required nuclear targeting, we compared the growth phenotypes of strains that overexpressed wild-type or NLS-deleted Btn2 or Cur1 (Figure 10E and Supplemental Figure S9C). Remarkably, deletion of the NLS motif completely abrogated the negative-growth phenotype of Cur1. Depletion of cytosolic Sis1 by overexpression of NLS-Sis1 likewise impaired yeast cell growth in the pre1-1 background (Supplemental Figure S9D). Thus we conclude that the growth-interfering activity of Cur1 is solely due to its ability to accumulate Sis1 in the nucleus.
On the basis of these observations, we theorized that the main function of Cur1 is to control the cytosolic availability of Sis1. Given that Sis1 is required for sorting to the juxtanuclear compartment, this suggested that Cur1 could regulate the partitioning of misfolded proteins between the nucleus and the periphery. In agreement with this idea, we noticed that the size of the juxtanuclear compartment was increased in Cur1-deficient cells (Supplemental Movies S1 and S2). Moreover, when we overexpressed Cur1 in yeast cells that expressed Hsp104 as an in situ marker for protein aggregates, we observed the formation of small peripheral foci (Figure 10F). Of importance, the formation of these deposition sites was not due to an induction of Hsp42 (Supplemental Figure S9E). Similar results were obtained with cells that expressed NLS-Sis1 (Figure 10E). Thus cytosolic depletion of Sis1 by Cur1 is sufficient to divert misfolded proteins to the periphery.
Taken together, these findings strongly argue for the possibility that the spatial PQC machinery is also active in cells growing under normal growth conditions. They also imply that the default direction of sorting is toward the nucleus. In summary, we conclude that misfolded proteins are not randomly distributed in the cytoplasm but are shuttled to specific sites through the concerted action of a network of stress-inducible protein-sorting factors and molecular chaperones.
DISCUSSION
Mounting evidence indicates that spatial PQC pathways are an integral part of the defense arsenal of eukaryotic organisms against environmental stress and the proteotoxicity that is associated with aging and disease (Kaganovich et al., 2008; Tyedmers et al., 2010; Specht et al., 2011). In this work, we identified Hsp42, Btn2, Cur1, and Sis1 as major players that regulate the sorting and sequestration of misfolded proteins in yeast cells that are exposed to mild heat stress. Together, our findings argue for the following molecular model (Figure 11).
FIGURE 11:
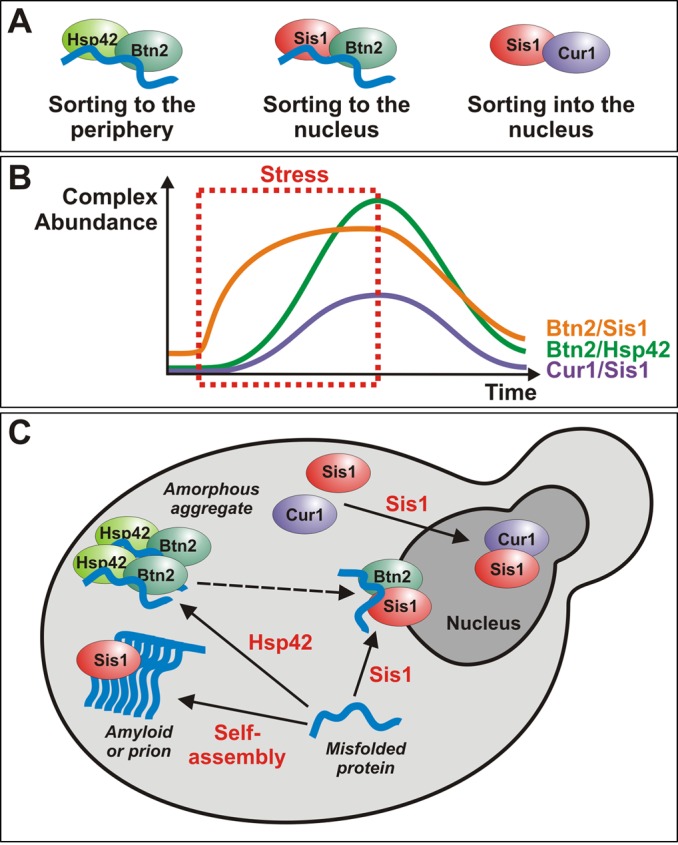
A model for the coordinated action of sorting factors and molecular chaperones during acute heat stress. (A) Schematic representation of the different sorting complexes that were identified in this work. A bound misfolded protein is depicted in blue. Note that only the complexes containing Btn2 are involved in aggregate sorting, whereas the Cur1 complex mediates sorting of Sis1 to the nucleus. (B) Schematic representation of the changes in complex abundance that happen before during and after a stress stimulus. (C) A model that summarizes the role of Btn2, Cur1, Sis1, and Hsp42 in the sorting of misfolded proteins to deposition sites. The events that induce sorting (association with a factor or assembly into an amyloid structure) are indicated in red. Arrows indicate the direction of sorting. The dashed arrow indicates that it is unclear whether misfolded proteins can directly be sorted from the peripheral to the juxtanuclear compartment.
Previous studies proposed that juxtanuclear and peripheral compartments serve different functions (Kaganovich et al., 2008; Specht et al., 2011). Substrate proteins in the juxtanuclear sites are more likely to be ubiquitylated and display a higher mobility. This led to the hypothesis that juxtanuclear substrates could be selectively eliminated by proteasomal degradation. In contrast, the peripheral compartment was suggested to protect the cell from proteotoxicity or promote the clearance of aggregates by autophagy. Our study now makes the important contribution that sorting is strongly dependent on whether a misfolded protein is able to associate with Hsp42 or Sis1. This suggests that changes in the expression level of these proteins and their cytosolic availability are important determinants for aggregate sorting. In agreement with this notion, the subcellular organization of protein aggregates is highly dynamic and changes with the duration of the stress (Figure 11).
On the basis of our observations (Supplemental Movie S1) and those of other groups (Kaganovich et al., 2008; Specht et al., 2011), the distribution of aggregates during mild heat stress follows a fixed temporal and spatial pattern: on sudden heat stress, yeast cells first form a juxtanuclear site, but the peripheral compartment rapidly catches up. When the stress stimulus persists, the peripheral sites become more abundant and grow in size. In the recovery phase, both compartments slowly disintegrate. We propose that these changes are predominantly determined by temporal changes in the relative cytosolic concentrations of Sis1 and Hsp42. At the onset of a stress stimulus, cytosolic Sis1 levels are high. Thus, in the early phase, Btn2 is able to team up with Sis1 to target misfolded proteins to the juxtanuclear site. Over time, however, induction of Cur1 leads to a gradual depletion of Sis1 from the cytosol. As a consequence, substrate flux to the juxtanuclear compartment decreases. This change is accompanied by an increased aggregate transport to the periphery caused by induction of HSP42. In the recovery phase, Sis1 reenters the cytosol and can engage with substrate proteins that are released from the disintegrating peripheral compartment.
Several lines of evidence support this model. First, Sis1 levels are only weakly up-regulated during stress (Luke et al., 1991), whereas Hsp42 levels are strongly induced (Haslbeck et al., 2004). Second, the main function of Cur1 during stress seems to be the depletion of Sis1 from the cytosol and not aggregate sorting. Third, yeast cells that lack or overexpress Cur1 display alterations in the partitioning of misfolded proteins. Finally, although Sis1 is recruited to peripheral aggregates, it is not required for targeting misfolded proteins to the periphery (see Figures 5E and 8F). Instead, it seems likely that peripheral Sis1 is involved in recruiting Ssa1 and Hsp104 to disengage misfolded proteins from Hsp42. Association of Sis1 with Btn2 could then facilitate the transfer of released proteins to the nucleus. Thus temporal changes in the cytosolic concentration of chaperones and protein-sorting factors determine whether aggregated proteins are stored in the periphery or shuttled to a juxtanuclear site, most likely to facilitate their degradation by the proteasome. Detailed quantitative measurements of the cytosolic concentrations of the involved proteins with high temporal and spatial resolution will be required to determine whether the proposed model holds true or whether other proteins and mechanisms are involved. In fact, it is very likely that additional sorting systems are activated when yeast cells are exposed to more severe heat stress or other stress stimuli such as oxidative stress (Molin et al., 2011).
The model outlined here requires that the expression levels of the involved proteins are exquisitely fine tuned. Indeed, transcription of BTN2 and CUR1 is strongly induced by stress (Gasch et al., 2000). Moreover, Btn2 and Cur1 are very unstable proteins that are rapidly turned over by the proteasome. However, when the activity of the proteasome is reduced during stress, they accumulate. Thus the unstable nature of Btn2 and Cur1 creates a dynamic feedback mechanism that ensures that the sorting machinery is only turned on when the capacity of the proteasome is exceeded by a high load of misfolded substrate proteins. The same hallmarks, namely inducibility by heat and rapid turnover by the proteasome, have recently been reported for Lsb2, a short-lived, actin-associated protein that affects the maintenance of a different yeast prion (Chernova et al., 2011). Thus the ubiquitin/proteasome system could have a general role in creating a time window of expression for proteins that are involved in stress-induced aggregate sorting.
How does Btn2 promote the sorting of misfolded proteins? The fact that Btn2 is a homologue of mammalian Hook proteins suggests that it could associate with the cytoskeleton to facilitate the transfer of cargo proteins. In agreement with this idea, a recent study reported that an intact actin network is absolutely required for aggregate sorting (Specht et al., 2011). Although our work provides evidence for an essential function in aggregate sorting, it remains to be seen whether Btn2 is also required for protein sequestration. Btn2-deficient cells are still able to sequester proteins in a peripheral location, albeit with reduced efficiency. This indicates that protein retention in the periphery is predominantly mediated by Hsp42, as previously suggested (Specht et al., 2011). However, Btn2 could have an important role in concentrating proteins at the nuclear envelope. This is suggested by our finding that juxtanuclear VHL aggregates are undetectable in Btn2-deficient cells. However, additional proteins might be required. Sis1, for example, could play an important role because Hsp40s can coassemble with misfolded proteins and oligomerize in a manner that is similar to small HSP (sHSP) aggregation (Kampinga and Craig, 2010).
Nuclear accumulation of heat shock proteins is a well-known phenomenon during stress, but it is generally believed to be associated with an essential function in the nucleus. Our findings, however, suggest that chaperone targeting to the nucleus can also have important regulatory functions in the cytosol. Btn2 and Cur1 are only able to enter the nucleus when they are complexed with Sis1. This mechanism ensures the time-dependent accumulation of Sis1 in the nucleus and thus its removal from cytosolic sorting pathways. Importantly, nuclear import of Sis1 requires NLS motifs in Btn2 and Cur1 and the nuclear import factor Srp1. The fact that Srp1 is able to recognize the NLS of Btn2 and Cur1 in vitro in the absence of Sis1 (Supplemental Figure S4F), however, suggests that association with Srp1 is not sufficient to induce nuclear targeting. Thus the nuclear targeting step could be subject to another layer of regulation in the cellular environment. A possible scenario is that association with Sis1 induces a conformational change that makes the NLS motifs more accessible. Alternatively, Sis1 could itself interact with additional nuclear targeting factors that are required for nuclear import. These and other open questions will require a thorough analysis of the structure/function relationships of the involved proteins and their reconstitution into active complexes in vitro.
Our study demonstrates that the complexity of aggregate sorting is much greater than previously anticipated. Yeast cells have at least two distinct compartments in the periphery—one for soluble and one for amyloidogenic proteins. This indicates that structural aspects of misfolded proteins are the most important determinants for aggregate sorting. In contrast to soluble proteins such as VHL, amyloidogenic proteins can self-assemble. Hence, the ability to form a replication-competent structure could be a key requirement for the localization to amyloid- and prion-containing sites. Such a self-aggregation mechanism is in contrast to the factor-assisted aggregation that we observed for soluble misfolded proteins such as VHL. Why yeast cells actively promote the aggregation of these misfolded proteins during stress needs to be determined. The most likely explanation, however, is that protein deposition decreases the substrate load of the PQC system. An additional mechanism is suggested by our observation that Sis1-positive foci are selectively retained in the mother cell when yeast cells reenter the cell cycle (Supplemental Movie S3). This asymmetric partitioning of aggregated proteins during cell division could be a mechanism to rejuvenate the progeny after heat stress and thus reduce aging.
Do higher organisms possess factors that have similar functions as Sis1, Hsp42, Btn2, and Cur1? Some mammalian Hsp40s shuttle between the nucleus and the cytosol and could therefore act analogously to Sis1 (Cheng et al., 2008; Zhang et al., 2008). Moreover, the Hook2 protein, a homologue of Btn2 and Cur1, was shown to promote the formation of mammalian aggresomes (Szebenyi et al., 2007). Association with aggresomes has also been described for mammalian sHSPs (Ito et al., 2002). Wickner and colleagues addressed the question of conservation by expressing Hook1 in yeast cells but did not find a prion curing effect (Kryndushkin et al., 2008). However, as we have shown, prion loss is largely due to changes in the availability of Sis1. It is therefore likely that mammalian Hook1 does not recognize Sis1. These data suggest that certain aspects of these proteins, such as their transport function, may have been conserved from yeast to human.
Many facts remain to be learned about protein homeostasis in eukaryotic cells. An entirely new level of complexity has recently been added by the discovery of diverse spatial PQC pathways. We have provided important insight into the molecular underpinnings of the spatial PQC machinery of the model eukaryote S. cerevisiae. Our study provides a starting point for future studies in yeast and other model organisms that will reveal additional aspects about how cells manage misfolded and aggregation-prone proteins. Undoubtedly, these studies will provide important insights into the causes and consequences of protein-misfolding diseases and aging.
MATERIALS AND METHODS
Cloning procedures
Cloning procedures were performed as described previously (Alberti et al., 2007, 2009; Halfmann et al., 2011). The variant version of Sis1 was generated synthetically as described in the Supplemental Information.
Yeast techniques, strains, and media
The media used were standard synthetic (SD or SGal) media or rich media containing 2% d-glucose (YPD) or 2% d-galactose (YPGal). The yeast strain backgrounds used in this study were W303 ADE+ (leu2-3112; his3-11,-15; trp1-1; ura3-1; can1-100; [psi-]; [PIN+]) or BY4741 (his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; [psi-]; [PIN+]). See Alberti et al. (2009) for details on the generation of the [NRP1C+] prion strain. Construction of the temperature-sensitive proteasome mutant strains Pre1-1 and Cim3-1 was described previously (Ghislain et al., 1993; Gerlinger et al., 1997). Srp1-31 strain construction was reported elsewhere (Tabb et al., 2000). A list of the strains used in this study can be found in Supplemental Table S3. Yeast cells were treated with the proteasome inhibitor MG132 as described previously (Liu et al., 2007).
Yeast gene deletions were performed using a PCR-based approach (Gueldener et al., 2002). The plasmids pUG6 or pUG27 were used for construction of gene deletion cassettes. Occasionally, the plasmid pAG32 was used (Goldstein and McCusker, 1999). C-terminal tagging of yeast genes was performed as described previously (Sheff and Thorn, 2004).
For analysis of yeast cell growth, cells were transformed with low- or high-copy constructs. The cells were grown overnight in glucose-containing media lacking uracil and/or leucine and washed once in water before the preparation of fivefold serial dilutions in water. Subsequently, the cells were spotted to either glucose-containing (repressing) or galactose-containing (inducing) media lacking uracil and/or leucine and incubated at different temperatures.
Microscopy and image analysis
General fluorescence microscopy (Figures 3, B and C, 5E, 6, A and B, 8, C and F, and 10F and Supplemental Figures S2E, S4D, S6, A and B, S7B, and S8, B and C) was performed using an Olympus BX61 microscope with a 100× oil immersion objective and MetaVue 5.0 software (Universal Imaging Corporation, West Chester, PA). All other images and time-lapse movies were acquired using a Deltavision microscope system with softWoRx 4.1.2 software (Applied Precision, Issaquah, WA). The system was based on an Olympus IX71 microscope, which we used with a 100×/1.4 numerical aperture (NA; or 150×/1.45 NA) objective. The images were collected with a Cool SnapHQ camera (Photometrics, Tucson, AZ) as 352 × 352 pixel (or 384 × 384 pixel) files using 1 × 1 (or 2 × 2) binning. All images were deconvolved using standard softWoRx deconvolution algorithms (enhanced ratio, high-to-medium noise filtering). Images acquired with the Deltavision setup were maximum-intensity projections of at least 20 individual images. Representative cells are shown in all figures, and each experiment was performed independently three times. We performed several control experiments with GFP or mCherry alone at different temperatures (unpublished data). In these experiments, we did not observe coalescence of GFP or mCherry into fluorescent foci or association with PQC components. In addition, GFP and mCherry did not colocalize with foci-forming proteins such as Hsp42 or Sis1.
Image quantification was performed using Fiji image analysis software. A square of fixed size that approximately corresponded to the area of one-fifth of a yeast nucleus was selected, and the average pixel intensity in the cytoplasm or nucleoplasm was measured using nondeconvolved images. To avoid irregularities or bright fluorescent foci distorting the measured overall pixel intensity, we placed the square in areas with diffuse and uniform fluorescence signals. The ratio of the average pixel intensity was obtained by dividing the nucleoplasmic intensity by the cytoplasmic intensity. For quantification of the size and intensity of juxtanuclear and peripheral foci, the total integrated pixel intensity of a two-dimensional object was determined from nondeconvolved images. Unless otherwise indicated, 50 values from different cells were averaged. All error bars designate the SE of the mean.
For quantification of the frequencies of the juxtanuclear and peripheral compartments, cells were arbitrarily divided into two categories (Figure 5C). Cells that exhibited only a juxtanuclear compartment or a juxtanuclear site that was larger than the peripheral site were assigned to category J. Cells with peripheral signals that were equal or larger than the juxtanuclear signal or cells that only had a peripheral compartment were assigned to category P.
Coimmunoprecipitation of yeast proteins from cell lysate
Yeast cells were transformed with a low-copy plasmid for the expression of a FLAG-tagged or hemagglutinin (HA)-tagged bait protein. The transformants were grown in selective liquid medium overnight, harvested by centrifugation, and resuspended in IP buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 5 mM EDTA; 1% Triton X-100) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF], 1.25 mM benzamidine, 10 μg/ml pepstatin, 10 μg/ml chymostatin, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml E-64). The resuspended cells were transferred to a tube containing glass beads. The cells were lysed using a TissueLyser II instrument (Qiagen, Valencia, CA) for 25 min at 25 Hz. The cell lysate was centrifuged for 5 min at 10,000 rpm in a tabletop centrifuge, and the supernatant was transferred to a new precooled tube. A 3-μl portion of the anti-FLAG M2 or anti-HA antibody was added, and the samples were rotated end over end for 2 h. We added 40 μl of protein G–Sepharose slurry, and the samples were rotated for an additional hour. The beads were collected by centrifugation and washed three to five times with 1 ml of cold IP buffer. Finally, the beads were resuspended in sample buffer and boiled for 10 min before loading onto a SDS–PAGE gel.
Protein purification and protein-binding assays
Btn2, Btn2ΔNLS, Cur1, Cur1ΔNLS, and EGFP were expressed as GST fusions in Escherichia coli BL21 DE3. Proteins were purified from bacterial lysates using glutathione–Sepharose 4B beads (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. Srp1 and Sis1 were expressed as His fusions and purified from E. coli BL21 DE3 cell lysates on HisTrap HP columns (GE Healthcare) using a gradient elution profile according to the manufacturer's protocol. For protein-binding studies, GST and His fusion proteins were incubated in equimolar amounts with glutathione–Sepharose beads (GE Healthcare) in phosphate-buffered saline (PBS) for 2 h at 4°C. The beads were collected by centrifugation at 9000 × g and washed three times with PBS containing 1% Tween 20. The proteins were eluted in a two-step elution by resuspending the beads in PBS containing 0.1 M glutathione, followed by 15 min of incubation at room temperature. The pooled eluates were precipitated with 12.5% trichloroacetic acid and analyzed by immunoblotting with specific antibodies.
Semidenaturing detergent–agarose gel electrophoresis
Semidenaturing detergent–agarose gel electrophoresis was performed as described (Alberti et al., 2010).
Gel filtration of yeast cell lysate
Cells were harvested in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1.0% Triton X-100, 0.4 mM PMSF, 8 mM N-ethylmaleimide, 1.25 mM benzamidine, 10 μg/ml pepstatin, 10 μg/ml chymostatin, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml E-64) and lysed using glass beads (600 μm; Sigma-Aldrich, St. Louis, MO) for 20 min in a TissueLyser II instrument (Qiagen) at 25 Hz. Beads were sedimented by centrifugation (2 min, 900 × g, 4°C), and the supernatant was applied onto Spin-X Centrifuge Tube filters (0.22 μm; Corning, Corning, NY) to be cleared of cellular debris (10,000 × g, 5 min, 4°C). Samples were resolved on a Superose 6HR 10/30 column (GE Healthcare), and 500-μl fractions were analyzed using the MiniFold DotBlot System (Whatman, GE Healthcare).
Supplementary Material
Acknowledgments
We thank C. Norden, R. Ernst, M. Coelho, and other members of the Max Planck Institute of Molecular Cell Biology and Genetics for critical reading of the manuscript. In addition, we thank S. Lindquist, D. Wolf, K. Mann, J. Frydman (through Addgene, Cambridge, MA), M. Nomura, and M. Route for reagents. B. Borgonovo and D. White are acknowledged for expert technical assistance. We are grateful to the Max Planck Society and the Dresden International Graduate School for Biomedicine and Bioengineering program for funding.
Abbreviations used:
- HSP
heat shock protein
- IPOD
insoluble protein deposit
- JUNQ
juxtanuclear protein quality control
- NLS
nuclear localization sequence
- PQC
protein quality control
- PrD
prion domain
- sHSP
small heat shock protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0194) on June 20, 2012.
*These authors contributed equally to this work
REFERENCES
- Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Halfmann R, Lindquist S. Biochemical, cell biological, and genetic assays to analyze amyloid and prion aggregation in yeast. Methods Enzymol. 2010;470:709–734. doi: 10.1016/S0076-6879(10)70030-6. [DOI] [PubMed] [Google Scholar]
- Allen KD, Wegrzyn RD, Chernova TA, Muller S, Newnam GP, Winslett PA, Wittich KB, Wilkinson KD, Chernoff YO. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchsenius AS, Wegrzyn RD, Newnam GP, Inge-Vechtomov SG, Chernoff YO. Yeast prion protein derivative defective in aggregate shearing and production of new “seeds.”. EMBO J. 2001;20:6683–6691. doi: 10.1093/emboj/20.23.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Douglas MG. Characterization of YDJ1: a yeast homologue of the bacterial dnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Pearce DA. Interaction with Btn2p is required for localization of Rsglp: Btn2p-mediated changes in arginine uptake in Saccharomyces cerevisiae. Eukaryotic Cell. 2002;1:606–612. doi: 10.1128/EC.1.4.606-612.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Belshan M, Ratner L. Hsp40 facilitates nuclear import of the human immunodeficiency virus type 2 Vpx-mediated preintegration complex. J Virol. 2008;82:1229–1237. doi: 10.1128/JVI.00540-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO. Stress and prions: lessons from the yeast model. FEBS Lett. 2007;581:3695–3701. doi: 10.1016/j.febslet.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chernova TA, et al. Prion induction by the short-lived, stress-induced protein lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol Cell. 2011;43:242–252. doi: 10.1016/j.molcel.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger UM, Guckel R, Hoffmann M, Wolf DH, Hilt W. Yeast cycloheximide-resistant crl mutants are proteasome mutants defective in protein degradation. Mol Biol Cell. 1997;8:2487–2499. doi: 10.1091/mbc.8.12.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Alberti S, Krishnan R, Lyle N, O'Donnell CW, King OD, Berger B, Pappu RV, Lindquist S. Opposing effects of glutamine and asparagine govern prion formation by intrinsically disordered proteins. Mol Cell. 2011;43:72–84. doi: 10.1016/j.molcel.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J. Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J. 2004;23:638–649. doi: 10.1038/sj.emboj.7600080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci USA. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Kamei K, Iwamoto I, Inaguma Y, Garcia-Mata R, Sztul E, Kato K. Inhibition of proteasomes induces accumulation, phosphorylation, and recruitment of HSP27 and alphaB-crystallin to aggresomes. J Biochem. 2002;131:593–603. doi: 10.1093/oxfordjournals.jbchem.a003139. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kama R, Robinson M, Gerst JE. Btn2, a Hook1 ortholog and potential Batten disease-related protein, mediates late endosome-Golgi protein sorting in yeast. Mol Cell Biol. 2007;27:605–621. doi: 10.1128/MCB.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti V, Kama R, Gerst JE. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol Biol Cell. 2011;22:1648–1663. doi: 10.1091/mbc.E10-11-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Shewmaker F, Wickner RB. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 2008;27:2725–2735. doi: 10.1038/emboj.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Fan CY, Younger JM, Ren H, Cyr DM. Identification of essential residues in the type II Hsp40 Sis1 that function in polypeptide binding. J Biol Chem. 2002;277:21675–21682. doi: 10.1074/jbc.M111075200. [DOI] [PubMed] [Google Scholar]
- Liu C, Apodaca J, Davis LE, Rao H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques. 2007;42:158, 160, 162. doi: 10.2144/000112389. [DOI] [PubMed] [Google Scholar]
- Lopez N, Aron R, Craig EA. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+] Mol Biol Cell. 2003;14:1172–1181. doi: 10.1091/mbc.E02-09-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke MM, Sutton A, Arndt KT. Characterization of SIS1, a Saccharomyces cerevisiae homologue of bacterial dnaJ proteins. J Cell Biol. 1991;114:623–638. doi: 10.1083/jcb.114.4.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin M, Yang J, Hanzen S, Toledano MB, Labarre J, Nystrom T. Life span extension and H(2)O(2) resistance elicited by caloric restriction require the peroxiredoxin Tsa1 in Saccharomyces cerevisiae. Mol Cell. 2011;43:823–833. doi: 10.1016/j.molcel.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Newnam GP, Birchmore JL, Chernoff YO. Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol. 2011;408:432–448. doi: 10.1016/j.jmb.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba M. Modulation of intracellular protein degradation by SSB1-SIS1 chaperon system in yeast S. cerevisiae. FEBS Lett. 1997;409:307–311. doi: 10.1016/s0014-5793(97)00535-8. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Langseth SX, Serio TR. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:e24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B, Lee S, Cyr DM. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure. 2000;8:799–807. doi: 10.1016/s0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht S, Miller SB, Mogk A, Bukau B. Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol. 2011;195:617–629. doi: 10.1083/jcb.201106037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebenyi G, Wigley WC, Hall B, Didier A, Yu M, Thomas P, Kramer H. Hook2 contributes to aggresome formation. BMC Cell Biol. 2007;8:19. doi: 10.1186/1471-2121-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb MM, Tongaonkar P, Vu L, Nomura M. Evidence for separable functions of Srp1p, the yeast homolog of importin alpha (Karyopherin alpha): role for Srp1p and Sts1p in protein degradation. Mol Cell Biol. 2000;20:6062–6073. doi: 10.1128/mcb.20.16.6062-6073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for Sis1 in delivering substrates to Hsp104. Mol Cell. 2008;32:584–591. doi: 10.1016/j.molcel.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Craig EA. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol. 1999;19:7751–7758. doi: 10.1128/mcb.19.11.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang Z, Cao Y, Zhang S, Li H, Huang Y, Ding YQ, Liu X. The Hsp40 family chaperone protein DnaJB6 enhances Schlafen1 nuclear localization which is critical for promotion of cell-cycle arrest in T-cells. Biochem J. 2008;413:239–250. doi: 10.1042/BJ20071510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.