Through an RNAi-based modifier screen, we identified the nucleoporin Nup205/NPP-3 as a negative regulator of mitotic onset in Caenorhabditis elegans. Strikingly, NPP-3 is lost from the nuclear envelope at mitotic onset in an AIR-1– and centrosome-dependent manner. We propose a model whereby centrosomes and AIR-1 promote timely mitosis by locally removing NPP-3.
Abstract
Regulation of mitosis in time and space is critical for proper cell division. We conducted an RNA interference–based modifier screen to identify novel regulators of mitosis in Caenorhabditis elegans embryos. Of particular interest, this screen revealed that the Nup205 nucleoporin NPP-3 can negatively modulate the timing of mitotic onset. Furthermore, we discovered that NPP-3 and nucleoporins that are associated with it are lost from the nuclear envelope (NE) in the vicinity of centrosomes at the onset of mitosis. We demonstrate that centrosomes are both necessary and sufficient for NPP-3 local loss, which also requires the activity of the Aurora-A kinase AIR-1. Our findings taken together support a model in which centrosomes and AIR-1 promote timely onset of mitosis by locally removing NPP-3 and associated nucleoporins from the NE.
INTRODUCTION
In eukaryotes, the genetic material is separated from the rest of the cell by the nuclear envelope (NE). To ensure genome stability upon cell division, the genetic material must be distributed faithfully to the two daughter cells. For this to be the case, chromosome segregation is coordinated with other mitotic events, including reorganization of the microtubule network and, in most eukaryotes, nuclear envelope breakdown (NEBD). How such coordination is achieved in time and space, in particular in a developing organism, is not fully understood.
In interphase, the NE constitutes a physical boundary that allows transport of molecules in and out of the nucleus through gates that are embedded in the NE and called nuclear pore complexes (NPCs) (reviewed in Hoelz et al., 2011). NPCs are large proteinaceous structures composed of nucleoporins that associate in distinct subcomplexes. There are approximately 30 nucleoporins in vertebrates (Antonin et al., 2008). Homologues for 23 of these have been identified in Caenorhabditis elegans and are referred to as NPP-1 to NPP-23 (Galy et al., 2003; Rodenas et al., 2012) (Table S1). Depletion of most nucleoporins in C. elegans affects nuclear morphology and/or permeability, as well as nuclear reassembly following mitosis (Galy et al., 2003, 2006; Schetter et al., 2006; Rodenas et al., 2009).
NPCs undergo dramatic reorganization and disassembly upon entry into mitosis. In human cells, live imaging with fluorescently tagged fusion proteins established that Nup98 is the first nucleoporin to dissociate from the NE in prometaphase (Dultz and Ellenberg, 2010). This is followed by the early and rapid departure from the NE of members of the Nup93 complex, as well as of the Nup107–160 and Nup214 complexes. Finally, the transmembrane nucleoporin POM121 dissociates from the NE and redistributes to the endoplasmic reticulum (ER); this occurs concomitantly to the disassembly of the nuclear lamina that underlies the NE (Beaudouin et al., 2002; Dultz et al., 2008). This sequence of events mirrors observations in starfish oocytes, where the nucleoporins Nup98, Nup153, Nup214, and POM121 depart sequentially from the NE (Lenart et al., 2003).
Phosphorylation events play an important role in the mitotic dynamics of nucleoporins. For instance, phosphorylation of Nup98 by Cdk1 and members of the NIMA-related kinase family of kinases in human cells promotes Nup98 removal from NPCs and facilitates their disassembly, as well as nuclear permeabilization at the onset of mitosis (Guttinger et al., 2009; Laurell et al., 2011). In C. elegans, three kinases are known to regulate mitotic entry and NEBD: the Cdk1 homologue NCC-1, the Polo-like kinase PLK-1, and the Aurora-A kinase AIR-1 (Boxem et al., 1999; Chase et al., 2000; Hachet et al., 2007; Portier et al., 2007). Depletion of these components by RNA interference (RNAi) either prevents (NCC-1 and PLK-1) or strongly delays (AIR-1) entry into M phase and NEBD. It is probable that one or several of these kinases impinge on given nucleoporins, for instance the transmembrane nucleoporins GP210/NPP-12, the depletion of which leads to a block in lamina disassembly (Galy et al., 2008). However, the potential link between these kinases and specific nucleoporins is not clear. More generally, the mechanisms governing NPC disassembly in C. elegans embryos are not fully understood.
RESULTS
An RNAi-based modifier screen for novel mitotic regulators
The one-cell-stage C. elegans embryo is well suited for analyzing the onset of mitosis and the execution of NEBD, which can be monitored with exquisite spatial and temporal resolution (Figure 1A). Using embryos in which the male and female pronuclei remain apart due to defective pronuclear migration, we found previously that such separated pronuclei undergo asynchronous NEBD (Figure 1B) in a manner that is dependent on centrosomes and on AIR-1 (Hachet et al., 2007).
FIGURE 1:
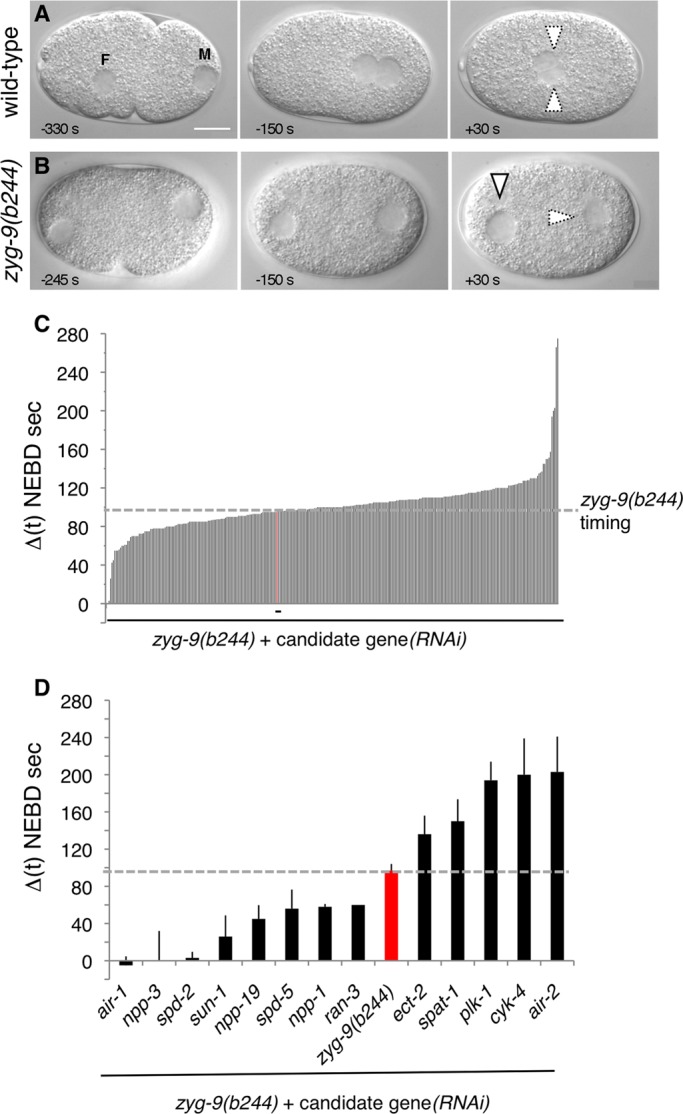
RNAi-based modifier screen for modulators of mitotic entry. (A and B) Images from time-lapse DIC microscopy of wild-type (A) and zyg-9(b244) (B) one-cell-stage embryos. In all figures, anterior is to the left, posterior to the right; F and M designate the female and male pronuclei, respectively. Arrowheads with thick lines: intact pronucleus; arrowheads with dashed lines: pronucleus undergoing NEBD. Scale bars, 10 μm. The onset of NEBD, as defined by the loss of the smooth line corresponding to the NE is defined as time 0. (C and D) Average time difference between NEBD in the male and female pronuclei in zyg-9(b244) + candidate gene(RNAi) in the initial screen (C; see also Table S2) or selected after the confirmation round using a threshold of ± 35 s as compared with the zyg-9(b244) timing (D). The time corresponding to zyg-9(b244) alone is shown in red. The following candidate genes that were selected in the initial screen did not give a reproducible phenotype in the confirmation round: T01C3.1, C03C10.3, ZC308.1a, ZK1037.5, nsh-1, T2308.9, T20B12.8, F53G12.5, ZK1127.7, and T13F2.7. In addition, glucosamine (F07A11.2) and sqv-4 (F29F11.1) were not retained because RNAi-treated embryos exhibited severe pleiotropic phenotypes, precluding a proper analysis of NEBD. Finally, the following genes could not be analyzed for their impact on asynchronous NEBD as their depletion rescued pronuclear migration to some extent: cyb-1, cyb-3, cye-1, ncc-1, cdc25.1, F09G2.4, gpr-2, rga-3, C03C10.3, F59B2.6, and hmg-3.
To uncover novel genes modulating mitotic entry, we used this assay to design an RNAi-based modifier screen to identify components contributing to the asynchrony normally observed when the two pronuclei are separated. We anticipated this screen to identify positive and negative regulators of mitotic entry. In principle, such regulators could act in a centrosome-dependent manner and thus exhibit alterations in the timing of the male pronucleus, which is the only one associated with centrosomes in this setting. Alternatively, such regulators may function in a centrosome-independent manner, in which case they might exhibit also, or perhaps only, alterations in the timing of the female pronucleus.
We selected a set of genes to screen using two criteria. First, we chose ∼1400 genes that, based on a compendium of microarray experiments, are coexpressed with known mitotic regulators, including ncc-1, plk-1, and air-1 (Kim et al., 2001). Second, we selected genes that are embryonic lethal when inactivated by RNAi, thus reducing the number of genes to test to 360 (Table S2). These 360 genes were inactivated singly by RNAi in the background of zyg-9(b244) mutant embryos, which exhibit defective pronuclear migration and asynchronous NEBD (Figure 1B). In each case, initially three to five embryos were analyzed by time-lapse differential interference contrast (DIC) microscopy, and the average time difference between the two pronuclei was determined (Figure 1C). A subsequent confirmation round, during which more embryos were analyzed, was performed with candidate genes identified in the initial screen.
In this manner, we identified five genes whose inactivation clearly increases the asynchrony between the separated pronuclei (Figure 1D). These genes encode PLK-1, which was only partially inactivated by RNAi in this experiment (see Materials and Methods), the Bora homologue SPAT-1, which regulates Aurora-A and functions with PLK-1 to regulate cell-cycle progression (Hutterer et al., 2006; Noatynska et al., 2010), two modulators of the Rho GTPase, the RhoGEF ECT-2 and the RhoGAP CYK-4, as well as the Aurora-B kinase AIR-2. The latter three genes are essential for cytokinesis, including during the meiotic divisions, and we hypothesize that the apparent increased asynchrony in these cases reflects delayed formation of the female pronucleus following impaired meiotic divisions. These five components are not the focus of this study and thus will not be discussed further.
We also identified eight genes whose inactivation clearly decreases the asynchrony between the separated male and female pronuclei (Figure 1D). The corresponding proteins include four that were expected from earlier work: AIR-1, SPD-2 and SPD-5 (both required for centrosome integrity [ Hamill et al., 2002; Kemp et al., 2004]), as well as SUN-1 [required to link centrosomes to the male pronucleus (Malone et al., 2003)]. Another protein whose inactivation decreases asynchrony is RAN-3, the Ran GTPase GEF RCC1. Interestingly, the three remaining proteins in this category are the nucleoporins Nup205/NPP-3, Nup54/NPP-1, and Nup35/NPP-19, which in other systems are known to interact with each other (Grandi et al., 1995; Kosova et al., 1999; Hawryluk-Gara et al., 2005).
These results led us to analyze systematically the impact on timely NEBD of depleting the nucleoporins NPP-1 to NPP-20, because some of them were not included in the initial set of 360 genes. The more recently described NPP-21 to NPP-23 were not analyzed. As shown in Figure S1 and Table S3, we found that the depletion of another nucleoporin, the Nup93 homologue NPP-13, which in other systems is also part of the same subcomplex as Nup205/NPP-3 (Kosova et al., 1999; Hawryluk-Gara et al., 2005), also significantly reduces the asynchrony between the two pronuclei. Moreover, we found that NE formation is severely compromised in npp-8(RNAi), npp-9(RNAi), and npp-20(RNAi) embryos (Galy et al., 2003), precluding a thorough analysis of a potential contribution to timely mitotic entry. The same is true of most npp-19(RNAi) embryos (Rodenas et al., 2009), despite the fact that some embryos were analyzable in the initial screen, presumably owing to incomplete RNAi-mediated inactivation (see Figure 1D).
Given that the depletion of NPP-3 has the most striking impact on asynchrony (Figure 1D), we focused further investigations on this component.
NPP-3 as a novel negative regulator of mitotic onset
Time-lapse DIC recordings of a larger number of embryos confirmed that the onset of NEBD is synchronous in zyg-9(b244) embryos following NPP-3 depletion (Figures 2, A–D, and S1, Table S3, and Movies M1 and M2). We also assayed NEBD in live embryos using yellow fluorescent protein (YFP)-lamin to visualize the lamina underlying the NE. The onset of lamina disassembly is synchronous in the two pronuclei in the wild type, whereas it is asynchronous in zyg-9(RNAi) embryos (Figure 2, E and F, Movie M3). As anticipated, we found that lamina disassembly is synchronous in the two pronuclei of zyg-9(RNAi) npp-3(RNAi) embryos (Figure 2, G and H, and Movie M4).
FIGURE 2:
NPP-3 depletion abolishes asynchronous NEBD of separated male and female pronuclei. (A and C) Images from time-lapse DIC microscopy of zyg-9(b244) (A) and zyg-9(b244) npp-3(RNAi) (C) one-cell-stage embryos. See also corresponding Movies M1 and M2. (E and G) Time-lapse fluorescence microscopy of zyg-9(RNAi) (E) and zyg-9(RNAi) npp-3(RNAi) (G) one-cell-stage embryos expressing YFP-lamin. See also corresponding Movies M3 and M4. Lamina disassembly is apparent by loss of the smooth appearance characteristic of earlier stages. Note that the 6% neutral density filter was removed in panel G (see also legends of Movies M3 and M4). Similar results were obtained with zyg-9(b244) npp-3(RNAi). (B, D, F, and H) Average time difference between NEBD onset in the male and female pronuclei in seconds ± SEM in zyg-9(b244) (B) or zyg-9(RNAi) (F), as well as zyg-9(b244) npp-3(RNAi) (D) or zyg-9(RNAi) npp-3(RNAi) (H) one-cell-stage embryos, monitored by DIC (B and D) or YFP-lamin (F and H). Numbers of embryos and statistical analysis are given in Table S3 for B and D. (I and J) Monitoring NCC-1 activation in the male and female pronuclei of zyg-9(b244) (I) and zyg-9(b244) npp-3(RNAi) (J) one-cell-stage embryos by staining for Cdk1P-Tyr15 (shown alone on the right and in red in the merged images), α-tubulin (green), and DNA (blue). Histograms of pixel intensities of the male (red) and female (blue) pronucleus from the two genotypes are depicted on the right. (K) Time separating the exit of meiosis II from NEBD of the male and female pronuclei in seconds ± SEM, determined from DIC time-lapse recordings. Extrusion of the second polar body was used as a reference for the exit from meiosis (time 0 s). Embryos were timed until NEBD in one-cell-stage embryos. The number of embryos analyzed (n) and p values (Student's t test, time from polar body extrusion until female NEBD, compared with the wild type) are as follows: wild type (n = 13); p zyg-9(b244) = 0.0025 (n = 14); p zyg-9(b244) npp-3(RNAi) = 0.74 (n = 14).
We next addressed whether the synchrony observed when scoring the onset of NEBD upon NPP-3 depletion reflects a more general impact on the timing of mitotic entry, in which case it should be accompanied by synchronous changes of Cdk1 activity in the two pronuclei. To address this question, we used a Cdk1P-Tyr15 antibody that recognizes specifically the inactive form of the NCC-1 kinase. As reported previously (Hachet et al., 2007), when NCC-1 is inactive during interphase, the Cdk1P-Tyr15 signal is high in both separated pronuclei. During prophase, the Cdk1P-Tyr15 signal diminishes earlier in the male pronucleus than in the female pronucleus in the majority of zyg-9(b244) embryos, indicative of earlier Cdk1 activation in the male pronucleus (Figure 2I; 76%, N = 25 embryos) (Hachet et al., 2007). As shown in Figure 2J, we found by contrast that the Cdk1P-Tyr15 signal diminishes earlier in the male pronucleus only in a minority of zyg-9(b244) npp-3(RNAi) embryos (Figure 2J; 43%, N = 42 embryos, binomial two-tail test calculation, p = 3.88 × 10−7). This result suggests that NPP-3 depletion has a more general effect on the timing of mitotic entry, although the impact on the onset of NEBD is most readily detectable.
In principle, synchronous mitotic entry in zyg-9(b244) npp-3(RNAi) embryos could be due to a delay of the male pronucleus or, instead, to an acceleration of the female pronucleus compared with zyg-9(b244) alone. To distinguish between these possibilities, we determined the overall duration of the first cell cycle by monitoring live embryos since the exit from meiosis II. Previous work established that the female pronucleus is delayed compared with the wild type in zyg-9(b244) embryos (Hachet et al., 2007). Here we found that the two separated pronuclei in zyg-9(b244) npp-3(RNAi) embryos enter mitosis approximately at the same time as do the two joined pronuclei in the wild type (Figure 2K). Therefore the synchrony in zyg-9(b244) npp-3(RNAi) embryos reflects an acceleration of mitotic entry in the female pronucleus, which is devoid of centrosomes in this setting.
These findings led us to conclude that NPP-3 acts as a negative regulator of mitotic onset in zyg-9 mutant embryos, the contribution of which is masked by the presence of centrosomes next to the male pronucleus. We then addressed whether NPP-3 depletion also accelerates mitotic onset in embryos lacking centrosomes, in which both centrosomes enter mitosis with wild-type timing (Hachet et al., 2007). To this end, we analyzed the overall duration of the first cell cycle in spd-5(or213) npp-3(RNAi) embryos, and found it to be statistically indistinguishable from that in spd-5(or213) embryos (mean, SD, and N: 940, 46, 10 vs. 920, 38, 13; p = 0.51, Student's t test). This finding indicates that NPP-3 depletion cannot advance mitotic onset further, presumably because another rate-limiting component is not active as of yet.
Nuclear permeability defects do not correlate with decreased asynchrony
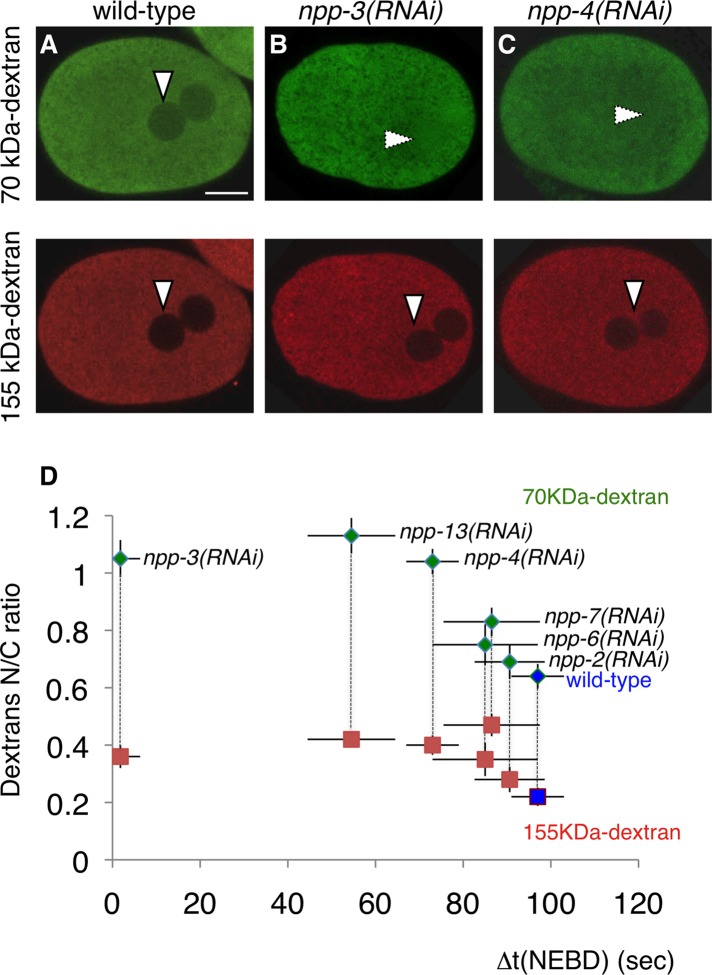
We considered whether general permeability defects in the NE could explain synchronous entry into mitosis in zyg-9(b244) npp-3(RNAi) embryos. Therefore we monitored nuclear permeability defects by monitoring the ability of pronuclei to exclude fluorescently labeled 70- and 155-kDa dextrans. Consistent with previous observations (Galy et al., 2003), we found that the size exclusion limit of the nucleus is defective in npp-3(RNAi) embryos, because 70-kDa dextran is not excluded from the pronuclei, whereas 155-kDa dextran is correctly excluded (Figure 3, A–D).
FIGURE 3:
Defective nuclear permeability does not drastically alter timing of mitotic entry. (A–C) Nuclear exclusion of fluorescently labeled dextrans of 70 kDa (green) and 155 kDa (red). Confocal images of wild-type (A), npp-3(RNAi) (B), and npp-4(RNAi) (C) live embryos are shown. Vertical arrowheads indicate nuclear exclusion; horizontal arrowheads indicate lack of exclusion. (D) Nuclear-to-cytoplasmic (N/C) ratio of fluorescently labeled dextrans (70 kDa, green; 155 kDa, red) plotted against the average time difference between NEBD in the male and female pronuclei (Δt (NEBD)). See also Figure S2.
Although this result could be compatible with general NE permeability defects being responsible for accelerated entry into mitosis of the female pronucleus in zyg-9(b244) npp-3(RNAi) embryos, we addressed this possibility further by investigating whether NE permeability defects always cause synchrony. To this end, we investigated the exclusion from the pronuclei of 70- and 155-kDa dextrans in embryos depleted of other nucleoporins. As shown in Figures 3D and S2, A–E, we found that embryos depleted of Nup93/NPP-13 or Nup45/58/NPP-4 exhibit nuclear permeability defects indistinguishable from those in embryos depleted of NPP-3. Despite this, zyg-9(b244) npp-13(RNAi) [and even more so, zyg-9(b244) npp-4(RNAi)] embryos retain substantial asynchrony between the two pronuclei (Figure 3D and Table S3). As a further means to compare permeability defects in npp-3(RNAi) and npp-4(RNAi) embryos, we monitored the ability of pronuclei to exclude green fluorescent protein (GFP)-β-tubulin, which was similarly compromised in embryos depleted of either NPP-3 or NPP-4, as well as of NPP-1 (Figure S2, F–I). Analogous conclusions were reached when comparing the distribution of GFP-β-tubulin upon NPP-3 and NPP-4 depletion in zyg-9(b244) embryos (Movies 5–7).
Overall, these findings indicate that bulk permeability defects, although potentially being a contributing factor, do not alone explain synchronous onset of NEBD and mitotic entry in zyg-9(b244) npp-3(RNAi) embryos. Instead, we postulate that NPP-3 can negatively impact timely mitotic entry when centrosomes are not in the vicinity of the NE by ensuring the proper nuclear/cytoplasmic distribution of a specific cell-cycle regulator. Upon NPP-3 depletion in a zyg-9(b244) mutant background, this as-of-yet unidentified component may gain premature access inside the female pronucleus and thus lead to synchronous mitotic entry of the two pronuclei.
Decreased asynchrony correlates with diminished NPP-3 at the NE
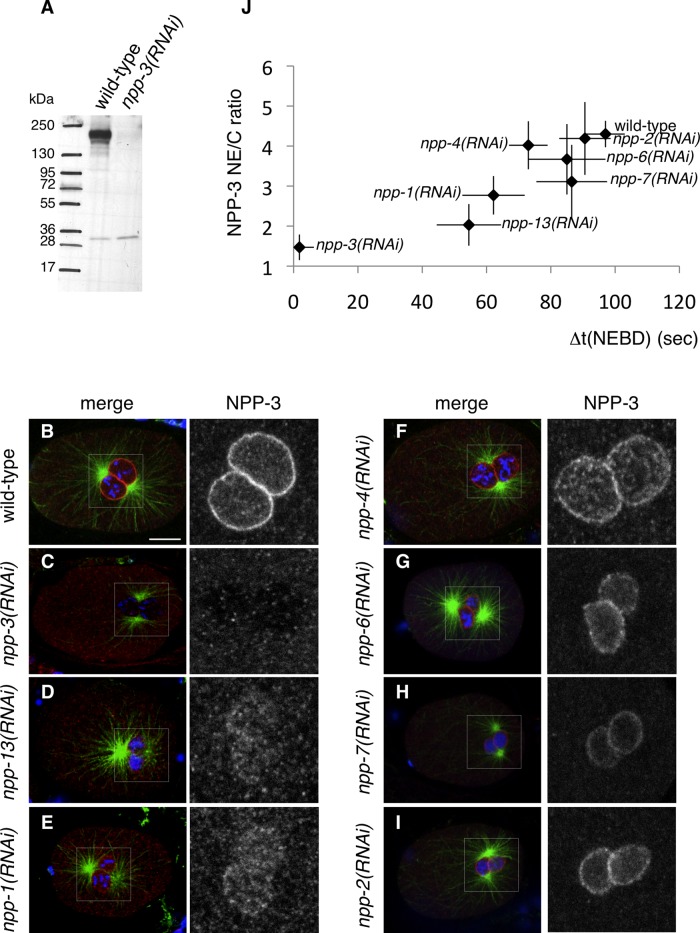
We proceeded to analyze the subcellular distribution of NPP-3. We raised and affinity-purified polyclonal antibodies against NPP-3, which recognize a single specific band at the expected size in wild-type embryonic extracts (Figure 4A). Immunofluorescence analysis of wild-type, one-cell-stage embryos probed with these antibodies demonstrates that NPP-3 localizes primarily at the NE (Figure 4, B and C).
FIGURE 4:
Decreased asynchrony correlates with loss of NPP-3 at the NE. (A) Western blot of extracts from wild-type or npp-3(RNAi) embryos probed with NPP-3 antibodies. (B–I) NPP-3 distribution at the NE in embryos of the indicated genotypes stained for NPP-3 (shown alone in the insets on the right and in red in the merged images), α-tubulin (green), and DNA (blue). Note that depletion of some nucleoporins affects nuclear size as previously observed (Galy et al., 2003). (J) NE-to-cytoplasmic (NE/C) ratio of NPP-3 signal plotted against the average time difference between NEBD in the male and female pronuclei in seconds. See also Table S4.
We set out to investigate whether alterations in NPP-3 distribution at the NE could explain the partial loss of asynchrony observed upon depletion of other nucleoporins in zyg-9(b244) embryos. We observed a strong reduction of NPP-3 at the NE upon depletion of Nup93/NPP-13 or of Nup54/NPP-1, the two NPPs that had the most significant impact on asynchrony after NPP-3 itself (Figure 4, D, E, and J, and Table S4). In contrast, we found that depletion of Nup45/Nup58/NPP-4, Nup160/NPP-6, Nup153/NPP-7, or Nup85/NPP-2, which all have a minor impact on asynchrony, does not strikingly alter NPP-3 levels at the NE (Figure 4, F–J, and Table S4).
Overall, these findings establish that lower levels of NPP-3 at the NE correlate with accelerated entry into mitosis in the female pronucleus in zyg-9(b244) embryos, indicating that the presence of NPP-3 at the NE negatively impacts mitotic entry.
NPP-3 local removal from the NE at mitosis
While investigating the distribution of NPP-3 across the cell cycle, we discovered that the protein is no longer detected at the NE in the vicinity of centrosomes, starting at the end of prophase and most strikingly in prometaphase (Figure 5, A–D). This is unlikely because of epitope masking, because polyclonal antibodies raised against a fusion protein were used. Moreover, a similar local loss is observed with antibodies against NPPs that are part of the same subcomplex (see later in the text). The lamina has been reported to disassemble initially in the vicinity of centrosomes in C. elegans embryos (Lee et al., 2000). By conducting double-labeling experiments, we found that NPP-3 local loss precedes lamina disassembly (Figure 5E). We conclude that NPP-3 local loss from the NE is an early event of NEBD.
FIGURE 5:
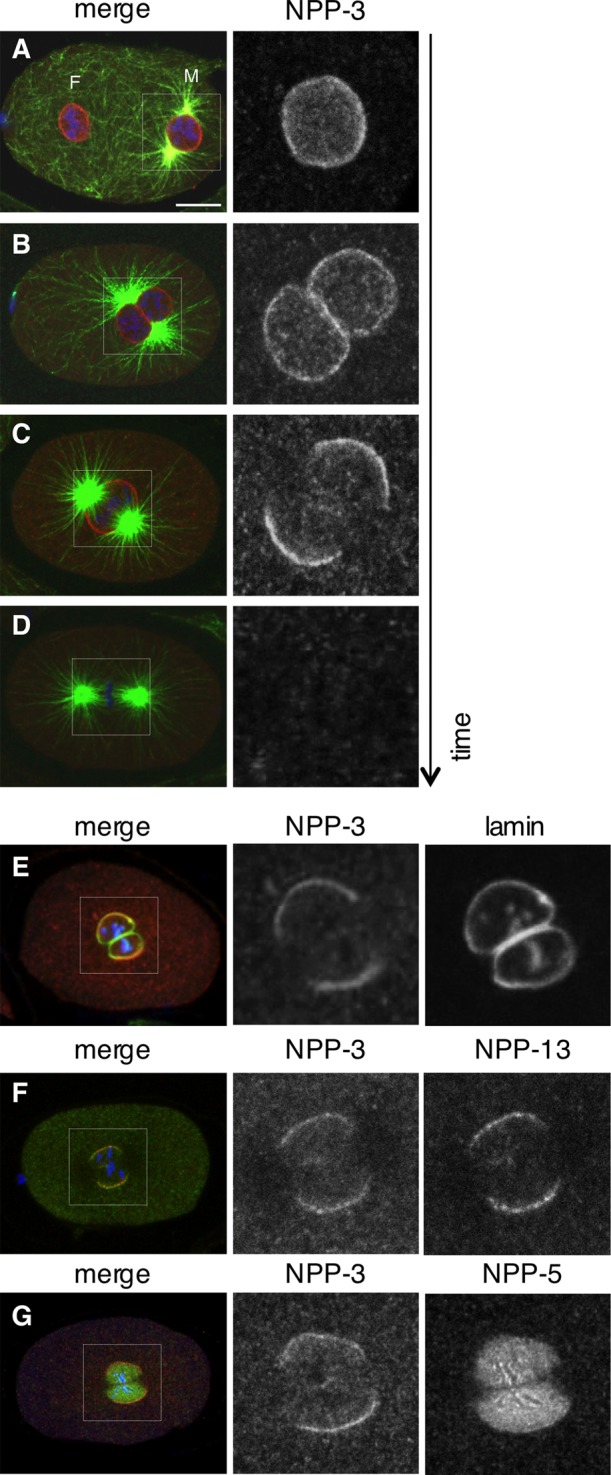
NPP-3 localization at mitotic onset. (A–D) Wild-type, one-cell-stage embryos in prophase (A and B), prometaphase (C), or metaphase (D) stained for NPP-3 (shown alone in the insets on the right and in red in the merged images), α-tubulin (green), and DNA (blue). (E–G) Wild-type, one-cell-stage embryos stained for NPP-3 (shown alone in the insets in the middle and in red in the merged images), lamin, NPP-13, or NPP-5, as indicated (shown alone in the insets on the right and in green in the merged images). DNA is shown in blue.
We next investigated whether such local loss is also exhibited by other nucleoporins. We first tested whether this is the case for the NPP-3–associated Nup93/NPP-13. To this end, we raised and affinity-purified polyclonal antibodies against NPP-13, which decorate the NE of wild-type embryos (Figure S3, A–D). Importantly, we found that NPP-13 also exhibits local loss near centrosomes at the onset of mitosis, concomitant with that of NPP-3 (Figure 5F). Similar results were obtained with embryos expressing GFP–NPP-19 (Figure S3, E and F). In contrast, we found that Nup107/NPP-5, which resides on the outer part of the NPC, as well as Nup98/NPP-10 and Nup153/NPP-7, which reside on the inner part of the NE, all disappear in a uniform manner from the NE and redistribute to kinetochores in mitosis (Figure 5G, Figure S3, G and H, and data not shown; Belgareh et al., 2001; Rodenas et al., 2009). To investigate whether the overall integrity of the NE is compromised at the time of NPP-3 local loss, we analyzed the distribution of the inner nuclear membrane components GFP-emerin and GFP–LEM-2, as well as that of the GFP-SP12 fusion that marks ER-localized proteins and thus also labels the outer nuclear membrane (Lee et al., 2000; Poteryaev et al., 2005). We found that these three fusion proteins do not exhibit local loss at the onset of mitosis (Figure S3I and data not shown), indicating that NPP-3 local removal does not result from an overall loss of membrane integrity.
Overall, we conclude that NPP-3, NPP-13, and NPP-19, which are all members of the Nup93 subcomplex, exhibit local loss from the NE in the vicinity of centrosomes at the onset of mitosis.
Centrosomes and AIR-1 kinase activity direct NPP-3 local removal from the NE
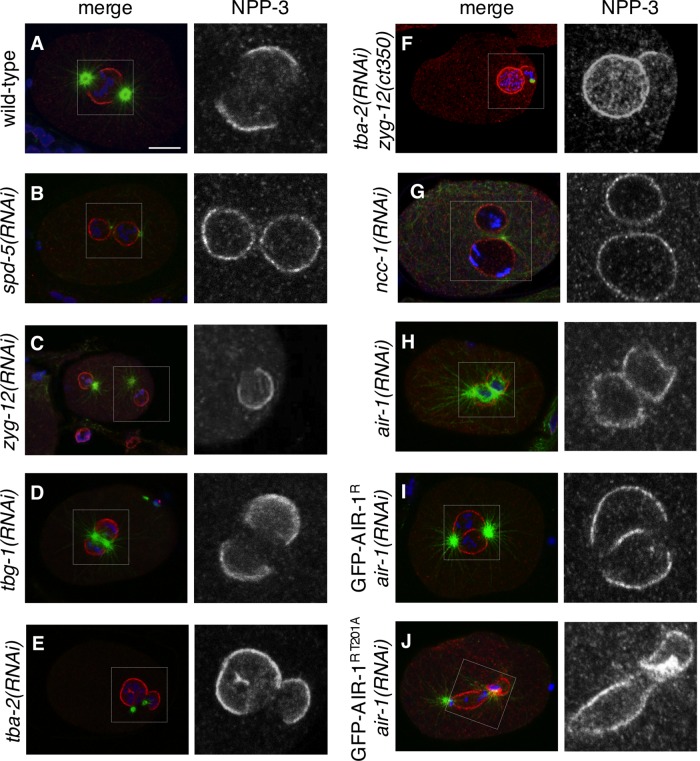
We set out to address the mechanisms that result in local loss of NPP-3 and associated NPPs from the NE in the vicinity of centrosomes at the onset of mitosis. To address whether centrosomes are necessary, we disrupted centrosome integrity through depletion of SPD-5 and found that this prevents NPP-3 local loss (Figure 6, A and B). To address whether centrosomes are sufficient, we depleted ZYG-12 such that the two centrosomes detach from the male pronucleus early in the cell cycle and move freely in the cytoplasm until they each contact stochastically one of the two separated pronuclei. We found that NPP-3 is invariably lost from the NE in the vicinity of the centrosome in such embryos (Figure 6C). We conclude that centrosomes are both necessary and sufficient for NPP-3 local loss from the NE at mitotic onset.
FIGURE 6:
NPP-3 local loss is centrosome- and AIR-1–dependent. (A–J) One-cell-stage embryos of the indicated genotypes stained for NPP-3 (shown alone in the insets on the right and in red in the merged images), α-tubulin (green), and DNA (blue). Note in (E) and (F) that the female pronucleus is much larger and located next to the male pronucleus, as reported previously for tba-2(RNAi) embryos (Sonneville and Gönczy, 2004). The numbers of embryos analyzed, which all exhibited the phenotype illustrated in the panels, except when mentioned otherwise, are as follows: spd-5(RNAi): 17; zyg-12(RNAi): 8; tbg-1(RNAi): 15, 1 of which did not exhibit NPP-3 local loss; tba-2(RNAi): 16; tba-2 (RNAi) zyg-12(ct350): 7; ncc-1(RNAi): 12; air-1(RNAi): 11; GFP-AIR-1R: 9; GFP-AIR-1R T201A: 19.
We next investigated whether microtubules nucleated from the centrosomes are involved. As shown in Figure 6D, we found that NPP-3 local loss is not altered upon depletion of the γ-tubulin component TBG-1, which significantly decreases centrosomal microtubules (Hannak et al., 2002). Moreover, NPP-3 local loss is also unaffected when microtubules are essentially absent following RNAi-mediated depletion of the α-tubulin gene tba-2 (Figure 6E). The same is true of embryos simultaneously depleted of ZYG-12 and TBA-2 function, in which residual short microtubules that could remain on tba-2(RNAi) alone are expected not to contact the NE (Figure 6F). We conclude that centrosomes appear to dictate NPP-3 local loss independently of microtubules.
We then verified whether NCC-1, which is known to be essential for NEBD and mitotic entry (Boxem et al., 1999), affects NPP-3 local loss, and found this to be the case indeed (Figure 6G). Next we tested whether the centrosomal kinase AIR-1, the depletion of which also leads to synchronous NEBD of separated pronuclei (Hachet et al., 2007; Portier et al., 2007), regulates NPP-3 local loss. As shown in Figure 6H, we found that NPP-3 local loss does not occur in air-1(RNAi) embryos. To address whether AIR-1 kinase activity is needed, we analyzed embryos depleted of endogenous air-1 and expressing an RNAi-resistant transgene encoding either wild type or a kinase-inactive version of GFP-tagged AIR-1. As illustrated in Figure 6, I and J, we found that NPP-3 local loss occurs in embryos expressing wild-type AIR-1, but not the corresponding kinase-inactive version.
These results demonstrate that centrosomes and AIR-1 kinase activity dictate local loss of NPP-3 from the NE at the onset of mitosis. Together with our finding that NPP-3 can negatively regulate the timing of mitotic onset, these findings lead us to propose a model whereby centrosomes and AIR-1 promote timely onset of mitosis by leading to the local removal of NPP-3 and associated nucleoporins from the NE (Figure 7).
FIGURE 7:
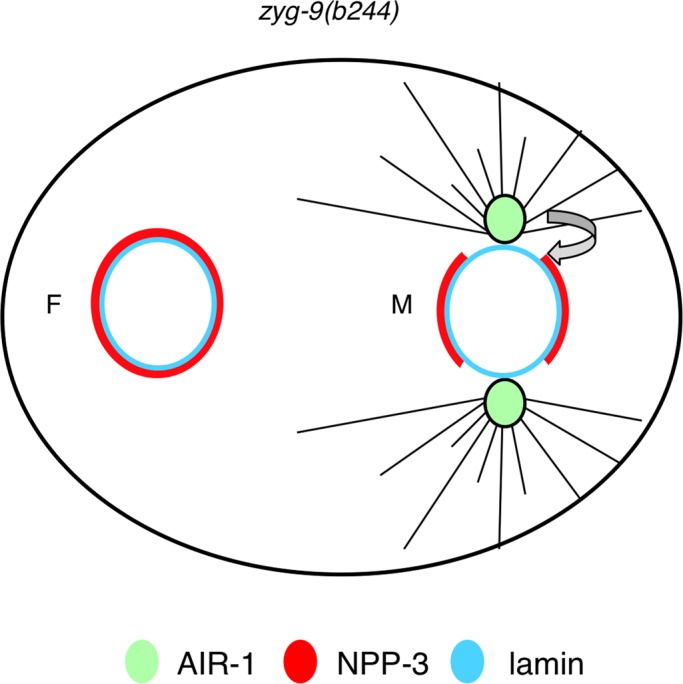
Model for the role of centrosomes and AIR-1 in promoting NPP-3 local loss and timely mitotic entry in C. elegans embryos. Green circle: AIR-1 at centrosomes. Note that cytoplasmic AIR-1 is not depicted in this figure but likely contributes to overall AIR-1 activity. Red: NPP-3; light blue: lamina; gray lines: microtubules; filled line: intact NPP-3 signal; dashed line: disrupted NPP-3 signal. Centrosomes and AIR-1 promote NPP-3 local loss from the NE, potentially facilitating NEBD and timely mitotic entry.
DISCUSSION
Our work demonstrates that NPP-3 can modulate the onset of mitosis in C. elegans embryos. This novel function for the nucleoporin Nup205/NPP-3 echoes recent results that revealed unsuspected roles for nucleoporins during mitosis, outside of their well-characterized role in NPC architecture and nucleocytoplasmic transport. For instance, the Nup107–160 subcomplex localizes to kinetochores and the spindle, and depletion of members of this subcomplex in human cells and C. elegans results in defective chromosome segregation, spindle assembly, and cytokinesis (reviewed in Loiodice et al., 2004; Chatel and Fahrenkrog, 2011; reviewed in Nakano et al., 2011; Rodenas et al., 2012). Moreover, recent studies unraveled a novel function for some nucleoporins in regulating gene expression (Capelson et al., 2010; Kalverda et al., 2010), whereas depletion of Nup133/NPP-15 in the mouse embryo reveals that this nucleoporin coordinates cell differentiation events (Lupu et al., 2008; D'Angelo et al., 2012).
Nup205/NPP-3 as a negative regulator of entry into mitosis
We focused our analysis on a subset of nucleoporins, the depletion of which decreases the asynchrony between the two separated pronuclei, with Nup205/NPP-3 exhibiting the most striking effect. We found that the synchrony observed in npp-3(RNAi) zyg-9(b244) embryos reflects an acceleration of mitotic entry in the female pronucleus. We propose that the impact on the female pronucleus is revealed because of the absence of centrosomes in its vicinity in zyg-9(b244) embryos. In essence, RNAi-mediated depletion of NPP-3 in this background alleviates the requirement for centrosomes in dictating mitosis onset. In such embryos, the female pronucleus could be considered to be an equivalent of the male pronucleus, thus resulting in synchronous mitotic entry of the two pronuclei, irrespective of the presence of centrosomes. Given that NPP-3 depletion does not further advance mitotic onset in embryos lacking centrosomes, our work suggests in addition that another rate-limiting factor contributes to the timing of mitotic onset in otherwise wild-type embryos. Furthermore, examination of cell-cycle duration in two-cell-stage npp-3(RNAi) embryos revealed an acceleration of mitotic onset in the AB cell, but an atl-1–independent delay of this process in P1 (data not shown). Together, these observations indicate that the contribution of NPP-3 to the timing of mitotic entry is complex and differs in distinct cellular contexts.
What are the mechanisms that lead to an acceleration of mitotic entry in the female pronucleus upon NPP-3 depletion in zyg-9 mutant embryos? Our findings suggest that defects in the permeability barrier caused by NPP-3 depletion do not alone explain the impact on mitotic onset, because depletion of other nucleoporins such as NPP-4 results in seemingly similar exclusion defects but hardly impairs mitotic onset. Instead, we consider the following two possibilities to explain the impact of NPP-3 depletion on mitotic entry.
First, it is formally possible that depletion of NPP-3 and associated subcomplex members could somehow generally impair the structure of NPCs, perhaps rendering them more fragile and thereby facilitating mitotic entry. In this scenario, phosphorylation of NPP-3 and associated NPPs by AIR-1 or other mitotic kinases might lead to the destabilization of NPCs and thereby facilitate mitotic entry. A second possibility is that depletion of NPP-3 and associated components affect more specifically the distribution of a critical mitotic regulator. We did not detect apparent defects in the steady-state distribution of CDC-25.1, Cyclin B3, AIR-1, and PLK-1 in npp-3(RNAi) embryos (unpublished observations). However, we cannot exclude that the dynamics or the activity of one of these components is affected. Alternatively, NPP-3 may govern the proper distribution of another mitotic regulator, the identity of which remains to be determined.
Mechanisms governing the local loss of NPP-3
We found that the distribution of NPP-3 is regulated in time and space, with the protein being lost from the NE in the vicinity of centrosomes at mitotic onset, before local nuclear lamina disassembly. Even though NPP-3 local removal occurs before local lamina disassembly, it does not appear to be the earliest nucleoporin to leave the NE. Thus, careful comparison of time-lapse recordings indicates that the uniform loss of GFP–NPP-5, which belongs to the NPP-160/NPP-6 subcomplex, precedes the local loss of GFP–NPP-19, which belongs to the Nup205/NPP-3 complex (unpublished observations). The early and uniform departure of Nup98/NPP-10 complex members from the NE is in line with observations in human cells and starfish oocytes demonstrating that Nup98 is the first component known to leave the NE (Lenart et al., 2003; Dultz et al., 2008). It will be interesting to carefully investigate the kinetics of the disassembly of Nup205 and associated subcomplex components in other systems to address whether they are also lost locally in the vicinity of centrosomes. Perhaps local loss is so rapid that it may be overlooked unless specifically looked for. Alternatively, it could be that the pathway resulting in such local loss is particularly active in C. elegans but less so in other systems.
What mediates the local loss of Nup205/NPP-3 and associated nucleoporins in C. elegans? Such local loss is dependent on AIR-1 activity, raising the possibility that phosphorylation of NPP-3 or of an associated NPP by this kinase is important. Conceivably, this could affect nuclear permeability in a spatially restricted manner and facilitate mitotic onset and NEBD. A conceptually analogous situation is encountered in human cells in which phosphorylation of the nucleoporin Nup98/NPP-10 by multiple kinases is important for NPC disassembly during mitotic entry (Laurell et al., 2011). Our findings taken together lead us to propose a model whereby centrosomes concentrate mitotic kinases, including AIR-1, thus enabling efficient phosphorylation of nucleoporins such as NPP-3 and other members of the Nup93 subcomplex. In turn, this induces local loss of this subcomplex from the NE, thus facilitating mitotic onset (Figure 7).
MATERIALS AND METHODS
Nematode strains and RNAi
Transgenic worms expressing GFP-AIR-1R (RNAi resistant), GFP-AIR-1R T201A, GFP–LEM-2, GFP–emerin, GFP–SP12, and GFP–NPP-5 were grown at 24°C (Lee et al., 2000; Poteryaev et al., 2005; Rodenas et al., 2009; Toya et al., 2011). Homozygous recessive mutant animals of genotypes zyg-9(b244) (Wood et al., 1980) and spd-5(or213) (Hamill et al., 2002) were grown at 15°C and shifted before analysis to the restrictive temperature for the duration indicated below.
Bacterial RNAi feeding strains for air-1, tbg-1, npp-12, and npp-13 were described (Bellanger and Gönczy, 2003). RNAi was performed by feeding L3 animals as follows: tbg-1(RNAi) and air-1(RNAi): 24–38 h at 24°C; zyg-9(RNAi) npp-3(RNAi): 16–24 h at 25°C; zyg-9(b244) npp-1(RNAi), zyg-9(b244) npp-4(RNAi), zyg-9(b244) npp-13(RNAi), and zyg-9(b244) npp-12(RNAi): 24 h at 16°C, then 16–20 h at 25°C; all combinations of zyg-9(b244) with other specific nucleoporins: 16–20 h at 25°C.
Visual screen and microscopy
For the RNAi modifier screen, bacterial feeding strains were obtained from either the C. elegans ORFeome RNAi library (Rual et al., 2004) or the Ahringer library (Kamath et al., 2003). RNAi was performed using three different conditions depending on the strength of the RNAi phenotype: 20 h at 25°C (condition A; e.g., spd-5), 24 h at 16°C followed by 20 h at 25°C (condition B; e.g., ect-2), or 8 h at 25°C (condition C; for genes classified as osmosensitive, e.g., for plk-1).
Embryos were analyzed at ∼23°C by time-lapse DIC microscopy (Gönczy et al., 1999) or dual-time fluorescence and DIC microscopy (Brauchle et al., 2003).
Antibody production
Antibodies against NPP-3 and NPP-13 were raised against recombinant glutathione S-transferase (GST) fusion proteins (aa 262–597 for NPP-3 and 1–360 for NPP-13). These fusion proteins were expressed in bacteria, purified using glutathione beads, and injected into rabbits (Eurogentec, Fremont, CA). Antibodies were subsequently strip-purified against the corresponding maltose binding protein (MBP) fusion proteins and eluted with glycine pH 2.5 and then neutralized with Tris-HCl, pH 8.0.
Immunofluorescence
Fixation and indirect immunofluorescence were performed essentially as described (Gönczy et al., 1999). The primary mouse α-tubulin antibody DM1A (Sigma, St. Louis, MO) was used at 1:200, whereas primary rabbit antibodies were used as follows: NPP-3 (1:200; this study), NPP-13 (1:400; this study), NPP-10 (1:250; gift from Iain Mattaj, EMBL, Heidelberg, Germany), AIR-1(1:1000; Hannak et al., 2001), and phosphoDetect anti-Cdk1 (pTyr15, 1:100; Calbiochem, San Diego, CA). GFP was monitored using anti-mGFP (1:100; Chemicon/Upstate/Millipore, Billerica, MA) or goat polyclonal antibody against GFP labeled with FITC (1:200; Abcam, Cambridge, MA). The secondary goat anti-mouse antibody coupled to Alexa488 (Molecular Probes. Eugene, OR) was used at 1:500. Slides were counterstained with Hoechst 33258 at ∼1 mg/ml (Sigma) to label DNA. Confocal microscopy was performed with a Leica TCS SP2 AOBS (Leica Microsystems, Wetzlar, Germany) using a 63X oil objective and a pinhole aperture of 114 μm (1 Airy Unit). For Figure 2, I and J, maximum intensity projections are shown. For the two embryos shown in Figure 2, I and J, the distribution of pixel intensities in each nucleus (red, M PN; blue, F PN) was measured using Matlab (MathWorks, Natick, MA).
Injection of fluorescently labeled dextrans
Fluorescently labeled dextrans (Sigma) of 70 kDa (coupled to fluorescein) and 155 kDa (coupled to rhodamine) were spun for 10 min at 12,000 rpm in a tabletop centrifuge using a nanosep 10K column (Pall, Port Washington, NY) to remove free dye and then injected into the gonad of adult hermaphrodite (2 mg/ml each in injection buffer: 20 mM KPO4, pH 7.5; 3 mM K Citrate, pH 7.5; 2% polyethylene glycol 6000). Injected worms were incubated at 25°C for 4 h before dissection. Their embryos were analyzed by confocal microscopy using a Leica TCS SP2 AOBS equipped with a 63X oil objective with a pinhole aperture of 114 μm (1 Airy Unit).
Supplementary Material
Acknowledgments
We thank Mattyas Gorjanacz and Iain Mattaj for the GFP-LEM 2 and GFP-emerin strains, as well as for NPP-10 antibodies, and Anthony Hyman for AIR-1 antibodies. We are grateful to Paul Guichard and Aitana Neves for help with image processing, to Christian Gentili for his work on cyk-4 and ect-2 as potential regulators of mitosis, as well as to Fernando Romero Balestra for critical reading of the manuscript. V.H. was supported by a Roche postdoctoral fellowship (Mkl/stm 120-2007) and by an MHV postdoctoral fellowship from the Swiss National Science Foundation (PMPD33-118694). Additional support was provided by the Swiss Cancer League (grant KLS 2160-02-2008 to P.G.). Work in the laboratory of P.A. was funded by the Spanish Ministry of Science and Innovation (BFU2010-15478).
Abbreviations used:
- DIC
differential interference contrast
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- NE
nuclear envelope
- NEBD
nuclear envelope breakdown
- NPC
nuclear pore complex
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0204) on June 27, 2012.
REFERENCES
- Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002;108:83–96. doi: 10.1016/s0092-8674(01)00627-4. [DOI] [PubMed] [Google Scholar]
- Belgareh N, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger JM, Gönczy P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol. 2003;13:1488–1498. doi: 10.1016/s0960-9822(03)00582-7. [DOI] [PubMed] [Google Scholar]
- Boxem M, Srinivasan DG, van den Heuvel S. The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development. 1999;126:2227–2239. doi: 10.1242/dev.126.10.2227. [DOI] [PubMed] [Google Scholar]
- Brauchle M, Baumer K, Gönczy P. Differential activation of the DNA replication checkpoint contributes to asynchrony of cell division in C. elegans embryos. Curr Biol. 2003;13:819–827. doi: 10.1016/s0960-9822(03)00295-1. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase D, Serafinas C, Ashcroft N, Kosinski M, Longo D, Ferris DK, Golden A. The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis. 2000;26:26–41. doi: 10.1002/(sici)1526-968x(200001)26:1<26::aid-gene6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Chatel G, Fahrenkrog B. Nucleoporins: leaving the nuclear pore complex for a successful mitosis. Cell Signal. 2011;23:1555–1562. doi: 10.1016/j.cellsig.2011.05.023. [DOI] [PubMed] [Google Scholar]
- D'Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, Mattaj I. A role for gp210 in mitotic nuclear-envelope breakdown. J Cell Sci. 2008;121:317–328. doi: 10.1242/jcs.022525. [DOI] [PubMed] [Google Scholar]
- Galy V, Askjaer P, Franz C, Lopez-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Curr Biol. 2006;16:1748–1756. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–5115. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, Schnabel R. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J Cell Biol. 1999;144:927–946. doi: 10.1083/jcb.144.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- Hachet V, Canard C, Gonczy P. Centrosomes promote timely mitotic entry in C. elegans embryos. Dev Cell. 2007;12:531–541. doi: 10.1016/j.devcel.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/s1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J Cell Biol. 2001;155:1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E, Oegema K, Kirkham M, Gönczy P, Habermann B, Hyman AA. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Shibuya EK, Wozniak RW. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol Biol Cell. 2005;16:2382–2394. doi: 10.1091/mbc.E04-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- Hutterer A, Berdnik D, Wirtz-Peitz F, Zigman M, Schleiffer A, Knoblich JA. Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev Cell. 2006;11:147–157. doi: 10.1016/j.devcel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/s1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Kosova B, Pante N, Rollenhagen C, Hurt E. Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane. J Biol Chem. 1999;274:22646–22651. doi: 10.1074/jbc.274.32.22646. [DOI] [PubMed] [Google Scholar]
- Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–550. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Lee KK, Gruenbaum Y, Spann P, Liu J, Wilson KL. C. elegans nuclear envelope proteins emerin, MAN1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol Biol Cell. 2000;11:3089–3099. doi: 10.1091/mbc.11.9.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart P, Rabut G, Daigle N, Hand AR, Terasaki M, Ellenberg J. Nuclear envelope breakdown in starfish oocytes proceeds by partial NPC disassembly followed by a rapidly spreading fenestration of nuclear membranes. J Cell Biol. 2003;160:1055–1068. doi: 10.1083/jcb.200211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell. 2008;14:831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Nakano H, Wang W, Hashizume C, Funasaka T, Sato H, Wong RW. Unexpected role of nucleoporins in coordination of cell cycle progression. Cell Cycle. 2011;10:425–433. doi: 10.4161/cc.10.3.14721. [DOI] [PubMed] [Google Scholar]
- Noatynska A, Panbianco C, Gotta M. SPAT-1/Bora acts with Polo-like kinase 1 to regulate PAR polarity and cell cycle progression. Development. 2010;137:3315–3325. doi: 10.1242/dev.055293. [DOI] [PubMed] [Google Scholar]
- Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, Oegema K. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev Cell. 2007;12:515–529. doi: 10.1016/j.devcel.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell. 2005;16:2139–2153. doi: 10.1091/mbc.E04-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenas E, Gonzalez-Aguilera C, Ayuso C, Askjaer P. Dissection of the NUP107 nuclear pore subcomplex reveals a novel interaction with spindle assembly checkpoint protein MAD1 in C. elegans. Mol Biol Cell. 2012;23:930–944. doi: 10.1091/mbc.E11-11-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenas E, Klerkx EP, Ayuso C, Audhya A, Askjaer P. Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev Biol. 2009;327:399–409. doi: 10.1016/j.ydbio.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Rual JF, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetter A, Askjaer P, Piano F, Mattaj I, Kemphues K. Nucleoporins NPP-1, NPP-3, NPP-4, NPP-11 and NPP-13 are required for proper spindle orientation in C. elegans. Dev Biol. 2006;289:360–371. doi: 10.1016/j.ydbio.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R, Gönczy P. zyg-11 and cul-2 regulate progression through meiosis II and polarity establishment in C. elegans. Development. 2004;131:3527–3543. doi: 10.1242/dev.01244. [DOI] [PubMed] [Google Scholar]
- Toya M, Terasawa M, Nagata K, Iida Y, Sugimoto A. A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat Cell Biol. 2011;13:708–714. doi: 10.1038/ncb2242. [DOI] [PubMed] [Google Scholar]
- Wood WB, Hecht R, Carr S, Vanderslice R, Wolf N, Hirsh D. Parental effects and phenotypic characterization of mutations that affect early development in Caenorhabditis elegans. Dev Biol. 1980;74:446–469. doi: 10.1016/0012-1606(80)90445-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.