Abstract
Nu61, a radiation-resistant human tumor xenograft, was selected from a parental radiosensitive tumor SCC-61 by eight serial cycles of passage in athymic nude mice and in vivo irradiation. Replicate DNA array experiments identified 52 genes differentially expressed in nu61 tumors compared with SCC-61 tumors. Of these, 19 genes were in the IFN-signaling pathway and moreover, 25 of the 52 genes were inducible by IFN in the nu61 cell line. Among the genes involved in IFN signaling, STAT1α and STAT1β were the most highly overexpressed in nu61 compared to SCC-61. STAT1α and STAT1β cDNAs were cloned and stably transfected into SCC-61 tumor cells. Clones of SCC-61 tumor cells transfected with vectors expressing STAT1α and STAT1β demonstrated radioprotection after exposure to 3 Gy (P < 0.038). The results indicate that radioresistance acquired during radiotherapy treatment may account for some treatment failures and demonstrate an association of acquired tumor radioresistance with up-regulation of components of the IFN-related signaling pathway.
Keywords: IFN signaling, acquired tumor radioresistance, microarrays, HSV-1 replication in tumors
Curative treatment of malignant tumors with ionizing radiation (IR) was introduced ≈80 years ago (1). During ensuing years, the failure to cure tumors by radiation therapy has been ascribed to intrinsic tumor cell radioresistance and tumor microenvironmental factors such as hypoxia, which favor tumor cell survival after IR (1, 2). Basic principals of signal transduction and DNA repair after IR have been investigated in simple genetic model systems and in cells derived from radiosensitive patients with genetic disorders such as Ataxia telangiectasia (3-5). Although these investigations have elucidated important basic principles and pathways of the response of eukaryotic cells to radiation damage, these studies have had little impact on improving clinical radiotherapy. There are several potential explanations for the difficulty in translating these findings to radiotherapy. (i) IR is delivered in multiple doses that are relatively small (100-300 cGy/day) compared with the larger single doses used in investigations of mechanisms of DNA repair and checkpoint control. (ii) Cells in an organized tissue or tumor respond differently to IR than cells in single cell suspension in vitro (6-9). To determine the mechanisms underlying resistance to IR in vivo, it is essential to compare genes differentially expressed from isogenic radiosensitive and radioresistant tumors. We selected radioresistant cells after sequential irradiation of tumors resulting from implantation of radiosensitive tumor cells. The key, totally unexpected, finding was that IR-resistant tumors overexpress a significant number of genes related to IFN pathways or inducible by IFN. Comparative analysis of these genes revealed that STAT1 transcription factor involved in the IFN signaling is overexpressed in the radioresistant tumor. Transfection of the isoforms of STAT1, STAT1α and STAT1β, conferred radioresistance on the recipient radiosensitive SCC-61 cells. These data implicate the IFN-signaling pathway and, specifically, isoforms of STAT1 as mediators of acquired tumor radioresistance.
Materials and Methods
Cell and Viruses. Human head and neck squamous cell carcinoma line SCC-61, the serial tumor explants, and the selected cell line nu61-8-4000 were grown in DMEM/F-12 (1:1) (GIBCO)/20% FBS (Intergen, Purchase, NY)/1% penicillin-streptomycin/0.4 μg/ml hydrocortisone at 37°C and 7% CO2. R3616 is a recombinant virus derived by genetic engineering of herpes simplex virus 1 strain F [HSV-1(F)]. The mutant virus lacks both copies of the γ134.5 gene (10). Virus stocks were made and titered in Vero cells.
Selection of Radioresistant SCC-61 Tumors. A total of 107 SCC-61 cells were injected into the right hind limb of female athymic nude mice (Frederick Cancer Research Center, Frederick, MD). Tumors were grown to 250 mm3 and irradiated at 5Gy by using a Pantak X-ray generator operating at 150 Kv and 25 mA with a dose rate of 192 cGy/min. Seven days later, the tumors were excised, minced, and explanted in 100-mm3 tissue culture dishes. Explants were incubated at 37°C, 7% CO2, and 100% humidity in culture media (see above). Contaminating fibroblast cells were removed by incubation with EDTA and rinsing. After four serial passages in vitro, the amplified cells were pooled and implanted into athymic nude mice as described above. Cycle was repeated eight times. The total dose of ionizing radiation was 4,000 cGy. The cell line derived from the final passage through nude mice was designated nu61-8-4000, hereafter called nu61.
In Vivo Radiation Experiments. In vivo radiation experiments were done as described elsewhere (11).
Measurements of Virus Replication in Tumors. SCC-61 and nu61 xenografts were generated as described above and allowed to grow to a volume of 150-200 mm3. At that time, 107 plaque-forming units (pfu) of the HSV-1 recombinant virus R3616 were injected intratumorally. The tumors were harvested 72 h after infection, homogenized, freeze-thawed, sonicated, and plated on monolayers of Vero cells. The results represent the average of two experiments with a total of eight sample tumors for each cell line.
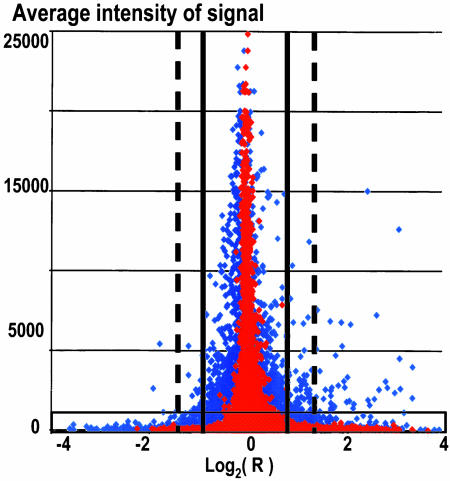
DNA Array Data Analysis. All experiments were performed with Afffymetrix U133 DNA chips, carrying probes for ≈22,000 human genes. Data were acquired and normalized (scaled) with the mas 5.0 software and exported into Microsoft excel for the further transformations. Our analysis was based on the pairwise comparison of replicated arrays. We used multistep filtration of data. First, we removed the data for which the signal intensity was called absent by the mas 5.0 based on the comparison of perfect match with mismatched probes. We next applied t tests for the selection of genes significantly different in nu61 relative to SCC-61 tumors. As a cutoff level, we used a P value ≤0.05. Second, we used receiver operating characteristic (ROC)-analysis for intensity-based filtration, to remove low-intensity signals as described in ref. 12. Third, we filtered the data by ratios by measuring standard deviations for the reference sets. Reference sets (same-to-same hybridizations) were based on the comparison of the hybridizations with parallel samples. Ratios, corresponding to ±2 SD (95.5% stringency) and ±3 SD (99.7% stringency) of the reference sets, varied depending on experiment (9). A graphic representation of this strategy is shown in Fig. 1.
Fig. 1.
Scheme of multistep filtration (MSF) of DNA array data. Red diamonds corresponds to the same-to-same hybridizations (nu61/nu61 and SCC-61/SCC-61), and blue diamonds correspond to the same-to-different hybridizations (nu61/SCC-61). Light blue region on the bottom of the histogram corresponds to the genes with intensity signals below cutoff level, estimated by the receiver operating characteristic (ROC) analysis. Note, that genes in this region possess the highest ratios of response producing false positive data (see ref. 12). Solid vertical bars, range of ±2 SD of the reference set, corresponding to ratios of ± 1.919. Dashed vertical bars, range of ± 3 SD of the reference set, corresponding to the ratios of ±2.657. Shown are data not filtrated by P values and CV.
Cloning of STAT1α and STAT1β was based on the full-size cDNA clones, obtained from Invitrogen (IMAGE clone ID 2452685, GenBank accession no. AI923499) and ATCC (IMAGE clone ID 3627218, GenBank accession no. BC002704, respectively). All clones were sequence verified. Synthetic AttB (13, 14) were annealed and cloned into MluI/SpeI-digested pIRES-Hyg3 (Clontech) to generate pAttb. Stat1α was released by using EcoRV and cloned into NaeI-digested pAttb to produce pAttbStat1α. The STAT1β coding sequence was released by using BamHI and BglII and cloned into BamHI-digested pIres-Hyg3 to produce pStat1-Hyg3-4. The STAT1β coding sequence was then transferred from pStat-Hyg3-4 into pAttb-Hyg3 by using AflII and BsiWI to produce pAttbStat1β. Resulted plasmids were sequence verified. pAttBStat1α, pAttBStat1β or pAttB were cotransfected into SCC-61 with pCMV-INT, kindly provided by Michele Calos (Department of Genetics, Stanford University School of Medicine, Stanford, CA). Cells were selected in the Hyg-containing media, and selected monoclones were further propagated for experiments. As the control, we used pAttB cotransfected with pCMV-INT as described above. Viability of clones after irradiation at 3 and 10 Gy was determined by the MTS assay, as described elsewhere (15).
Transient Transfection. For transient transfections, we used pAttBStat1α, pAttBStat1β, or pAttB-empty vector. Cells were treated with the TransFast reagent (Promega), using a charge ratio of 1:1 (1 μg of DNA plus 3 μl of TransFast reagent), in the presence of pAttBStat1α, pAttBStat1β, or pAttB plasmids. Cells were incubated for1hwiththe transfection complexes, complete media was then added, and incubation was continued for 24 h before irradiation with 3 Gy. Surviving cells were scored by the Trypan Blue exclusion assay 24 h after irradiation (16). Data were presented as percent of surviving cells, where 100% corresponded to the unirradiated control for each transfectant.
Western Blots. Total cellular protein was extracted in RIPA buffer with protease inhibitors added (1× PBS/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/1 mM Na3VO4/2 μg/ml aprotinin/1 mM PMSF). Proteins were separated on SDS/7.5% PAGE gels and transferred to nitrocellulose membranes. Stat1α and -β were detected with the anti-Stat1 p84/p91 (E-23) antibody (Santa Cruz Biotechnology).
Results
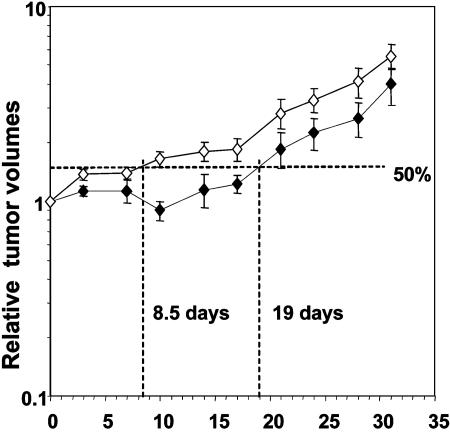
Selection of Radioresistant Tumors. We selected a radioresistant variant (nu61) of the relatively radiosensitive head and neck cancer cell line, SCC-61 (17), by injection of tumor cells into nude mice. Tumors were exposed to 5 Gy and excised, and tumor and stromal cells were separated in vitro and reinjected to form tumors. The cycle of irradiation and reexcision was repeated eight times. After eight cycles and a cumulative total of 4,000 cGy of in vivo irradiation, the tumor cells were excised and established as a cell line designated nu61-8-4000 (nu61). For quantitative estimation, we determined the time it took for the tumors to increase 1.5-fold (or 50%) in relative tumor volume after six 5-Gy irradiations with each dose delivered daily. As illustrated in Fig. 2, whereas the parent SCC-61 tumors required 19 days to reach a volume 1.5-fold greater than the starting volume, the nu61 tumors required only 8.5 days. Therefore, the estimated difference in the radiation induced growth delay was reduced ≈2.2-fold. Moreover, the doubling time for the nonirradiated nu61 tumors was 8.5 days compared to 14.6 days for the SCC-61 tumors (data not shown). The decrease in radiation induced growth delay and accelerated growth of nu61 tumors reflects acquisition of radioresistance.
Fig. 2.
Relative increase in SCC-61 and nu61 tumors as a function of time after irradiation. Tumors were permitted to grow to a volume of 250-300 mm3. At that time (day 0) the tumors were subjected to six 500-cGy irradiations as described in Materials and Methods. Measurements of relative tumor volumes were performed during 32 days. Growth delay was defined as the time, necessary for irradiated tumor to increase relative volume by 50%, compared to the volume on day 0. Error bars are SEM.
Genes in the IFN-Signaling Pathway Are Differentially Expressed in nu61 Tumors. Identification of the genes differentially expressed in nu61 relative to SCC-61 tumors was done as follows. In the first experiment, we profiled RNAs pooled from three animals for each array. In the second independent experiment, we profiled RNA from a single animal for each array. In each experiment, three arrays were used for each experimental point. At the 95.5% stringency (mean ± 2 SD, see Fig. 1), 276 genes were differentially expressed between SCC-61 and nu61 in the first experiment, 119 genes were differentially expressed in the second experiment, and 52 genes were differentially expressed in both experiments. These genes are presented in Table 1. Investigation of the list of these 52 genes (see Table 1) showed that 19 genes are known to be components of the IFN inducible pathway, including isoforms of 2′,5′-oligoadenylate synthetase and MHC class I (18, 19). To confirm the association of genes, differentially expressed in nu61 with IFN-inducible genes, we exposed nu61 tumor cell line for 5 h to IFNα, -β, and -γ (50, 50, and 5 ng/ml, respectively, corresponding to the IC50 for each in nu61). After IFN exposure, we identified 343 responding genes (see Materials and Methods). Of this number, 25 genes overlap with the 52 genes listed in Table 1. These genes include IRF7, genes conferring protection from double-stranded RNA and viruses (OAS1, OAS3, and MX1), and genes in the IFN-signaling pathway (Stat1α and Stat1β). Microarray data were confirmed by quantitative RT-PCR (see Table 1).
Table 1. Genes, differentially expressed in nu61 relative to SCC-61 and common between experiments 1 and 2.
| ID | Name | Symbol | N/S R, experiment 1 | N/S R, experiment 2 | IFN/UTC R, experiment 3 |
|---|---|---|---|---|---|
| 202869_at | 2′,5′-oligoadenylate synthetase 1, 40/46kDa | OAS1*† | 3.286 | 2.282 | 3.922 |
| 205552_s_at | 2′,5′-oligoadenylate synthetase 1, 40/46kDa | OAS1*† | 4.506 | 2.491 | 4.290 |
| 218400_at | 2′-5′-oligoadenylate synthetase 3, 100kDa | OAS3*† | 3.624 | 2.318 | 3.113 |
| 201641_at | Bone marrow stromal cell antigen 2 | BST2*† | 10.343 | 5.196 | 5.744 |
| 204533_at | IFNγ-inducible protein 10 | CXCL10* | 10.319 | 2.820 | 10.619 |
| 204439_at | Chromosome 1 ORF 29 | Clorf29*† | 33.479 | 45.942 | 4.793 |
| 205899_at | Cyclin A1 | CCNA1* | 2.172 | 2.376 | 2.443 |
| 218986_s_at | Hypothetical protein FLJ20035 | FLJ20035* | 8.787 | 6.389 | 3.798 |
| 219352_at | Hypothetical protein FLJ20637 | FLJ20637* | 7.315 | 3.441 | 4.096 |
| 208436_s_at | IFN regulatory factor 7 | IRF7* | 3.254 | 2.713 | 4.814 |
| 204415_at | IFN, α-inducible protein | G1P3* | 3.401 | 5.341 | 5.451 |
| 205483_s_at | IFN, α-inducible protein | G1P2* | 8.500 | 6.452 | 4.967 |
| 202411_at | IFN, α-inducible protein 27 | IFI27*† | 5.425 | 9.462 | 4.231 |
| 209417_s_at | IFN-induced protein 35 | IFI35*† | 4.528 | 2.653 | 4.652 |
| 214059_at | IFN-induced protein 44 | IFI44*† | 5.654 | 3.212 | 3.976 |
| 214453_s_at | IFN-induced protein 44 | IFI44*† | 8.102 | 4.187 | 4.151 |
| 204747_at | IFN-induced protein with tetratricopeptide repeats | IFIT4*† | 6.479 | 2.781 | 6.701 |
| 203153_at | IFN-induced protein with tetratricopeptide repeats 1 | IFIT1*† | 8.606 | 9.055 | 6.124 |
| 202086_at | IFN-inducible protein p78 | MX1*† | 6.204 | 6.327 | 4.240 |
| 205569_at | Lysosomal-associated membrane protein 3 | LAMP3*† | 7.084 | 3.025 | 11.535 |
| 219209_at | Melanoma differentiation-associated protein 5 | MDA5* | 2.617 | 2.761 | 5.238 |
| 202446_s_at | Phospholipid scramblase 1 | PLSCR1*† | 3.498 | 2.428 | 2.398 |
| 200887_s_at | Signal transducer and activator of transcription 1 | STAT1α*† | 2.370 | 2.578 | 2.430 |
| 209969_s_at | Signal transducer and activator of transcription 1 | STAT1β*† | 4.094 | 2.272 | 4.192 |
| 201110_s_at | Thrombospondin 1 | THBS1* | 2.496 | 3.542 | 2.837 |
| 200798_x_at | Myeloid cell leukemia sequence 1 (BCL2-related) | MCL1** | 2.146 | 2.496 | - |
| 200923_at | Lectin | LGALS3BP** | 2.226 | 2.288 | - |
| 201150_s_at | Tissue inhibitor of metalloproteinase 3 | TIMP3** | 2.419 | 2.498 | - |
| 201601_x_at | IFN induced transmembrane protein 1 (9-27) | IFITM1**† | 4.298 | 3.036 | - |
| 201893_x_at | Decorin | DCN** | 0.415 | 0.315 | - |
| 202145_at | Lymphocyte antigen 6 complex, locus E | LY6E** | 4.124 | 2.630 | - |
| 202409_at | Putative insulin-like growth factor 2-associated protein | NA**‡ | 0.046 | 0.377 | - |
| 202736_s_at | LSM4 homolog | LSM4** | 2.336 | 3.756 | - |
| 203148_s_at | Tripartite motif-containing 14 | TRIM14** | 3.270 | 3.175 | - |
| 204019_s_at | Likely ortholog of mouse Src homology 3 domain YSC-like 1 | SH3YL1** | 0.218 | 0.253 | - |
| 204417_at | Galactosylceramidase | GALC** | 2.190 | 2.968 | - |
| 204470_at | Chemokine (C-X-C motif) ligand 1 | CXCL1** | 3.093 | 2.470 | - |
| 204614_at | Serine (or cysteine) proteinase inhibitor | SERPINB2** | 0.358 | 0.428 | - |
| 205623_at | Aldehyde dehydrogenase 3 family | ALDH3A1** | 0.325 | 0.317 | - |
| 205829_at | Hydroxysteroid (17-β) dehydrogenase 1 | HSD17B1** | 3.428 | 4.291 | - |
| 206376_at | Homolog of rat orphan transporter v7-3 | NTT73** | 2.102 | 2.322 | - |
| 208282_x_at | Deleted in azoospermia 2 | DAZ2** | 2.138 | 3.192 | - |
| 208729_x_at | MHC, class 1, B | HLA-B** | 2.377 | 2.371 | - |
| 209301_at | Carbonic anhydrase II | CA2** | 2.655 | 2.480 | - |
| 211529_x_at | MHC, class 1, A | HLA-A** | 2.683 | 2.583 | - |
| 212463_at | CD59 | CD59** | 0.516 | 0.447 | - |
| 213194_at | Roundabout, axon guidance receptor, homolog 1 | ROBO1**† | 5.562 | 4.970 | - |
| 213294_at | Hypothetical protein FLJ38348 | FLJ38348** | 2.401 | 2.276 | - |
| 214657_s_at | Trophoblast-derived noncoding RNA | TncRNA** | 0.505 | 0.405 | - |
| 215239_x_at | Zinc finger protein 273 | ZNF273** | 0.463 | 0.414 | - |
| 216922_x_at | Deleted in azoospermia | DAZ** | 2.320 | 2.968 | - |
| 218669_at | RAP2C, member of RAS oncogene family | RAP2C** | 0.520 | 0.070 | - |
Shown are 52 genes, common between two independent experiments in vivo. Numbers represent the ratios of expression of genes in nu61 relative to SCC-61 (columns 5 and 6) or ratios of gene expression in nu61 exposed to IFNs relative to the untreated cells (column 7). Each target was counted as separate gene or potential gene isoform. OAS1, IFI44, and Stat1 are represented by two targets each. Stat1 targets correspond to α (200887_s_at) and β (209969_s_at) isoforms. Asterisks represent 25 genes overlapped with experiment 3 (response to IFN in vitro). Double asterisks represent 27 genes, not inducible by IFN in experiment 3. Daggers indicate the genes confirmed by the real-time PCR analysis. Not included here but also confirmed by the real-time PCR genes are: ISG15 (IFN-stimulated protein, 15 kDa), MX2 (IFN-regulated resistance GTP-binding protein MxB), TRIM22 (tripartite motif-containing 22), USP18 (ubiquitin-specific protease 18), CLCA4 (chloride channel, calcium-activated, member 4), HAIK1 (type 1 intermediate filament cytokeratin), KLK7 (kallikrein 7), PPP1R3C (protein phosphatase 1, regulatory subunit 3C), IGF2 (insulin-like growth factor 2), and RIL (LIM domain protein). Quantitative RT-PCR was performed with the pooled RNA samples, obtained from the first experiment (see text), and genes not shown here were selected based on the levels of expression, exceeding ±3 SD cutoff level (see Materials and Methods and Fig. 1) and producing primers, able to generate positive products in quantitative RT-PCR amplification reactions. Overall correlation between DNA array data and quantitative RT-PCR data, measured as Pearson's correlation coefficient, was 0.894.
NA, not available
Functional Competence of the IFN-Inducible System in nu61. Xeno-grafts of nu61 or SCC-61 tumors growing in athymic nude mice were each inoculated with the R3616 mutant of HSV-1(F). The tumors were harvested 3 days after inoculation and processed as described in Materials and Methods. The recovery of infectious virus from SCC-61 and nu61 tumors was 2.0 × 105 ± 1.5 × 105 and 0.5 × 104 ± 2.8 × 104, respectively. Significantly (P = 0.001, as estimated by Mann-Whitney test; see ref. 20) more infectious virus was recovered from SCC-61 than nu61. In light of the heightened sensitivity of this mutant to IFN, the restricted proliferation of R3616 in nu61 cells could reflect activation of the IFN pathway in these tumors as compared to SCC-61 tumors.
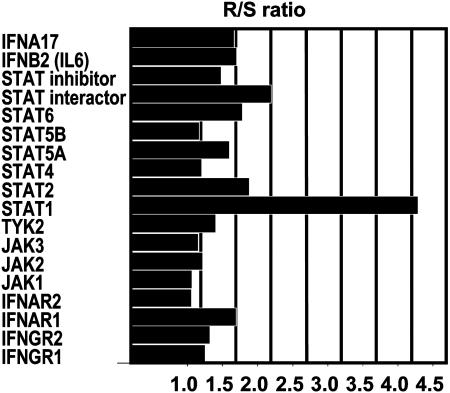
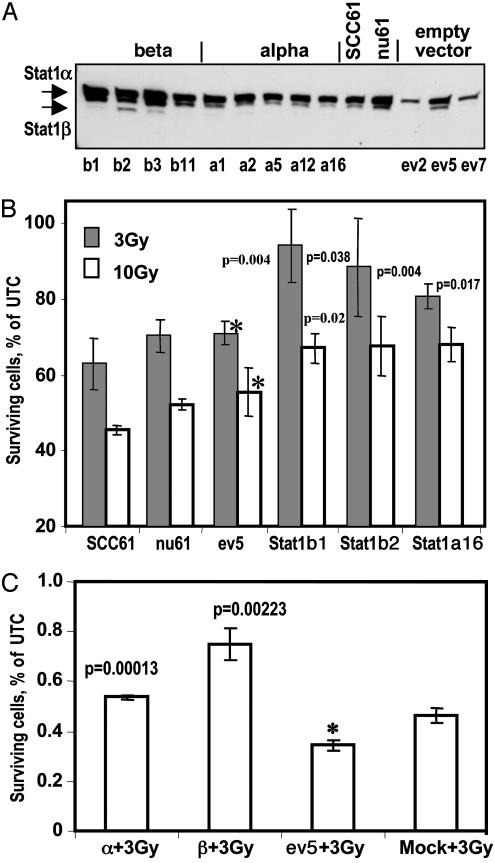
STAT1 as Potential Mediator of Radioresistance in nu61 Tumors. The results of the microarray analyses indicate that STAT1 is the most highly up-regulated gene among the genes in the IFN-signaling pathway listed in Table 1. Stat1 is an upstream mediator of IFN signaling. The question arose whether Stat1 was the sole upstream mediator of IFN signaling up-regulated in these studies. To answer this question, we compared the ratios of Stat1 expression in nu61 relative to SCC-61 with the ratios of other genes involved in IFN signaling contained in the DNA array database of the first experiment even if they were not up-regulated as defined by our criteria. Fig. 3 demonstrates that, for nu61, Stat1β is the most highly differentially expressed gene in the IFN-signaling pathway. We further investigated the effects of irradiation on SCC-61 tumor cells stably transfected with pAttBStat1α, pAttBStat1β, and pAttB as described in Materials and Methods. Fig. 4A shows protein expression of Stat1α and Stat1β in selected stably transfected clones. Fig. 4B demonstrates that stable transfectants of SCC-61 by STAT1 expression vectors show an increase in radioresistance compared with pAttB (empty vector) transfectants as measured by MTS viability assay (see Materials and Methods). For the Stat1β1 clone, the surviving fraction increased by 32.8% (P = 0.00377) after exposure to 3 Gy and 21.3% after 10 Gy (P = 0.020). For the Stat1β2 clone, the increase in survival was 24.6% (P = 0.03815) after 3 Gy and 22.1% (P = 0.050) after 10 Gy. For the Stat1α16 clone, the increase in survival after 3 Gy was 13.9%, (P = 0.00431) and 23.1% (P = 0.017) after 10 Gy. The results indicate that both α and β isoforms of STAT1 protect SCC-61 cells from ionizing radiation-mediated death. Because stable transfectants are clonal lines that may have acquired secondary mutations during the selection process, SCC-61 cells were exposed to 3 Gy 24 h after transient transfection with the same expression vectors, encoding Stat1α and Stat1β as noted above. The surviving cells were counted by using the Trypan Blue exclusion assay. As shown in Fig. 4C, transient transfection with pAttBStat1α led to a 1.55-fold increase (P = 0.00013) in survival. Transfection with pAttBStat1β led to a 2.2-fold increase (P = 0.00223) in survival (radioprotection). Thus, the results of transient and stable transfection experiments demonstrate that the isoforms of STAT 1 confer enhanced survival of tumor cells after radiation.
Fig. 3.
Stat1 is overexpressed in the nu61 compared with SCC-61 more than other genes, involved in the IFN signaling and presented on U133A DNA Chip. Data were taken from the first experiment with pooled RNA samples. Databases, obtained with U133A arrays were screened manually to identify genes, involved in IFN signaling. Shown are the ratios of the gene expression in the nu61 cells relative to the SCC-61 parental cells. Stat1 (β isoform, target ID 209969_s_at) is overexpressed more than other IFN-signaling genes.
Fig. 4.
Stable transfection of SCC-61 parental cell line by Stat1α and Stat1β cDNAs leads to increased radioprotection of the transfected clones. (A) Protein expression of Stat1 in stably transfected clones. pAttBStat1α, pAttBStat1β, or pAttB were cotransfected into SCC-61 with pCMV-INT, kindly provided by Michele Calos. Cells were selected in the Hyg-containing media, and selected monoclones were further propagated for experiments (see Materials and Methods). Several clones, transfected by pAttBStat1β, pAttBStat1α, or pAttB (empty vector), were checked by PCR of genomic DNA for appropriate integration and checked by Western analysis for expression of α and β isoforms of Stat1. b1, b2, b3, and b11 correspond to the clones stably transfected by pAttBStat1β. a1, a2, a5, a12, and a16 correspond to the clones stably transfected by pAttBStat1α. ev2, ev5, and ev7 correspond to the clones, stably transfected by pAttB and related as “empty vector.” For the further experiments we choose b1, b2, and a16 clones as Stat1 stable transfectants and ev5 clone as the “empty vector” control. (B) Stably transfected α and β clones protect parental cell line from ionizing radiation. SCC-61, nu61, and b1, b2, a16, and ev5 clones were plated in the 96-well plates and either irradiated at 3 or 10 Gy (see Materials and Methods) or left untreated. All experiments were made in triplicates. Viability of irradiated and untreated cells was assessed by the MTS assay 72 h after irradiation (see Materials and Methods). Fraction of surviving cells was calculated as percent of viable cells after irradiation to unirradiated control for the each type of the cells. For the stable transfectants of the SCC-61, significance of survival was calculated relative to the ev5 clone, marked by asterisks. P values were calculated as the two-tailed, unpaired t test and are indicated in B above each corresponding bar. Error bars are standard deviations of triplicate experiments. (C) Transient transfection of the parental cell line SCC-61 by plasmids, carrying Stat1β and Stat1α leads to the increased radioprotection. SCC-61 cells were transfected with the pAttBStat1α, pAttBStat1β or pAttB plasmids (see above and Materials and Methods). As the mock we used SCC-61, treated in the same way as transfected cells but without exogenous plasmids. Twenty-four hours after transfection, cells were irradiated at 3 Gy, and 24 h after irradiation viable cells were scored by the Trypan Blue assay (see Materials and Methods). Surviving fraction was calculated as the percent of viable cells after irradiation to unirradiated control for the each type of the cells. Significance was calculated relative to the ev5-transfected cells (marked by asterisk), using two-tailed unpaired t test. P values are indicated above the appropriate columns. Error bars are standard deviations of the triplicate experiments.
Discussion
The salient features of this report and their significance are as follows.
(i) The emergence of tumors after radiotherapy has been attributed to repopulation of tumors with cells surviving irradiation. The conditions of the tumor microenvironment that favor tumor cell survival after IR include hypoxia and secretion of γ radiation protective cytokines and growth factors that promote the growth and survival of tumor tissue (2, 21, 22). Also, a number of candidate genes have been implicated in the response of eukaryotic cells to IR. These include a number of cell cycle, checkpoint, and DNA repair genes as well as mediators of apoptosis such as p53, Bax, Bcl-2, etc. (23-26). Expression or repression of these genes is associated with cell survival or death in simple model systems but shed no light on intercellular events in in situ tumors or clinical outcome of radiotherapy. An important yet unanswered question in radiotherapy is whether radioresistant tumor cells exist de novo or whether they develop during a course of fractionated radiotherapy. The studies described in this report were designed to develop a system in which many of the questions regarding the resistance of tumors to IR in situ can be answered at a molecular level. We report the isolation of an ionizing radiation-resistant tumor by serial passage of tumor cells in mice coupled with irradiation of the tumors in situ. To our knowledge, this is the first report of the isolation of a radiation-resistant tumor by multiple exposures to IR of a sensitive parental tumor in situ. The derivation of such tumors opens the way for genetic analysis of radiation resistance in situ. Moreover, our results support the hypothesis that resistance to ionizing radiation may be selected after fractionated radiation of tumors, a common practice in clinical radiotherapy.
(ii) Comparison of gene expression of radioresistant and radiosensitive tumors in situ led to the identification of 52 genes whose transcription was significantly up-regulated in radioresistant tumors in two independent experiments. Of these, 25 genes were induced by interferons in the nu61 cell line and 19 are known components of IFN-inducible pathways (see Table 1). These results led to the association of overexpression of the IFN pathway and a radioresistant phenotype. These results are supported by previous reports that exposure to IFN enhances cell survival after radiation (27-29).
(iii) To test the functional competence of the IFN-inducible system, we inoculated the tumors with a highly attenuated, IFN-sensitive HSV-1 mutant R3616 lacking both copies of the γ134.5 genes (30). The basis of attenuation of the R3616 mutant is its inability to block the effects of the activated IFN-signaling pathway (31, 32). In knockout mice lacking components of the pathway, R3616 becomes as virulent as its wild-type parent (33). The results indicated that the R3616 mutant replicated significantly better in the SCC-61 tumors than in the nu61 tumors. The results indicate a dissociation of the effects of IFN on tumor growth from the effects of the IFN pathway on viral replication
(iv) STAT1 is an upstream mediator of IFN-signaling. As noted in Fig. 3, STAT1 is the most highly expressed gene in the IFN-signaling pathway in nu61 compared with SCC-61 tumors among related genes presented on the U133A arrays. Our experiments with stably and transiently transfected SCC-61 showed that both isoforms of STAT1 (α and β) increased radioresistance of transfected cells (see Fig. 4 B and C). STAT1 expression is associated with a tumor suppressor and proapoptotic functions in some experimental systems (34). STAT proteins, including STAT1, are multifunctional and associated with diverse cellular functions (35). For example, STAT1 overexpression has been demonstrated in several human cancers, including head and neck cancer (36). STAT1 protein has also been shown to be overexpressed during lung tumor progression (37). Intriguingly, STAT1 is induced by radiation in association with epidermal growth factor receptor (EGFR) (38). The EGFR-signaling pathway has been associated with tumor radio-resistance. We propose that isoforms of STAT1 in some cellular contexts may transduce survival/growth signals that enhance tumor survival after x-irradiation under some conditions.
Here we directly compare the expression of cellular genes in radiosensitive and radioresistant tumors in situ. The availability of such tumor lines provides a means for analyses of the acquired tumor phenotype at the molecular level. Results from these experiments may affect the delivery of radiotherapy. For example, if radioresistant tumor cells evolve during therapy, then a few large doses may be a more favorable delivery schedule than the multiple smaller doses currently used. Also, specific molecular targets in addition to DNA repair/checkpoint genes are identified by these experiments. However, much remains to be done to assess the impact of the individual genes identified in this report on the development of resistance to radiotherapy.
Acknowledgments
We are grateful to Dr. Samuel Hellman (Department of Radiation and Cellular Oncology, The University of Chicago, Chicago) for his fruitful discussion of the manuscript. These studies were aided by National Cancer Institute Grants CA71933 and CA78766 and the U.S. Public Health Service.
Abbreviations: IR, ionizing radiation; HSV, herpes simplex virus.
References
- 1.Hall, E. J. (2000) Radiobiology for Radiologist (Lippincott Williams and Wilkins, Philadelphia), 5th Ed., pp 5-17.
- 2.Harris, A. L. (2002) Nat. Rev. Cancer 2, 38-47. [DOI] [PubMed] [Google Scholar]
- 3.Taylor, A. M. (1998) Int. J. Radiat. Biol. 73, 365-371. [DOI] [PubMed] [Google Scholar]
- 4.Young, B. R. & Painter, R. B. (1989) Hum. Genet. 82, 113-117. [DOI] [PubMed] [Google Scholar]
- 5.Valerie, K. & Povirk, L. F. (2003) Oncogene 22, 5792-5812. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Barros, M., Paris, F., Cordon-Cardo, C., Lyden, D., Rafii, S, Haimovitz-Friedman, A., Fuks, Z. & Kolesnick, R. (2003) Science 300, 1155-1159. [DOI] [PubMed] [Google Scholar]
- 7.Weaver, V. M., Lelievre, S., Lakins, J. N., Chrenek, M. A, Jones, J. C., Giancotti, F., Werb, Z. & Bissell, M. J. (2002) Cancer Cell. 2, 205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, S. K., Karlsson, M. C., Ravetch, J. V. & Kerbel, R. S. (2002) Cancer Res. 62, 6891-6900. [PubMed] [Google Scholar]
- 9.Khodarev, N. N., Park, J. O., Yu, J., Gupta, N., Nodzenski, E., Roizman, B. & Weichselbaum, R. R. (2001) Proc. Natl. Acad. Sci. USA 98, 12665-12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roizman, B. (1999) Acta Virol. 43, 75-80. [PubMed] [Google Scholar]
- 11.Hallahan, D. E., Mauceri, H. J., Seung, L. P., Dunphy, E. J., Wayne, J. D., Hanna, N. N., Toledano, A., Hellman, S., Kufe, D. W. & Weichselbaum, R. R. (1995) Nat. Med. 1, 786-791. [DOI] [PubMed] [Google Scholar]
- 12.Khodarev, N. N., Park, J., Kataoka, Y., Nodzenski, E., Hellman, S., Roizman, B., Weichselbaum, R. R. & Pelizzari, C. A. (2003) Genomics 81, 202-209. [DOI] [PubMed] [Google Scholar]
- 13.Groth, A. C., Olivares, E. C., Thyagarajan, B. & Calos, M. P. (2000) Proc. Natl. Acad. Sci. USA 97, 5995-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thyagarajan, B., Olivares, E. C., Hollis, R. P., Ginsburg, D. S. & Calos, M. P. (2001) Mol. Cell. Biol. 21, 3926-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pederson, L. C., Buchsbaum, D. J., Vickers, S. M., Kancharla, S. R., Mayo, M. S., Curiel, D. T. & Stackhouse, M. A. (1997) Cancer Res. 57, 4325-4332. [PubMed] [Google Scholar]
- 16.Khodarev, N. N., Yu, J., Labay, E., Darga, T., Brown, C. K., Mauceri, H. J., Yassari, R., Gupta, N. & Weichselbaum, R. R. (2003) J. Cell Sci. 116, 1013-1022. [DOI] [PubMed] [Google Scholar]
- 17.Quiet, C. A., Weichselbaum, R. R. & Grdina, D. J. (1991) Int. J. Radiat. Oncol. Biol. Phys. 20, 733-738. [DOI] [PubMed] [Google Scholar]
- 18.Samuel, C. E. (2001) Clin. Microbiol. Rev. 14, 778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi, T., Ogasawara, K., Takaoka, A. & Tanaka, N. (2001) Annu. Rev. Immunol. 19, 623-655. [DOI] [PubMed] [Google Scholar]
- 20.Townend, J. (2002) Practical Statistics for the Environmental and Biological Scientists (Wiley, New York), pp. 189-191.
- 21.Hallahan, D. E., Haimovitz-Friedman, A., Kufe, D. W., Fuks, Z. & Weichselbaum, R. R. (1993) Important Adv. Oncol., 71-80. [PubMed]
- 22.McBride, W. H. & Dougherty, G. J. (1995) Nat. Med. 1, 1215-1217. [DOI] [PubMed] [Google Scholar]
- 23.Amundson, S. A., Bittner, M. & Fornace, A. J., Jr. (2003) Oncogene 22, 5828-5833. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed, M. M., Alcock, R. A., Chendil, D., Dey, S., Das, A., Venkatasubbarao, K., Mohiuddin, M., Sun, L., Strodel, W. E. & Freeman, J. W. (2002) J. Biol. Chem. 277, 2234-2246. [DOI] [PubMed] [Google Scholar]
- 25.Wahl, G. M. & Carr, A. M. (2001) Nat. Cell Biol. 12, E277-E286. [DOI] [PubMed] [Google Scholar]
- 26.Haas-Kogan, D. A., Kogan, S. S., Yount, G., Hsu, J., Haas, M., Deen, D. F. & Israel, M. A. (1999) Int. J. Radiat. Oncol. Biol. Phys. 43, 399-403. [DOI] [PubMed] [Google Scholar]
- 27.Kita, K., Sugaya, S., Zhai, L., Wu, Y. P., Wano, C., Chigira, S., Nomura, J., Takahashi, S., Ichinose, M. & Suzuki, N. (2003) Radiat Res. 160, 302-308. [DOI] [PubMed] [Google Scholar]
- 28.Savoldi-Barbosa, M. & Sakamoto-Hojo, E. T. (2001) Teratog. Carcinog. Mutagen. 21, 417-429. [DOI] [PubMed] [Google Scholar]
- 29.Sirota, N. P., Bezlepkin, V. G., Kuznetsova, E. A., Lomayeva, M. G., Milonova, I. N., Ravin, V. K., Gaziev, A. I. & Bradbury, R. J. (1996) Radiat. Res. 146, 100-105. [PubMed] [Google Scholar]
- 30.Chou, J., Kern, E. R., Whitley, R. J. & Roizman, B. (1990) Science 250, 1262-1266. [DOI] [PubMed] [Google Scholar]
- 31.Chou, J., Chen, J.-J., Gross, M. & Roizman, B. (1995) Proc. Natl. Acad. Sci. USA 92, 10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He, B., Gross, M. & Roizman, B. (1997) Proc. Natl. Acad. Sci. USA 94, 843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leib, D. A., Harrison, T. E., Kathleen, M. L., Machalek, M. A., Moorman, N. J. & Virgin, H. W. (1999) J. Exp. Med. 189, 663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy, D. E. & Darnell, J. E. (2002) Nat. Rev. Mol. Cell Biol. 3, 651-662. [DOI] [PubMed] [Google Scholar]
- 35.O'Shea, J. (1997) Immunity 7, 1-11. [DOI] [PubMed] [Google Scholar]
- 36.Buettner, R., Mora, L. B. & Jove, R. (2002) Clin. Cancer Res. 8, 945-954. [PubMed] [Google Scholar]
- 37.Yao, R., Wang, Y., Lubet, R. A. & You, M. (2002) Oncogene 21, 5814-5821. [DOI] [PubMed] [Google Scholar]
- 38.Amorino, G. P., Hamilton, V. M., Valerie, K., Dent, P., Lammering, G. & Schmidt-Ullrich, R. K. (2002) Mol. Biol. Cell. 13, 2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]