OCA2 is used as a model melanosome cargo protein to define primary sequence elements required for acidic dileucine–motif binding to adaptors AP-1 and AP-3. OCA2 must bind to AP-3 for melanosome localization. BLOC-1 is also required and thus can cooperate with either adaptor for cargo delivery to lysosome-related organelles.
Abstract
Cell types that generate unique lysosome-related organelles (LROs), such as melanosomes in melanocytes, populate nascent LROs with cargoes that are diverted from endosomes. Cargo sorting toward melanosomes correlates with binding via cytoplasmically exposed sorting signals to either heterotetrameric adaptor AP-1 or AP-3. Some cargoes bind both adaptors, but the relative contribution of each adaptor to cargo recognition and their functional interactions with other effectors during transport to melanosomes are not clear. Here we exploit targeted mutagenesis of the acidic dileucine–based sorting signal in the pigment cell–specific protein OCA2 to dissect the relative roles of AP-1 and AP-3 in transport to melanosomes. We show that binding to AP-1 or AP-3 depends on the primary sequence of the signal and not its position within the cytoplasmic domain. Mutants that preferentially bound either AP-1 or AP-3 each trafficked toward melanosomes and functionally complemented OCA2 deficiency, but AP-3 binding was necessary for steady-state melanosome localization. Unlike tyrosinase, which also engages AP-3 for optimal melanosomal delivery, both AP-1– and AP-3–favoring OCA2 variants required BLOC-1 for melanosomal transport. These data provide evidence for distinct roles of AP-1 and AP-3 in OCA2 transport to melanosomes and indicate that BLOC-1 can cooperate with either adaptor during cargo sorting to LROs.
INTRODUCTION
Lysosome-related organelles (LROs) are cell type–specific organelles that share some characteristics with lysosomes but are distinguishable by their content of unique cargo proteins that confer distinct morphological and functional characteristics (Dell'Angelica et al., 2000; Raposo et al., 2007). Some cell types simultaneously harbor LROs and lysosomes and consequently must use unique sorting and trafficking pathways to divert specific cargo proteins from the general endocytic pathway and deliver them to maturing LROs (Dell'Angelica et al., 2000; Raposo and Marks, 2007; Raposo et al., 2007). For example, cargoes of melanosomes— LROs of melanocytes and ocular pigment cells in which melanin pigments are synthesized and stored (Marks and Seabra, 2001; Hearing, 2005)—are biosynthetically delivered from distinct domains of early endosomes (Raposo and Marks, 2007; Sitaram and Marks, 2012) from which other cargoes are destined for lysosomes or for endosomal recycling (Raposo et al., 2001; Delevoye et al., 2009). The mechanisms that regulate the differential delivery of cargoes toward melanosomes and away from classical endocytic organelles are only beginning to be understood.
Several studies have implicated two members of the heterotetrameric clathrin adaptor (AP) family, AP-1 and AP-3, in cargo sorting from endosomes toward melanosomes (Höning et al., 1998; Huizing et al., 2001; Theos et al., 2005; Chapuy et al., 2008; Delevoye et al., 2009). AP complexes facilitate sorting by coupling coat formation to clustering of target integral membrane cargoes that bear short cytoplasmically exposed sorting signals, to which APs directly bind (Boehm and Bonifacino, 2001; Bonifacino and Traub, 2003; Robinson, 2004). One class of AP-binding signals is the acidic dileucine (aLL) motif, with the consensus [D/E]xxxL[L/I] (Bonifacino and Traub, 2003). The melanosomal enzymes tyrosinase (TYR) and tyrosinase-related protein 1 (TYRP1) bear aLL-based sorting signals that are required for their steady-state localization to melanosomes in melanocytes or to late endosomes/lysosomes in other cell types (Vijayasaradhi et al., 1995; Calvo et al., 1999; Simmen et al., 1999; Huizing et al., 2001; Theos et al., 2005; Setty et al., 2007; Delevoye et al., 2009) and are capable of binding in vitro to AP-1 and AP-3 (TYR) or only AP-1 (TYRP1; Höning et al., 1998; Theos et al., 2005; although note that the acidic residue, glutamic acid, in the TYRP1 signal is at position −5 relative to the dileucine rather than −4 in almost all other signals). Consistently, both TYR and TYRP1 localize to AP-1–coated endosomal buds and tubules that accumulate near melanosomes in melanocytic cells, whereas only TYR is found in morphologically similar AP-3–coated structures (Raposo et al., 2001; Theos et al., 2005; Delevoye et al., 2009). This segregation of tyrosinase with AP-3 and TYRP1 with AP-1 has been replicated in a reconstituted budding assay (Chapuy et al., 2008). In AP-3–deficient melanocytes, TYR is largely mislocalized to early endosomes and multivesicular late endosomes, suggesting that AP-3 is required to efficiently divert TYR from the classical endosomal degradation pathway and toward melanosomes (Huizing et al., 2001; Richmond et al., 2005; Theos et al., 2005). However, a small cohort of TYR in AP-3–deficient cells is delivered to melanosomes, correlating with an increased cohort of TYR present in AP-1–coated buds; this suggested that AP-1 can partially compensate for AP-3 in melanosomal delivery (Theos et al., 2005). By contrast to TYR, TYRP1 largely localizes correctly to melanosomes in AP-3–deficient cells (Huizing et al., 2001; Setty et al., 2007) but requires AP-1 for efficient egress from vacuolar endosomal domains via recycling endosome-derived tubules (Delevoye et al., 2009). Thus AP-1 and AP-3 each function in the delivery of distinct cargoes from endosomes toward melanosomes, likely via distinct pathways (Raposo and Marks, 2007; Sitaram and Marks, 2012). However, it is not understood how AP-1 and AP-3 discriminate among cargo proteins, cooperate in the delivery of cargoes that are recognized by both complexes, or interface with other components of the trafficking machinery. Of importance, while the molecular details of the interaction between the α and σ chains of the AP-2 core complex and an aLL-like signal from CD4 are known (Kelly et al., 2008) and some of the molecular details are shared during aLL signal binding by AP-1 and AP-3 (Mattera et al., 2011), the features of aLL signals that distinguish between AP-1 and AP-3 binding have not been clearly identified.
The genes encoding two AP-3 subunits, β3A and δ, are among the 15 genes targeted by mutation in different Hermansky–Pudlak syndrome (HPS) subtypes and mouse models (Kantheti et al., 1998; Dell'Angelica et al., 1999; Feng et al., 1999). HPS comprises a group of genetic diseases that result from impaired LRO biogenesis in multiple cell types, including pigment cells and platelets (Di Pietro and Dell'Angelica, 2005; Huizing et al., 2008). In several HPS models, including those with mutations in AP-3 subunits, impaired LRO function has been linked to defective membrane dynamics required for cargo delivery to nascent LROs (Di Pietro and Dell'Angelica, 2005; Wei, 2006). HPS models with the most severe defects in melanosome biogenesis are those with mutations in subunits of the Biogenesis of Lysosome-Related Organelles Complex 1 (BLOC-1). The rod-like, eight-subunit BLOC-1 (Lee et al., 2012) has no known molecular function but is absolutely required for the melanosomal delivery of TYRP1 and the copper transporter, ATP7A (Di Pietro et al., 2006; Setty et al., 2007, 2008). By contrast, tyrosinase localization to melanosomes is only modestly impaired in BLOC-1–deficient cells (Di Pietro et al., 2006; Setty et al., 2007, 2008). On the basis of this differential sensitivity to perturbations in AP-3 and BLOC-1 expression and on the differential enrichment of AP-3 and BLOC-1 on buds and tubular domains of early endosomes, respectively (Di Pietro et al., 2006; Setty et al., 2007), it has been proposed that melanosomal cargoes follow one of two pathways from early endosomes to melanosomes—one controlled by AP-3 and another by BLOC-1 (Raposo and Marks, 2007; Setty et al., 2007). Moreover, the similarity of the cellular defects caused by AP-1 knockdown and BLOC-1 deficiency has led to the additional proposal that AP-1 and BLOC-1 function together to promote TYRP1 trafficking to melanosomes via recycling endosomal tubules (Delevoye et al., 2009).
Although a model in which AP-1 and BLOC-1 cooperate in one dedicated cargo delivery pathway to melanosomes and AP-3 functions in a separate pathway is consistent with genetic data in melanocytes, it fails to account for several published observations. First, a robust physical interaction has been detected between AP-3 and BLOC-1 in detergent extracts of membranes from several cell types (Di Pietro et al., 2006; Salazar et al., 2006; Gokhale et al., 2012), including melanocytes (S. Gangi Setty, unpublished observation), and each complex regulates the association of the other with membranes (Di Pietro et al., 2006). By contrast, despite several efforts, no physical interaction has been observed between AP-1 and BLOC-1 (Salazar et al., 2009; and unpublished observations). Second, AP-3 and BLOC-1 cooperate in regulating the dynamics of a set of cargoes in neurons (Newell-Litwa et al., 2009; Salazar et al., 2009; Larimore et al., 2011) and in the stable incorporation of a putative transporter, SLC35D3, in platelets (Meng et al., 2012), and BLOC-1 and AP-3 are coisolated upon immunoaffinity purification from neurons (Newell-Litwa et al., 2009; Salazar et al., 2009; Gokhale et al., 2012) and from clathrin-coated vesicles from HeLa cells (Borner et al., 2006). These data suggest that BLOC-1 and AP-3 might function together under certain circumstances. Moreover, it is unclear how signals such as those in tyrosinase that are capable of binding to both AP-3 and AP-1 “choose” between the two distinct pathways.
To further clarify the relationships between AP-1, AP-3, and BLOC-1 in melanosomal protein sorting, we exploited an additional melanosomal cargo protein, OCA2 (also known as pink-eyed dilution or P protein; Rinchik et al., 1993). OCA2 is a 12–transmembrane domain, pigment cell–restricted protein that bears homology to a superfamily of transporters (Rinchik et al., 1993; Lee et al., 1995). Mutations in the OCA2 gene underlie oculocutaneous albinism (OCA) type 2, the most common form of OCA worldwide (King, 1998), as well as phenotypic changes in skin hue and eye color (Lao et al., 2007; Norton et al., 2007; Sulem et al., 2007; Eiberg et al., 2008; Mengel-From et al., 2010). We previously showed that OCA2 localizes to melanosomes by virtue of an aLL sorting signal in the cytoplasmic N-terminal domain (Sitaram et al., 2009). Like the TYR sorting signal, the OCA2 sorting signal binds in vitro to both AP-1 and AP-3. Here we sought to define those features of the OCA2 aLL sorting signal that confer the ability to bind AP-1 or AP-3 and then to exploit mutants that only bind to one of these complexes to dissect the respective role of each adaptor in OCA2 melanosome transport. Our results define sequence features of aLL sorting signals that distinguish between AP-1 and AP-3 binding, show that OCA2 absolutely requires AP-3 interaction for stable localization to melanosomes, and provide evidence for two distinct AP-3–dependent melanosomal transport pathways in melanocytes—one that is independent of BLOC-1 and one that requires BLOC-1. Together our data suggest that AP-1 and AP-3 play different roles in melanosome transport and that BLOC-1 can cooperate with multiple adaptors to effect cargo delivery to cell type-specific LROs.
RESULTS
The OCA2 acidic dileucine–based melanosome-targeting signal binds to AP-1 and AP-3 hemicomplexes in a yeast three-hybrid assay
The N-terminal cytoplasmic domain of human OCA2 contains three consensus aLL motifs termed LL1, LL2, and LL3 (Figure 1a). We previously showed that substitution of the two critical leucines of LL1 by alanine (AA1N) abolished steady-state localization to melanosomes, implicating LL1 as the critical sorting signal for melanosome delivery. Moreover, a fusion protein consisting of the OCA2 N-terminal cytoplasmic domain fused to glutathione S-transferase (GST) was capable of binding to both AP-1 and AP-3 from human melanoma or HeLa cell cytosol, and this interaction required intact LL1 (Sitaram et al., 2009). We first sought to determine those features of LL1 that were required for its interaction with AP-1 and/or AP-3 and thus distinguished it from the weak LL2 and completely inactive LL3. To more rapidly assay in parallel the interaction between OCA2 mutants and AP complexes, we adapted a yeast three-hybrid (Y3H) assay that was previously used to study acidic dileucine motifs in the melanogenic enzyme tyrosinase and the HIV protein Nef (Janvier et al., 2003; Theos et al., 2005; Chaudhuri et al., 2007, 2009). In this assay, Gal4 fusion proteins of the test cytoplasmic domain are coexpressed with hemicomplexes consisting of the small subunit (σ1, σ2, or σ3A) and corresponding large subunit (γ, α, or δ) of AP-1, -2, and -3, respectively, which comprise the AP surface that engages aLL-sorting motifs (Janvier et al., 2003; Doray et al., 2007; Kelly et al., 2008; note that the σ4/ε hemicomplex of AP-4 has not been demonstrated to bind an aLL signal). The complete cytoplasmic N-terminal domain of human OCA2 (amino acids 1–173) was fused to the C-terminus of the yeast Gal4 DNA-binding domain. One of the three σ subunits was expressed from the MET5 promoter within a second transcription unit in the same plasmid, and the γ, α, or δ subunit was fused to the C-terminus of the Gal4 activation domain in a separate plasmid. The Saccharomyces cerevisiae strain HF7c was cotransformed with both plasmids and grown on methionine-deficient selective medium to express all three proteins. An interaction between the OCA2 cytoplasmic domain and an AP hemicomplex activates Gal4-dependent expression of HIS3 and allows for growth on histidine (His)-deficient medium. The cytoplasmic domain of human tyrosinase (from amino acid 499 through the C-terminus), previously shown to interact with all three AP hemicomplexes in this assay (Janvier et al., 2003; Chaudhuri et al., 2009), served as a positive control.
FIGURE 1:
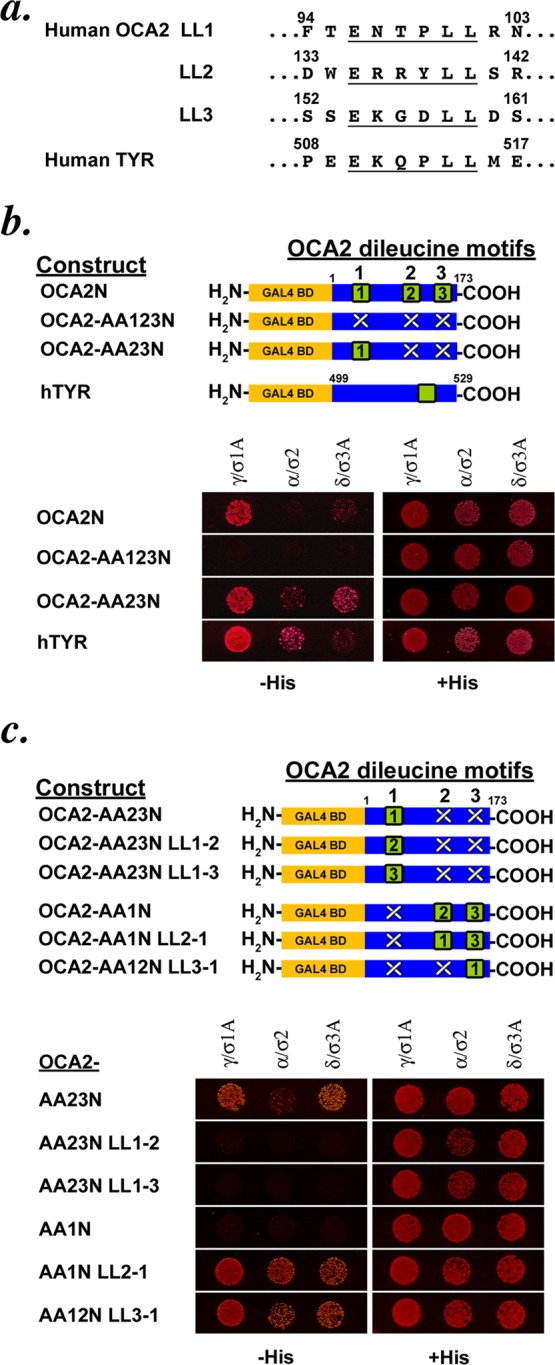
The cytoplasmic domain of OCA2 shows dileucine-dependent interaction with adaptor proteins in a yeast three-hybrid system. (a) Sequences of the three acidic dileucine sorting motifs in the cytoplasmic N-terminus of human OCA2 and the single acidic dileucine motif in the cytoplasmic C-terminus of human tyrosinase. (b) Schematic of OCA2 N-terminal-domain Gal4 fusion constructs used in the assay. GAL4 BD, Gal4-binding domain; green square, intact dileucine motif; white X, disrupted dileucine motif. Black numbers within the green square indicate whether the primary sequence of the LL1, LL2, or LL3 motif is inserted at the indicated position within the domain. Below, a yeast three-hybrid assay of various OCA2 constructs coexpressed with hemicomplexes of the AP-1 (γ/σ1A), AP-2 (α/σ2), and AP-3 (δ/σ3A) complexes. A protein–protein interaction leads to expression of HIS3, allowing growth on His-deficient medium. All transformed yeast grow on His-containing control medium. (c) Schematic of OCA2 constructs used in the assay. Below, yeast three-hybrid assay of the OCA2 constructs in which dileucine motif sequences are repositioned within the OCA2 cytoplasmic domain.
As expected, the tyrosinase cytoplasmic domain interacted with hemicomplexes from AP-1, AP-2, and AP-3, as indicated by growth of all three sets of transformants on His-deficient medium (Figure 1b). The full-length human OCA2 cytoplasmic N-terminal domain also interacted with all three AP hemicomplexes, confirming previous results obtained using a GST-cytoplasmic domain fusion pull-down assay (Sitaram et al., 2009); consistent with previous results, the strength of interaction with the AP-2 α/σ2 hemicomplex varied among experiments (data not shown, but see later figures). Also consistently, the interaction was ablated when all three acidic dileucine motifs were mutagenized to substitute the three dileucine pairs for dialanines (OCA2-AA123N). An intact critical LL1 motif alone (OCA2-AA23N) was sufficient to restore the interaction with the AP hemicomplexes. These data confirm that the Y3H assay robustly recapitulates the ability of LL1 to interact with AP complexes.
Acidic dileucine motif position does not affect AP binding or sorting activity
Our previous analyses of transiently transfected immortalized melanocytes that express full-length OCA2 variants revealed that neither the second nor third acidic dileucine motif (LL2 or LL3) alone could support steady-state localization of OCA2 to melanosomes in wild-type melanocytes or fully restore melanin synthesis when expressed in OCA2-deficient melanocytes (Sitaram et al., 2009). Moreover, cytoplasmic domain fusion proteins in which LL1 was ablated by mutagenesis showed impaired binding to AP complexes using a GST pull-down assay. To determine why these motifs were inactive in melanosome sorting and AP binding, we considered two parameters: position within the cytoplasmic domain and sequence of the intervening “x” residues within the [D/E]xxxL[L/I] consensus.
Because proximity to the membrane has previously been reported to affect the ability of a consensus sorting motif to function and to bind adaptors (Rohrer et al., 1996; Geisler et al., 1998; White et al., 1998), we used the Y3H assay to determine whether the inability of the LL2 or LL3 motif to bind APs was due to its placement within the cytoplasmic domain. We made variants of the OCA2-AA23N construct in which the sequence of the active LL1 signal was replaced in situ by that of LL2 (OCA2-AA23N LL1-2) or LL3 (OCA2-AA23N LL1-3; Figure 1c). Whereas all three AP hemicomplexes interact with OCA2-AA23N bearing intact LL1, no AP interacted with either the LL1-2 or LL1-3 mutant, indicating that the sequences of the LL2 and LL3 motifs were inherently inactive even when placed in the normal position for LL1. We then performed the complementary experiment in which the LL1 sequence was placed in the position of the second or third motif (in the context of an inactivated LL1 motif at position 1 in construct OCA2-AA1N). Whereas the disruption of the LL1 motif in OCA2-AA1N ablates the interaction with all three AP hemicomplexes, reintroduction of the LL1 sequence into the second position (OCA2-AA1N LL2-1) or the third position (OCA2-AA12N LL3-1) within the cytoplasmic domain restores this interaction. These data indicate that the LL1 motif is active in AP binding regardless of its position within the cytoplasmic domain, implicating specific sequence elements as critical to its AP-binding activity.
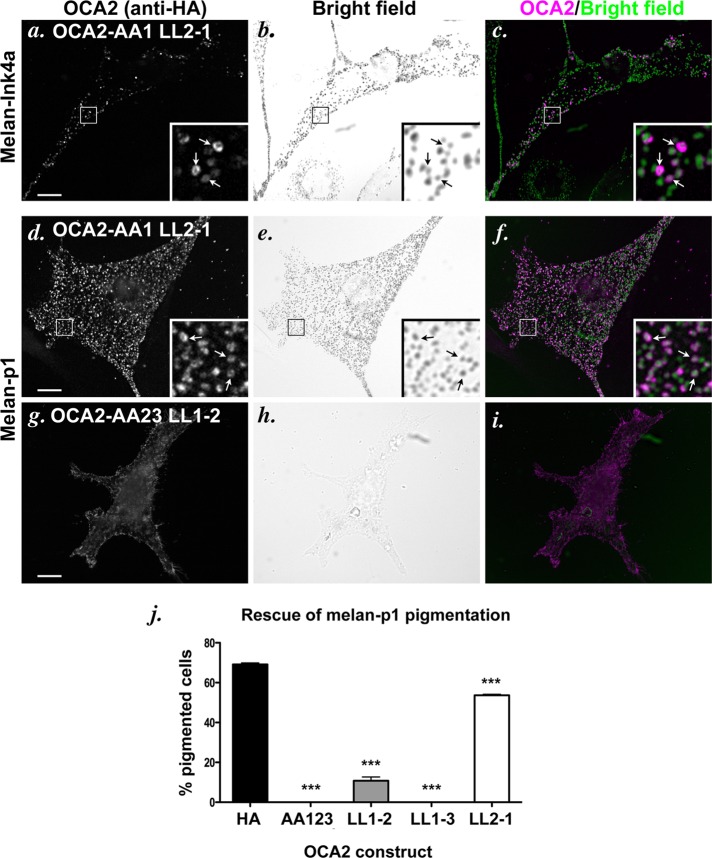
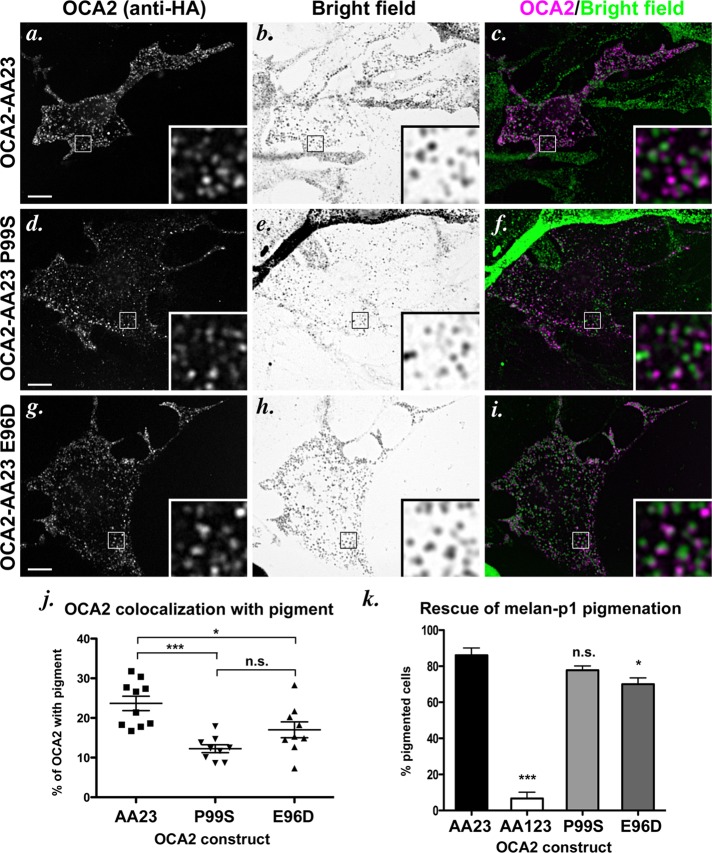
To determine whether motif position affected the activity of the sorting signal, we engineered the aforementioned mutations into the full-length, hemagglutinin (HA)-tagged human OCA2 and transiently expressed the mutants in wild-type pigmented melan-Ink4a mouse melanocytes. Indirect immunofluorescence microscopy (IFM) analyses in these cells showed that OCA2-AA1 LL2-1—in which LL1 is placed in position 2—distributes largely in HA-positive rings that surround pigmented structures (Figure 2, a–c), similar to the staining pattern seen with expression of wild-type HA-tagged or native OCA2 (Sitaram et al., 2009). To determine whether this construct retained function, we expressed it in nonpigmented, OCA2-deficient melan-p1 mouse melanocytes; we previously showed that OCA2 function correlates with melanosomal localization, since expression of human OCA2 constructs that localize to melanosomes but not ones that are restricted to the cell surface restores pigment synthesis in melan-p1 melanocytes (Sviderskaya et al., 1997; Sitaram et al., 2009). Consistently, transiently expressed OCA2-AA1N LL2-1 localized to vesicular structures in melan-p1 cells as in wild-type cells (Figure 2, d–f) and rescued pigmentation in 54% of transfected cells, compared with 69% of cells expressing wild-type OCA2 (Figure 2j). By contrast, full-length OCA2 constructs bearing the LL1-2 and LL1-3 mutations were mislocalized to the cell surface when expressed in wild-type melanocytes (Figure 3j and unpublished data) or melan-p1 cells (Figure 2, g–i) and were incompetent to rescue melanin synthesis in melan-p1 cells (Figure 2, g–j, and unpublished data). Together these data confirm that the LL2 and LL3 sequences are intrinsically poor at interacting with AP complexes and at effecting melanosome localization, even when placed in a “favorable” position within the protein. Conversely, the LL1 sequence is intrinsically competent to bind AP complexes in a relatively position-independent manner.
FIGURE 2:
Localization and function of OCA2 dileucine-motif position mutants. (a–i) IFM analysis of melan-Ink4a cells (a–c) or melan-p1 cells (d–i) expressing selected mutant OCA2 variants. The melan-Ink4a melanocytes were transfected with OCA2-AA1 LL2-1 (a–c), and the melan-p1 cells were transfected with OCA2-AA1 LL2-1 (d–f) or OCA2-AA23 LL1-2 (g–i). Transgenes were visualized with anti-HA antibodies (a, d, g), and melanosomes were visualized by bright-field microscopy (b, e, h). c, f, and i are merged OCA2-HA (magenta) and inverted bright-field (green) images. All insets show 5× magnified images of the boxed region. Arrows point to regions of overlap of OCA2 constructs with melanosomes. Bar, 10 μm. (j) Dileucine mutants were expressed in OCA2-deficient melan-p1 cells, and transfected cells were visually inspected for the presence of pigmented melanosomes. Shown is the percentage of transfected cells expressing each indicated construct that contained pigmented melanosomes (percentage pigmented cells). All columns were significantly different from rescue by OCA2-HA. ***, p < 0.001.
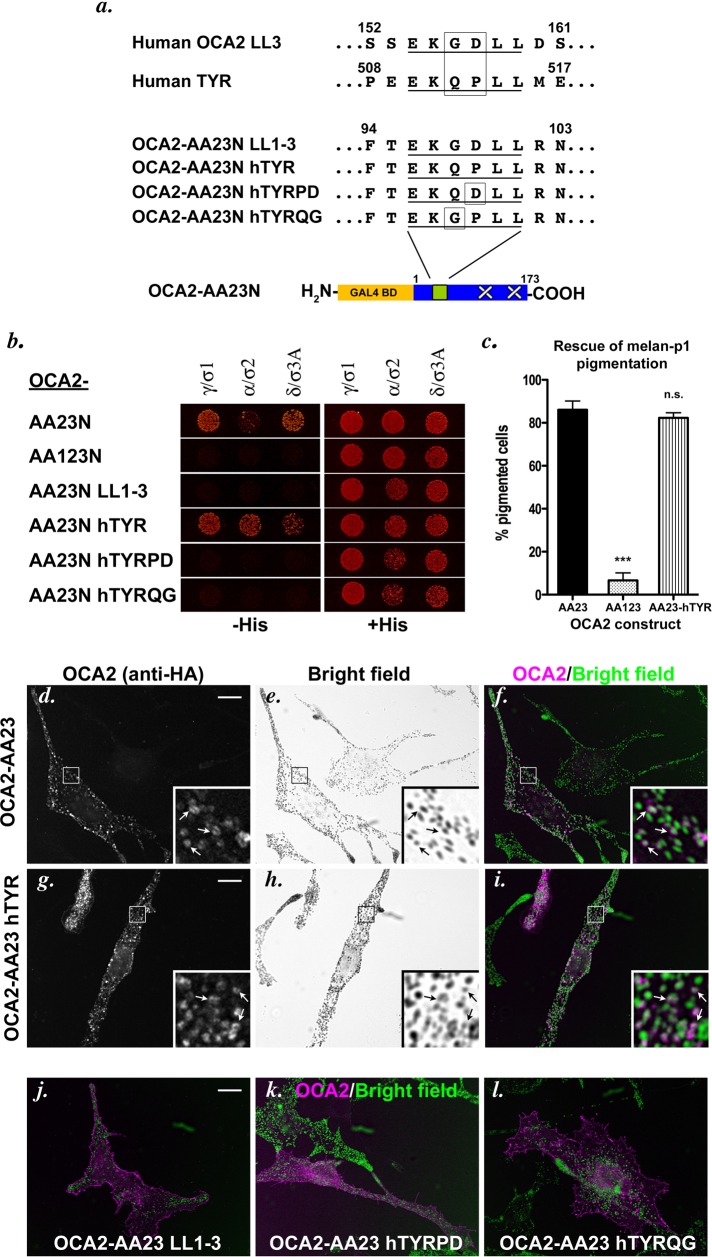
FIGURE 3:
Internal sorting signal residues affect AP complex interaction. (a) Comparison of the LL3 acidic dileucine motif from human OCA2 and the sole acidic dileucine motif in human tyrosinase. OCA2-AA23N hTYR constructs were made in which the first acidic dileucine motif of the OCA2 cytoplasmic domain was replaced with the sequence from tyrosinase, with or without the indicated amino acid substitutions. (b) Yeast three-hybrid analyses of AP hemicomplex interaction with the OCA2-AA23N hTYR constructs. (c) Melan-p1 rescue assay performed as in Figure 2j. n.s., not significant. ***, p < 0.001. (d–l) Melan-Ink4a mouse melanocytes were transiently transfected with full-length, HA-tagged OCA2-AA23 (d–f), OCA2-AA23 hTYR (g–i), OCA2-AA23 LL1-3 (j), OCA2-AA23 hTYRPD (k), or OCA2-AA23 hTYRQG (l) constructs bearing the identical mutations used in the yeast three-hybrid assay. Cells were stained with anti-HA antibodies (d, g; colored magenta in merged pictures, f, i–l) and subjected to indirect immunofluorescence microscopy. Bright-field images (e, h) were inverted and colored green in merged pictures. Arrows point to regions of overlap between OCA2 constructs and melanosomes. Bar, 10 μm.
Acidic dileucine motif sequence requirements for AP-binding and melanosome-targeting activity
To begin to dissect those primary sequence features of the OCA2 dileucine motifs that conferred AP binding and sorting activity, we compared the sequence of the inactive LL3 motif in OCA2 to that of active acidic dileucine sorting signals. In particular, the sole acidic dileucine sorting motif in human TYR, EKQPLL, differs at only two positions from the LL3 motif in human OCA2, EKGDLL (Figure 3a). Whereas AP-1, -2, and -3 hemicomplexes did not interact with the OCA2-AA23N LL1-3 mutant, in which the LL3 sequence in position 1 is the only intact motif (Figures 1c and 3b), all three AP hemicomplexes interacted with an OCA2-AA23N construct bearing the EKQPLL motif of human TYR at position 1 (OCA2-AA23N hTYR; Figure 3b). However, constructs bearing either of the two single amino acid substitutions that change the middle residues of the TYR signal to those in the inactive LL3 motif (OCA2-AA23N TYRPD or OCA2-AA23N TYRQG) did not interact with any AP hemicomplexes (Figure 3b). The AP-binding ability of the motifs correlated completely with sorting activity, since full-length OCA2-AA23 bearing endogenous LL1 (Figure 3, d–f) and OCA2-AA23N hTYR bearing the intact tyrosinase signal localized to melanosomes at steady state when expressed in melan-Ink4a melanocytes (Figure 3, d–i) and stimulated melanin synthesis in a high proportion of transfected melan-p1 cells (Figure 3c), but neither of the two “LL3-like” single-point-mutant chimeric constructs did. Instead, these mutants localized to the plasma membrane (Figure 3, k and l). Taken together, our results indicate that the “x” amino acids of the [D/E]xxxL[L/I] consensus greatly influence AP binding and sorting activity in the context of OCA2, and that neither glycine at position −2 nor aspartate at position −1 (relative to the first leucine) can support either activity.
OCA2 interactions with AP-1 and AP-3 are separable
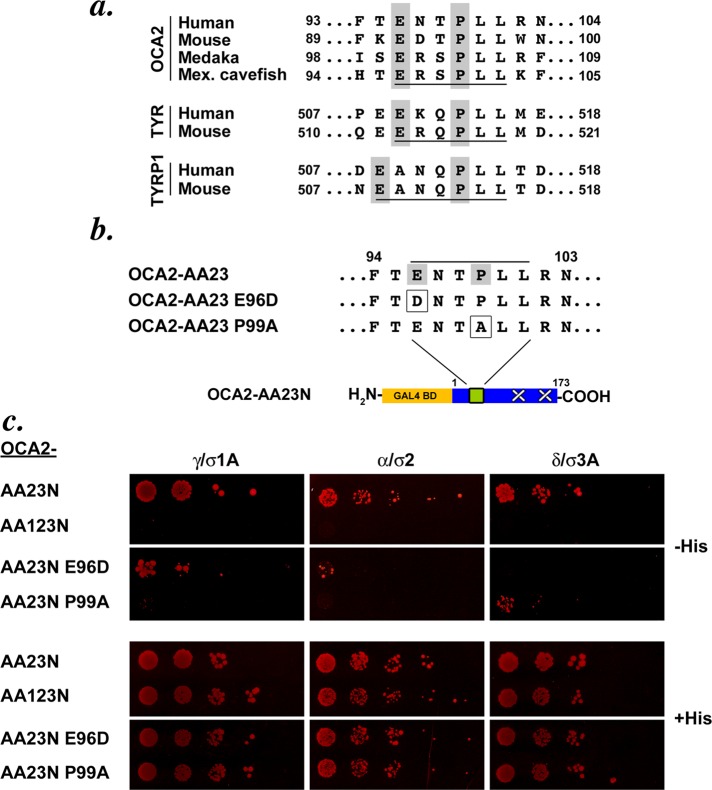
To further define the sequence requirements for AP binding and for melanosome targeting, we compared LL1 to acidic dileucine–based sorting signals from other melanocytic proteins. The LL1 motif in human OCA2 is the only one of the three human motifs that has a counterpart in OCA2 homologues in the mouse, Japanese medaka fish, or Mexican cavefish. Although the general consensus for acidic dileucine motifs is [D/E]xxxL[L/I], the three nonhuman OCA2 homologues (and indeed nearly all vertebrate homologues; Supplemental Table S1) and the LL1 (but not LL2 or LL3) motif of human OCA2 share a more restricted consensus of ExxPLL (Figure 4a). Moreover, the sequences of the cytoplasmic sorting motifs found in TYR and TYRP1 also correspond to the more restricted motif, suggesting the specific importance of a −4 glutamate (or −5 for the unusually extended TYRP1 motif) and −1 proline for sorting to melanosomes. We tested the effect of a conservative Glu-to-Asp mutation of the OCA2 LL1 motif (OCA2-AA23N E96D) and of a nonconservative Pro-to-Ala mutation (OCA2-AA23N P99A; Figure 4b). The E96D mutant ablated the interaction of OCA2-AA23N with AP-3 hemicomplexes by Y3H assay (Figure 4c). Its interactions with AP-1 and AP-2 hemicomplexes were reduced but not eliminated; this manifested as variability in yeast growth over several trials (e.g., compare Figure 4c to Supplemental Figure S1b). Conversely, the P99A mutation abolished interaction of OCA2-AA23N with AP-1 hemicomplexes but consistently retained an interaction with AP-3 hemicomplexes (albeit reduced). The P99A mutation also consistently impaired the interaction with AP-2 hemicomplexes (data not shown). These data suggest that AP-1 but not AP-3 is tolerant of an aspartate residue at position −4, whereas AP-3 but not AP-1 is tolerant of an alanine at position −1.
FIGURE 4:
AP-1 and AP-3 interaction with the OCA2 LL1 motif are separable. (a) Comparison of sequences of acidic dileucine motifs in OCA2 isoforms from human, mouse, Japanese medaka fish, and Mexican cavefish. All motifs are underlined, and gray boxes highlight conserved elements in the OCA2 motifs, as well as in motifs from human and mouse tyrosinase and TYRP1. (b) Single amino acid substitutions were made in the OCA2-AA23N construct to test for changes in interaction with AP complexes. (c) Yeast three-hybrid analyses of AP interaction with OCA2 sorting motif mutants. Transformants were spotted in fivefold serial dilutions. E96D and P99A show reciprocal interaction with AP-1 and AP-3.
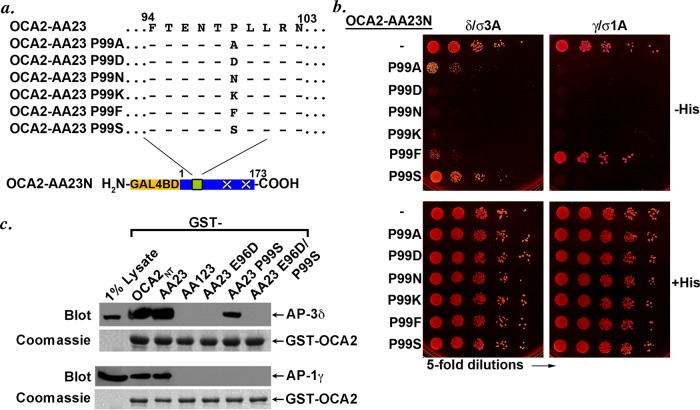
To determine what features at position −1 were necessary for binding to AP-1 and AP-3 hemicomplexes, we further mutagenized the −1 Pro residue to several different amino acids to test the suitability of a range of side groups for the residue in that position (Figure 5a). Of Ala, Asp, Asn, Lys, Phe, and Ser, only the serine substitution interacted with AP-3 hemicomplexes at a level that was comparable to that of the wild-type OCA2 LL1 motif; of importance, like the alanine substitution, the serine substitution did not support interaction with AP-1 hemicomplexes (Figure 5b). The phenylalanine substitution also interacted variably with both AP-1 and AP-3 hemicomplexes. As predicted by the individual mutations, incorporation of both the E96D and P99S mutations together within the LL1 motif of OCA2-AA23N ablated the interaction with either AP-1 or AP-3 hemicomplexes (Supplemental Figure S1, a and b; see also Figure 5c). Although each individual mutant retained the ability to bind to the AP-2 hemicomplex, the combined mutant also ablated this interaction (Supplemental Figure S1b). Taken together, these data suggest that interaction of an acidic dileucine motif with AP-1 requires a proline or perhaps a large hydrophobic amino acid residue at position −1, whereas AP-3 binding can tolerate small, uncharged residues such as serine or alanine at this position.
FIGURE 5:
AP-3 is more tolerant than AP-1 of substitutions at the −1 position in the OCA2 LL1 sorting motif. (a) Schematic of OCA2-AA23N constructs containing a range of amino acid substitutions at the conserved proline of the LL1 sorting motif of human OCA2. (b) Yeast three-hybrid analyses of the interaction of OCA2-AA23N proline substitution constructs with AP-3 or AP-1. (c) GST pull-down assay. Fusion proteins consisting of GST fused to the full-length OCA2 N-terminal cytoplasmic domain (GST-OCA2NT) or to the indicated mutants were generated by expression in E. coli, bound to glutathione–Sepharose beads and incubated with detergent lysates of the human melanoma, MNT-1. Bound proteins were recovered by centrifugation, fractionated by SDS–PAGE, and analyzed by immunoblotting for AP-3 (with anti–δ-adaptin antibody) or AP-1 (with anti–γ-adaptin antibody). Bottom, Coomassie stain of the same gels showing equal loading of GST-OCA2 cytoplasmic domains.
To confirm the results of the Y3H assay, we tested for the ability of the E96D, P99S, and the combined E96D/P99S variants to bind to AP-1 and AP-3 using a GST pull-down assay previously used to demonstrate the binding of the OCA2 LL1 motif to these adaptors (Sitaram et al., 2009). Detergent lysates of MNT-1 human melanoma cells were incubated with GST fused to the OCA2 N-terminal cytoplasmic domain with or without mutations in the LL1, LL2, and LL3 motifs, and bound AP-3 or AP-1 complexes were detected by immunoblotting with antibodies to δ- and γ-adaptin, respectively (Figure 5c). As shown previously, both the intact OCA2 cytoplasmic domain (OCA2NT) and a variant lacking LL2 and LL3 (AA23) bound to AP-3 and AP-1 from cell lysates, but a variant lacking all three aLL motifs (AA123) did not. Consistent with the Y3H data, an AA23 variant harboring the P99S mutation bound to AP-3 effectively (30.1 ± 14% relative to AA23) but not variants harboring the E96D mutation alone or in tandem with P99S. Consistent with the weak and variable binding of the E96D variant to AP-1 γ/σ1 hemicomplexes by Y3H, no binding above background was detected for this variant to either AP-1 or AP-3. Taken together, these data support the conclusions drawn from Y3H that the P99S variant favors binding to AP-3 and the E96D variant binds with low affinity, if at all, to AP-1.
OCA2 interaction with AP-3 is required for melanosomal localization but not function
Because we had isolated OCA2 constructs that showed contrasting preferences for AP-1 or AP-3 binding—E96D weakly for AP-1 and P99S or P99A for AP-3—we next tested the role of each complex in melanosomal sorting by determining whether the changes in interactions with AP complexes affected OCA2 localization. Each sorting motif mutation was placed into full-length, HA-tagged human OCA2-AA23 (in which the dileucine residues of LL2 and LL3 are replaced by alanine, such that the only active motif is the wild-type or modified LL1) and transiently expressed in melan-Ink4a melanocytes. In these cells, most of the visualized “wild-type” OCA2-AA23 localized to rings, and ∼49% of these structures surrounded pigmented melanosomes (Figures 3, d–f, and 6g). Consistently, by immuno–electron microscopy analyses of immunogold-labeled ultrathin cryosections, OCA2-AA23 localized largely to melanosomes with some additional labeling of the Golgi and occasional small vesicles (Supplemental Figure S2). By comparison, the AP-3–biased OCA2-AA23 P99S or P99A constructs consistently showed increased steady-state labeling at the cell surface but also localized properly to rings in most cells (Figure 6, a–c; note, however, that in some cells, only cell surface labeling was observed). About 40% of the intracellular rings enclosed pigmented structures (Figure 6g). The increased cell surface localization but near-wild-type distribution of the intracellular pool of the P99A and P99S variants to melanosomes suggests that OCA2 interaction with AP-1 and AP-2 might facilitate more efficient intracellular retention of OCA2 but might not be absolutely required for melanosomal trafficking. By contrast, the AP-1–biased OCA2-AA23 E96D variant localized to structures that were localized eccentrically to one side of pigmented melanosomes (Figure 6, d–f). This eccentric localization of the E96D variant led to a decrease in the degree of overlap with melanin (∼ 25%) relative to wild-type OCA2-AA23 (p < 0.01; although also reduced relative to the P99A or P99S variants, the reduction was not statistically significant due to cell-to-cell variability of localization measurements). This eccentric distribution would be consistent with localization to perimelanosomal recycling endosome-derived transport carriers (Delevoye et al., 2009). The data thus suggest that AP-1 binding is sufficient for initial targeting steps toward melanosomes but that AP-3 binding is necessary for a late sorting step to target OCA2 to melanosomes. As expected for a mutant that was unable to bind to any of the adaptors, full-length OCA2-AA23N bearing the double E96DP99S mutation localized to the cell surface when expressed in wild-type melan-Ink4a melanocytes (Supplemental Figure S1c).
FIGURE 6:
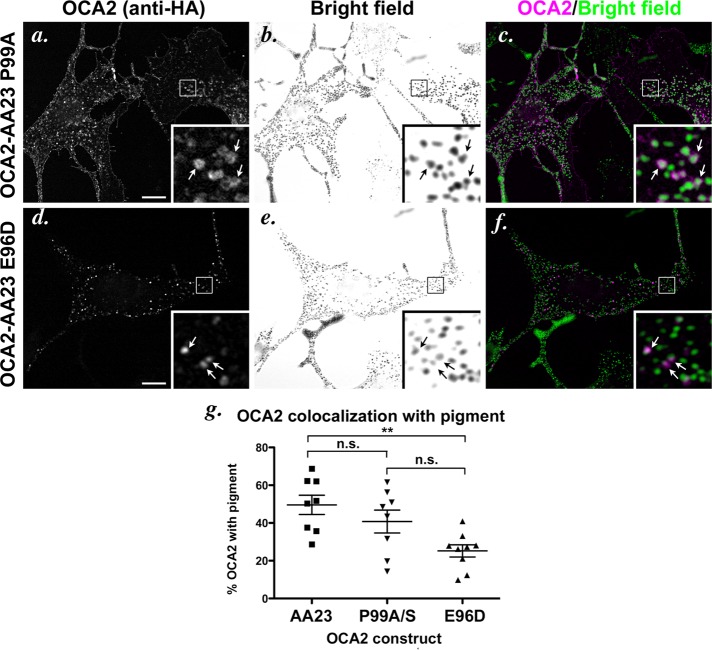
Localization of dileucine-motif point mutants in wild-type melanocytes. (a–f) IFM analysis of melan-Ink4a cells expressing selected mutant OCA2 variants. Melan-Ink4a melanocytes were transfected with OCA2-AA23 P99A (a–c) or OCA2-AA23 E96D (d–f). Transgenes were visualized with anti-HA antibodies (a, d), and melanosomes were visualized by bright-field microscopy (b, e). c and f are merged OCA2-HA (magenta) and inverted bright-field (green) images. All insets show 5× magnified images of the boxed region. Arrows point to regions of overlap of OCA2 constructs with melanosomes. Bar, 10 μm. (g) Deconvolved IFM images of each mutant were converted to binary images and analyzed for marker overlap. Cells transfected with OCA2-AA23 P99A or OCA2-AA23 P99S were combined in the analysis. Each data point represents one cell. Shown is the percentage of punctate/vesicular HA staining that overlapped with pigment in each cell (percentage of OCA2 with pigment). Each pair of mutants was compared. n.s., not significant. **, p < 0.01.
To confirm the role of AP-3 in melanosomal trafficking of OCA2, we also expressed the constructs in AP-3–deficient melan-pe melanocytes, which are derived from the HPS type 2 mouse model, pearl (Theos et al., 2005). A large cohort of tyrosinase is mislocalized in these cells, but the cells are significantly pigmented (Theos et al., 2005), and melanosomal localization of other cargoes, such as TYRP1 and ATP7A, is largely intact (Theos et al., 2005; Setty et al., 2007, 2008). We observed two different phenotypes in different cultures of melan-pe cells. One phenotype resembled that observed originally, in which tyrosinase localization and melanosome ultrastructure have been well characterized (Theos et al., 2005). In these cells, like the AP-1–preferring E96D mutant in wild-type cells, transiently expressed OCA2-AA23 localized to punctate structures that were near or adjacent to pigmented melanosomes (Figure 7, a–c), and only 25% of these structures overlapped with pigment (Figure 7j). This result is consistent with a model in which AP-3 is necessary for melanosomal delivery of OCA2 from adjacent endosomal structures. Both the E96D and P99S constructs were also detected on some structures that abutted pigmented melanosomes in melan-pe cells (Figure 7, d–i) but to a lesser degree than OCA2-AA23 (Figure 7j). Thus in all cases, the lack of AP-3 appeared to prevent efficient melanosomal delivery of OCA2. Moreover, partially or severely reduced affinity for AP-1, as in E96D and P99S, respectively, correlated with a decrease in localization to perimelanosomal structures, suggesting that AP-1 might be required for effective accumulation in these structures.
FIGURE 7:
Localization of dileucine-motif point mutants in AP-3–deficient melanocytes. (a–i) IFM analysis of AP-3–deficient cells expressing selected mutant OCA2 variants. Melan-pe melanocytes were transfected with OCA2-AA23 (a–c), OCA2-AA23 P99S (d–f), or OCA2-AA23 E96D (g–i). Transgenes were visualized with anti-HA antibodies (a, d, g), and melanosomes were visualized by bright-field microscopy (b, e, h). c, f, and i are merged OCA2-HA (magenta) and inverted bright-field (green) images. All insets show 5× magnified images of the boxed region. Bar, 10 μm. (j) Deconvolved IFM images of each mutant were converted to binary images and analyzed for marker overlap. Each data point represents one cell. Shown is the percentage of punctate/vesicular HA staining that overlapped with pigment in each cell (percentage of OCA2 with pigment). The two point mutants were significantly different from OCA2-AA23 but not from each other. *, p < 0.05; ***, p < 0.001. (k) OCA2-deficient melan-p1 melanocytes were rescued as in Figure 2j. All columns were compared with OCA2-AA23. n.s., not significant. *, p < 0.05; ***, p < 0.001.
In independent cultures of melan-pe cells, we observed a distinct phenotype. In these cells, melanosomes were dramatically enlarged, highly pigmented, and abnormally distributed near the nucleus. In these cultures, all OCA2 mutants, including the AP-3–favoring P99A, were localized to the enlarged melanosomes (Supplemental Figure S3, a–c and g–l). However, a number of additional proteins were also unexpectedly localized to these aberrant melanosomes, including the early endosomal soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) syntaxin 13 (Setty et al., 2007), the late endosome/lysosome SNARE syntaxin 7 (data not shown), and OCA2-AA1, which was not detected on melanosomes in wild-type cells (Sitaram et al., 2009; Supplemental Figure S3, d–f); none of these cargoes was detected on pigment granules in the more modestly pigmented cultures shown in Figure 7. We therefore interpret these observations as the result of indirect, adverse effects of long-term AP-3 depletion on endosomal trafficking and the maintenance of the distinct identities of endosomes and melanosomes. By contrast, the results with the less pigmented cultures are consistent with observations of aLL signal variants in wild-type melanocytes and thus more likely reflect direct consequences of AP-3 loss on cargo transport.
We next tested whether changes in steady-state localization of the mutants affected function by using the melan-p1 pigmentation rescue assay. In this set of experiments, expression of OCA2-AA23 rescued pigmentation in 86% of transfected melan-p1 cells, whereas the expression of the surface-localized OCA2-AA123 and OCA2-AA23 E96DP99S mutants rescued pigmentation in very few cells (Figure 7k and Supplemental Figure S1d). Consistent with the similar degree of localization to melanosomes in wild-type melanocytes, the AP-3–biased P99S variant stimulated pigment synthesis as well as OCA2-AA23. Surprisingly, the AP-1–biased E96D variant was only slightly less efficient than OCA2-AA23 in rescuing pigmentation in melan-p1 cells, despite the reduced localization to melanosomes in wild-type cells (Figure 7k). The high degree of rescue effected by the E96D variant suggests that OCA2 might be able to fulfill its function in melanogenesis from within structures that are adjacent to melanosomes.
OCA2 trafficking to melanosomes is dependent on BLOC-1
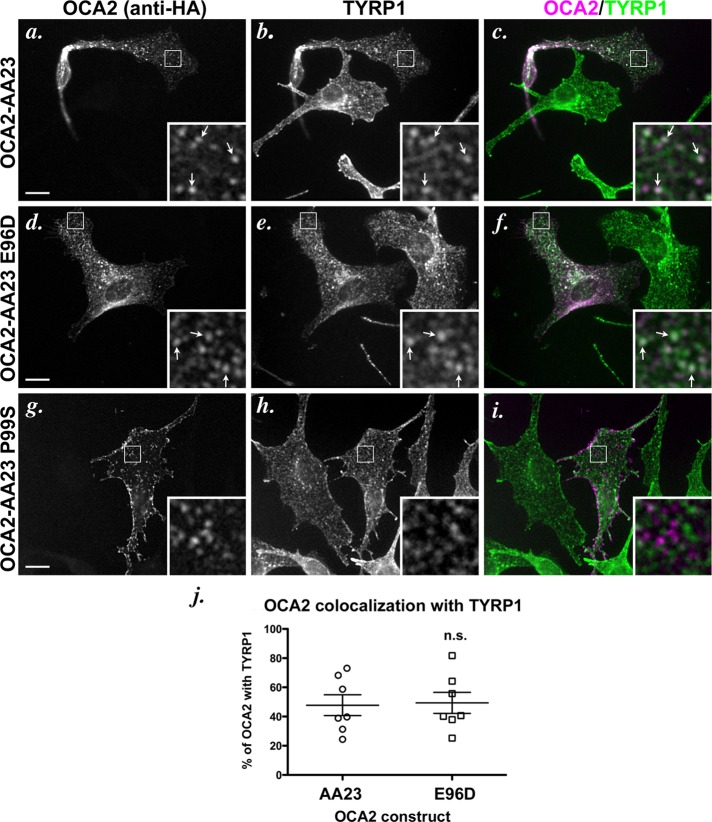
The foregoing data indicate that AP-1 might facilitate an early step in melanosomal trafficking of OCA2 but that AP-3 is required for ultimate efficient accumulation in melanosomes. This would tend to suggest that OCA2 follows a transport pathway from endosomes to melanosomes that is similar to TYR but dissimilar from TYRP1, which is sorted to melanosomes in an AP-3–independent manner. To further test this model, we probed the requirement for BLOC-1 in melanosomal transport of OCA2. Melanosomal delivery of TYR and TYRP1 shows different levels of dependence on BLOC-1, such that TYRP1 accumulation absolutely requires BLOC-1, but TYR accumulation is only modestly reduced in BLOC-1–deficient cells (Setty et al., 2007). To test whether melanosomal delivery of OCA2 variants requires BLOC-1, we examined their localization after transient expression in melan-pa1 melanocytes derived from the BLOC-1–deficient pallid mouse. We previously showed that TYRP1 in these cells is mislocalized largely to vacuolar endosomes, as a result of the failure of BLOC-1–dependent siphoning toward melanosomes, and to the cell surface, as a result of “default” increased endosomal recycling (Setty et al., 2007). By IFM, ∼50% of transiently expressed OCA2-AA23 colocalized with the internal pool of TYRP1 in melan-pa1 cells, and there was also a substantial pool of OCA2-AA23 at the plasma membrane (Figure 8, a–c and j). Moreover, OCA2-AA23 did not overlap at all with PMEL, a marker for the nonpigmented melanosomes in these cells (unpublished data), supporting the interpretation that OCA2-AA23 is excluded from melanosomes in these cells. These data indicate that melanosomal trafficking of OCA2 is similarly BLOC-1 dependent. The AP-1–biased E96D variant overlapped with TYRP1 to a similar degree (Figure 8, d–f and j), supporting the view that BLOC-1 and AP-1 cooperate in cargo delivery. Of interest, the AP-3–biased P99S variant was nearly completely mislocalized to the cell surface in melan-pa1 cells; indeed, cell surface localization of P99S was more extensive than that for TYRP1, and those puncta that were observed showed little overlap with intracellular TYRP1 and likely represent surface projections (Figure 8, g–i). This result might be explained by a combination of increased endosomal recycling to the cell surface, as has been observed for TYRP1 in BLOC-1–deficient cells (Setty et al., 2007), and decreased endocytosis as a consequence of the decreased interaction of the P99S variant with AP-2 relative to the other OCA2 constructs (unpublished data). Taken together, these data suggest that OCA2 requires both AP-3 and BLOC-1 for effective delivery to melanosomes.
FIGURE 8:
Localization of dileucine-motif point mutants in BLOC-1–deficient melanocytes. (a–i) IFM analysis of BLOC-1–deficient cells expressing selected mutant OCA2 variants. Melan-pa1 melanocytes were transfected with OCA2-AA23 (a–c), OCA2-AA23 E96D (d–f), or OCA2-AA23 P99S (g–i). Transgenes were visualized with anti-HA antibodies (a, d, g), and endogenous TYRP1 was visualized in b, e, and h. c, f, and i are merged OCA2-HA (magenta) and TYRP1 (green) images. All insets show 5× magnified images of the boxed region. Bar, 10 μm. (j) Deconvolved IFM images of OCA2-AA23 and OCA2-AA23 E96D were converted to binary images and analyzed for marker overlap. The P99S mutant was not analyzed. Each data point represents one cell. Shown is the percentage of punctate/vesicular HA staining that overlapped with TYRP1 in each cell (percentage of OCA2 with TYRP1). E96D was not significantly different (n.s.) from OCA2-AA23.
As discussed, TYR requires AP-3 but not BLOC-1 for efficient accumulation in melanosomes. To test whether the lack of requirement for BLOC-1 is a property of the aLL-sorting signal, we expressed OCA2-AA23 hTYR bearing the aLL motif from human tyrosinase in BLOC-1–deficient melan-pa1 cells. As shown earlier, this substituted aLL motif conferred wild-type localization and function to OCA2. Consistently, OCA2-AA23 hTYR was unable to traffic to melanosomes in melan-pa1 cells, as shown by a lack of colocalization with PMEL (Supplemental Figure S4). This result indicates that the transplantable aLL motif from tyrosinase cannot confer BLOC-1 independence on OCA2. Taken together, the data suggest that AP-3 can function in either a BLOC-1–independent pathway or a BLOC-1–dependent pathway and that the choice of pathway is not dictated by the properties of the aLL motif alone.
DISCUSSION
On the basis of the behavior of model cargo proteins studied to date, we previously proposed two routes by which cargoes are delivered from endosomes toward maturing melanosomes in melanocytic cells: one mediated by AP-3 and used largely by TYR (Huizing et al., 2001; Theos et al., 2005), and the other mediated by AP-1 and BLOC-1 and used by TYRP1 (Setty et al., 2007; Delevoye et al., 2009), the copper transporter ATP7A (Setty et al., 2008), and perhaps a cohort of TYR (Theos et al., 2005). By analyzing the behavior of wild-type OCA2 and variants that bind selectively to AP-1 or AP-3, we now extend this model by providing evidence for a pathway that requires both BLOC-1 and AP-3. Our data obtained using a Y3H assay confirm previous findings that OCA2 can bind to both AP-1 and AP-3 via a critical [D/E]xxxL[L/I]-consensus aLL sorting motif but show for the first time that the identity of the “x” residues within the core motif—but not the placement of the motif within the cytoplasmic domain— underlies binding specificity for each adaptor. The distinct intracellular localization of mutants that interact preferentially with either AP-1 or AP-3 provides evidence that AP-3 mediates a final step in the delivery of OCA2 to melanosomes, highlighting an expanded role for this complex in LRO biogenesis.
AP-1, -2, and -3 are well known for their role in coupling clathrin coat recruitment to cargo selection via recognition of cytoplasmic sorting motifs and consequent cargo clustering into buds for directed vesicular trafficking within the endosomal system (Bonifacino and Traub, 2003; Robinson, 2004). The cytoplasmic domains of a number of melanosomal proteins, including TYR and TYRP1, contain aLL-based sorting signals that have been shown to be necessary for melanosomal transport (Vijayasaradhi et al., 1995; Calvo et al., 1999; Simmen et al., 1999). Human OCA2 has three consensus aLL motifs, but only the most membrane-distal motif, LL1, confers melanosomal localization and full function (Sitaram et al., 2009). Unlike aLL signals in other melanosomal and lysosomal proteins, which tend to be positioned within 6-11 residues of the transmembrane domain, the LL1 motif of human OCA2 is located 70 amino acid residues from the transmembrane domain. Motifs at such a distance tend to be limited for use in internalization rather than targeting to lysosomes or LROs (reviewed in Bonifacino and Traub, 2003), as seems to be the case for an aLL signal in the distal cytoplasmic domain of PMEL (Lepage and Lapointe, 2006; Theos et al., 2006; Robila et al., 2008), but in this case it is likely that protein:protein or protein:lipid interactions bring the motif close enough to the membrane to engage AP-1 and AP-3. Nevertheless, the data presented here demonstrate that the LL1 signal functions in a position-independent manner—at least within the most membrane-proximal 100 amino acid residues of the OCA2 N-terminal domain—and is capable of interacting with AP complexes and effecting melanosome targeting when moved to sites that normally harbor inactive LL2 and LL3 motifs. We conclude that the primary determinant of aLL-based targeting activity is ultimately a property of its sequence, supporting conclusions drawn from an analysis of differential binding of wild-type and mutant adaptor complexes to natural aLL signals from tyrosinase, LIMPII, and HIV Nef (Mattera et al., 2011). In this regard, the LL1 sequence is different from other aLL motifs such as a CD4/CD3-γ chimera (Geisler et al., 1998), the distal motif of Iip31 (Pond et al., 1995), and the motif of Vam3p (Darsow et al., 1998), all of which were affected by the residue immediately upstream of the acidic residue. It may be that OCA2 folds in such a way that only the core motif is presented in the correct orientation for AP binding, without the engagement of nearby residues. Of interest, the LL1 sequence is exactly conserved in nearly all of the predicted mammalian homologues of OCA2, with the exception of mouse and rat (Supplemental Table S1), suggesting that the sequence may have evolved to optimize function.
Although the adjacent residues do not seem to be important for LL1 interaction with AP complexes, the x residues within the core [D/E]xxxL[L/I] motif are, and signal activity is extraordinarily sensitive to changes in these residues. In our comparison of the human tyrosinase motif with the similar but nonfunctional OCA2 LL3 motif, the mutation of either the −2 Gln or the −1 Pro in the TYR motif to the Gly or Asp of the LL3 motif, respectively (OCA2-AA23 hTYRQG or OCA2-AA23 hTYRPD), completely abolished the interaction with all three AP complexes and caused mislocalization of the full-length protein to the cell surface, despite the fact that this motif still conforms to the ExxxLL consensus. Moreover, the hTYRQG sequence conforms to the more specific ExxPLL consensus found in other melanosomal proteins. Because the motifs were all placed in the same position within the OCA2 domain, the difference in AP interaction between the EKQPLL motif and the EKGPLL motif must reflect differences in the direct interaction between the core motif and the AP complexes. There is not yet a crystal structure of an aLL motif in complex with the core domain of AP-1 or AP-3, but there is one for AP-2 with the motif from CD4 (Kelly et al., 2008). The side chains for residues at the −2 and −3 positions of the motif point into the solvent in vitro and thus do not contribute significantly to binding affinity to the AP-2 core. The large effect on AP binding that we observed upon mutation in the −2 position of the TYR aLL motif suggests that the orientation of these residues might be more restricted when binding to AP-1 or AP-3. AP-2 is used for endocytosis of a wide variety of proteins from the cell surface, and a less rigid binding orientation is believed to contribute to its broad specificity as a cargo adaptor (Kelly et al., 2008). It may be that AP-1 and AP-3 have more rigid binding surfaces that are more sensitive to the identities of the x residues than AP-2. Indeed, yeast three-hybrid analyses by Mattera et al. (2011) suggested that aLL motifs bind to the analogous surfaces of the AP-1 and AP-3 complexes. Furthermore, fine specificity of AP complex binding was shown to depend on variations in AP complex subunit composition, as well as the sequence of cargo-sorting motifs, in agreement with our results.
The E96D mutation in OCA2 greatly reduced but did not eliminate AP-1 interaction and had less of an effect on AP-2 interaction, consistent with mutagenesis results with the distal aLL motif of LRP9 (Doray et al., 2007). Our data additionally show that the conservative E96D mutation eliminates AP-3 interaction. Acidic residues upstream of the dileucine pair are known to be necessary for AP-3 binding to LIMP-II (Höning et al., 1998) and Vam3p (Darsow et al., 1998), but to our knowledge this study represents the first report of specificity for the identity of the acidic residue. In contrast to the E96D mutant, the P99A and P99S mutants of OCA2 eliminated AP-1 interaction and greatly reduced AP-2 interaction but maintained a significant level of AP-3 interaction. Proline has been specifically noted as a common −1 residue in several aLL motifs (Pond et al., 1995; Darsow et al., 1998; Höning et al., 1998; Bennett and Lamoreux, 2003; Doray et al., 2007). Consistent with the importance of the −1 Pro for AP-1 and AP-3 interaction, mutagenesis of a −1 Pro in the distal motif of LRP9 eliminates binding to AP-1 and severely reduces its interaction with AP-2, and the cation-independent mannose 6-phosphate receptor can acquire the ability to interact with AP-1 and AP-2 from a heterologous motif that includes a −1 Pro (Doray et al., 2008). The −1 Pro does not seem to be required for AP-3 interaction since the same position could accommodate alanine (with a small hydrophobic side chain), serine (polar uncharged side chain), and phenylalanine (bulky hydrophobic side chain) in our Y3H assay. However, neither OCA2 LL1 (Figure 5) nor the OCA2-AA23 hTYRPD chimera (Figure 3) was able to tolerate an acidic residue (aspartate) in this position. In the AP-2 crystal structure the −4 acidic residue binds in a pocket that has some flexibility, the two leucine residues bind in a separate hydrophobic pocket, and the −1 position protrudes into the solvent. It is again tempting to speculate that the differential requirements for the identity of the acidic residue or the −1 Pro will be reflected in structural differences between AP-1 and AP-3 aLL motif–interacting surfaces. The aLL motif of Nef, ENTSLL, is identical to the OCA2 P99S variant, yet Nef does interact with AP-1 in a yeast three-hybrid assay (Mattera et al., 2011). However it has also been suggested that residues outside of the sorting motif in Nef contribute to AP binding (Chaudhuri et al., 2009; Mattera et al., 2011). The aLL motif of mouse tyrosinase, ERQPLL, interacts in a three-hybrid assay with both AP-1 and AP-3 (Theos et al., 2005), and the identical sequence in the lipid kinase PI4K2A mediates coimmunoprecipitation from cell lysates of PI4K2A with AP-3 but less so with AP-1 (Craige et al., 2008). Whether this difference in AP-binding specificity reflects a difference in cargo protein conformation or important extra-motif residues or simply a difference between the two assays is unknown.
Although the E96D and P99S variants show opposite preferences for AP interaction, full-length OCA2 bearing either mutation was found to localize to postendosomal structures and to confer nearly complete function to OCA2-deficient melanocytes. This indicates that either AP-1 or AP-3 alone is sufficient to direct proteins into the endosomal system and into endosomal domains bound for melanosomes. However, steady-state melanosomal localization of OCA2 seemed to critically depend on an OCA2/AP-3 interaction because 1) the AP-1–biased E96D variant localized to endosomal structures closely apposed to melanosomes in wild-type melanocytes rather than to melanosomal “rings” and 2) no construct expressed in the majority of AP-3–deficient melan-pe melanocytes localized properly to melanosomes. The unusual structures to which OCA2 localized in the absence of AP-3 or a strong AP-3 binding signal were adjacent to and partially overlapping with melanin. This pattern of localization resembles that of AP-1 and recycling endosomal markers in a human pigmented melanoma cell line and correlates with tubular transport intermediates that physically connect recycling endosomal domains with melanosomes (Delevoye et al., 2009). We thus speculate that OCA2 accumulates in such tubular extensions when it is capable of binding to AP-1 but not to AP-3 (Figure 9). Consistently, by double immunogold labeling and immuno–electron microscopy analysis of ultrathin cryosections of transiently transfected wild-type melanocytes, we detected OCA2 in small vesicles that also contained AP-1 (Supplemental Figure S2, insets) and that are morphologically similar to intermediates in MNT-1 melanoma cells that harbor melanosome-bound TYRP1 (Raposo et al., 2001; Delevoye et al., 2009). The likely interpretation that the E96D variant localized to endosomal tubules that are physically continuous with melanosomes in wild-type cells and rescued melanin synthesis in OCA2-deficient cells nearly as well as wild-type OCA2 raises the intriguing possibility that OCA2 might not need to be present specifically on melanosomes in order to function. OCA2 is believed to function as a transporter, and thus either OCA2 might in fact function from the tubules themselves or mislocalization of OCA2 itself to the tubules might be overcome by diffusion of its putative transport substrate into melanosomes via passage through the tubules.
FIGURE 9:
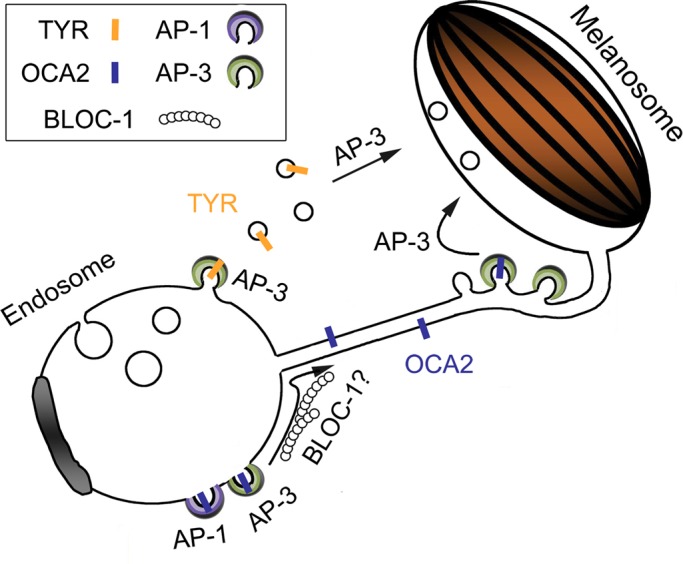
Model for role of AP-3, AP-1 and BLOC-1 in transport of OCA2 from endosomes to melanosomes. Shown is a model based on a related model from Delevoye et al. (2009) and the data from this article. In vacuolar endosomes, OCA2 encounters either AP-1 or AP-3, and either is sufficient to target OCA2 to melanosome-bound transport carriers that emerge from these domains. BLOC-1 also functions in an as-yet-unknown capacity during this step. From the transport carriers, binding to AP-3 is required to effect a subsequent transport step that results in stable association with melanosomes.
The dependence of OCA2 on AP-3 distinguishes its trafficking from that of TYRP1. Yet, like TYRP1, all OCA2 constructs were mislocalized in BLOC-1–deficient melanocytes; indeed, OCA2 in these cells largely overlapped with TYRP1, which is largely entrapped in early endosomal vacuoles (Setty et al., 2007), and the OCA2 constructs did not colocalize with the melanosomal marker PMEL. This dependence on BLOC-1 differentiates OCA2 trafficking from that of TYR, for which a substantial cohort can be found in the nonpigmented melanosomes in BLOC-1–deficient cells (Setty et al., 2007, 2008). Thus, our data define a pathway of endosome-to-melanosome trafficking that is both BLOC-1 dependent and AP-3 dependent. Of interest, this pathway is consistent with analyses of enhanced coat color dilution in mice homozygous for mutations in OCA2 and HPS complexes AP-3, BLOC-1, or a related complex BLOC-2, which suggested that OCA2 functionally interacts with each of these trafficking complexes in the context of coat pigment generation (Hoyle et al., 2011). This pathway furthermore aligns our previous model of distinct BLOC-1–mediated and AP-3–mediated melanosomal trafficking pathways with observations in neurons and platelets that suggest that BLOC-1 and AP-3 work together in cargo distribution (Newell-Litwa et al., 2009; Salazar et al., 2009; Larimore et al., 2011; Meng et al., 2012), although cell-type specificity of trafficking mediated by the ubiquitously expressed AP-3 may also stem from interactions with LRO-specific Rabs (Bultema et al., 2012). Finally, our model provides a critical biological context in a LRO-generating cell type for a previously described physical interaction between AP-3 and BLOC-1 (Di Pietro et al., 2006; Salazar et al., 2006). We speculate that these distinct sorting functions for AP-3 are mediated from distinct domains of early-sorting and/or recycling endosomes. Consistently, we and others (Peden et al., 2004; Theos et al., 2005; unpublished observations) detect AP-3 in two different pools in melanocytes and other cell types. OCA2 sorting by AP-3 might occur from the tubular recycling endosomal domains from which TYRP1 is delivered, whereas TYR sorting by AP-3 might occur from distinct domains of sorting endosomes (Figure 9). Further analyses will be required to test this model.
If AP-3 mediates cargo sorting from two distinct domains, what determines whether cargoes enter the BLOC-1–dependent or BLOC-1–independent pathway? This decision point likely reflects some feature of the cargo other than the AP-interacting sorting signal. The failure of OCA2-AA23 hTYR, bearing the sorting motif of the largely BLOC-1–independent tyrosinase, to localize to immature melanosomes in BLOC-1–deficient melanocytes suggests that the aLL motif alone cannot determine BLOC-1 dependence. Instead, other features are likely critical. Unlike tyrosinase, which has only a single membrane-spanning domain, OCA2 has 12 membrane-spanning domains, and its oligomeric state is unknown. We speculate that the 12 membrane-spanning domains preclude entry of OCA2 either into early-sorting endosomal domains from which tyrosinase accesses AP-3 for the BLOC-1–independent pathway or into vesicles that likely serve as the transport intermediates from these domains (Peden et al., 2004; Theos et al., 2005). By contrast, OCA2 might readily enter the tubular transport intermediates that have been observed between recycling endosomal domains and melanosomes and through which TYRP1 likely enters melanosomes (Delevoye et al., 2009). Definitive identification of the determining feature will have to await ultrastructural analyses of OCA2 mutant and chimeric protein localization in HPS model melanocytes.
MATERIALS AND METHODS
Chemicals
All chemicals were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise specified.
Cell culture and transgene expression
All culture reagents were purchased from Invitrogen (Carlsbad, CA) unless stated otherwise. All cells were grown at 37°C and 10% CO2. Immortalized mouse melanocyte cell lines, including melan-Ink4a from C57BL/6 Ink4a-Arf−/− (wild-type) mice (Sviderskaya et al., 2002), melan-p1 from C3H-pcp/p25H (pink-eyed dilute) mice (Sviderskaya et al., 1997), melan-pa1 from B6.Cg-PldnPaInk4a-Arf−/− (pallid) mice (Setty et al., 2007), and melan-pe from B6Pin.C3-Ap3b1pe/Ap3b1pe (pearl) mice (Theos et al., 2005), were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 200 nM phorbol 12-myristate-13-acetate. Melan-pe growth medium was additionally supplemented with 200 pM cholera toxin. Transfections in mouse melanocytes were performed using FuGENE-6 (Roche Diagnostics, Indianapolis, IN) according to manufacturer's instructions with 1–2 μg of DNA. MNT-1, a pigmented human melanoma cell line (Cuomo et al., 1991), was cultured as previously described (Raposo et al., 2001).
Construction of OCA2 expression plasmids and glutathione S-transferase fusion proteins
The construction of pCR3/OCA2-HA containing the coding sequence of 3xHA-tagged, full-length human OCA2 and derivative constructs pCR3/OCA2-AA1, pCR3/OCA2-AA23, pCR3/OCA2-AA12, and pCR3/OCA2-AA123 (Sitaram et al., 2009) has been previously described. The pBridge plasmids containing the AP σ1A, σ2, or σ3A construct and the cytoplasmic domain of tyrosinase (Theos et al., 2005) and the pGADT7 plasmids containing fusions of mouse AP γ1, rat αC, and human δ to yeast Gal4-AD (Janvier et al., 2003; Chaudhuri et al., 2007) have been previously described. Each full-length OCA2 dileucine mutant presented here was constructed by site-directed mutagenesis using two-step PCR amplification (Higuchi et al., 1988) with GoTaq (Promega, Madison, WI) and the appropriate pCR3/OCA2-HA variant as the template. For example, the OCA2-AA23 LL1-2 fragment was synthesized by mutagenic two-step amplification of the pCR3/OCA2-AA23 template. Mutagenized fragments were then subcloned into the BamHI and BsiWI sites of the appropriate pCR3/OCA2-HA variant. The corresponding yeast three-hybrid pBridge constructs were made by PCR amplification of the N-terminal portion of the appropriate pCR3/OCA2-containing plasmid with primers that added a 5′ EcoRI site and 3′ SalI site. The products were subcloned into the same restriction sites downstream of yeast Gal4-BD in pBridge plasmids containing human σ1A, rat σ2, or human σ3A in the second transcription unit. GST-OCA2 fusion proteins containing full-length OCA2 N-terminal cytoplasmic domain or the AA23 and AA123 variants were as described (Sitaram et al., 2009). GST fusions to OCA2 dileucine variants were generated by two-step PCR amplification of the coding region for the entire 173 N-terminal cytoplasmic domain of the full-length pCR3/OCA2 dileucine variant using primers that added a 5′ BamHI site and a 3′ SalI site to the PCR product. The amplification products were then subcloned into pGEX-5X-1 (GE Healthcare, Little Chalfont, United Kingdom). All recombinant plasmids were verified by automated sequencing by the University of Pennsylvania Cell Center or the Nucleic Acid/Protein Research Core Facility at the Children's Hospital of Pennsylvania. Details of PCR primers, sequences, and conditions will be provided upon request.
Antibodies
Antibodies used were as follows: rat anti-HA 3F10 was from Roche Diagnostics; mouse anti-HA 16B12 and rabbit anti-HA.11 were from Covance Research Products (Princeton, NJ); TA99 (Mel-5) anti-TYRP1 was from the American Type Culture Collection (Manassas, VA); HMB45 anti-human PMEL was from LabVision (Fremont, CA); SA4 anti-AP-3δ was from Developmental Studies Hybridoma Bank (Iowa City, IA) and a kind gift from Victor Faundez (Emory University, Atlanta, GA); and anti–γ-adaptin to AP-1 was from BD Biosciences (San Jose, CA). Goat anti–mouse immunoglobulin G (IgG) secondary antibodies conjugated to Alexa 488 and Alexa 594 were from Invitrogen. Donkey anti–rat IgG antibodies conjugated to DyLight 488, unconjugated donkey anti–mouse IgG antibodies, and donkey anti–mouse IgG antibodies conjugated to horseradish peroxidase or alkaline phosphatase were from Jackson ImmunoResearch Laboratories (West Grove, PA). Conjugation of Alexa 488 to monoclonal antibodies was performed using a protein labeling kit from Invitrogen.
Transfections and immunofluorescence microscopy
Mouse melanocytes were seeded at 1 × 105 per well in six-well dishes or at 0.5 × 105 per well in 12-well dishes on coverslips coated with Matrigel (BD Biosciences) according to manufacturer's instructions. The next day, cells were transfected with 1–2 μg of the relevant OCA2 construct by using FuGENE-6 reagent. Two days later, cells were fixed for 30 min in phosphate-buffered saline (PBS)/2% formaldehyde (Thermo Fisher Scientific, Waltham, MA), stained with primary and fluorochrome-conjugated secondary antibodies, and mounted onto glass slides as described previously (Calvo et al., 1999). Slides were analyzed on a DM IRBE microscope (Leica, Wetzlar, Germany) equipped with an Orca (Hamamatsu, Bridgewater, NJ) or a Retiga Exi Fast 1394 digital camera (Qimaging, Surrey, Canada). Images were captured and manipulated using OpenLab software (Improvision, Lexington, MA) with the volume deconvolution package. Final images were prepared using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Yeast culture, transformation, and three-hybrid assays
The S. cerevisiae strain HF7c (Clontech) was maintained on complete yeast extract/peptone/dextrose plates. Cotransformation with pBridge and pGADT7 plasmids was performed by a modification of the lithium acetate procedure as described in the Yeast Protocols Handbook from Clontech. HF7c transformants were selected by spreading on plates lacking leucine, tryptophan, and methionine. For colony growth assays, HF7c transformants were pooled and spotted once or in fivefold serial dilutions on plates lacking leucine, tryptophan, methionine, and histidine and allowed to grow at 30°C for 3–5 d.
Glutathione S-transferase pull-down assays
GST pull-down assays were conducted essentially as described (Starcevic and Dell'Angelica, 2004; Sitaram et al., 2009) with slight buffer modifications. Briefly, GST-OCA2 fusion proteins were expressed in Escherichia coli (BL21(DE) star; Invitrogen) by induction with 10 μM isopropyl β-d-1-thiogalactopyranoside for 3 h at 30ºC, and bacterial cell lysates in 1% Triton X-100 were flash frozen and stored at −80ºC. MNT-1 cells (4 × 107 per pull down) were harvested with 0.5 mM EDTA in PBS, resuspended (2 × 107 cells/ml) in lysis buffer (100 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 1 mM NaF, 0.2 mM GTPγS, protease inhibitors [Roche], and 4% NP-40), and lysed by Dounce homogenization on ice. Lysates were clarified by centrifugation at 20,000 × g for 15 min and precleared by incubation under rotation with 15 μl of packed glutathione–Sepharose 4B beads (Amersham, GE Healthcare) for 1 h at 4ºC. Cleared lysates were incubated with equal quantities (20 μg) of GST-OCA2 fusion proteins prebound to 15 μl of packed glutathione–Sepharose beads for 1 h at 4ºC. Beads were then washed four times in ice-cold buffer (100 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM NaF, and 0.1% NP-40), resuspended in 50 μl of 2× reducing SDS–PAGE loading dye, and heated at 65ºC for 5 min. A 5-μl portion of each sample was fractionated by SDS–PAGE (12% gels) and stained with Coomassie blue R-250; 45 μl of each sample was fractionated by SDS–PAGE (10% gels) and analyzed by immunoblotting for AP-1 or AP-3 using ECL Plus enhanced chemiluminescence (GE Healthcare) and exposure to film. Film images were scanned and adjusted using Adobe Photoshop CS4, version 11.02, software and quantified using ImageQuant Software (GE Healthcare).
Statistical analysis
Quantification of melanin synthesis rescue was performed as follows. Transfected melan-p1 cells were identified by positive labeling with anti-HA antibody by immunofluorescence microscopy and then assessed for the presence of pigmented melanosomes by bright-field microscopy. Within each experiment, the percentage of transfected cells (≥100 cells/experiment) that were positive for pigment rescue was calculated. Graphs represent rescue by each construct averaged over three independent experiments. Note that the results varied with different thaws of melan-p1 cells and different batches of transfection reagents; each graph represents a group of samples that were analyzed at the same time using the same cells and reagents. An initial repeated measures analysis of variance was performed on the matched sets, followed by Dunnett's multiple comparison test to compare rescue by each mutant to rescue by wild-type OCA2. To quantify overlap of OCA2 dileucine mutants with pigment or with TYRP1, deconvolved paired images were rendered binary in ImageJ (National Institutes of Health, Bethesda, MD) by density slicing, and the total area of overlap between them was calculated for objects containing >5 pixels, excluding the densely labeled perinuclear area. Seven to 10 cell profiles were quantified for each pairwise comparison. To compare colocalization with pigment, a one-way analysis of variance was performed on the unmatched sets, followed by Bonferroni's multiple comparison test. To compare the colocalization of OCA2-AA23 or OCA2-AA23 E96D with TYRP1, we performed an unpaired two-tailed t test. All statistical analyses were performed using GraphPad Prism (GraphPad Software, San Diego, CA).
Supplementary Material
Acknowledgments
We thank Victor Faundez and Andrew Peden for generous gifts of antibodies. This work was supported by National Institutes of Health Grant R01 EY015625 from the National Eye Institute (to M.S.M.), Training Grants T32 GM007229 and T32 HL007971 (for A.S.) and K12 GM081259 (for M.K.D.), the Institut Curie and Centre National de la Recherche Scientifique (for G.R.), the Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (for J.S.B.), and Wellcome Trust Grant 078327 (to E.V.S. and D.C.B.).
Abbreviations used:
- aLL
acidic dileucine
- AP
adaptor protein
- BLOC
biogenesis of lysosome-related organelles complex
- ECL
enhanced chemiluminescence
- GST
glutathione S-transferase
- HA
hemagglutinin
- HIV
human immunodeficiency virus
- HPS
Hermansky–Pudlak syndrome
- IFM
indirect immunofluorescence microscopy
- IgG
immunoglobulin G
- LROs
lysosome-related organelles
- OCA
oculocutaneous albinism
- TYR
tyrosinase
- TYRP1
tyrosinase-related protein-1
- Y3H
yeast three hybrid
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0509) on June 20, 2012.
*Present address: Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore 560 012, India.
REFERENCES
- Bennett DC, Lamoreux ML. The color loci of mice—a genetic century. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- Boehm M, Bonifacino JS. Adaptins: the final recount. Mol Biol Cell. 2001;12:2907–2920. doi: 10.1091/mbc.12.10.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Borner GH, Harbour M, Hester S, Lilley KS, Robinson MS. Comparative proteomics of clathrin-coated vesicles. J Cell Biol. 2006;175:571–578. doi: 10.1083/jcb.200607164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultema JJ, Ambrosio AL, Burek CL, Di Pietro SM. BLOC-2, AP-3, and AP-1 function in concert with Rab38 and Rab32 to mediate protein trafficking to lysosome-related organelles. J Biol Chem. 2012;287:19550–19563. doi: 10.1074/jbc.M112.351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo PA, Frank DW, Bieler BM, Berson JF, Marks MS. A cytoplasmic sequence in human tyrosinase defines a second class of di-leucine-based sorting signals for late endosomal and lysosomal delivery. J Biol Chem. 1999;274:12780–12789. doi: 10.1074/jbc.274.18.12780. [DOI] [PubMed] [Google Scholar]
- Chapuy B, Tikkanen R, Muhlhausen C, Wenzel D, von Figura K, Höning S. AP-1 and AP-3 mediate sorting of melanosomal and lysosomal membrane proteins into distinct post-Golgi trafficking pathways. Traffic. 2008;9:1157–1172. doi: 10.1111/j.1600-0854.2008.00745.x. [DOI] [PubMed] [Google Scholar]
- Chaudhuri R, Lindwasser OW, Smith WJ, Hurley JH, Bonifacino JS. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J Virol. 2007;81:3877–3890. doi: 10.1128/JVI.02725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R, Mattera R, Lindwasser OW, Robinson MS, Bonifacino JS. A basic patch on alpha-adaptin is required for binding of human immunodeficiency virus type 1 Nef and cooperative assembly of a CD4-Nef-AP-2 complex. J Virol. 2009;83:2518–2530. doi: 10.1128/JVI.02227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B, Salazar G, Faundez V. Phosphatidylinositol-4-kinase type IIα contains an AP-3-sorting motif and a kinase domain that are both required for endosome traffic. Mol Biol Cell. 2008;19:1415–1426. doi: 10.1091/mbc.E07-12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo M, Nicotra MR, Apollonj C, Fraioli R, Giacomini P, Natali PG. Production and characterization of the murine monoclonal antibody 2G10 to a human T4-tyrosinase epitope. J Invest Dermatol. 1991;96:446–451. doi: 10.1111/1523-1747.ep12470092. [DOI] [PubMed] [Google Scholar]
- Darsow T, Burd CG, Emr SD. Acidic di-leucine motif essential for AP-3-dependent sorting and restriction of the functional specificity of the Vam3p vacuolar t-SNARE. J Cell Biol. 1998;142:913–922. doi: 10.1083/jcb.142.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C, et al. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J Cell Biol. 2009;187:247–264. doi: 10.1083/jcb.200907122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC, Mullins C, Caplan S, Bonifacino JS. Lysosome-related organelles. FASEB J. 2000;14:1265–1278. doi: 10.1096/fj.14.10.1265. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the b3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Dell'Angelica EC. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic. 2005;6:525–533. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, Dell'Angelica EC. BLOC-1 interacts with BLOC-2 and the AP-3 complex to facilitate protein trafficking on endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B, Knisely JM, Wartman L, Bu G, Kornfeld S. Identification of acidic dileucine signals in LRP9 that interact with both GGAs and AP-1/AP-2. Traffic. 2008;9:1551–1562. doi: 10.1111/j.1600-0854.2008.00786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray B, Lee I, Knisely J, Bu G, Kornfeld S. The gamma/sigma1 and alpha/sigma2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol Biol Cell. 2007;18:1887–1896. doi: 10.1091/mbc.E07-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiberg H, Troelsen J, Nielsen M, Mikkelsen A, Mengel-From J, Kjaer KW, Hansen L. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum Genet. 2008;123:177–187. doi: 10.1007/s00439-007-0460-x. [DOI] [PubMed] [Google Scholar]
- Feng L, et al. The beta3A subunit gene (Ap3b1) of the AP-3 adaptor complex is altered in the mouse hypopigmentation mutant pearl, a model for Hermansky-Pudlak syndrome and night blindness. Hum Mol Genet. 1999;8:323–330. doi: 10.1093/hmg/8.2.323. [DOI] [PubMed] [Google Scholar]
- Geisler C, Dietrich J, Nielsen BL, Kastrup J, Lauritsen JP, Odum N, Christensen MD. Leucine-based receptor sorting motifs are dependent on the spacing relative to the plasma membrane. J Biol Chem. 1998;273:21316–21323. doi: 10.1074/jbc.273.33.21316. [DOI] [PubMed] [Google Scholar]
- Gokhale A, Larimore J, Werner E, So L, Moreno-De-Luca A, Lese-Martin C, Lupashin VV, Smith Y, Faundez V. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J Neurosci. 2012;32:3697–3711. doi: 10.1523/JNEUROSCI.5640-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing VJ. Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J Dermatol Sci. 2005;37:3–14. doi: 10.1016/j.jdermsci.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höning S, Sandoval IV, von Figura K. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J. 1998;17:1304–1314. doi: 10.1093/emboj/17.5.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle DJ, Rodriguez-Fernandez IA, Dell'Angelica EC. Functional interactions between OCA2 and the protein complexes BLOC-1, BLOC-2, and AP-3 inferred from epistatic analyses of mouse coat pigmentation. Pigment Cell Melanoma Res. 2011;24:275–281. doi: 10.1111/j.1755-148X.2010.00815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, Gahl WA. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet. 2008;9:359–386. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Sarangarajan R, Strovel E, Zhao Y, Gahl WA, Boissy RE. AP-3 mediates tyrosinase but not TRP-1 trafficking in human melanocytes. Mol Biol Cell. 2001;12:2075–2085. doi: 10.1091/mbc.12.7.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janvier K, Kato Y, Boehm M, Rose JR, Martina JA, Kim BY, Venkatesan S, Bonifacino JS. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 gamma-sigma1 and AP-3 delta-sigma3 hemicomplexes. J Cell Biol. 2003;163:1281–1290. doi: 10.1083/jcb.200307157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantheti P, et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Kelly BT, McCoy AJ, Spate K, Miller SE, Evans PR, Höning S, Owen DJ. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RA. Albinism. In: Nordlund JJ, Boissy RE, Hearing VJ, King RA, Ortonne J-P, editors. The Pigmentary System: Physiology and Pathophysiology. New York: Oxford University Press; 1998. pp. 553–575. [Google Scholar]
- Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet. 2007;71:354–369. doi: 10.1111/j.1469-1809.2006.00341.x. [DOI] [PubMed] [Google Scholar]
- Larimore J, et al. The schizophrenia susceptibility factor dysbindin and its associated complex sort cargoes from cell bodies to the synapse. Mol Biol Cell. 2011;22:4854–4867. doi: 10.1091/mbc.E11-07-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Nemecek D, Schindler C, Smith WJ, Ghirlando R, Steven AC, Bonifacino JS, Hurley JH. Assembly and architecture of Biogenesis of Lysosome-related Organelles Complex-1 (BLOC-1) J Biol Chem. 2012;287:5882–5890. doi: 10.1074/jbc.M111.325746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ST, Nicholls RD, Jong MT, Fukai K, Spritz RA. Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics. 1995;26:354–363. doi: 10.1016/0888-7543(95)80220-g. [DOI] [PubMed] [Google Scholar]
- Lepage S, Lapointe R. Melanosomal targeting sequences from gp100 are essential for MHC class II-restricted endogenous epitope presentation and mobilization to endosomal compartments. Cancer Res. 2006;66:2423–2432. doi: 10.1158/0008-5472.CAN-05-2516. [DOI] [PubMed] [Google Scholar]
- Marks MS, Seabra MC. The melanosome: membrane dynamics in black and white. Nat Rev Mol Cell Biol. 2001;2:738–748. doi: 10.1038/35096009. [DOI] [PubMed] [Google Scholar]
- Mattera R, Boehm M, Chaudhuri R, Prabhu Y, Bonifacino JS. Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J Biol Chem. 2011;286:2022–2030. doi: 10.1074/jbc.M110.197178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng R, et al. SLC35D3 delivery from megakaryocyte early endosomes is required for platelet dense granule biogenesis and differentially defective in Hermansky-Pudlak syndrome models. Blood. 2012;120:404–414. doi: 10.1182/blood-2011-11-389551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel-From J, Borsting C, Sanchez JJ, Eiberg H, Morling N. Human eye colour and HERC2, OCA2 and MATP. Forensic Sci Int Genet. 2010;4:323–328. doi: 10.1016/j.fsigen.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Newell-Litwa K, Salazar G, Smith Y, Faundez V. Roles of BLOC-1 and adaptor protein-3 complexes in cargo sorting to synaptic vesicles. Mol Biol Cell. 2009;20:1441–1453. doi: 10.1091/mbc.E08-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 2007;24:710–722. doi: 10.1093/molbev/msl203. [DOI] [PubMed] [Google Scholar]
- Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond L, Kuhn LA, Teyton L, Schutze MP, Tainer JA, Jackson MR, Peterson PA. A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J Biol Chem. 1995;270:19989–19997. doi: 10.1074/jbc.270.34.19989. [DOI] [PubMed] [Google Scholar]
- Raposo G, Marks MS. Melanosomes—dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr Opin Cell Biol. 2007;19:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J Cell Biol. 2001;152:809–824. doi: 10.1083/jcb.152.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond B, Huizing M, Knapp J, Koshoffer A, Zhao Y, Gahl WA, Boissy RE. Melanocytes derived from patients with Hermansky-Pudlak syndrome types 1, 2, and 3 have distinct defects in cargo trafficking. J Invest Dermatol. 2005;124:420–427. doi: 10.1111/j.0022-202X.2004.23585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, Spritz RA, Avidano KM, Jong MT, Nicholls RD. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993;361:72–76. doi: 10.1038/361072a0. [DOI] [PubMed] [Google Scholar]
- Robila V, Ostankovitch M, Altrich-Vanlith ML, Theos AC, Drover S, Marks MS, Restifo N, Engelhard VH. MHC class II presentation of gp100 epitopes in melanoma cells requires the function of conventional endosomes and is influenced by melanosomes. J Immunol. 2008;181:7843–7852. doi: 10.4049/jimmunol.181.11.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Rohrer J, Schweizer A, Russell D, Kornfeld S. The targeting of Lamp1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J Cell Biol. 1996;132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G, et al. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 cargoes. Mol Biol Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G, Zlatic S, Craige B, Peden AA, Pohl J, Faundez V. Hermansky-Pudlak syndrome protein complexes associate with phosphatidylinositol 4-kinase type II alpha in neuronal and non-neuronal cells. J Biol Chem. 2009;284:1790–1802. doi: 10.1074/jbc.M805991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 2008;454:1142–1146. doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, et al. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen T, Schmidt A, Hunziker W, Beermann F. The tyrosinase tail mediates sorting to the lysosomal compartment in MDCK cells via a di-leucine and a tyrosine-based signal. J Cell Sci. 1999;112:45–53. doi: 10.1242/jcs.112.1.45. [DOI] [PubMed] [Google Scholar]
- Sitaram A, Marks MS. Mechanisms of protein delivery to melanosomes in pigment cells. Physiology (Bethesda) 2012;27:85–99. doi: 10.1152/physiol.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A, Piccirillo R, Palmisano I, Harper DC, Dell'Angelica EC, Schiaffino MV, Marks MS. Localization to mature melanosomes by virtue of cytoplasmic dileucine motifs is required for human OCA2 function. Mol Biol Cell. 2009;20:1464–1477. doi: 10.1091/mbc.E08-07-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2 and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- Sviderskaya EV, Bennett DC, Ho L, Bailin T, Lee ST, Spritz RA. Complementation of hypopigmentation in p-mutant (pink-eyed dilution) mouse melanocytes by normal human P cDNA, and defective complementation by OCA2 mutant sequences. J Invest Dermatol. 1997;108:30–34. doi: 10.1111/1523-1747.ep12285621. [DOI] [PubMed] [Google Scholar]
- Sviderskaya EV, Hill SP, Evans-Whipp TJ, Chin L, Orlow SJ, Easty DJ, Cheong SC, Beach D, DePinho RA, Bennett DC. p16(Ink4a) in melanocyte senescence and differentiation. J Natl Cancer Inst. 2002;94:446–454. doi: 10.1093/jnci/94.6.446. [DOI] [PubMed] [Google Scholar]
- Theos AC, et al. Dual loss of ER export and endocytic signals with altered melanosome morphology in the silver mutation of Pmel17. Mol Biol Cell. 2006;17:3598–3612. doi: 10.1091/mbc.E06-01-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos AC, et al. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol Biol Cell. 2005;16:5356–5372. doi: 10.1091/mbc.E05-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayasaradhi S, Xu Y, Bouchard B, Houghton AN. Intracellular sorting and targeting of melanosomal membrane proteins: identification of signals for sorting of the human brown locus protein, gp75. J Cell Biol. 1995;130:807–820. doi: 10.1083/jcb.130.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ML. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 2006;19:19–42. doi: 10.1111/j.1600-0749.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- White S, Hatton SR, Siddiqui MA, Parker CD, Trowbridge IS, Collawn JF. Analysis of the structural requirements for lysosomal membrane targeting using transferrin receptor chimeras. J Biol Chem. 1998;273:14355–14362. doi: 10.1074/jbc.273.23.14355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.