Summary
Oct4 is one of the most important transcription factors required to maintain an undifferentiated state (self-renewal) and pluripotency of human and mouse embryonic stem (ES) cells as well as early embryonic cells. In addition, Oct4 is the only known transcription factor that has to be exogenously introduced into differentiated cells to make induced pluripotent stem (iPS) cells. Therefore, it is of great importance to understand how Oct4 transcription is regulated in ES cells and embryos and how it becomes activated during iPS cell formation. In this article, we will review the regulation of the mouse Oct4 gene from the viewpoint of DNA methylation, binding of orphan nuclear receptors, histone modifications and synergistic effects with other pluripotency factors. We will also raise several key questions that need to be addressed in future work to improve our understanding of Oct4 gene regulation and its essential role in self-renewal and pluripotency.
Keywords: ES cells, Epigenetics, Oct4, Pluripotency
Introduction
Oct4, Sox2 and Nanog constitute a triad of transcription factors that are critical for self-renewal and pluripotency of human and mouse embryonic stem (ES) cells and blastocyst inner cell mass (ICM) cells (Pesce and Scholer, 2001; Chambers and Smith, 2004; Boiani and Scholer, 2005; Niwa, 2007). The Oct4 protein contains a POU domain that binds to the octamer sequence, ATGCAAAT, or its inverse complement DNA located in the promoter or enhancer regions of its target genes. Oct4 is specifically expressed in pluripotent cells, including mouse and human ICM cells, ES cells, germ cells, embryonic carcinoma cells and embryonic germ cells. The ICM cells in Oct4−/− mouse embryos lose pluripotency and tend to differentiate into the trophoblast lineage (Nichols et al., 1998). The concentration of Oct4 within a cell needs to be tightly regulated to allow for self-renewal of ES cells because even a twofold increase of the Oct4 protein induces differentiation toward primitive endoderm and mesoderm and a 50% decrease causes differentiation into trophectoderm (Niwa et al., 2000). Although little is known about how the protein level of Oct4 is precisely controlled in ES cells, there should be a highly sensitive sensor mechanism that is capable of detecting the level of the Oct4 protein and sending suppression signals for its expression as the first step in a negative feedback loop.
The importance of Oct4 for pluripotency has been further highlighted by the recent invention of induced pluripotent stem (iPS) cell technology (Takahashi and Yamanaka, 2006; Maherali and Hochedlinger, 2008). This technology creates pluripotent ES-like cells by introducing up to four transcription factors, including Oct4, into differentiated cells. Among the several combinations of transcription factors needed to make iPS cells, Oct4 is the only one that is always required to be introduced from the outside. The majority of iPS protocols do not require exogenous Nanog, which is activated by Oct4 and other introduced factors. Exogenous Sox2 can be also omitted if target cells, such as neural stem cells, already express this protein (Kim et al., 2009). A large number of original articles and review papers have been published in the past few years on the molecular mechanism of self-renewal and pluripotency; however, large pieces are still missing from our understanding of Oct4 gene regulation. This article will review the regulatory mechanism of Oct4 transcription, focusing on chromatin modifications and other proteins directly bound to the Oct4 locus.
Cis-regulatory elements and DNA methylation of the Oct4 locus
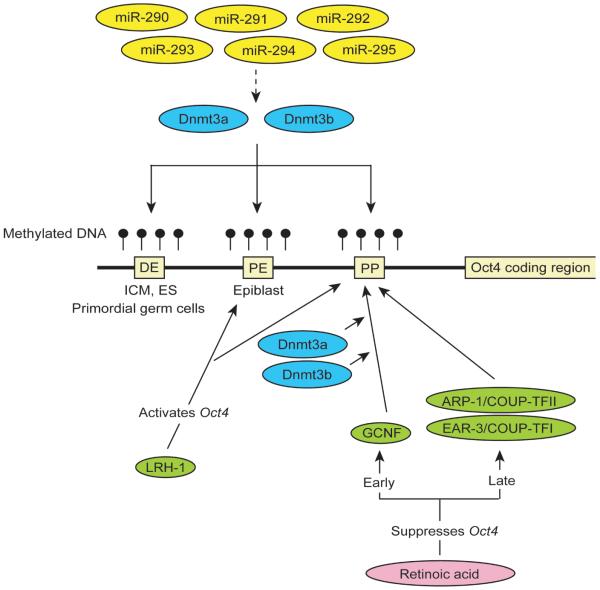
The upstream of the transcriptional initiation site of the mouse Oct4 gene contains three regulatory elements for its transcription, including distal enhancer (DE), proximal enhancer (PE) and proximal promoter (PP) (Yeom et al., 1996; Niwa, 2007) (Fig. 1). The two enhancers are differentially active depending on the developmental stage of the mouse embryo. The DE drives Oct4 expression in the ICM, ES cells and primordial germ cells, while the PE activates Oct4 expression in epiblast cells. These three elements serve as landmarks for protein binding and DNA methylation. DNA methylation of these three elements reflects the transcriptional status of the Oct4 gene. While they are unmethylated in ES cells, these elements are methylated in somatic cells which do not express Oct4 (Hattori et al., 2004). The de novo DNA methyltransferases Dnmt3a and Dnmt3b are responsible for the methylation during differentiation of ES cells (Li et al., 2007). The transcription of Dnmt3a and Dnmt3b is indirectly facilitated by ES cell-specific microRNAs, miR-290 through miR-295, which have been proposed to suppress a transcriptional repressor for Dnmt3a and Dnmt3b, such as Rbl2 (Sinkkonen et al., 2008).
Fig. 1.
Regulation of Oct4 transcription through DNA methylation and binding of orphan nuclear receptors. During differentiation of mouse ES cells, miR-290 through mir-295 indirectly facilitate transcription of Dnmt3a and Dnmt3b, which in turn methylate the DNA in the three regulatory regions of the Oct4 gene. As a separate mechanism, retinoic acid induces GCNF (early period) and ARP-1/COUP-TFII and EAR-3/COUP-TFII (late period), both of which bind to the PP. GCNF recruits Dnmt3a and Dnmt3b to the PP, contributing to suppression of the Oct 4 gene. In contrast, LRH-1 binds to the PE and the PP and activates Oct4 in undifferentiated ES cells through an uncharacterized mechanism.
When somatic cells are dedifferentiated and acquire pluripotency, their Oct4 locus is expected to become active, accompanied by DNA demethylation. Thus, DNA demethylation of the Oct4 locus has been routinely monitored in several models of cell or nuclear dedifferentiation. For instance, the Oct4 locus of somatic cells undergoes demethylation when the cells are fused with ES cells and reprogrammed to a pluripotent state (Kimura et al., 2004). Demethylation is also observed upon injection of somatic nuclei into oocytes (Byrne et al., 2003) and upon incubation of somatic nuclei in embryonal carcinoma cell extract (Freberg et al., 2007). The demethylation of the three regulatory regions is commonly examined to evaluate the similarity and difference between iPS cells and ES cells. The first protocol used to selectively grow iPS cells demonstrated only partial demethylation of the Oct4 regulatory regions (Takahashi and Yamanaka, 2006), which was one of the challenges to improving the protocol. Subsequent protocols enabled selection of iPS cells with fully demethylated Oct4 and other pluripotency genes (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007).
Orphan nuclear receptors that regulate Oct4 transcription
Several nuclear orphan receptors bind to the aforementioned three regulatory elements to activate or suppress Oct4 transcription in mouse ES cells (Mullen et al., 2007) (Fig. 1). The best characterized orphan nuclear receptor involved in the regulation of Oct4 transcription is germ cell nuclear factor (GCNF). This protein is rapidly upregulated during differentiation of ES cells by retinoic acid and suppresses Oct4 transcription through binding to the PP and recruiting Dnmt3a and Dnmt3b (Gu et al., 2005b; Sato et al., 2006). The suppressive role of GCNF is physiologically important to restrict Oct4 expression in the germ cell lineage in mouse embryos (Fuhrmann et al., 2001). Two other orphan receptors, ARP-1/COUP-TFII and EAR-3/COUP-TFI, also bind to the PP and suppress Oct4 transcription (Schoorlemmer et al., 1994; Sylvester and Scholer, 1994; Ben-Shushan et al., 1995). These two proteins are also increased upon treatment of embryonal carcinoma cells with retinoic acid. However, they may not be the primary regulators of Oct4 suppression since they are expressed after Oct4 has already begun to be downregulated by retinoic acid (Ben-Shushan et al., 1995; Mullen et al., 2007). Another orphan receptor LRH-1, which stands for liver receptor homolog 1, activates Oct4 transcription through binding to the PP and the PE but the mechanism of gene activation remains unknown (Gu et al., 2005a). Consistent with its specific binding to the PE, disruption of the LRH-1 gene leads to the loss of Oct4 expression in the epiblast but not in the ICM. In addition, ES cells derived from the ICM of LRH-1−/− blastocysts can remain undifferentiated. However, their expression of Oct4 decreases more rapidly than that of wild-type ES cells when leukemia inhibitory factor (LIF), a key cytokine for self-renewal, is removed from the culture medium. Collectively, several orphan nuclear receptors have been validated for their roles in regulating Oct4 expression but their connections with epigenetic modifications, in particular histone modifications and binding of other chromatin proteins, have not been fully elucidated.
Histone modifications and Oct4 regulation
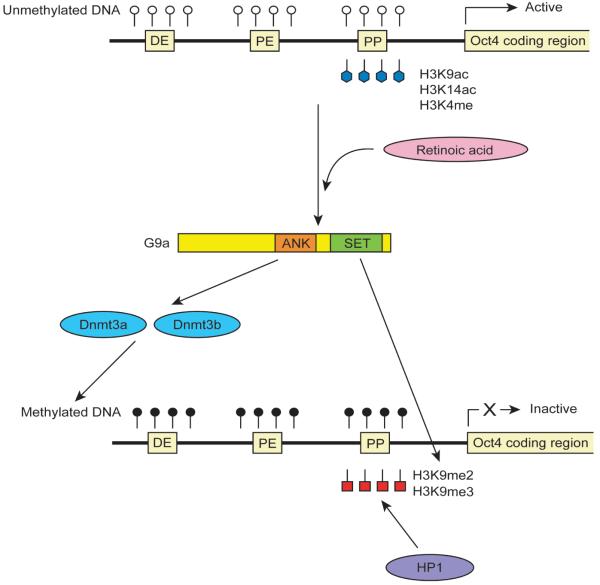
Several histone modifications take place at the PP when Oct4 becomes inactive during early differentiation of mouse ES cells and embryonal carcinoma cells (Fig. 2). Acetylation of lysine 9 and 14 and methylation of lysine 4 on histone H3 are generally markers of transcriptionally active genes, and these histone modifications begin to be removed within 24 hrs after retinoic acid is added to the culture medium to induce differentiation (Feldman et al., 2006). In contrast, methylation of lysine 9 on histone H3, which is a marker of inactive genes, increases during the same period. DNA methylation of the regulatory elements occurs later, between 72 and 96 hrs after addition of retinoic acid, and is therefore thought to stabilize already inactivated Oct4.
Fig. 2.
G9a independently induces DNA methylation and histone H3 methylation of the Oct4 gene through its two domains. Addition of retinoic acid to embryonal carcinoma cells induces DNA methylation of the Oct4 gene through the ankyrin repeat (ANK) of G9a as well as di- and trimethylation of lysine 9 on histone H3 (H3K9me2 and H3K9me3) through the SET domain of G9a. H3K9me2 and H3K9me3 recruit HP1 and trigger heterochromatin formation at the Oct4 locus. During these processes, acetylation of lysine 9 and lysine 14 on histone H3 (H3K9ac and H3K14ac) and methylation of lysine 4 on histone H3 (H3K4me) are lost, which is independent of G9a.
The best characterized histone modifying enzyme in the context of Oct4 suppression is the methyltransferase G9a, which is necessary for di- and trimethylation of histone H3 at the PP (Tachibana et al., 2002; Feldman et al., 2006). G9a directly induces primarily dimethylation of lysine 9; therefore, the trimethylation of lysine 9 could be an indirect effect through recruitment of another unidentified histone methyltransferase to the PP. These lysine methylations attract the heterochromatin protein HP1, resulting in heterochromatin formation and silencing of Oct4. G9a also induces DNA methylation at the regulatory elements but this is independent of lysine 9 methylation. This is because G9a with an inactivated methyltransferase domain (SET domain) can still induce DNA methylation by recruiting Dnmt3a and Dnmt3b to the regulatory elements depending on the ankyrin repeat sequence on G9a (Epsztejn-Litman et al., 2008). The two independent functions of G9a have been also reported with retrotransposons and other target genes of G9a (Dong et al., 2008; Tachibana et al., 2008).
When LIF is removed from the culture medium of mouse ES cells, the cells become differentiated and Oct4 becomes inactive. But Oct4 can be readily reactivated in differentiated cells derived from G9a−/− ES cells once LIF is added back to the culture medium, unlike wild-type ES cells (Feldman et al., 2006). In addition, G9a−/− ES cell nuclei are more easily reprogrammed than wild-type nuclei to support early development in somatic cell nuclear transfer experiments (Epsztejn-Litman et al., 2008). These in vivo findings indicate a key role of G9a in suppressing Oct4 and potentially other pluripotency genes.
Mechanistic link between cytokine signaling and Oct4 transcription
LIF is the best characterized cytokine essential for self-renewal of mouse ES cells (Burdon et al., 1999; Chambers and Smith, 2004). LIF, a member of the IL-6 family of cytokines, binds to its receptor and induces dimerization of the receptor with the membrane protein gp130. This heterodimer activates receptor-associated Janus kinases (JAKs), which phosphorylate tyrosine 705 of STAT3. Phosphorylated STAT3 then enters the nucleus and binds to promoters of its target genes to regulate their expression. Phosphorylation of STAT3 is sufficient for self-renewal of mouse ES cells without LIF but this requires other components derived from serum (Niwa et al., 1998; Matsuda et al., 1999). Currently, target DNA sequences for phosphorylated STAT3 in mouse ES cell are not well characterized. It is also unknown whether phsophorylated STAT3 is directly involved in the regulation of Oct4 transcription. Given the central role of LIF and Oct4 for self-renewal and pluripotency of mouse ES cells, the missing link between these two molecules appears to be one of the most important unanswered questions in the field. Even less clear is the connection between Oct4 transcription and two other cytokine signaling pathways, the BMP pathway and the Wnt pathway, whose contributions to self-renewal and pluripotency are poorly characterized compared to the LIF pathway in mouse ES cells (Chambers and Smith, 2004; Boiani and Scholer, 2005).
Although many differences may exist in Oct4 regulation between mouse and human ES cells, there is clear evidence of such differences in cytokine signaling. In stark contrast to mouse ES cells, the LIF-STAT3 signaling pathway, though active in human ES cells, is not necessary for self-renewal of these cells (Daheron et al., 2004; Humphrey et al., 2004). Virtually nothing is known as to why this is the case. Self-renewal of human ES cells is more dependent on other cytokines, such as basic fibroblast growth factor (Chase and Firpo, 2007). How different cytokine signaling pathways converge on the same transcriptional activation of Oct4 is also one of the fundamental questions in ES cell biology.
The Paf1 complex and Oct4 activation
Little was known until recently about how active Oct4 transcription is maintained in ES cells. This was partly due to the lack of an effective in vitro model to study Oct4 activation as a counterpart to the well-established inactivation model by addition of retinoic acid and removal of LIF. With this background in mind, the recent discovery of the Paf1 complex as an activator of Oct4 is highly significant to efforts to uncover how active Oct4 transcription is maintained in ES cells (Ding et al., 2009). The Paf1 complex binds to the PP of the Oct4 locus but is lost during differentiation of mouse ES cells, which appears to be due to the rapid downregulation of each of the five subunits of the complex. Knockdown of each subunit decreases Oct4 mRNA and induces differentiation of ES cells. In a complementary approach, overexpression of a subunit sustains Oct4 expression and blocks differentiation of ES cells. These findings establish a key role for the Paf1 complex in Oct4 activation through direct interaction with its promoter.
The Paf1 complex was originally identified as an RNA polymerase II-binding complex in Saccharomyces cerevisiae (Shi et al., 1997). This complex is ubiquitously expressed and co-localized with RNA polymerase II at the promoters and coding regions, regulating initiation and elongation of transcription (Pokholok et al., 2002). Knockout mice of Hrpt2, encoding the Cdc73 subunit of the complex, die before embryonic day 6.5 (Wang et al., 2008). Blastocysts of these mice do not hatch in vitro and do not form ES cells due to apoptosis. The roles of the Paf1 complex appear to be highly specific to selected genes since depletion of Cdc73 upregulates or downregulates only less than 100 genes each in mouse embryonic fibroblasts (Wang et al., 2008).
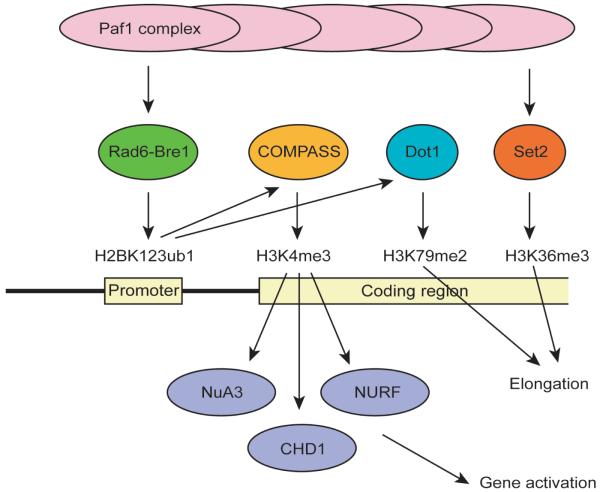
The discovery of a connection between the Paf1 complex and Oct4 locus is significant because the Paf1 complex serves as a platform to organize multiple histone modifications during the initiation and elongation of transcription as demonstrated mainly in S. cerevisiae (Sims et al., 2004; Shilatifard, 2006) (Fig. 3). First, the Paf1 complex promotes monoubiquitination of lysine 123 on histone H2B (H2BK123ub1) at promoters through the Rad6-Bre1 complex. This ubiquitination then results in trimethylation of lysine 4 on histone H3 (H3K4me3) through the COMPASS complex and dimethylation of lysine 79 on histone H3 (H3K79me2) by Dot1. In addition, the Paf1 complex facilitates trimethylation of lysine 36 on histone H3 (H3K36me3) by Set2, which is independent of H2BK123ub1. Among these three histone methylations, H3K4me3 has been most extensively investigated (Sims and Reinberg, 2006; Ruthenburg et al., 2007). H3K4me3 is primarily localized at the 5′ end of active genes and recruits a number of downstream effectors during gene activation. The recruited molecules include the chromatin remodeling ATPases CHD1 and NURF and the acetyltransferase complex NuA3, all of which lead to gene activation (Berger, 2007). H3K79me2 and H3K36me3 are mainly localized in the coding region of transcriptionally active genes and are implicated in elongation (Guenther et al., 2007). These recent advances have enriched the tools available to investigate how transcriptional activation of the Oct4 gene is maintained in ES cells and how it is downregulated during differentiation from the point of view of histone modifying enzymes.
Fig. 3.
The Paf1 complex induces a series of histone modifications at the target genes. The Paf1 complex binds to the promoters and coding regions of its target genes. At the promoter it induces H2BK123ub1, which subsequently recruits COMPASS and Dot1. These two enzymes induce H3K4me3 and H3K79me2, respectively. H3K4me3 triggers binding of chromatin modifying enzymes, such as NuA3, CHD1 and NURF, for gene activation. The Paf1 complex also recruits Set2 to induce H3K36me3. These cascades have been mainly studied in Saccharomyces cerevisiae and have not been proven to exist at the Oct4 locus in ES cells.
Synergistic Oct4 activation with other pluripotency factors
Oct4, Nanog and Sox2 synergistically regulate transcription of their target genes. These three factors respectively occupy 3%, 9% and 7% of the promoter regions of approximately 18,000 genes in human ES cells (Boyer et al., 2005). A similar finding was reported with mouse ES cells (Loh et al., 2006). Importantly, at least two of the three factors co-occupy the promoters of more than 300 genes, including the promoters or enhancers of the Oct4, Nanog and Sox2 genes themselves, and activate these genes (Chew et al., 2005; Kuroda et al., 2005; Okumura-Nakanishi et al., 2005; Rodda et al., 2005). This notion of co-occupancy is supported by the presence of adjacent Sox2- and Oct4-binding elements located within enhancers of their target genes. Furthermore, Oct4, Nanog and Sox2 can be present within the same protein complexes (Wang et al., 2006; Liang et al., 2008). These findings suggest that the three transcription factors function in close collaboration with each other and form an autoregulatory feedback loop to sustain their expression level for self-renewal (Jaenisch and Young, 2008).
Recent identification of additional proteins that interact with Oct4 or bind to the Oct4 locus has expanded the potential members involved in the feedback loop. For example, Oct4 and Nanog can form several different complexes that contain histone deacetylase HDAC2 (Wang et al., 2006; Liang et al., 2008), which might be involved in the suppression of Oct4 transcription when it is overexpressed. The zinc-finger transcription factor Sall4 binds to the DE and activates Oct4 (Zhang et al., 2006). T-cell factor-3 (Tcf3), a terminal component of the Wnt signaling pathway, and Oct4 co-occupy more than 1,000 loci, including the PP, and suppresses Oct4 transcription (Cole et al., 2008). Although these factors have been identified as synergistic regulators of Oct4, their precise mechanism for Oct4 regulation remains unclear. Their functions are expected to come down to epigenetic modifications of the Oct4 locus, meaning that there must be connections with histone- and DNA-modifying proteins.
Conclusion
Recent progress in research on pluripotency in ES cells has rapidly expanded our understanding of the transcriptional regulation of the Oct4 gene; however, several key questions remain unanswered concerning the mechanistic links between Oct4 gene regulation and ES cell self-renewal and pluripotency. One of the most important unanswered questions is how LIF signaling is related to Oct4 transcription. Another fundamental question relates to how different cytokine signaling pathways converge on the transcriptional activation of Oct4 and how this may differ between species, namely human and mouse. Although the protein level of Oct4 needs to be precisely controlled in ES cells to maintain self-renewal, little is known about the feedback mechanism responsible for detecting the level of Oct4 mRNA or protein and regulating the level accordingly. Finally, our view of the regulation of Oct4 transcription has become more complex, and it is now apparent that many pluripotency factors are involved in the synergistic regulation of the Oct4 gene. This raises important questions about the combined effects of these regulators on epigenetic modifications of the Oct4 locus. Addressing each of these questions is important for the sake of improving our understanding of the basic science of pluripotency and early development of mouse and human embryos. The discovery of iPS technology, however, has made it even more important to answer these questions in order to improve the efficiency and thus practical usefulness of this highly promising technology.
Acknowledgements
We thank Meri Firpo and Michael Franklin for critical reading of the manuscript. The work in the authors’ laboratory has been supported by the Faculty Seed Grant of the Academic Health Center, Leukemia Research Fund and the NIH grant R01 GM068027 to N.K.
References
- Ben-Shushan E, Sharir H, Pikarsky E, Bergman Y. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol. Cell. Biol. 1995;15:1034–1048. doi: 10.1128/mcb.15.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Youg RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Chambers I, Stracey C, Niwa H, Smith A. Signaling mechanisms regulating self-renewal and differentiation of pluripotent embryonic stem cells. Cells Tissues Organs. 1999;165:131–143. doi: 10.1159/000016693. [DOI] [PubMed] [Google Scholar]
- Byrne JA, Simonsson S, Western PS, Gurdon JB. Nuclei of adult mammalian somatic cells are directly reprogrammed to Oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 2003;13:1206–1213. doi: 10.1016/s0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chase LG, Firpo MT. Development of serum-free culture systems for human embryonic stem cells. Curr. Opin. Chem. Biol. 2007;11:367–372. doi: 10.1016/j.cbpa.2007.06.421. [DOI] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daheron L, Opitz SL, Zaehres H, Lensch MW, Andrews PW, Itskovitz-Eldor J, Daley GQ. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 2004;22:770–778. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, Hubner N, Doss MX, Sachinidis A, Hescheler J, Iacone R, Anastassiadis K, Stewart AF, Pisabarro MT, Caldarelli A, Poser I, Theis M, Buchholz F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schubeler D, Tachibana M, Shinkai Y, Lorincz MC. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell. Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Freberg CT, Dahl JA, Timoskainen S, Collas P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol. Biol. Cell. 2007;18:1543–1553. doi: 10.1091/mbc.E07-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann G, Chung AC, Jackson KJ, Hummelke G, Baniahmad A, Sutter J, Sylvester I, Scholer HR, Cooney AJ. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev. Cell. 2001;1:377–387. doi: 10.1016/s1534-5807(01)00038-7. [DOI] [PubMed] [Google Scholar]
- Gu P, Goodwin B, Chung AC, Xu X, Wheeler DA, Price RR, Galardi C, Peng L, Latour AM, Koller BH, Gossen J, Kliewer SA, Cooney AJ. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol. Cell. Biol. 2005a;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, LeMenuet D, Chung AC, Mancini M, Wheeler DA, Cooney AJ. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol. Cell. Biol. 2005b;25:8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Nishino K, Ko YG, Ohgane J, Tanaka S, Shiota K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- Humphrey RK, Beattie GM, Lopez AD, Bucay N, King CC, Firpo MT, Rose-John S, Hayek A. Maintenance of pluripotency in human embryonic stem cells is STAT3 independent. Stem Cells. 2004;22:522–530. doi: 10.1634/stemcells.22-4-522. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, Meyer J, Hübner K, Bernemann C, Ortmeier C, Zenke M, Fleischmann BK, Zaehres H, Schöler HR. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kimura H, Tada M, Nakatsuji N, Tada T. Histone code modifications on pluripotential nuclei of reprogrammed somatic cells. Mol. Cell. Biol. 2004;24:5710–5720. doi: 10.1128/MCB.24.13.5710-5720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol. Cell. Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, Xu GL. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell. Biol. 2007;27:8748–8759. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell. Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Guidelines and techniques for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;3:595–605. doi: 10.1016/j.stem.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, Heike T, Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen EM, Gu P, Cooney AJ. Nuclear receptors in regulation of mouse ES cell pluripotency and differentiation. PPAR. Res. 2007;2007:61563. doi: 10.1155/2007/61563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes. Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- Pesce M, Scholer HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Sato N, Kondo M, Arai K. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem. Biophys. Res. Commun. 2006;344:845–851. doi: 10.1016/j.bbrc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J, van Puijenbroek A, van Den Eijnden M, Jonk L, Pals C, Kruijer W. Characterization of a negative retinoic acid response element in the murine Oct4 promoter. Mol. Cell. Biol. 1994;14:1122–1136. doi: 10.1128/mcb.14.2.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Sylvester I, Scholer HR. Regulation of the Oct-4 gene by nuclear receptors. Nucleic Acids Res. 1994;22:901–911. doi: 10.1093/nar/22.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wang P, Bowl MR, Bender S, Peng J, Farber L, Chen J, Ali A, Zhang Z, Alberts AS, Thakker RV, Shilatifard A, Williams BO, Teh BT. Parafibromin, a component of the human PAF complex, regulates growth factors and is required for embryonic development and survival in adult mice. Mol. Cell. Biol. 2008;28:2930–2940. doi: 10.1128/MCB.00654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, Ng HH, Lufkin T, Robson P, Lim B. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]