Abstract
Interleukins (ILs) are key mediators of the immune response and inflammatory process. Plasma levels of IL-10, IL-1Ra, and IL-6 are associated with metabolic conditions, show large inter-individual variations, and are under strong genetic control. Therefore, elucidation of the genetic variants that influence levels of these ILs provides useful insights into mechanisms of immune response and pathogenesis of diseases. We conducted a genome-wide association study (GWAS) of IL-10, IL-1Ra, and IL-6 levels in 707 non-diabetic African Americans using 5,396,780 imputed and directly genotyped single nucleotide polymorphisms (SNPs) with adjustment for gender, age, and body mass index. IL-10 levels showed genome-wide significant associations (p<5×10−8) with eight SNPs, the most significant of which was rs5743185 in thePMS1 gene (p=2.30×10−10). We tested replication of SNPs that showed genome-wide significance in 425 non-diabetic individuals from West Africa, and successfully replicated SNP rs17365948 in the YWHAZ gene (p=0.02). IL-1Ra levels showed suggestive associations with two SNPs in the ASB3 gene (p=2.55×10−7), 10 SNPs in the IL-1 gene family (IL1F5, IL1F8, IL1F10, and IL1Ra, p=1.04×10−6 to 1.75×10−6), and 23 SNPs near the IL1A gene (p=1.22×10−6 to 1.63×10−6). We also successfully replicated rs4251961 (p=0.009); this SNP was reported to be associated with IL-1Ra levels in a candidate gene study of Europeans. IL-6 levels showed genome-wide significant association with one SNP (RP11-314E23.1; chr6:133397598; p=8.63×10−9). To our knowledge, this is the first GWAS on IL-10, IL-1Ra, and IL-6 levels. Follow-up of these findings may provide valuable insight into the pathobiology of IL actions and dysregulations in inflammation and human diseases.
Keywords: interleukin, interleukin-10, interleukin-1Ra, interleukin-6, genome-wide association study, African American
Background
Interleukins (ILs) are cytokines that regulate the immune response, and play a crucial role in inflammation. ILs are the key mediators and markers of the inflammatory process in metabolic conditions such as obesity, insulin resistance, type 2 diabetes (T2D), cardiovascular diseases (CVDs), and associated complications (Juge-Aubry et al. 2005; Wellen and Hotamisligil 2005; Shoelson et al. 2006; King 2008; Donath and Shoelson 2011). Of the wide set of ILs produced in humans, we were primarily interested in three: IL-10 (predominantly anti-inflammatory), IL-1Ra (anti-inflammatory), and IL-6 (inflammatory) that have been shown to be associated with several metabolic disorders and CVDs (Charles et al. 2011; Juge-Aubry et al. 2003; Bluher et al. 2005; Cartier et al. 2009; Doumatey et al. 2009; Lai et al. 2009; Doumatey et al. 2010; Fisman and Tenenbaum 2010).
The plasma levels of these three ILs show large inter-individual variations, and there is evidence that their circulating levels are under significant genetic influence. Heritability estimates of the IL levels based on twin studies ranges from 50 to 75% for IL-10 (Westendorp et al. 1997; Reuss et al. 2002; de Craen et al. 2005), 53% for IL-1Ra (de Craen et al. 2005), and 57% for IL-6 (de Craen et al. 2005). Despite the strong heritability estimated for the ILs, there have been few attempts at identifying the specific genetic variants that influence their circulating levels. A few candidate gene studies have tested association with variants in genes encoding the ILs. These studies reported associations of single base change polymorphisms in the IL10 gene with IL-10 levels (Gibson et al. 2001; Reuss et al. 2002; Hohaus et al. 2009), in the IL1Ra and IL1Rβ genes with IL-1Ra levels (Hurme and Santtila 1998; Di Renzo et al. 2007; Rafiq et al. 2007; Reiner et al. 2008), and in the IL6 gene with IL-6 levels (Fishman et al. 1998; Ferrari et al. 2003; Ferrari et al. 2004). A genome-wide linkage analysis in the Quebec Family Study did not find any robustly linked loci for IL-6 levels (Ruchat et al. 2008).
In general, association studies between IL-10, IL-1Ra, and IL-6 levels and polymorphisms in the genes encoding these ILs have reported inconsistent results (Smith and Humphries 2009). More importantly, the identified polymorphisms explain only a small fraction of the genetic contribution; hence the postulation that the production and levels of ILs may be influenced by genes outside their primary coding loci (Reuss et al. 2002). The unbiased approach of the genome-wide association study (GWAS) is a useful strategy for identifying such variants. However and to our knowledge, no GWAS has been conducted on IL-10, IL-1Ra, or IL-6 levels. Here, we report the first GWAS of these three ILs in a population-based sample of African Americans that are disproportionately affected by metabolic conditions (e.g., obesity, T2D, and CVDs) in which ILs are key mediators (Crawford et al. 2010).
Methods
Study population
Individuals included in this study were participants in the Howard University Family Study (HUFS), a population based cohort of families and unrelated African Americans enrolled from the Washington, DC metropolitan area. The study population, data collection procedures, and ethical process have been described in detail elsewhere (Adeyemo et al. 2009). Ethical approval was obtained from the Howard University Institutional Review Board, and written informed consent was obtained from each participant. Data collected from each participant included anthropometric measures, blood pressure, and an overnight fasting blood sample. IL-10, IL-1Ra, and IL-6 levels were measured in plasma using Quantikine ELISA kits (R&D Systems, Minneapolis, Minnesota, USA). For this GWAS, subjects with T2D were excluded.
Genotyping, imputation, and data quality control
Genotyping was done using the Affymetrix Genome-Wide Human SNP Array 6.0 (McCarroll et al. 2008). Data quality filter was performed using PLINK v.1.07 (Purcell et al. 2007). SNPs that had a call rate <95% across all individuals (n=41,885), a minor allele frequency ≤ 0.01 (n=19,154), or a Hardy-Weinberg equilibrium test p < 0.001 (n=6,317) were excluded. A total of 808,465 autosomal SNPs passed quality control filters. The average call rate for the remaining SNPs was 99.5%.
To increase the number of SNPs for association analysis, imputation was done with the program MACH 1.0 (Li, Willer et al. 2010) using both the Yoruba (YRI) and Northern and Western European ancestry (CEU) population data from the 2010 release of the 1000 Genomes Project (http://www.1000genomes.org/) as reference data. The detailed imputation procedure has been described elsewhere (Shriner et al. 2010). We obtained 8,642,787 SNPs consisting of 808,465 directly genotyped and 7,834,322 imputed SNPs. Further quality filters resulted in the exclusion of 50 SNPs based on Hardy-Weinberg equilibrium (p <0.001), 8 SNPs with MAF < 0.01, and 3,245,949 SNPs for which >10% subjects had missing genotypes. Finally, 5,396,780 autosomal SNPs that passed quality control filters were analyzed in 707 unrelated individuals. Of these individuals, 704 had measured values for plasma levels of IL-10, 706 for IL-1Ra, and 697 for IL-6. Power of the study was calculated using the GWAPower statistical program designed for quantitative trait GWAS (Feng et al. 2011). Assuming LD (r2) between a SNP and causative genetic variant of 0.5, estimated “heritability” (i.e., IL level variation explained by genetic variants) of 0.04, and 20% variance explained by the five covariates, the power of the GWAS was estimated to be ~80% (Supplementary Figure 1).
Population stratification was assessed using a series of procedures. First, the AWClust Algorithm (Gao and Starmer 2008) was employed to investigate clustering of genotypes. All study subjects formed a single cluster; seven individuals that were outliers to this cluster were excluded. Second, the genomic control method was used to examine inflation of type I error rate. The genomic inflation factor was 0.996 for IL-10, 1.015 for IL-1Ra, and 1.003 for IL-6 indicating that population stratification had minimal effect. Third, the quantile-quantile (QQ) plots of distribution of the test statistic were examined and showed minimal population stratification in the sample (Supplementary Figure 2). Principal components (PCs) were computed using the eigenstrat method (Price et al. 2006).The first two PCs were retained and used as covariates during association analysis to adjust for any potential residual population stratification.
Statistical analysis
Normality of IL-10, IL-1Ra, and IL-6 levels was assessed using descriptive statistics and distribution plots (histograms and scatter plots) in SPSS Statistics v.17 program (Supplementary Figure 3). None of the ILs was normality distributed; therefore, we log10-transformed them prior to genome-wide analysis. The analysis was performed in PLINK v.1.07 using linear regression models under the additive genetic model with adjustment for age, sex, BMI, and the first two PCs as covariates. Genome-wide significance was set as p< 5×10−8. The top scoring SNPs for each phenotype were further annotated to understand the genomic context for each significant finding (Ge et al. 2008).
We attempted to replicate our findings in an independent sample of 425 non-diabetic individuals from West Africa who participated in the Africa America Diabetes Mellitus Study (Rotimi et al. 2004). Seven of the eight SNPs that reached genome-wide significance with IL-10 levels were genotyped in the replication sample (Supplementary Table 1). Of these, three passed our quality control filters and were included in the final analysis with adjustment for age, sex, and BMI.
Finally, combined analysis (or meta-analysis) of the discovery and replication datasets was done for the SNPs typed in both cohorts using METAL software program (Willer et al. 2010). We combined the evidence of association in two sided p-values using a fixed-effects model. Sample size of each SNP was used as a weight, and the sign of the beta value (coefficient of the linear regression model adjusted for sex, age, and BMI) was used as the direction of association of the minor allele. Evidence for the between-study heterogeneity was tested using Cochran’s Q statistic and I2, and the p-value of the chi-square was used to declare significance (Higgins et al. 2003).
Results
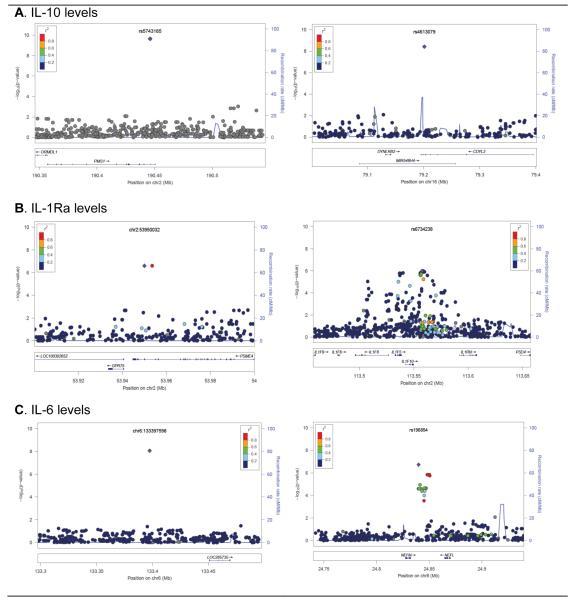
Characteristics of the study subjects are presented in Table 1. Women had higher BMI, IL-6 and IL-1Ra levels, and lower IL-10 levels than men. The relationships between IL levels and other variables (including age, gender, and BMI) are presented in Supplementary Table 2. Age, gender, and BMI were significantly associated with IL-6 and IL-1Ra levels in both univariate and multivariate analyses. Chromosomal distributions of the p-values (Manhattan plots) are shown in Supplementary Figure 4, and regional plots of loci highlighted in the study are shown in Figure 1. The top ranking SNPs with p<10−6 in the GWAS are presented in Table 2, and the top 30 SNPs for each IL are presented in Supplementary Table 3.
Table 1.
Characteristics of African American subjects included in this genome-wide association study.
| Mean ± s.d. |
||||
|---|---|---|---|---|
| Characteristics | Overall (n=707) | Men (n=307) | Women (n=400) | p valuea |
| Age (years) | 42.9 ± 10.7 | 43.1 ± 11.0 | 42.7 ± 10.5 | 0.555 |
| BMI(Kg/m2) | 29.9 ± 8.0 | 28.2 ± 6.9 | 31.2 ± 8.6 | <0.0001 |
| IL-10(pg/ml) | 10.4±3.4 | 10.7±3.2 | 10.2±3.4 | 0.052 |
| IL-1Ra(pg/ml) | 399.2±256.3 | 358.2±222.0 | 430.6±276.0 | <0.0001 |
| IL-6(pg/ml) | 1.8±1.8 | 1.5±1.5 | 2.0±2.0 | <0.0001 |
p value shows significance of mean difference between men and women
Figure 1. Regional plots of “top hits” identified in the GWAS.
SNPs are plotted by chromosomal position (in megabases, according to build hg18) on the x-axis against association (−10Log10 p value) with IL-10, IL-1Ra, and IL-6 levels on the y-axis. Two loci are shown for each of the interleukins. Light blue lines show recombination rates taken from HapMap YRI to reflect local LD structure around the associated SNP. The lower panel shows gene annotations taken from the UCSC genome browser. The purple diamond represents the SNP that shows the strongest association in the plotted region. The LD of other SNPs and the best ranking SNP in the plotted region is shown in a scale from minimum (blue) to maximum (red).
Table 2.
Top SNPs associated with IL-10, IL-1Ra and IL-6 plasma levels among African Americans with p value <10−6.
| IL | SNP | Chr | Position (bp) |
Closest gene | Distance to gene (bp) |
Minor allele |
MAF | β | p value |
|---|---|---|---|---|---|---|---|---|---|
| IL-10 | rs5743185 | 2 | 190737838 | PMS1 | 0 | T | 0.16 | −0.07 | 2.30 ×10−10 |
| rs4613079 | 16 | 80643957 | CDYL2 | 0 | T | 0.19 | −0.06 | 5.43 ×10−10 | |
| chr22:18133848 | 22 | 18133848 | BCL2L13 | 0 | G | 0.14 | −0.07 | 1.73 ×10−9 | |
| rs2238776 | 22 | 19757892 | TBX1 | 0 | A | 0.18 | −0.06 | 3.70 ×10−9 | |
| rs12279202 | 11 | 9432090 | IPO7 | 0 | A | 0.14 | −0.06 | 1.82 ×10−8 | |
| rs17365948 | 8 | 101956877 | YWHAZ | 0 | A | 0.13 | −0.06 | 2.56 ×10−8 | |
| rs16877320 | 6 | 15923026 | ARPC3P5 | −11987 | G | 0.15 | −0.06 | 2.82 ×10−8 | |
| rs11225547 | 11 | 102967852 | DCUN1D5 | −4908 | C | 0.05 | −0.08 | 4.24 ×10−8 | |
| rs13201744 | 6 | 6126845 | F13A1 | 17473 | A | 0.18 | −0.05 | 7.32 ×10−8 | |
| chr22:18130773 | 22 | 18130773 | BCL2L13 | 0 | C | 0.17 | −0.06 | 7.67 ×10−8 | |
| rs2238773 | 22 | 19752973 | TBX1 | 0 | T | 0.18 | −0.06 | 7.99 ×10−8 | |
| chr22:18132943 | 22 | 18132943 | BCL2L13 | 0 | C | 0.18 | −0.06 | 1.21 ×10−7 | |
| rs1199964 | 13 | 37632651 | FAM48A | 0 | A | 0.25 | 0.04 | 1.32 ×10−7 | |
| rs1211337 | 13 | 37617299 | FAM48A | 0 | A | 0.25 | 0.04 | 1.34 ×10−7 | |
| rs17054796 | 13 | 37570731 | ALG5 | 0 | C | 0.25 | 0.04 | 2.01 ×10−7 | |
| rs198284 | 7 | 24265343 | AC004485.3 | 0 | C | 0.04 | 0.09 | 2.26 ×10−7 | |
| chr13:36436587 | 13 | 36436587 | DCLK1 | 0 | C | 0.25 | 0.04 | 2.61 ×10−7 | |
| chr13:36435315 | 13 | 36435315 | DCLK1 | 0 | T | 0.26 | 0.04 | 3.73 ×10−7 | |
| rs17054831 | 13 | 37619600 | FAM48A | 0 | G | 0.23 | 0.04 | 3.81 ×10−7 | |
| chr13:36429834 | 13 | 36429834 | DCLK1 | 0 | T | 0.26 | 0.04 | 4.07 ×10−7 | |
| chr13:36463694 | 13 | 36463694 | DCLK1 | 0 | G | 0.26 | 0.04 | 4.08 ×10−7 | |
| rs8000672 | 13 | 37554345 | ALG5 | 0 | C | 0.25 | 0.04 | 4.27 ×10−7 | |
| rs7989205 | 13 | 37570182 | ALG5 | 0 | C | 0.31 | 0.04 | 4.54 ×10−7 | |
| chr13:36435082 | 13 | 36435082 | DCLK1 | 0 | A | 0.25 | 0.04 | 5.30 ×10−7 | |
| rs8001724 | 13 | 37549928 | ALG5 | 0 | G | 0.25 | 0.04 | 5.53 ×10−7 | |
|
| |||||||||
| IL-1Ra | chr2:53950032 | 2 | 53950032 | ASB3 | 0 | C | 0.02 | 0.23 | 2.55 ×10−7 |
| chr2:53953538 | 2 | 53953538 | ASB3 | 0 | T | 0.02 | 0.23 | 2.55 ×10−7 | |
| rs1917805 | 10 | 50805983 | CHAT | −11158 | G | 0.10 | 0.09 | 6.09 ×10−7 | |
| rs12452028 | 17 | 78212231 | SLC26A11 | 0 | C | 0.15 | 0.08 | 7.49 ×10−7 | |
|
| |||||||||
| IL-6 | chr6:133397598 | 6 | 133397598 | RP11-314E23.1 | −11621 | T | 0.01 | −0.52 | 8.63 ×10−9 |
| rs196854 | 8 | 24783980 | NEFM | 7373 | G | 0.07 | −0.17 | 1.86 ×10−7 | |
| rs7623146 | 3 | 63703985 | AC136289.1 | 23984 | A | 0.10 | −0.14 | 6.58 ×10−7 | |
| rs4473152 | 15 | 78381660 | SH2D7 | 0 | G | 0.14 | 0.12 | 8.15 ×10−7 | |
Eight SNPs were associated with IL-10 levels with genome-wide significant p-values (p<5×10−8). The SNPs that achieved genome-wide significance were rs5743185 (PMS1; p=2.30×10−10), rs4613079 (CDYL2; p=5.43 × 10−10), chr22:18133848 (BCL2L13; p=1.73×10−9), rs2238776 (TBX1; p= 3.70×10−9), rs12279202 (IPO7; p=1.82×10−8), rs17365948 (YWHAZ; p=2.56×10−8), rs16877320 (near ARPC3P5; p=2.82×10−8), and rs11225547 (near DCUN1D5; p=4.24×10−8 ).
No SNP reached genome-wide significance for IL-1Ra levels. The top two SNPs showing suggestive association with IL-1Ra levels (p=2.55×10−7) were in the ASB3 gene. Notable among the other top IL-1Ra associated SNPs was a cluster of 10 SNPs within a 21.8 kb interval on chromosome 2q14 that harbors the IL-1 gene family (IL1F5, IL1F8, IL1F10, and IL1Ra; p=1.04 ×10−6 to 1.75 ×10−6) (Figure 1). When we repeated the analysis conditioning on rs6734238, the best associated SNP in the cluster and the nearest SNP to the IL1Ra gene, the association with the other seven SNPs disappeared, indicating that the association is likely to be solely driven by this variant. A set of 23 SNPs spanning a 2.2kb region at a physical distance of 13-17kb from the IL1A gene also showed suggestive association with IL-1Ra levels (p=1.22×10−6 to 1.63×10−6). However, the association of the 22 SNPs disappeared when the analysis was repeated conditioning on the lead SNP at position chr2:113557312.
The best hit for IL-6 levels was on chromosome 6 (chr6:133397598; p=8.63×10−9). The magnitude of effect of the association with this variant was also the strongest; having one copy of the T allele of this variant resulted in lower log10-transformed IL-6 levels (beta = 0.52 Z-score units of log10-transformed IL-6 levels). Other suggestive associations include rs196854 (near NEFM; p=1.86×10−7) and rs4473152 (SH2D7; p=8.15×10−7).
In the West African replication dataset (refer to Supplementary Table 4 for descriptive statistics), IL-10 levels were significantly associated with rs17365948 (YWHAZ; p=0.02) and marginally associated with rs12279202 (IPO7; p=0.09). The direction of both associations was the same as that in the GWAS and the effect was stronger in the replication dataset compared to that observed in the discovery dataset (Supplementary Table 5). Both SNPs showed genome-wide significance in meta-analysis, with rs17365948 showing stronger evidence in the combined analysis compared to the GWAS (p combined= 6.91 ×10−9 vs. p discovery=2.56 ×10−8). Cochran’s Q statistic and I2 tests showed high heterogeneity between the individual datasets, especially marked for rs2238776 that showed no association in the replication dataset (Table 3).
Table 3.
Meta-analysis results of three SNPs associated with IL-10 levels in the combined sample of African Americans and West Africans.
| Discovery | Replication | Meta-analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||
| SNP | Nearest gene |
Chr | Position (bp) |
Minor allele |
MAF | p value | β |
p value |
β | Z- score |
p value | Dir | Het I2 | p het |
| rs17365948 | YWHAZ | 8 | 101956877 | T | 0.08 | 2.56 ×10−8 | −0.06 | 0.025 | −0.13 | −5.79 | 6.91 ×10−9 | - | 61.4 | 0.107 |
| rs12279202 | IPO7 | 11 | 9432090 | T | 0.09 | 1.82 ×10−8 | −0.06 | 0.095 | −0.33 | −5.50 | 3.88 ×10−8 | - | 76.8 | 0.038 |
| rs2238776 | TBX1 | 22 | 19757892 | A | 0.12 | 2.33 ×10−9 | −0.06 | 0.555 | +0.05 | −4.36 | 1.31 ×10−5 | − | 93.8 | 5.94 ×10−5 |
We also searched the literature for SNPs associated with IL-10, IL-1Ra, and IL-6 levels. One study focusing on the IL1Ra gene in subjects from Tuscany, Italy reported significant association of IL-1Ra levels with five SNPs (rs4251961, rs579543, rs2637988, rs315943, and rs1374281) (Rafiq et al. 2007). We successfully replicated association of IL-1Ra levels with the top associated SNP rs4251961 (p=0.009, β (C allele) =−0.04), that tagged all others in the reported study. Consistent with a candidate gene study that reported association between two IL6 gene SNPs (rs1800795 and rs1554606) with IL-6 plasma levels in Caucasians but not in African Americans (Walston et al. 2007), we did not observe significant association with these SNPs in our African American cohort.
Discussion
In this first GWAS for IL-10, IL-1Ra, and IL-6 plasma levels in African Americans, we found genome-wide significant associations for IL-10 and IL-6 and suggestive associations for IL-1Ra levels. The strength of association in most of the variants was similar after adjusting for covariates including age, sex, and BMI. This is in contrast to, but not directly comparable with, a previously reported genome-wide linkage study that showed suggestive evidence of linkage for IL-6 levels on chromosome 14q12 (D14S1280) which disappeared after adjustment for BMI (Ruchat et al. 2008). The finding that these associations were independent of BMI suggests that the influence of the identified variants on IL levels is direct, rather than being mediated through adiposity.
We identified genome-wide significant associations of IL-10 levels with several SNPs. We replicated one of these SNPs located in the YWHAZ gene using an independent sample from West Africa, and further observed that the evidence for association improved in a combined analysis of both datasets (i.e., African American and West African combined). The strongest association of IL-10 levels was with a SNP in the PMS1 gene that encodes a protein involved in the repair of DNA mismatches. This finding is consistent with that of two studies that showed a link between the PMS1 and IL10 molecules. A co-immunoprecipitation experiment in HeLa cell lines showed that in humans PMS1 binds with CYLD protein (Cannavo et al. 2007). CYLD is a deubiquitinating enzyme which attenuates NF-κB and c-Jun NH2-terminal kinase (JNK) signaling. In transgenic mouse CYLDex7/8 protein increased expression of mouse Il10 mRNA in cultured bone-marrow-derived dendritic cells (Bros et al. 2010). Further studies are needed to understand the functional roles of the genome-wide associated SNPs in PMS1, IPO7 (encodes a protein that mediates nuclear import of proteins with a classical nuclear localization signal), YWHA (encodes a protein that is a member of a highly conserved protein family that mediates signal transduction by binding to phosphoserine-containing proteins), and CDYL2 (encodes a protein involved in catalytic activity and protein binding).
Interestingly, common variants in PMS1, IPO7, YWHAZ, and CDYL2 genes that were associated with IL-10 levels in the present study have also been associated with systolic blood pressure (SBP) in a GWAS conducted in the same cohort (Adeyemo et al. 2009). The PMS1 gene was also associated with SBP and hypertension in the CHARGE consortium GWAS (Levy et al. 2009). The associations we identified between IL-10 levels and these variants were not modified with adjustment for SBP and/or hypertension status in the logistic regression models. Moreover, there is virtually no correlation between IL-10 levels and SBP in this cohort of African Americans (r=−0.04, p=0.318), suggesting that the observed IL-10 associations were not being driven by SBP. Therefore, we postulate that there may be a small set of loci that co-regulate both IL-10 and SBP. Animal model studies provide evidence that manipulation of IL-10 levels affects blood pressure levels. For example, treatment of hypertensive pregnant rats with exogenous IL-10 leads to lowering of SBP, normalization of plasma endothelin-1 levels, and vascular relaxation (Tinsley et al. 2010). Another study showed that IL-10 deficiency and hypoxia in a mouse model leads to excessive activation of anti-angiogenic and apoptotic pathways, resulting in severe preeclampsia-like disease in which hypertension is a defining characteristic (Lai, Kalkunte et al. 2011). Our findings are consistent with the above studies that imply a pathophysiological relationship between IL-10 levels and blood pressure, and suggest the possibility of a common genetic influence on both traits.
We found suggestive association and relatively strong effect size between IL-1Ra levels and two variants in the ASB3 gene. The protein encoded by the ASB3 gene is a member of the ankyrin repeat and suppressor of cytokine signaling (SOCS) box-containing (ASB) protein family that suppresses cytokine signaling by inhibiting the Janus kinase and signal transducers and activators of transcription (JAK-STAT) pathway (Kile et al. 2001). Our study showed that higher IL-1Ra levels are associated with the minor alleles of the two associated SNPs; this observation suggests that these variants may down-regulate expression of the ASB3 gene thereby modulating increased expression of the IL-1Ra cytokine. Several variants that were suggestively associated with IL-1Ra levels are located in the IL-1 gene family clustering near or in IL1F5, IL1F8, IL1F10, and IL1Ra genes. Our best scoring SNP in the cluster, rs6734238, is 3.8kb upstream of the IL1Ra gene that encodes the IL-1Ra protein. It is known that IL-1Ra modulates a variety of anti-inflammatory and immune responses by competing with IL-1 alpha and IL-1 beta for binding to the IL-1 receptor type I (Mulero et al. 1999). The IL-1 gene family that had multiple SNPs associated with IL-1Ra levels in our study is known to be largely conserved in mammals (Sims and Smith 2010). Our finding concurs with that of previous candidate gene studies which showed that IL-1Ra levels in healthy individuals are influenced by alleles of the IL1Ra and IL1β genes (Hurme and Santtila 1998; Rafiq et al. 2007). One of the associated SNPs in the IL-1 gene family, rs6758965, was located in the intron of the IL1F8 gene, a recently discovered member of the IL-1 gene family which encodes a protein known to have inflammatory effects in joint cells (Yang et al. 2010; Nicklin, et al. 2002).
We successfully replicated rs4251961, the SNP reported as most strongly associated with IL-1Ra levels among European populations (Rafiq et al. 2007). A cluster of SNPs around the IL1A gene were associated with IL-1Ra levels. IL1A is a member of the IL-1 cytokine family that encodes the pro-inflammatory cytokine IL-1α. Molecular studies have shown that IL-1α and IL-1Ra bind competitively to IL-1 receptors thereby inducing pro- and anti-inflammatory responses, respectively (Juge-Aubry et al. 2003). Our study provides additional evidence supporting the observation that variants of the IL1A and IL1Ra genes influence IL-1Ra levels. In all, we showed that genetic variants in the IL-1 gene complex are likely mediators of IL-1Ra plasma levels in humans.
The variant that showed genome-wide significant association with IL-6 levels was 11.6kb downstream from the RP11-314E23.1 gene the function of which has not been clearly described. Given the paramount role of IL-6 in various diseases, replication of the finding in an independent cohort and characterization of the locus may be helpful in understanding the pathogenesis of several diseases. We suspect that studies with larger sample sizes will likely improve the limitation of our study and detect additional loci with smaller effects for all three ILs. Although we included only apparently healthy subjects, we acknowledge that apart from adjusted factors such as age, sex, and BMI, a residual confounding effect of an underlying disease or socio-economic status cannot be completely ruled out. In fact, high levels of oxidative stress and inflammatory markers including cytokines have been reported in the African American population (Feairheller et al. 2011). Further studies are needed to understand the complex interaction of genetic and socio-environmental factors that are associated with chronic stress and low grade inflammation that in turn influence the level of ILs (Baltrus at al. 2010).
Generated by pathogen-driven selection, the extreme polymorphisms within IL genes modulate their biological functions and expression levels. Our study corroborates others’ that showed that the presence of such selection polymorphisms is more common in the IL-1 gene family than in IL6 and IL10 genes (Fumagalli et al. 2009). Unlike intragenic variations, the involvement of extragenic variations in the regulation of IL expression is not well studied but could be a novel evolutionary adaptation of cytokine regulation to the rapidly evolving mechanisms developed by pathogens to evade the classical immune system in humans. As a result of involvement of multiple loci, selective absence of one or more regulatory loci can be compensated by presence of others ensuring maintenance of the immune response (Sims and Smith 2010).
In conclusion, we observed significant associations between several genomic loci that may modulate circulating levels of IL-10, IL-1Ra, and IL-6 in African Americans. Several of the genes harboring variants associated with IL-10, IL-1Ra, and IL-6 levels are involved in inflammation, cell signaling, angiogenesis, viral infections, and different forms of cancer (http://www.genecards.org/). This observation is consistent with the hypothesis that ILs play key roles in the progression of several diseases, and points to the relevance of investigating possible common pathways mediated by these ILs to understand the pathogenesis of complex diseases.
Supplementary Material
Acknowledgements
The study was supported by grants S06GM008016-320107 to CR and S06GM008016-380111 to AA, both from the NIGMS/MBRS/SCORE Program. Participant enrollment was carried out at the Howard University General Clinical Research Center (GCRC), which is supported by grant number 2M01RR010284 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Additional support was provided by the Coriell Institute for Biomedical Sciences. This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, in the Center for Research in Genomics and Global Health (Z01HG200362). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Adeyemo A, Gerry N, Chen G, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus PT, Shim RS, Ye J, Watson L, Davis SK. Socioeconomic position, stress, and cortisol in relation to waist circumference in African American and white women. Ethn Dis. 2010;20:376–82. [PMC free article] [PubMed] [Google Scholar]
- Bluher M, Fasshauer M, Tonjes A, Kratzsch J, Schon MR, Paschke R. Association of interleukin-6, C-reactive protein, interleukin-10 and adiponectin plasma concentrations with measures of obesity, insulin sensitivity and glucose metabolism. Exp Clin Endocrinol Diabetes. 2005;113:534–7. doi: 10.1055/s-2005-872851. [DOI] [PubMed] [Google Scholar]
- Bros M, Dexheimer N, Besche V, Masri J, Trojandt S, Hövelmeyer N, Reissig S, Massoumi R, Grabbe S, Waisman A, Reske-Kunz AB. Mutated cylindromatosis gene affects the functional state of dendritic cells. Eur J Immunol. 2010;40:2848–57. doi: 10.1002/eji.200939285. [DOI] [PubMed] [Google Scholar]
- Cannavo E, Gerrits B, Marra G, Schlapbach R, Jiricny J. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J Biol Chem. 2007;282:2976–86. doi: 10.1074/jbc.M609989200. [DOI] [PubMed] [Google Scholar]
- Cartier A, Bergeron J, Poirier P, et al. Increased plasma interleukin-1 receptor antagonist levels in men with visceral obesity. Ann Med. 2009;41:471–8. doi: 10.1080/07853890903022801. [DOI] [PubMed] [Google Scholar]
- Charles BA, Doumatey A, Huang H, Zhou J, Chen G, Shriner D, Adeyemo A, Rotimi CN. The Roles of IL-6, IL-10, and IL-1RA in Obesity and Insulin Resistance in African-Americans. J Clin Endocrinol Metab epub. 2011:1945–7197. doi: 10.1210/jc.2011-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford AG, Cote C, Couto J, Daskiran M, Gunnarsson C, Haas K, Haas S, Nigam SC, Schuette R. Prevalence of obesity, type II diabetes mellitus, hyperlipidemia, and hypertension in the United States: findings from the GE Centricity Electronic Medical Record database. Popul Health Manag. 2010;13:151–61. doi: 10.1089/pop.2009.0039. [DOI] [PubMed] [Google Scholar]
- de Craen AJ, Posthuma D, Remarque EJ, van den Biggelaar AH, Westendorp RG, Boomsma DI. Heritability estimates of innate immunity: an extended twin study. Genes Immun. 2005;6:167–70. doi: 10.1038/sj.gene.6364162. [DOI] [PubMed] [Google Scholar]
- Di Renzo L, Bigioni M, Del Gobbo V, et al. Interleukin-1 (IL-1) receptor antagonist gene polymorphism in normal weight obese syndrome: relationship to body composition and IL-1 alpha and beta plasma levels. Pharmacol Res. 2007;55:131–8. doi: 10.1016/j.phrs.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11:98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- Doumatey AP, Lashley KS, Huang H, et al. Relationships among obesity, inflammation, and insulin resistance in African Americans and West Africans. Obesity (Silver Spring) 2010;18:598–603. doi: 10.1038/oby.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feairheller DL, Park JY, Rizzo V, Kim B, Brown MD. Racial differences in the responses to shear stress in human umbilical vein endothelial cells. Vasc Health Risk Manag. 2011;7:425–31. doi: 10.2147/VHRM.S22435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010;10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- Feng S, Wang S, Chen CC, Lan L. GWAPower: a statistical power calculation software for genome-wide association studies with quantitative traits. BMC Genet. 2011;12:12. doi: 10.1186/1471-2156-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari SL, Ahn-Luong L, Garnero P, Humphries SE, Greenspan SL. Two promoter polymorphisms regulating interleukin-6 gene expression are associated with circulating levels of C-reactive protein and markers of bone resorption in postmenopausal women. J Clin Endocrinol Metab. 2003;88:255–9. doi: 10.1210/jc.2002-020092. [DOI] [PubMed] [Google Scholar]
- Ferrari SL, Karasik D, Liu J, et al. Interactions of interleukin-6 promoter polymorphisms with dietary and lifestyle factors and their association with bone mass in men and women from the Framingham Osteoporosis Study. J Bone Miner Res. 2004;19:552–9. doi: 10.1359/JBMR.040103. [DOI] [PubMed] [Google Scholar]
- Fishman D, Faulds G, Jeffery R, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–76. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisman EZ, Tenenbaum A. The ubiquitous interleukin-6: a time for reappraisal. Cardiovasc Diabetol. 2010;9:62. doi: 10.1186/1475-2840-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Pozzoli U, Cagliani R, Comi GP, Riva S, Clerici M, Bresolin N, Sironi M. Parasites represent a major selective force for interleukin genes and shape the genetic predisposition to autoimmune conditions. J Exp Med. 2009;206:1395–408. doi: 10.1084/jem.20082779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Starmer JD. AWclust: point-and-click software for non-parametric population structure analysis. BMC Bioinformatics. 2008;9:77. doi: 10.1186/1471-2105-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge D, Zhang K, Need AC, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–3. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166:3915–22. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohaus S, Giachelia M, Massini G, et al. Clinical significance of interleukin-10 gene polymorphisms and plasma levels in Hodgkin lymphoma. Leuk Res. 2009;33:1352–6. doi: 10.1016/j.leukres.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Hurme M, Santtila S. IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1beta genes. Eur J Immunol. 1998;28:2598–602. doi: 10.1002/(SICI)1521-4141(199808)28:08<2598::AID-IMMU2598>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Juge-Aubry CE, Somm E, Giusti V, et al. Adipose tissue is a major source of interleukin-1 receptor antagonist: upregulation in obesity and inflammation. Diabetes. 2003;52:1104–10. doi: 10.2337/diabetes.52.5.1104. [DOI] [PubMed] [Google Scholar]
- Juge-Aubry CE, Henrichot E, Meier CA. Adipose tissue: a regulator of inflammation. Best Pract Res Clin Endocrinol Metab. 2005;19:547–66. doi: 10.1016/j.beem.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Kile BT, Nicola NA, Alexander WS. Negative regulators of cytokine signaling. Int J Hematol. 2001;73:292–8. doi: 10.1007/BF02981953. [DOI] [PubMed] [Google Scholar]
- King GL. The role of inflammatory cytokines in diabetes and its complications. J Periodontol. 2008;79:1527–34. doi: 10.1902/jop.2008.080246. [DOI] [PubMed] [Google Scholar]
- Lai S, Fishman EK, Lai H, Pannu H, Detrick B. Serum IL-6 levels are associated with significant coronary stenosis in cardiovascularly asymptomatic inner-city black adults in the US. Inflamm Res. 2009;58:15–21. doi: 10.1007/s00011-008-8150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Kalkunte S, Sharma S. A critical role of interleukin-10 in modulating hypoxia-induced preeclampsia-like disease in mice. Hypertension. 2011;57:505–14. doi: 10.1161/HYPERTENSIONAHA.110.163329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–87. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Kuruvilla FG, Korn JM, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–74. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- Mulero JJ, Pace AM, Nelken ST, et al. IL1HY1: A novel interleukin-1 receptor antagonist gene. Biochem Biophys Res Commun. 1999;263:702–6. doi: 10.1006/bbrc.1999.1440. [DOI] [PubMed] [Google Scholar]
- Nicklin MJ, Barton JL, Nguyen M, FitzGerald MG, Duff GW, Kornman K. A sequence-based map of the nine genes of the human interleukin-1 cluster. Genomics. 2002;79:718–25. doi: 10.1006/geno.2002.6751. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiq S, Stevens K, Hurst AJ, et al. Common genetic variation in the gene encoding interleukin-1-receptor antagonist (IL-1RA) is associated with altered circulating IL-1RA levels. Genes Immun. 2007;8:344–51. doi: 10.1038/sj.gene.6364393. [DOI] [PubMed] [Google Scholar]
- Reiner AP, Wurfel MM, Lange LA, et al. Polymorphisms of the IL1-receptor antagonist gene (IL1RN) are associated with multiple markers of systemic inflammation. Arterioscler Thromb Vasc Biol. 2008;28:1407–12. doi: 10.1161/ATVBAHA.108.167437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss E, Fimmers R, Kruger A, Becker C, Rittner C, Hohler T. Differential regulation of interleukin-10 production by genetic and environmental factors--a twin study. Genes Immun. 2002;3:407–13. doi: 10.1038/sj.gene.6363920. [DOI] [PubMed] [Google Scholar]
- Rotimi CN, Chen G, Adeyemo AA, et al. A genome-wide search for type 2 diabetes susceptibility genes in West Africans: the Africa America Diabetes Mellitus (AADM) Study. Diabetes. 2004;53:838–41. doi: 10.2337/diabetes.53.3.838. [DOI] [PubMed] [Google Scholar]
- Ruchat SM, Despres JP, Weisnagel SJ, Chagnon YC, Bouchard C, Perusse L. Genome-wide linkage analysis for circulating levels of adipokines and C-reactive protein in the Quebec family study (QFS) J Hum Genet. 2008;53:629–36. doi: 10.1007/s10038-008-0291-1. [DOI] [PubMed] [Google Scholar]
- Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriner D, Adeyemo A, Chen G, Rotimi CN. Practical considerations for imputation of untyped markers in admixed populations. Genet Epidemiol. 2010;34:258–65. doi: 10.1002/gepi.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev. 2009;20:43–59. doi: 10.1016/j.cytogfr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Tinsley JH, South S, Chiasson VL, Mitchell BM. Interleukin-10 reduces inflammation, endothelial dysfunction, and blood pressure in hypertensive pregnant rats. Am J Physiol Regul Integr Comp Physiol. 2010;298:R713–9. doi: 10.1152/ajpregu.00712.2009. [DOI] [PubMed] [Google Scholar]
- Walston JD, Fallin MD, Cushman M, et al. IL-6 gene variation is associated with IL-6 and C-reactive protein levels but not cardiovascular outcomes in the Cardiovascular Health Study. Hum Genet. 2007;122:485–94. doi: 10.1007/s00439-007-0428-x. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–9. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorp RG, Langermans JA, Huizinga TW, Verweij CL, Sturk A. Genetic influence on cytokine production in meningococcal disease. Lancet. 1997;349:1912–3. doi: 10.1016/s0140-6736(05)63910-4. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genome wide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Shen N, Ye DQ, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.