Summary
Remodeling of the peptidoglycan (PG) exoskeleton is intimately tied to the growth and division of bacteria. Enzymes that hydrolyze PG are critical for these processes, but their activities must be tightly regulated to prevent the generation of lethal breaches in the PG matrix. Despite their importance, the mechanisms regulating PG hydrolase activity have remained elusive. Here we investigate the control of cell division hydrolases called amidases (AmiA, AmiB, and AmiC) required for Escherichia coli cell division. Poorly regulated amiB mutants were isolated encoding lytic AmiB variants with elevated basal PG hydrolase activities in vitro. The structure of an AmiB ortholog was also solved, revealing that the active site of AmiB is occluded by a conserved alpha-helix. Strikingly, most of the amino acid substitutions in the lytic AmiB variants mapped to this domain and are predicted to disrupt its interaction with the active site. Our results therefore support a model in which cell separation is stimulated by the reversible relief of amidase auto-inhibition governed by conserved sub-complexes within the cytokinetic ring. Analogous conformational control mechanisms are likely to be part of a general strategy used to control PG hydrolases present within multi-enzyme PG remodeling machines.
Keywords: autolysin, cytokinesis, morphogenesis, murein, sacculus
Introduction
Most bacteria surround themselves with a tough cell wall exoskeleton made of peptidoglycan (PG). This structure is constructed from polysaccharide strands crosslinked to one another via attached peptides to form a three-dimensional meshwork that encapsulates the cytoplasmic membrane and protects it from osmotic rupture (Vollmer & Seligman, 2010; Vollmer et al, 2008a). Because the PG matrix is continuous, its cleavage and expansion is intimately tied to the growth and division of bacterial cells. Also, the unique and essential nature of the PG layer makes its biogenesis an ideal target for antibiotics. Indeed, assembly of the PG layer requires synthetic enzymes called penicillin binding proteins (PBPs), the primary targets of penicillin and related β-lactam drugs, one of our oldest and arguably most effective classes of antibiotics (Typas et al, 2012).
In addition to the PG synthases, bacterial cells also produce an array of hydrolytic enzymes that cleave PG bonds to promote cell wall remodeling events important for cell growth, cell division, and certain developmental processes (Vollmer et al, 2008b). To function in this capacity, however, PG hydrolases must be tightly regulated (Vollmer et al, 2008b; Typas et al, 2012; Uehara & Bernhardt, 2011). Otherwise, aberrant cleavage of the PG layer may result in cell lysis. While this fact has been appreciated for some time, the cellular mechanisms responsible for regulating PG hydrolases have remained largely mysterious (Vollmer et al, 2008b; Typas et al, 2012; Uehara & Bernhardt, 2011). Elucidating these mechanisms is important because they should reveal new ways to subvert the PG biogenesis process for antibiotic development.
Cell division in Escherichia coli has proven to be an excellent model system for studies of PG hydrolase regulation (Uehara et al, 2010; Yang et al, 2011). In most bacteria, the division process begins with the polymerization of the tubulin-like FtsZ protein into a ring structure (the Z-ring) just underneath the cytoplasmic membrane at the prospective site of division (Bi & Lutkenhaus, 1991; de Boer, 2010). This structure then promotes the recruitment of a number of essential and non-essential division proteins to the division site, ultimately organizing the formation of a trans-envelope molecular machine capable of driving cell constriction (de Boer, 2010). An important function of this so called septal ring or divisome machine is the highly localized synthesis of the new (septal) PG material that will ultimately fortify the poles of the daughter cells (Typas et al, 2012). Septal PG appears to be initially shared between the developing daughter cells and must be split by PG hydrolases to shape the poles and complete the division process (Uehara & Bernhardt, 2011). Septal PG splitting in E. coli depends critically on the activities of three LytC-type N-acetylmuramyl-L-alanine amidases (Pfam: Amidase_3): AmiA, AmiB and AmiC (Heidrich et al, 2001; Priyadarshini et al, 2007). Amidases are PG hydrolases that break crosslinks in the PG meshwork by cleaving bonds that link stem peptides to the glycan strands (Vollmer et al, 2008b). The three amidases appear to be largely redundant with mutants lacking individual enzymes dividing essentially normally. However, mutants lacking all three amidases fail to split septal PG and form long chains of cells with distinct cytoplasmic compartments connected by shared layers of PG and a partially constricted outer membrane layer (Heidrich et al, 2001; Priyadarshini et al, 2007).
We previously demonstrated that the cell separation amidases have low basal activities in vitro, and that in order to promote division, these enzymes must be activated by septal ring factors with LytM domains: EnvC and NlpD (Uehara et al, 2010; 2009). In vitro PG hydrolase assays and in vivo genetic analyses indicate that EnvC specifically activates AmiA and AmiB while NlpD specifically activates AmiC (Uehara et al, 2010). Accordingly, mutants lacking both EnvC and NlpD have a chaining phenotype that resembles a triple amidase mutant (Uehara et al, 2009). We recently further showed that EnvC activity is controlled by an ATP-binding cassette (ABC) transporter-like complex composed of the ATPase FtsE and transmembrane domain subunit FtsX (Yang et al, 2011). FtsEX directly recruits EnvC to the septal ring via an interaction between EnvC and a periplasmic loop of FtsX (Yang et al, 2011). Interestingly, FtsEX variants predicted to be ATPase defective still recruit EnvC to the septum but fail to promote cell separation (Yang et al, 2011). Our results therefore support a model in which amidase activation via EnvC in the periplasm is regulated by conformational changes in the FtsEX complex mediated by ATP hydrolysis in the cytoplasm. Importantly, a role for FtsEX in the control of septal PG splitting appears to be conserved as Sham et al. (Sham et al, 2011) have directly connected the essential cell separation factor PcsB (Ng et al, 2004) with FtsEX in Streptococcus pneumoniae.
Although the factors controlling amidase activity have been identified, it has remained unclear how amidase activity is inhibited prior to cell division and how this inhibition is relieved by the EnvC-FtsEX system. Here we use a combination of genetic and structural biology to address this problem. Employing a genetic enrichment strategy based on the release of plasmids from lysing cells, we isolated plasmids encoding poorly regulated (lytic) variants of AmiB. The amino acid substitutions in these AmiB variants led to elevated basal PG hydrolase activity in vitro and mapped to a ~50 residue domain that was found to be unique to cell separation amidases of gram-negative bacteria. When similar substitutions were engineered in related amidases, proper regulatory activity was also disrupted. Structural analysis of an AmiB ortholog from Bartonella henselae, the first structure of a cell separation amidase ortholog, revealed that the identified regulatory domain forms an alpha-helix that occludes the amidase active site. Strikingly, most of the amino acid substitutions in the lytic AmiB variants mapped to this domain and are predicted to disrupt its interaction with the active site. Our results therefore support a model in which amidase activity is blocked prior to cell division by an auto-regulatory domain. Upon assembly of the cytokinetic ring, inhibition is then relieved by a cascade of conformational changes, starting with FtsE-mediated ATP hydrolysis in the cytoplasm and culminating in the EnvC-stimulated release of a regulatory helix from the amidase active site on the opposite side of the membrane. Analogous auto-inhibition and conformational control mechanisms are likely to be part of a general strategy used by multi-enzyme PG remodeling machines to control PG hydrolase activity.
Experimental Procedures
Media, bacterial strains and plasmids
Cells were grown in LB [1% tryptone, 0.5% yeast extract, 0.5% NaCl] or minimal M9 medium (Miller, 1972) supplemented with 0.2% casamino acids and 0.2% sugar (glucose, arabinose or maltose as indicated). Unless otherwise indicated, antibiotics were used at 10 (chloramphenicol; Cm), 15 (ampicillin; Amp) or 20 (kanamycin; Kan) µg/ml. Bacterial strains used are listed in Table S2. All E. coli strains used in the reported experiments are derivatives of MG1655. Plasmids used in this study are listed in Table S3. Those used to identify and characterize the lytic amidase alleles were medium copy number vectors with a p15A origin of replication.
Mutagenesis, plasmid release enrichment, and growth curves
Two independent mutant plasmid libraries were constructed by specifically mutagenizing the amiB gene encoded in pDY187 [Para::amiB] using the GeneMorph II EZClone Domain Mutagenesis Kit from Agilent Technologies. A detailed description of the mutagenesis protocol and the methods used for plasmid release are presented in Supplementary Materials. Please see figure legends for details about growth conditions and sample preparation methods used for specific experiments.
Protein purification, crystallography, and biochemical methods
With the exception of BHAmiB, all proteins were overexpressed and purified with a 6xHis-SUMO (H-SUMO) tag fused to their N-termini (Mossessova & Lima, 2000; Marblestone et al, 2006). The tag was subsequently removed post purification as described previously (Bendezú et al, 2009). For structural characterization of the N-acetylmuramyl-L-alanine amidase catalytic domain, a part of the Bartonella Henselae str. Houston-1 amiB gene encoding the C-terminus of BHAmiB from residues S179 to L409 was cloned into vector pMCSG19B, generating the expression construct pAPC62366.1. Detailed purification and crystallographic methods are described in Supplemental Material. Crystallographic statistics are presented in Table S4. PG hydrolase activity was monitored using a dye-release assay as described previously (Uehara et al, 2010).
Results
Isolation of amiB mutants that induce cell lysis
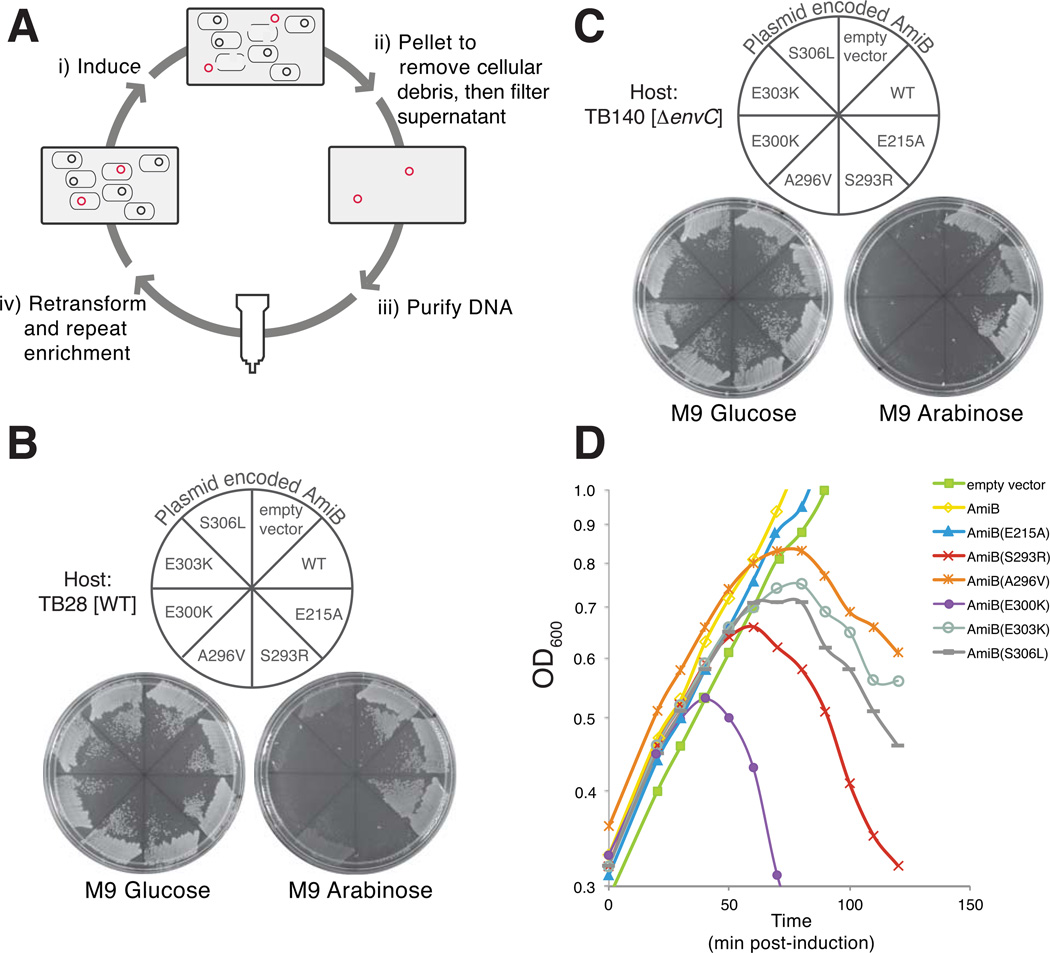
Although we recently showed that the cell separation amidases require activation by their cognate LytM factor to promote cell separation (Uehara et al, 2010), the mechanism of amidase activation has remained unclear. We hypothesized that the amidases are likely to be conformationally regulated and therefore exist in an “OFF” or an “ON” state. Thus, according to this model, the LytM proteins would activate the amidases by promoting the transition to or stabilizing the ON conformation. If this were true, we reasoned that we should be able to isolate amiB mutants that encode AmiB variants with an increased propensity to visit the ON conformation in the absence of activation. We further anticipated that, when exported to the periplasm, the poorly regulated activity of such mutants would create lesions in the PG matrix and induce cell lysis. A plasmid release enrichment strategy (Fig. 1A) (Delisle et al, 2006)(W. Roof and R. Young, unpublished data) was therefore initiated to isolate lytic amiB (lytamiB) mutants. To this end, we generated a library of plasmids encoding amiB under control of the arabinose promoter [Para::amiB] in which the amiB gene was mutagenized by error-prone PCR. The library was transformed into TB28 [WT] cells and the resulting transformants were pooled and grown in LB broth. Upon reaching an OD600 of 0.3, amiB was induced by the addition of arabinose to 0.2% and growth was continued for an additional 3 hrs. During the induction period, we presumed that rare plasmids encoding lytamiB mutants would cause cell lysis, promoting their released into the medium. Therefore, following removal of cells and cell debris, we purified plasmid DNA from the growth medium of the induced culture using Qiagen spin columns (see Experimental Procedures). Purified DNA was used to transform TB28 [WT] cells and the transformants were recovered on non-inducing medium. To identify transformants harboring lytamiB-containing plasmids, colonies from the primary transformation plate were screened for those that failed to grow or grew poorly on agar containing 0.2% arabinose (inducing conditions).
Figure 1. Plasmid release enrichment strategy for the isolation of lytic amiB (lytamiB) mutants.
(A) Overview of the plasmid release enrichment protocol. (i) Cells carrying mutagenized amiB-containing plasmid (black and red circles) are induced with 0.2% arabinose. (ii) Plasmids encoding rare lytamiB mutants (red circles) promote cell lysis and are released into the culture supernatant. Cells and cell debris are removed by centrifugation and filtration. (iii) Plasmid DNA is purified from the culture supernatants using Qiagen columns. (iv) Purified DNA can then be retransformed into the parental strain to repeat the enrichment protocol for as many rounds as necessary. (B) TB28 [WT] cells carrying an empty vector or expression constructs encoding AmiB (WT), AmiB(E215A) or the indicated LytAmiB variants were spread on M9-casamino acid (M9-CAA) agar containing either 0.2% glucose or 0.2% arabinose supplemented with 10 µg/ml chloramphenicol (Cm10). Plates were incubated overnight at 37°C and photographed. The AmiB(E215A) variant is a catalytically dead mutant used as a control (Uehara et al, 2010). (C) Same as (B) except the host strain was TB140 [ΔenvC]. (D) TB170 [ΔamiB] carrying an empty vector or expression constructs encoding AmiB (WT), AmiB(E215A) or the indicated LytAmiB variants were were grown at 37°C in M9-CAA-Cm10 supplemented with 0.2% maltose to an OD600 of about 0.3. At t = 0, arabinose was added to a final concentration of 0.2% and growth was monitored by following culture OD600. Samples for immunoblot analysis were taken just prior to lysis at t = 50 (see Fig. S1). Growth of cells harboring the empty vector or the AmiB (WT) or AmiB(E215A) control constructs continued without observable lysis.
Using two independently mutagenized plasmid libraries and one round of plasmid release for each enrichment, 33 out of 116 plasmid-release transformants showed differential growth on arabinose versus glucose media. Plasmid DNA was isolated from each of these candidates and the resident amiB genes were sequenced. As expected, a majority of the plasmids (29/33) were found to encode amiB mutants (Table S1). The three plasmids containing wild-type amiB were found to yield much more DNA than the parental vector following plasmid purification (data not shown). We assume these plasmids were released during the enrichment because their apparent copy number increase led to a level of AmiB overproduction that was sufficiently high to induce lysis. These isolates were not studied further.
To verify that the plasmid encoded amiB mutants isolated in the plasmid release enrichment were indeed lytamiB alleles, mutants from unique isolates were subcloned into the parental vector backbone. In each case, TB28 [WT] cells harboring the resulting plasmids were found to be inducer sensitive, indicating that the amiB mutants were toxic (Table 1 and Fig. 1B). Site directed mutagenesis was also used to generate constructs producing the AmiB variants: AmiB(H302P), AmiB(S306P), AmiB(D314V), and AmiB(A405V) because plasmids encoding variants with these individual substitutions were not isolated in the enrichment. AmiB(H302P) and AmiB(S306P) were found to be toxic whereas the others were not (Table 1). Finally, a subset of the mutant amiB isolates were confirmed to have lytic activity in liquid broth (Fig. 1D) and to produce equivalent levels of protein relative to wild-type amiB (Fig. S1). As expected for mutants thought to possess elevated basal activity, the AmiB variants were also found to be toxic in a strain lacking their cognate activator EnvC (Fig. 1C). We thus conclude that the lytamiB alleles isolated in the plasmid release enrichment procedure encode poorly regulated AmiB variants that induce cell lysis when overproduced in vivo.
Table 1.
AmiB variants isolated in the plasmid release enrichment.
| Variants from Enrichmenta |
Lethal Activityf | |
|---|---|---|
| WT | ΔenvC | |
| W8L, N263D, M295Ib | +++ | +++ |
| G33D, E300Kb | +++ | +++ |
| S87P, S306L | +++ | +++ |
| G170C, S293Rb | +++ | +++ |
| S173N, S293R | +++ | +++ |
| L229S, A296V, D314Vb | +++ | +++ |
| S293Ic, e | +++ | +++ |
| S293Re | +++ | +++ |
| M295Ic, e | +++ | +++ |
| A296Vc, e | +++ | +++ |
| E300Kc, e | +++ | +++ |
| H302P, A405Vb, e | +++ | +++ |
| E303Ke | +++ | +++ |
| S306Lc, e | +++ | +++ |
| S306Pd, e | +++ | +++ |
| Q333Le | + | +++ |
| R372Ce | + | +++ |
| Site-Directed Variants | ||
| H302P | +++ | +++ |
| S306P | +++ | +++ |
| D314V | − | − |
| A405V | − | − |
each row in the table indicates the substitution(s) found in a single variant
also contain silent mutation(s)
variant isolated from two independent libraries
variant contains a premature stop at codon 422
lytic phenotype phenotype was reproduced upon subcloning into the parental plasmid backbone. All others were not subcloned.
Plasmids encoding the indicated AmiB variants under control of the arabinose promoter were assessed for lethal activity by streaking transformants of TB28 [WT] or TB140 [ΔenvC] on M9-CAA-Cm10 agar containing either 0.2% glucose or 0.2% arabinose and visually determining if growth was significantly inhibited on the arabinose-containing plate.
Substitutions in the LytAmiB variants cluster within a predicted regulatory domain
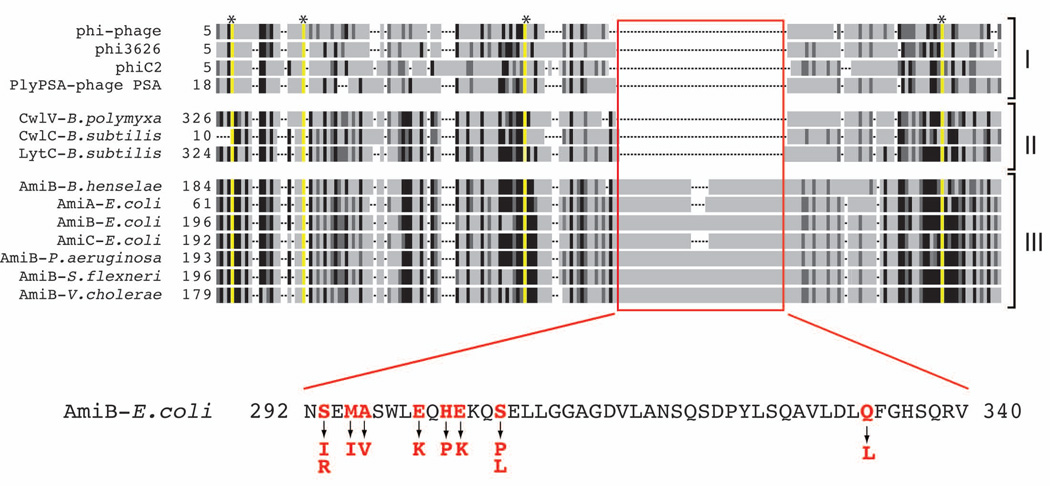
LytC-type amidases are widely distributed within the bacterial domain and are also encoded in the genomes of many bacteriophages (Finn et al, 2008). These enzymes can be grouped into at least three different functional classes: i) phage-encoded enzymes (endolysins) that are likely used to elicit host cell lysis at the end of an infection cycle (e.g. PLY-PSA) (Korndörfer et al, 2006), ii) enzymes encoded by sporulating bacteria that promote mother cell lysis for the release of mature spores (e.g. LytC) (Smith & Foster, 1995), and iii) enzymes encoded by gram-negative proteobacteria that are employed to promote cell separation (e.g. AmiB) (Heidrich et al, 2001). Interestingly, a sequence alignment of selected amidases from these three functional classes revealed that the known or predicted cell separation enzymes contain an ~50 amino acid insertion in their amidase domain that is absent in the lytic amidases (Fig. 2 and S2). Strikingly, 24/29 lytamiB candidates sequenced encoded proteins with at least one amino acid substitution within the N-terminal third of this domain (residues 293–306 of E. coli AmiB) (Table S1, Fig. 2). Those mutants encoding proteins with substitutions outside of this region (Q333L or R372C) were weak alleles with lower lytic activity (Table 1). Our results therefore implicate the ~50 amino acid insertion in the cell separation amidases as a regulatory domain.
Figure 2. Amino acid substitutions in the LytAmiB variants map to a potential regulatory domain.
Shown is a schematic representation of a multiple sequence alignment generated using amidase sequences from bacteria and phages. Identities and similarities are indicated by the black and dark grey regions, respectively. Residues essential for catalysis are highlighted in yellow and with an asterisk at the top of the alignment. Gaps in the alignment are indicated as dashed lines. The amidases are grouped into the following categories: (I) phage endolysins, (II) bacterial amidases involved in mother cell lysis following sporulation, and (III) cell separation amidases. The red box highlights what appears to be a ~50 amino acid insertion region found only in the cell separation amidases. The sequence of the insertion from E. coli AmiB is shown below the alignment. Residues in red were altered in LytAmiB variants. The identity of the substitutions are indicated by the arrows. See Figure S2 for the complete sequence alignment.
Structural analysis of an AmiB ortholog
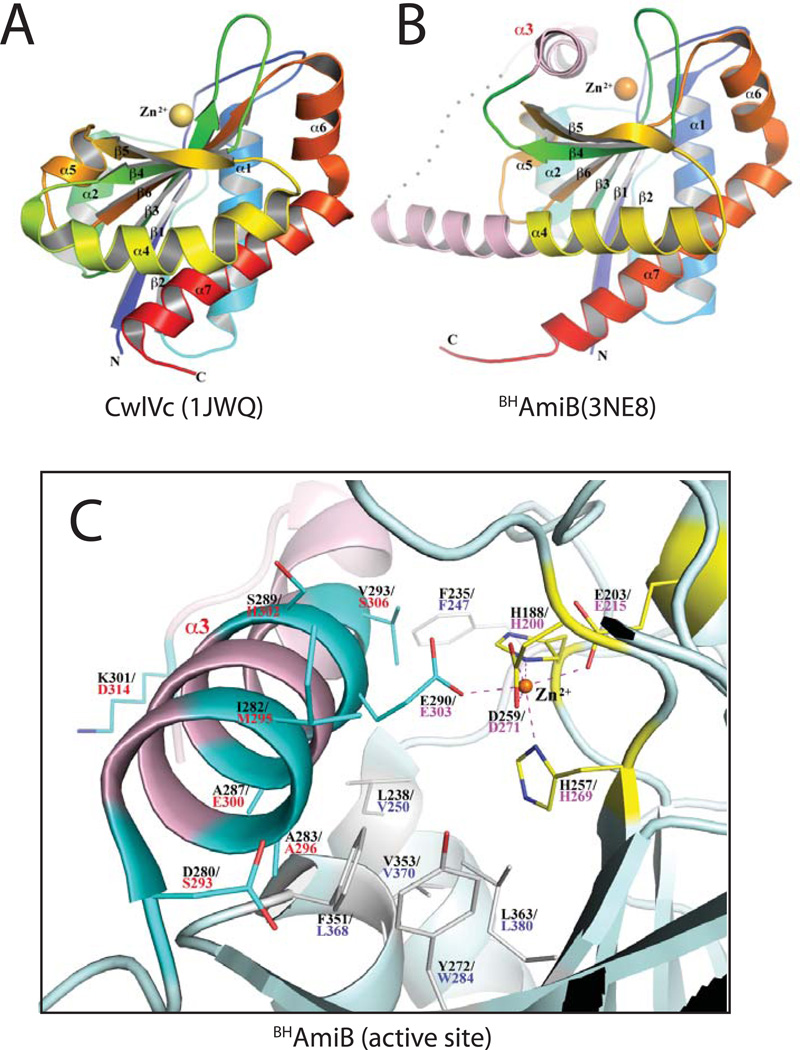
The structures of several phage or mother cell lysis amidases have been solved: CD27L, PLY-PSA, and CwlVc (Mayer et al, 2011; Korndörfer et al, 2006) (PDB: 3QAY, 1XOV, 1JWQ). They are metallo-enzymes and all contain a Zn2+-binding site in the middle of a cleft that is thought to be the catalytic center where the PG peptide substrate is bound and cleaved (Mayer et al, 2011; Korndörfer et al, 2006; Shida et al, 2001) (Fig. 3A). In the lytic amidases, the active site Zn2+ is coordinated octahedrally by three water or other small molecules and three amino acid side chains: two His and one Glu (Fig. S3A). These residues are highly conserved in LytC-type amidases and all of them have been demonstrated to be critical for catalysis (Shida et al, 2001). The active site of the lytic enzymes is open and the Zn2+ ion in these structures is solvent accessible as indicated by the coordinating water molecules (Mayer et al, 2011; Korndörfer et al, 2006) (Fig. 3A and S3A). Thus, the activity of the phage and mother cell lysis enzymes is unlikely to be autoregulated.
Figure 3. Crystal structure of Bartonella henselae AmiB.
(A–B) Shown are rainbow-colored cartoon diagrams of the CwlVc (A) and BHAmiB (B) structures with the N-termini in blue and the C-termini in red. The catalytic Zn2+ ion is drawn as a sphere. All secondary structures are labeled. In numbering α-helices, α3 is omitted from the numbering of CwlVc so that all common elements are numbered identically for CwlVc and BHAmiB. The regulatory region unique to cell separation amidases is colored pink in the BHAmiB structure with the structurally disordered loop region (E303-T308) represented by a dotted line. The unique α3 helix is labeled in pink. (C) Close-up view of the BHAmiB active site and regulatory helix. All residues are drawn in stick format and the Zn2+ ion is represented as an orange sphere. The Zn2+ ion is coordinated by H188, E203, H257, D259 (bidentate) and E290 with their carbon atoms drawn in yellow. Residue numbers for E.coli AmiB are provided in red underneath those for BHAmiB. Positions in the structure corresponding to amino acid substitutions in the LytAmiB variants are drawn with their carbon atoms in cyan. The residue corresponding to Q333 of E.coli AmiB (T315 of BHAmiB) is located in the N-terminal region of the α4 helix and they are out of view in this figure. Hydrophobic residues on the floor of binding cleft are also displayed in white.
Here we report the first structure of a cell separation amidase ortholog, AmiB from Bartonella henselae (BHAmiB). The core structure of BHAmiB is similar to that of the lytic amidases (Fig. 3A–B). A structural alignment between BHAmiB and CwlVc yields a root mean square deviation value of 1.4Å over 167 amino acids even though their sequences identity is less than 35%. As in other amidases, the LytC-domain of BHAmiB is an α/β fold with a highly twisted central β-sheet that has an order of 2-1-3-6-4-5, in which the strands 4 and 5 are antiparallel to the others (Fig. 3, S4). The β-sheet is flanked by four helices on one side, α1, consecutive α6 and α7, and an extensive α4. On the other side it is packed by two short parallel helices, α2 and α5. The packing of these two short helices helps create an extensive shallow cleft across the β-strands.
Strikingly, and in excellent agreement with the genetic analysis, the region of BHAmiB corresponding to the ~50 amino acid insertion identified in cell separation amidases is in part composed of a helical domain (α3, D280-L296) that completely buries the active site Zn2+ ion and occludes the active site cleft, thus presumably blocking access by substrate (Fig. 3, S4). The insertion domain continues from α3 to include an unstructured loop region and the first half of the extended α4 helix. This is in contrast to amidases without the insertion domain in which the β4 strand is directly linked to a much shorter α4 helix as observed, for example, in CwlVc (Fig. 3, S4). Not surprisingly, the buried nature of the active site ion Zn2+ results in an altered coordination sphere relative to the lytic amidases. It retains the octahedral-like configuration, but the water or other coordinating solute molecules observed in other structures are replaced by additional side-chain interactions. Notably, in addition to the coordinating residues typically observed in the LytC-type PG amidases (H188, E203, and H257), extra bonds are formed by two residues that are uniquely and highly conserved in the cell separation amidases (Fig. S2 and S3B). A bidentate bond is formed between the Zn2+ ion and D259 side-chain. This residue is typically a Asn in the lytic amidases and is oriented away from the metal. The other coordinating residue is E290 that extends into the active site from the occluding α3 helix.
The probable PG binding cleft of BHAmiB is rich in hydrophobic residues. Interestingly, the α3 helix is amphipathic with a polar and non-polar face. The non-polar face inserts into the active site cleft creating a predominantly hydrophobic interaction between the helix and the active site. The Zn2+ coordinating E290 residue is the only polar residue on the non-polar side of the α3 helix and its interaction with the metal ion appears to be the only conserved specific interaction that helps to position the helix within the cleft.
Importantly, mapping the amino acid substitutions found in the LytAmiB variants onto the BHAmiB crystal structure show that these residues are primarily located on helix α3 (Fig. 3C). Moreover, the majority of these residues are also found on the face of α3 that is interacting with the active site cleft (Fig. 3C). When modeled in the structure, the substituting residues, especially the positively charged lysines and arginine, likely cause α3 to fit poorly into the active site cleft and are predicted to destabilize the cleft-helix interaction. Additionally, in the case of the E303K substitution, the conserved glutamate that coordinates the active site Zn2+ is converted to a basic residue that is likely to have an unfavorable electrostatic interaction with the positively charged metal ion. Thus, in most cases, the LytAmiB variants are predicted to have a destabilized cleft-helix interaction. The tight correspondence between the genetic and structural data, as well as the conservation of the regulatory domain among predicted cell separation amidases, strongly suggests that these enzymes are autoinhibited by a regulatory helix “plug” analogous to α3 in BHAmiB. Furthermore, the results imply that the LytM-domain activators must somehow promote the release of the regulatory helix from the active site cleft to stimulate amidase activity (see Discussion).
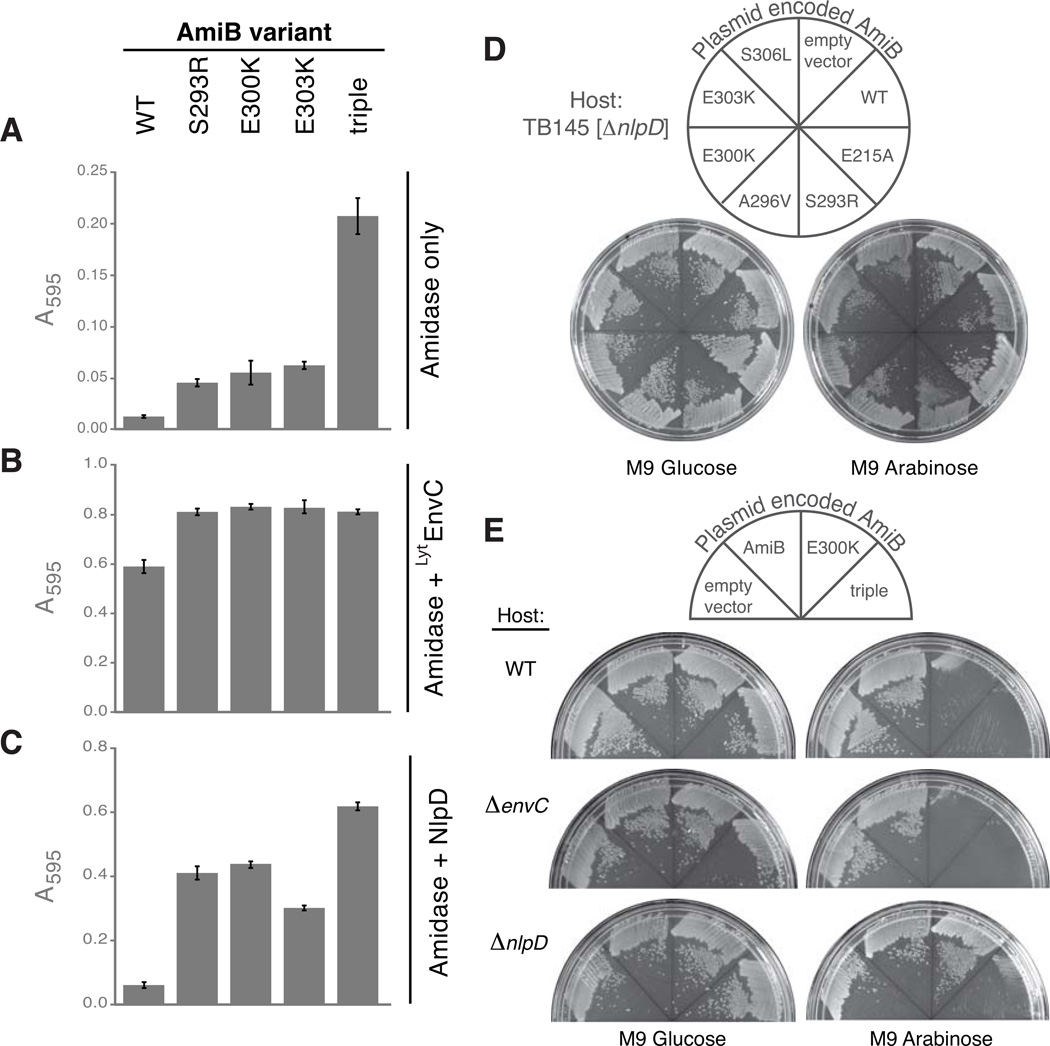
LytAmiB variants have elevated basal activity in vitro
The BHAmiB structure indicates that the interaction between the regulatory helix and the active site cleft is likely to be weakened in the E. coli LytAmiB variants. These enzymes should therefore have active sties that are more accessible and thus have higher basal PG hydrolase activities. To test this, we purified three of the most toxic LytAmiB variants [AmiB(S293R), AmiB(E300K), and AmiB(E303K)] as well as a variant containing all three substitutions [AmiB(triple)]. We then compared their basal activities to the wild-type enzyme using a dye-release assay for PG hydrolysis in which the liberation of dye from labeled PG is a reflection of PG hydrolase activity (Uehara et al, 2010). PG sacculi covalently labeled with Remazol Brilliant Blue (RBB) were incubated with the amidases (2 µM) at 37°C for 30 min. Undigested PG was pelleted by centrifugation and the absorbance of the supernatants was measured at 595 nm. As observed previously (Uehara et al, 2010), AmiB(WT) led to minimal dye release in the absence of activation (Fig. 4A). The LytAmiB variants with single substitutions in the regulatory helix were all found to have low but increased basal activity compared to AmiB(WT) (Fig. 4A). The effects of the single substitutions were additive with AmiB(triple) displaying roughly three times the basal activity of the LytAmiB variants identified in the enrichment (Fig. 4A). Basal PG hydrolytic activity is therefore inversely correlated with the apparent stability of the regulatory helix-active site interaction.
Figure 4. LytAmiB variants have elevated basal PG hydrolase activity.
(A) Dye-release assays measuring basal PG hydrolase activity of the LytAmiB variants relative to AmiB(WT). The variant referred to as “triple” is AmiB(S293R, E300K, E303K).
The indicated proteins (2 µM) were incubated with RBB-labeled PG at 37°C for 30 min. Undigested PG was pelleted by centrifugation and the absorbance of the supernatants was measured at 595 nm. Reactions were performed in triplicate and the error bars indicate the standard deviation. (B–C) Same as in (A) except LytEnvC (2 µM) or NlpD (2 µM) were combined with the amidases. Supernatants from control reactions containing LytEnvC only (2 µM), NlpD only (2 µM), lysozyme (4 µM) or buffer alone had the following A595 readings (average +/− std. dev.): 0.02 +/− 0.002, 0.04 +/− 0.008, 0.56 +/− 0.022 and 0.007 +/− 0.002. The A595 reading for the lysozyme reaction is a typical reading for a reaction that has gone to completion. (D) TB145 [ΔnlpD] cells carrying an empty vector or expression constructs encoding AmiB (WT), AmiB(E215A) or the indicated LytAmiB variants were spread on M9-CAA-Cm10 agar containing either 0.2% glucose or 0.2% arabinose. Plates were incubated overnight at 37°C and photographed. (E) TB28[WT], TB140[ΔenvC], or TB145[ΔnlpD] carrying an empty vector or expression constructs encoding AmiB (WT), AmiB(E300K), or AmiB(triple) were plated and photographed as described in (D).
All of the LytAmiB variants tested retained the ability to be activated by the LytM domain of EnvC [LytEnvC] and possessed slightly higher activity levels following activation than the wild-type enzyme (Fig. 4B). Strikingly, unlike AmiB(WT), which can only be activated by EnvC, the LytAmiB variants were also activated by the non-cognate LytM factor, NlpD (Fig. 4C). This observation suggests that the stability of the regulatory helix-active site interaction is an important determinant of the specificity of the amidases for their cognate LytM activators (see Discussion). The observed mis-activation appeared to be physiologically relevant as the LytAmiB variants with single substitutions required NlpD activity to elicit cell lysis (Fig. 4D and S5). AmiB(triple), on the other hand, was capable of inducing lysis of both WT and NlpD− cells when it was produced at levels similar to the single mutants (Fig. 4E,S5, and S6). Thus, a threshold level of mis-regulated amidase activity appears necessary for the induction of cell lysis. In WT cells this is accomplished by the aberrant activation of the LytAmiB variants by a non-cognate LytM activator. In the case of AmiB(triple), on the other hand, it appears that given enough basal activity, lysis can be induced independently of the activators. However, we were unable to definitively demonstrate this because of the significant growth defect displayed by a strain lacking all four LytM factors (Uehara et al, 2009).
The autoinhibition mechanism for amidase regulation is conserved
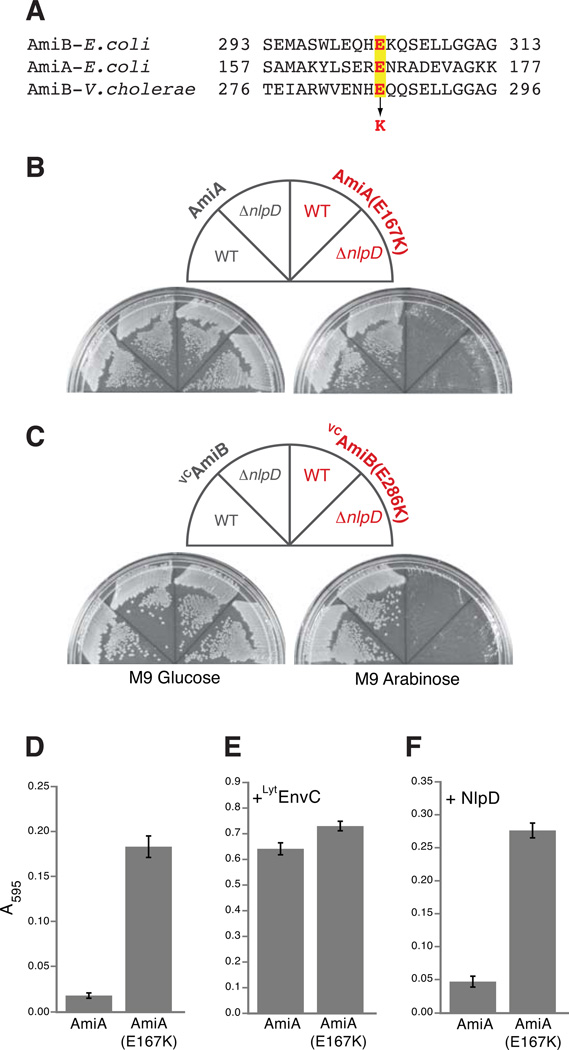
We tested the potential conservation of the amidase autoregulatory mechanism by mutating E. coli amiA and Vibrio cholerae amiB (VCamiB) so that they encoded enzymes with the equivalent substitution as the LytAmiB variant AmiB(E303K) (Fig. 5A). Production of the wild-type versions of each enzyme was not lethal. However, AmiA(E167K) and VCAmiB(E286K) were lethal in both WT and NlpD− cells (Fig. 5B–C). Purified AmiA(E167K) had elevated PG hydrolase activity in vitro relative to AmiA(WT) (Fig. 5D). As for the LytAmiB variants, AmiA(E167K) retained the ability to be activated by EnvC (Fig. 5E). It was also mis-activated by the non-cognate NlpD activator (Fig. 5F), underscoring the likely role of the regulatory helix-active site interaction in determining amidase-activator specificity. Interestingly, the basal activity of AmiA(E167K) approached that of AmiB(triple). This relatively high basal activity is probably responsible for the NlpD-independence of AmiA(E167K) toxicity in vivo. Unfortunately, solubility problems prevented us from purifying VCAmiB and VCAmiB(E286K) to test their basal activities. Nevertheless, their behavior in vivo, combined with the genetic, structural, biochemical, and bioinformatic results for the other amidases, strongly supports the idea that there is a conserved conformational control mechanism governing cell separation amidase activity at the cytokinetic ring of E. coli and related gram-negative bacteria.
Figure 5. Lytic and PG hydrolase activities of other amidases with substitutions in their regulatory domains.
(A) Sequence alignment of a portion of the regulatory domains of E. coli AmiB, E. coli AmiA, and V. cholerae AmiB (VCAmiB). The highlighted glutamic acid (E) residues in red were changed to lysines (K) in AmiA and VCAmiB to yield AmiA(E167K) and VCAmiB(E286K), respectively. (B–C) As indicated, TB28[WT] or TB145[ΔnlpD] cells carrying AmiA, AmiA(E167K), VCAmiB, or VCAmiB(E286K) were spread on M9-CAA-Cm10 agar containing either 0.2% glucose or 0.2% arabinose. Plates were incubated overnight at 37°C and photographed. (D) Dye-release assay measuring basal PG hydrolase activity for AmiA(E167K) relative to AmiA (WT). Assays were performed as described in Fig. 4 except that reactions contained 1µM amidase with or without an additional 1µM of purified LytM factor as indicated.
Discussion
Because of their potentially lethal activity, strict control mechanisms governing the activity of cellular PG hydrolases have long been postulated. Most of the regulatory strategies described to date, however, appear to be general mechanisms for adjusting the total level of cellular PG hydrolase activity in response to developmental programs or changes in growth conditions. These strategies involve the control of PG hydrolase gene expression or the global control of hydrolase activity through the production of hydrolase inhibitors or enzymes that modify the cell wall to block cleavage (Vollmer et al, 2008b; Dubrac et al, 2008; Chai et al, 2010; Cozy & Kearns, 2010; Yamamoto et al, 2008a; Moynihan & Clarke, 2011; Clarke et al, 2010; Laaberki et al, 2011). For controlling PG hydrolase activity needed for the proper expansion and division of the cell wall throughout the cell cycle, specific spatiotemporal mechanisms are likely to be critical. Despite their importance for cell growth and relevance to antimicrobial development, very little is known about these regulatory systems.
Spatiotemporal control of PG hydrolase activity
PG hydrolases from numerous organisms have been found to adopt specific subcellular localizations, implying that targeting mechanisms that guide PG hydrolases to their sites of action are important for their spatiotemporal regulation (Schlag et al, 2010; Yamada et al, 1996; Baba & Schneewind, 1998; Bernhardt & de Boer, 2003; Peters et al, 2011; Yamamoto et al, 2008b; Steen et al, 2003; Carballido-López et al, 2006). Indeed, in E. coli, AmiB and AmiC are both recruited to the divisome by N-terminal targeting domains (Bernhardt & de Boer, 2003; Peters et al, 2011). Curiously, however, cell separation is not strongly affected in mutants lacking AmiB and AmiC, indicating that AmiA alone can promote relatively efficient septal PG splitting (Chung et al, 2009). Since AmiA is not specifically recruited to the division site, this observation indicates that additional mechanisms for regulating the activity of PG hydrolases are likely working in concert with the localization systems to ensure that the amidases are only hydrolyzing PG at the appropriate time and place. Accordingly, we recently discovered that the amidases require activation by LytM factors at the cytokinetic ring to promote proper cell division (Uehara et al, 2010), and that one of these factors, EnvC, is itself controlled by the divisome-associated, ABC transporter-like FtsEX complex (Yang et al, 2011). What has remained unclear, however, is how amidase activity is kept off prior to the assembly of the septal ring and how the LytM factors stimulate PG hydrolysis when cell constriction is initiated.
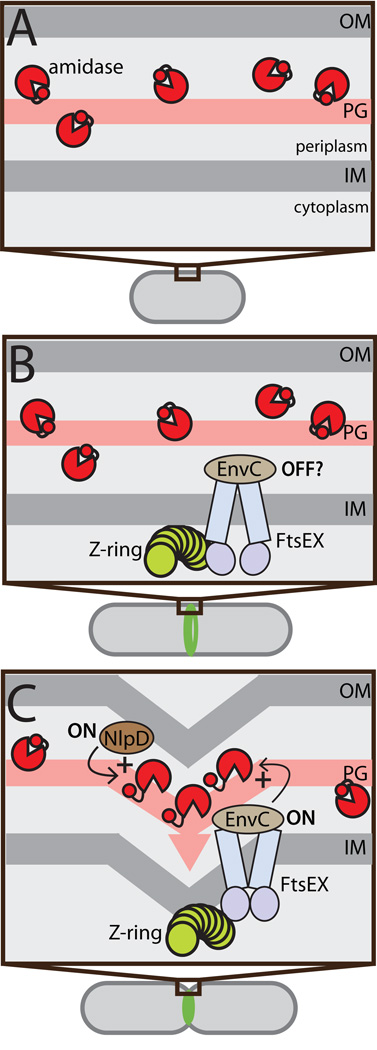
Here, we show that auto-inhibition is a critical feature of amidase regulation. We solved the the first structure of an ortholog of PG amidases required for cell division in gram-negative bacteria and discovered that the active site of this enzyme is occluded by a regulatory alpha-helix. Importantly, we combined this structural information with genetic and biochemical evidence that strongly supports a model in which cell separation amidases from gram-negative bacteria are controlled through the modulation of the regulatory helix-active site interaction. In the case the amidases from E. coli and related proteobacteria, this conformational control likely plays a significant role in restricting amidase activity to the septal ring when the machinery is engaged in cell constriction (Fig. 6). In newborn cells prior to division or in regions of the cell removed from the septal ring, amidase activity is blocked by auto-inhibition. Later in the cycle, Z-ring assembly promotes FtsEX and EnvC recruitment to midcell, while NlpD localization is delayed until the latter stages of the septal ring maturation process (Peters et al, 2011; Schmidt et al, 2004). It is not clear when the activation systems become active following their recruitment nor is it currently known what other septal ring component(s) might be responsible for their activation. Nevertheless, maximal amidase activation likely does not occur until after septal PG synthesis is initiated because AmiB and AmiC recruitment to the division site requires the activity of the essential septal PG synthase FtsI(PBP3) (Peters et al, 2011).
Figure 6. Conformational control of amidase activity during the cell cycle.
Shown is a schematic diagram illustrating the activation status of cell separation amidases through the cell cycle. (A) At early stages in the cell cycle prior to the formation of the Z-ring, periplasmic amidases (red pac-men) are likely to be largely inhibited by their regulatory helices (red circles). (B) FtsEX and EnvC are early recruits to the Z-ring, arriving well before the initiation of constriction (Peters et al, 2011). It is not known if the FtsEX-EnvC system is capable of amidase activation immediately following its recruitment, or if it requires further septal ring maturation. Even if it is active at this stage, it is unlikely to stimulate a high level of amidase activity because the amidase are not concentrated at midcell before the onset of cell constriction (Peters et al, 2011). (C) Once constriction is initiated NlpD, AmiB, and AmiC are recruited to the septal ring and both amidase activation systems are presumably activated to stimulate amidase activity (Peters et al, 2011). Our results indicate that amidase activation proceeds via the release of the regulatory helix from their active site. For simplicity, the three different amidases are not individually identified in the figure.
Given the relatedness of the amidases and their activators, EnvC and NlpD most likely stimulate the activity of their target amidases in the same way: by promoting the release of the regulatory helix from the active site. We do not presently understand how the LytM factor binding promotes this this conformational change, but we envision that it may do so by intervening in the baseline level of interconversions between the open and closed conformations in the amidases. We propose that the stability of the regulatory helix-active site interaction normally biases the equilibrium in favor of the closed (inactive) state. The LytM factors, therefore, may stabilize the open (active) state by binding directly to the regulatory helix or elsewhere in the ~50 residue regulatory domain (Fig. S7). In either case, the specificity of an amidase for a particular LytM factor is likely to be determined by a competition between LytM binding and the interaction between the regulatory alpha-helix and the amidase active site. Accordingly, our results suggest that weakening the regulatory helix-active site interaction in either AmiA or AmiB results in promiscuous amidases that can be activated by the non-cognate factor NlpD as well as their cognate factor EnvC. NlpD presumably has a lower affinity for these amidases than EnvC and thus can only compete with the regulatory helix-active site interaction when it is weakened by the amino acid substitutions in the LytAmiB variants.
For the EnvC-AmiA/B system, FtsEX is likely used to extend amidase conformational control to the cytoplasm where activation can be coupled with ATP hydrolysis by FtsE (Yang et al, 2011). We currently favor the idea that the amidases cycle between ON and OFF conformations in-tune with the ATPase cycle of FtsE as this would likely afford the septal ring with exquisite control over septal PG hydrolysis. However, we cannot rule out the alternative possibility that ATP binding by FtsE keeps the amidases in the ON conformation for multiple rounds of PG cleavage potentially even for the entire septal PG splitting process. Much less is known about the regulation of the NlpD-AmiC system within the septal ring. Since NlpD is a verified lipoprotein predicted to localize to the outer membrane (Ichikawa et al, 1994), its ability to activate AmiC is likely to be controlled by a mechanism distinct from EnvC, which is associated with the outer face of the inner membrane via its interaction with FtsX (Yang et al, 2011). Because they are associated with different membranes, an attractive hypothesis to explain the existence of these two partially redundant septal PG splitting systems is that they are optimized to couple the PG splitting process with the invagination of the membranes to which they localize: EnvC with the inner membrane and NlpD with the outer membrane.
A potentially general role for auto-inhibition in PG hydrolase regulation
The conserved role of FtsEX in cell separation in S. pneumoniae suggests that the ABC-system regulates PG hydrolysis in a diverse array of organisms by promoting a similar cascade of conformational changes that span the cytoplasmic membrane. Although the PG hydrolases subject to FtsEX regulation in other bacteria have yet to be uncovered, we suspect that they will also prove to be controlled by an auto-regulatory mechanism analogous to the one we have identified for the proteobacterial amidases. In fact, a growing number of PG hydrolases structures suggest that auto-inhibition is likely to be a general mode of activity regulation for these enzymes. The structures of S. aureus LytM, Listeria monocytogenes Auto, Mycobacterium tuberculosis RipA, and S. pneumoniae LytC all contain extra domains that occlude or restrict access to their active sites (Pérez-Dorado et al, 2010; Odintsov et al, 2004; Bublitz et al, 2009; Ruggiero et al, 2010; Böth et al, 2011). Consistent with a regulatory role for these domains, their removal by limited proteolysis in some cases has been shown to activate enzymatic activity (Firczuk et al, 2005; Ruggiero et al, 2010; Bublitz et al, 2009). The physiological relevance of these observations is unclear, however, as are the mechanisms by which these PG hydrolases are regulated in vivo. For example, it is not known whether mycobacterial RipA, itself a cell separation factor, is activated by proteolytic processing or a conformational change (Ruggiero et al, 2010; Böth et al, 2011). Also, in the case of LytM and Auto, the physiological functions of the crystallized PG hydrolases themselves are not even clear (Singh et al, 2010; Bublitz et al, 2009). Thus, further genetic and biochemical analysis of these and other structurally characterized PG hydrolases is warranted to determine the extent to which auto-regulatory domains are important for the in vivo control of PG cleavage and what other types of regulators might be used to activate auto-inhibited PG hydrolases at the time and place they are needed.
Mis-activation of PG hydrolases as an antibiotic target
Wild-type E. coli cells treated with β-lactams lyse rapidly through lesions at their division sites, and this rapid lysis requires an active septal PG splitting system (Uehara et al, 2009; Donachie & Begg, 1970; Lederberg, 1956; Schwarz et al, 1969; Staugaard et al, 1976). These PG synthase targeting drugs therefore appear to induce a lethal divisome malfunction by upsetting the balance between septal PG synthesis and hydrolysis. Our results suggest that another potential route for accomplishing this would be via small molecules that mis-activate the cell separation amidases, for example by stimulating the release of the regulatory helix from the amidase active site independently of the FtsEX-EnvC system or by locking the entire system in the ON state. Interestingly, the validity of the later mechanism is supported by a recent study investigating the antibacterial activity of the chemokines CXCL9 and CXCL10 (Crawford et al, 2011). Bacillus anthracis mutants resistant to the chemokines were isolated and found to have lesions in ftsX or lytE, a gene encoding a putative PG hydrolase with a modular structure similar to the FtsX-interacting partners EnvC (E. coli) and PcsB (S. pneumoniae) (Crawford et al, 2011). Thus, the chemokines may kill cells by aberrantly activating an analogous FtsEX-PG hydrolase regulatory system in Bacillus species. While future work is required to definitively demonstrate this mode of action, the chemokine results highlight the potential of the FtsEX-EnvC-amidase regulatory system as a therapeutic target for the development of new lytic antibiotics.
Supplementary Material
Acknowledgements
We thank all members of the Bernhardt lab past and present for helpful suggestions and comments as well as Renée Yang for help with the figures. The plasmid release method was originally developed by William D. Roof and Ryland F. Young to study bacteriophage-induced cell lysis. This work was supported by the Massachusetts Life Science Center, the Burroughs Wellcome Fund, and the National Institutes of Health (R01 AI083365-01). TGB holds a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund.
Abbreviations
- PG
peptidoglycan
- RBB
remazol brilliant blue
References
- Baba T, Schneewind O. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 1998;17:4639–4646. doi: 10.1093/emboj/17.16.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, Hale CA, Bernhardt TG, de Boer PAJ. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol. 2003;48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Böth D, Schneider G, Schnell R. Peptidoglycan Remodeling in Mycobacterium tuberculosis: Comparison of Structures and Catalytic Activities of RipA and RipB. J Mol Biol. 2011;413:247–260. doi: 10.1016/j.jmb.2011.08.014. [DOI] [PubMed] [Google Scholar]
- Bublitz M, Polle L, Holland C, Heinz DW, Nimtz M, Schubert W-D. Structural basis for autoinhibition and activation of Auto, a virulence-associated peptidoglycan hydrolase of Listeria monocytogenes. Mol Microbiol. 2009;71:1509–1522. doi: 10.1111/j.1365-2958.2009.06619.x. [DOI] [PubMed] [Google Scholar]
- Carballido-López R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell. 2006;11:399–409. doi: 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010;24:754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D. Rapid beta-lactam-induced lysis requires successful assembly of the cell division machinery. Proc Natl Acad Sci USA. 2009;106:21872–21877. doi: 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke CA, Scheurwater EM, Clarke AJ. The vertebrate lysozyme inhibitor Ivy functions to inhibit the activity of lytic transglycosylase. J Biol Chem. 2010;285:14843–14847. doi: 10.1074/jbc.C110.120931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozy LM, Kearns DB. Gene position in a long operon governs motility development in Bacillus subtilis. Mol Microbiol. 2010;76:273–285. doi: 10.1111/j.1365-2958.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford MA, Lowe DE, Fisher DJ, Stibitz S, Plaut RD, Beaber JW, Zemansky J, Mehrad B, Glomski IJ, Strieter RM, Hughes MA. Identification of the bacterial protein FtsX as a unique target of chemokine-mediated antimicrobial activity against Bacillus anthracis. Proc Natl Acad Sci USA. 2011;108:17159–17164. doi: 10.1073/pnas.1108495108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PAJ. Advances in understanding E. coli cell fission. Curr Opin Microbiol. 2010;13:730–737. doi: 10.1016/j.mib.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisle AL, Barcak GJ, Guo M. Isolation and expression of the lysis genes of Actinomyces naeslundii phage Av-1. Appl Environ Microbiol. 2006;72:1110–1117. doi: 10.1128/AEM.72.2.1110-1117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie WD, Begg KJ. Growth of the bacterial cell. Nature. 1970;227:1220–1224. doi: 10.1038/2271220a0. [DOI] [PubMed] [Google Scholar]
- Dubrac S, Bisicchia P, Devine KM, Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firczuk M, Mucha A, Bochtler M. Crystal structures of active LytM. J Mol Biol. 2005;354:578–590. doi: 10.1016/j.jmb.2005.09.082. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H, de Pedro MA, Höltje JV. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol Microbiol. 2001;41:167–178. doi: 10.1046/j.1365-2958.2001.02499.x. [DOI] [PubMed] [Google Scholar]
- Ichikawa J, Li C, Fu J, Clarke S. A gene at 59 minutes on the Escherichia coli chromosome encodes a lipoprotein with unusual amino acid repeat sequences. J Bacteriol. 1994;176:1630–1638. doi: 10.1128/jb.176.6.1630-1638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korndörfer IP, Danzer J, Schmelcher M, Zimmer M, Skerra A, Loessner MJ. The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls. J Mol Biol. 2006;364:678–689. doi: 10.1016/j.jmb.2006.08.069. [DOI] [PubMed] [Google Scholar]
- Laaberki M-H, Pfeffer J, Clarke AJ, Dworkin J. O-Acetylation of peptidoglycan is required for proper cell separation and S-layer anchoring in Bacillus anthracis. J Biol Chem. 2011;286:5278–5288. doi: 10.1074/jbc.M110.183236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J. Bacterial Protoplasts Induced by Penicillin. Proc Natl Acad Sci USA. 1956;42:574–577. doi: 10.1073/pnas.42.9.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marblestone JG, Edavettal SC, Lim Y, Lim P, Zuo X, Butt TR. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MJ, Garefalaki V, Spoerl R, Narbad A, Meijers R. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J Bacteriol. 2011;193:5477–5486. doi: 10.1128/JB.00439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Moynihan PJ, Clarke AJ. O-Acetylated peptidoglycan: Controlling the activity of bacterial autolysins and lytic enzymes of innate immune systems. Int J Biochem Cell Biol. 2011;43:1655–1659. doi: 10.1016/j.biocel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Ng W-L, Kazmierczak KM, Winkler ME. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol Microbiol. 2004;53:1161–1175. doi: 10.1111/j.1365-2958.2004.04196.x. [DOI] [PubMed] [Google Scholar]
- Odintsov SG, Sabala I, Marcyjaniak M, Bochtler M. Latent LytM at 1.3A resolution. J Mol Biol. 2004;335:775–785. doi: 10.1016/j.jmb.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Peters NT, Dinh T, Bernhardt TG. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J Bacteriol. 2011;193:4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Dorado I, González A, Morales M, Sanles R, Striker W, Vollmer W, Mobashery S, García JL, Martínez-Ripoll M, García P, Hermoso JA. Insights into pneumococcal fratricide from the crystal structures of the modular killing factor LytC. Nat Struct Mol Biol. 2010;17:576–581. doi: 10.1038/nsmb.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, de Pedro MA, Young KD. Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. J Bacteriol. 2007;189:5334–5347. doi: 10.1128/JB.00415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiero A, Marasco D, Squeglia F, Soldini S, Pedone E, Pedone C, Berisio R. Structure and functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation. Structure. 2010;18:1184–1190. doi: 10.1016/j.str.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Götz F. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol. 2010;75:864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, Phillips GJ, Weiss DS. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol. 2004;186:785–793. doi: 10.1128/JB.186.3.785-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz U, Asmus A, Frank H. Autolytic enzymes and cell division of Escherichia coli. J Mol Biol. 1969;41:419–429. doi: 10.1016/0022-2836(69)90285-x. [DOI] [PubMed] [Google Scholar]
- Sham L-T, Barendt SM, Kopecky KE, Winkler ME. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci USA. 2011;108:E1061–E1069. doi: 10.1073/pnas.1108323108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Hattori H, Ise F, Sekiguchi J. Mutational analysis of catalytic sites of the cell wall lytic N-acetylmuramoyl-L-alanine amidases CwlC and CwlV. J Biol Chem. 2001;276:28140–28146. doi: 10.1074/jbc.M103903200. [DOI] [PubMed] [Google Scholar]
- Singh VK, Carlos MR, Singh K. Physiological significance of the peptidoglycan hydrolase, LytM, in Staphylococcus aureus. FEMS Microbiol Lett. 2010;311:167–175. doi: 10.1111/j.1574-6968.2010.02087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Foster SJ. Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J Bacteriol. 1995;177:3855–3862. doi: 10.1128/jb.177.13.3855-3862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staugaard P, van den Berg FM, Woldringh CL, Nanninga N. Localization of ampicillin-sensitive sites in Escherichia coli by electron microscopy. J Bacteriol. 1976;127:1376–1381. doi: 10.1128/jb.127.3.1376-1381.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen A, Buist G, Leenhouts KJ, Khattabi El M, Grijpstra F, Zomer AL, Venema G, Kuipers OP, Kok J. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J Biol Chem. 2003;278:23874–23881. doi: 10.1074/jbc.M211055200. [DOI] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Bernhardt TG. More than just lysins: peptidoglycan hydrolases tailor the cell wall. Curr Opin Microbiol. 2011;14:698–703. doi: 10.1016/j.mib.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29:1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Seligman SJ. Architecture of peptidoglycan: more data and more models. Trends in Microbiology. 2010;18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008a;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008b;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, Matsumoto A, Suginaka H. An autolysin ring associated with cell separation of Staphylococcus aureus. J Bacteriol. 1996;178:1565–1571. doi: 10.1128/jb.178.6.1565-1571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Hashimoto M, Higashitsuji Y, Harada H, Hariyama N, Takahashi L, Iwashita T, Ooiwa S, Sekiguchi J. Post-translational control of vegetative cell separation enzymes through a direct interaction with specific inhibitor IseA in Bacillus subtilis. Mol Microbiol. 2008a;70:168–182. doi: 10.1111/j.1365-2958.2008.06398.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, Sekiguchi J. The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL-endopeptidase LytF in Bacillus subtilis. Mol Microbiol. 2008b;70:297–310. doi: 10.1111/j.1365-2958.2008.06397.x. [DOI] [PubMed] [Google Scholar]
- Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M, Bernhardt TG. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA. 2011;108:E1052–E1060. doi: 10.1073/pnas.1107780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.