Abstract
Papaya Mosaic Virus (PapMV) is a filamentous plant virus that belongs to the Alphaflexiviridae family. Flexible filamentous viruses have defied more than two decades of effort in fiber diffraction, and no high-resolution structure is available for any member of the Alphaflexiviridae family. Here we report our structural characterization of PapMV by X-ray crystallography and cryo-EM 3D reconstruction. We found that PapMV is 135 Å in diameter with a helical symmetry of ~ 10 subunits per turn. Crystal structure of the C-terminal truncated PMV coat protein reveals a novel all helix fold with seven α-helices. Thus the PMV CP structure is different from the four-helix bundle fold of Tobacco Mosaic Virus (TMV) in which helix bundling dominates the subunit interface in TMV and conveys rigidity to the rod virus. PapMV coat protein was crystallized as an asymmetrical dimer in which one protein lassoes the other by the N-terminal peptide. Mutation of residues critical to the inter-subunit lasso interaction abolishes coat protein polymerization. The crystal structure suggests that PMV may polymerize via the consecutive N-terminal loop lassoing mechanism. The structure of PapMV will be useful for rational design and engineering of the PapMV nanoparticles into innovative vaccines.
Keywords: Plant virus, Alphaflexiviridae, protein structure, cryo-EM, X-ray crystallography
Introduction
Papaya mosaic virus (PapMV) is a filamentous plant virus that belongs to the genus of Potexvirus and the family of Alphaflexiviridae. The virus contains 6656-base positive sense single strand RNA that encodes five proteins. The virus coat protein (CP, locus:NP_044334.1), the only structural protein of the virus, is a 215-residue peptide that self assembles around the viral RNA into a flexible helical sheath of 530 n min length. This flexible property is in stark contrast with another major group of rod-shaped plant virus with rigid rod morphology. Fiber diffraction of the oriented sols of the filamentous viruses was highly successful in revealing the atomic structures of several rigid rod tobamoviruses, including the tobacco mosaic virus (TMV) at 2.9 Å resolution, the cucumber green mottle mosaic virus at 3.4 Å resolution, and the ribgrass mosaic virus at 2.9 Å resolution. Most recently, several groups have determined the structures of TMV by cryo-EM at 4.7 Å, 4.6 Å resolution, and at 3.3 Å resolution. The tobamoviruses are homologous in sequence identity (40 ~ 50%) and share the same four α-helix bundle fold.
In contrast to the rich structural information on the rigid rod tobamoviruses, high-resolution structural information for the flexible filamentous virus has been lacking despite decades of structural effort. PapMV was initially studied by fiber diffraction, which provided basic geometric information. Vibrational circular dichroism studies showed that the coat proteins of the filamentous flexible viruses Potato virus X (PVX), PapMV, and Narcissus mosaic virus (NMV) are dominated by helical structures but with noticeable structural differences from the rigid rod TMV. Low-resolution cryo-EM structures of the flexible filamentous PVX and soybean mosaic virus (SMV) were reported recently. The cryo-EM maps revealed that both PVX and SMV are helical structures with a diameter of 130 ~ 140 Å and a central channel of ~ 30 Å. PVX and SMV also had a similar helical symmetry of approximately nine subunits per turn. Efforts directed towards obtaining high-resolution structures of flexible filamentous viruses have been going on for decades, the closest being a preliminary crystallization report of PapMV coat protein, however no crystal structures have been reported so far. The challenge for determining the crystal structure of CP of any filamentous virus has been to produce a low molecular weight CP that is not prone to aggregation. We have previously shown that the PapMV CP and modified versions of the protein can be produced in bacteria where they self assemble into nanoparticles in E. coli that resemble the wild type PapMV virus purified from infected plants. This expression system was used for biochemical investigation of the PapMV CP and also for engineering chimeric recombinant nanoparticles that may be used as adjuvant or as a vaccine platform. PapMV nanoparticles appear to be perceived by the innate immune system as a pathogen associated molecular pattern. This property makes them excellent immunomodulatory molecules for improving the seasonal flu vaccine or the typhoid fever candidate vaccine. By systematic mutagenesis, we found that Phe13 of PapMV CP is critical for self assembly. We also found that Lys97 is critical for binding of PapMV CP subunits to the viral RNA, and that Glu128 improves the length of nanoparticles produced in bacteria, possibly through an improved affinity for the viral RNA.
We combined cryo-EM of the intact PapMV and X-ray crystallography of the CP to characterize the structure of the filamentous flexible virus. Our studies provide structural information that may be useful for engineering and refining the vaccine platform.
Results and Discussion
Limited Proteolysis and Mass Spectroscopy
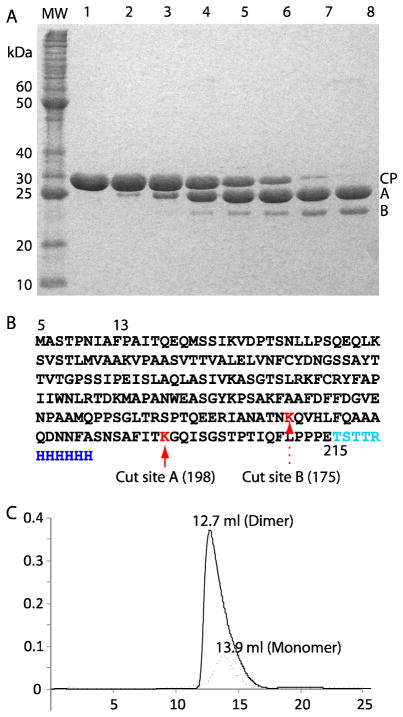
The full-length PapMV CP contains 215 residues and has a theoretical mass of 23.8 kDa. We cloned and expressed in E. coli PapMV CP missing the N-terminal five residues (6 - 215). An Ala2 was added to accommodate cloning into the NcoI site. We showed previously that the N-terminal 26 residues of CP are required for self-assembly, but removing the N-terminal five residues does not prevent the protein from self assembly to form the virus-like particle (VLP). Unfortunately, this protein cannot be concentrated sufficiently for crystallization because it tends to aggregate into heterogeneous large molecular weight complexes. Therefore, we carried out a limited trypsin digestion of purified PapMV CP (6-215)to generate a more soluble core for crystallization; two new bands(A and B) were observed in SDS-PAGE (Fig. 1A). Overnight trypsin treatment at higher enzyme to protein ratio (1:140 and 1:70) converted most of the protein into the lower band B (Data not shown). Tandem mass spectroscopy identified the two bands as the proteolysis products of PapMV CP cleaved at Lys198(Band A) and Lys175 (Band B), respectively (Fig. 1B). The final cleavage product is a 170-residue peptide corresponding to Met6 to Lys175 of PapMV CP. This protein covers ~ 80% of the wild type PapMV CP sequence. We further purified the overnight trypsin-treated products by gel filtration. We found that the digested and concentrated protein eluted from the Superdex 200 gel filtration column earlier (12.7 ml) than the original PapMV CP(13.9 ml), indicating dimerization of the cleavage product (Fig. 1C). The cleavage product was still aggregation-prone at concentrations higher than 10 mg/ml, thus we concentrated the protein to 5 mg/ml and setup crystallization plates immediately after purification and before the onset of aggregation.
Fig 1. Limited proteolysis of PapMV CP.
(A) SDS-PAGE of PapMV CP after trypsin digestion at a protein to enzyme ratio of 1:280. Lane 1 through 8 show digestion products at time point 0, 2,5, 10, 15, 20, 30, and 50 min, respectively. Protein digestion was terminated with a trypsin inhibitor cocktail. Band A and B are proteolytic products of CP. (B) Full amino acid sequence of the N-terminal truncated and C-terminal 6x His tagged PapMV construct. The proteolytic sites are determined by mass spectrometry. (C). Size exclusion chromatograms(Superdex 200 10/300 GL) of the undigested (dotted curve) and the digested CP (from residue 6 to 175) (continuous curve). The earlier elution volume and the asymmetrical elution profile indicate that the digested CP product dimerizes at higher concentration.
Crystal Structure of PapMV CP
The best crystals, with a size of ~10μm, were obtained within two weeks after setting up the plates at 4 °C with the hanging drop method in solution containing 100 mM Hepes, pH 7.6, 9% PEG 6000. Although they were tiny, these crystals diffracted X-rays strongly at the National Synchrotron Light Source (NSLS) beamline X25. We searched for heavy atom derivatives by blue native gel electrophoresis of the purified protein incubated with a panel of heavy atom compounds at 2 mM concentration, and found that the protein band was visibly shifted by Hg(Ac)2. We then soaked the crystals in freshly-prepared mother liquor with 2 mM Hg(Ac)2, and collected SAD data. The crystal structure of PapMV CP was solved at 2.7-Å resolution (See Table 1 for data collection and crystallographic statistics). The N-terminal five residues and the C-terminal Lys175 of the protein were disordered, so 164 residues from Ile11 to Asn174 were modeled in the crystal structure.
Table 1.
Data reduction and Crystallographic statistics of PapMV CP.
| Native | Hg2+ derivative peak | |
|---|---|---|
| Data collection | ||
| Space group | P65 | P65 |
| Cell dimensions | ||
| a × b × c (Å) | 72.94, 72.94, 133.33 | 73.20,73.20, 132.86 |
| α×β×γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 45-2.70 (2.75-2.7) | 25.0-3.20(3.26-3.20) |
| Wavelength (Å) | 1.100 | 1.0082 |
| Rsym* | 12.9 (44.6) | 18.4(46.8) |
| Completeness(%)* | 99.8 (99.0) | 99.9(99.6) |
| Redundancy* | 8.5(8.3) | 5.8(5.6) |
| I/σ(I)* | 16.7(4.2) | 7.7(2.9) |
| f′ (Hg) | −15.21 | |
| f″ (Hg) | 9.62 | |
| No. reflections | 93,946 | 76,543 |
| Unique reflections | 11,087 | 13,141 |
| Refinement Statistics | ||
| Resolution (Å) | 30-2.7 | |
| Rwork/Rfree(%)*,** | 16.8 (22.8) | |
| No. of Atoms | 2,621 | |
| Protein | 2,468 | |
| H2O | 153 | |
| Rms deviations | ||
| Bond length(Å) | 0.019 | |
| Bond angle(°) | 1.710 | |
| Overall B-factor (Å2) | 29.9 | |
| Ramachandran statistics | ||
| favored(%) | 96.0 | |
| allowed(%) | 4.0 | |
| Disallowed (%) | 0.0 | |
| MolProbity | ||
| Clashscore, all atoms | 4.67[100th]# | |
| Molprobity Score | 2.10[97th] | |
Numbers in brackets refer to highest resolution shell.
A random 5% of the reflection data was omitted in the refinement and used to calculate Rfree.
Numbers in square brackets are the percentiles among structures of comparable resolution. 100th percentile is the best; 0th percentile is the worst.
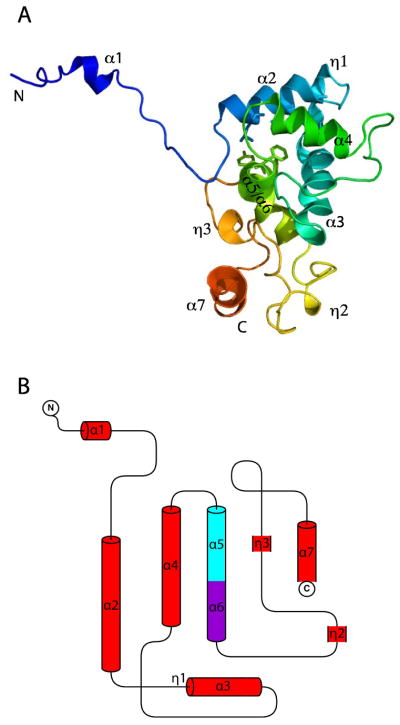
The CP structure consists of an all-helix fold with seven α-helices (Fig. 2A and B). Helix 5 (H5) and H6 are actually from one helix, broken into two segments by a ~ 60° kink at Pro112. No similar structure was found in the Protein Data Bank (PDB) and DALI database server. Therefore, the PMV coat protein structure is a novel fold.
Fig 2. Crystal structure of the PapMV CP monomer.
(A) One CP structure is shown in cartoon view. The structure is colored in rainbow scheme from blue at the N-terminus to red at the C-terminus. (B) Topology of the seven α-helices of the PapMV CP.
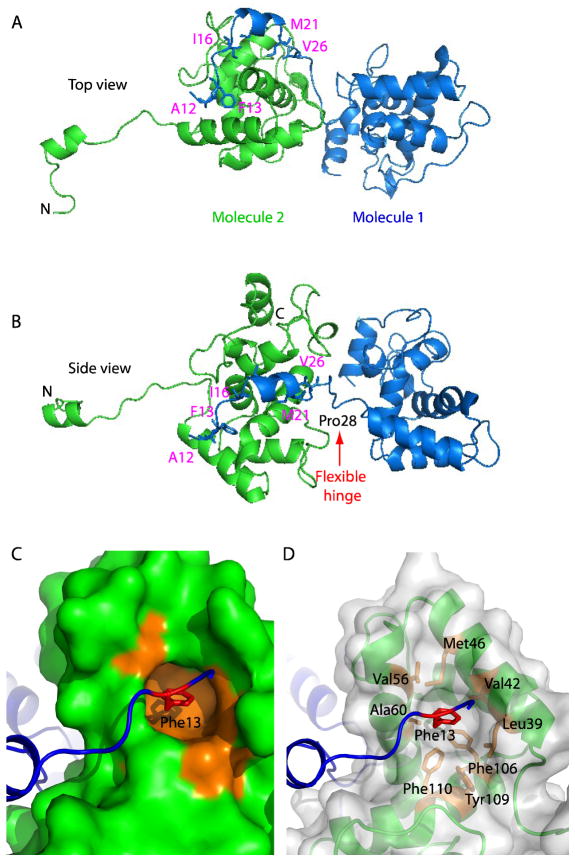
The truncated PMV CP is crystallized as an asymmetrical dimer. In the crystal structure, one protein molecule interacted with its neighbor via the extensive N-terminal peptide contacts(Fig. 3A and B). The subunit-subunit interface is formed via hydrophobic interaction, with a long groove in one protein occupied by the N-terminal peptide 11–26 of its neighbor. Important residues in the resolved 16-residue N-terminal peptide include Ala12, Phe13, Ile16, Met21, Ile24, and Val26 (Fig. 3A and B). The protruding N-terminal peptide is linked to the core domain via a flexible hinge loop centered at Pro28 between H1 and H2(the red arrow in Fig 3B). We previously mutated one of these residues, Phe13, and found that such mutation abolished self-assembly, implying a crucial role for this aromatic residue in the interaction between the subunits of the nanoparticles. In the crystal structure, Phe13 dips deeply into and fits snugly within a hydrophobic pocket in the neighboring molecule formed by Leu39, Val42, Met46, Val56, Ala60, Phe106, Tyr109, and Phe110 (Fig. 3C and D). This, coupled with our previous demonstration that deletion of the N-terminal 26 residues or 13 residues or mutation of Phe13 to Ala13 entirely abolishes the subunit-subunit interaction, rendering the protein assembly-incompetent, strongly suggests that hydrophobic interaction between the N-terminal peptide and the neighboring deep hydrophobic groove is the underlying mechanism of polymerization of the filamentous plant virus.
Fig 3. Crystal structure of the a symmetrical PMV CP dimer.
(A and B) Crystal dimer structure of the PapMV is shown in cartoon view. Note that the N-terminal peptide of the green molecule wraps around the neighboring magenta molecule via a domain swapping mechanism and that a flexible hinge region, marked by a red arrow, links the two molecules. (C) Phe13 of the N-terminal peptide fits snugly inside the hydrophobic pocket of the neighbor CP. (D) Residues contributing to the hydrophobic interaction are labeled and shown as sticks.
Domain swapping is shown in a number of systems as one mechanism for protein oligomerization. A dimer forms if domain swapping is reciprocal: protein A donates a domain to protein B and vice versa. Larger oligomers form if the swapping is not immediately reciprocated and is open ended, i.e., protein A donates a domain to B, and B to C, and so on. An oligomeric ring results if the last protein donates the domain to the first protein A. However, along polymer can result if the domain swapping propagates forward ad infinitum. Based on the asymmetrical dimer crystal structure, we propose an open-ended N-terminal loop swapping PMV assembly mechanism by which the CP subunits in the helical virus are likely linked together at the lateral interfaces via the protruding N-terminal peptides. Such a mechanism is compatible with the biochemical observation that the CP alone is prone to aggregation, and is capable of forming VLP in the absence of RNA. This mechanism is also supported by our previous mutation study indicating the crucial role of Phe13 in the domain-swapped N-peptide in polymerization. We further suggest that the flexible hinge loop may partially account for the flexible nature of the large family of filamentous plant viruses.
Cryo-EM of Intact PapMV
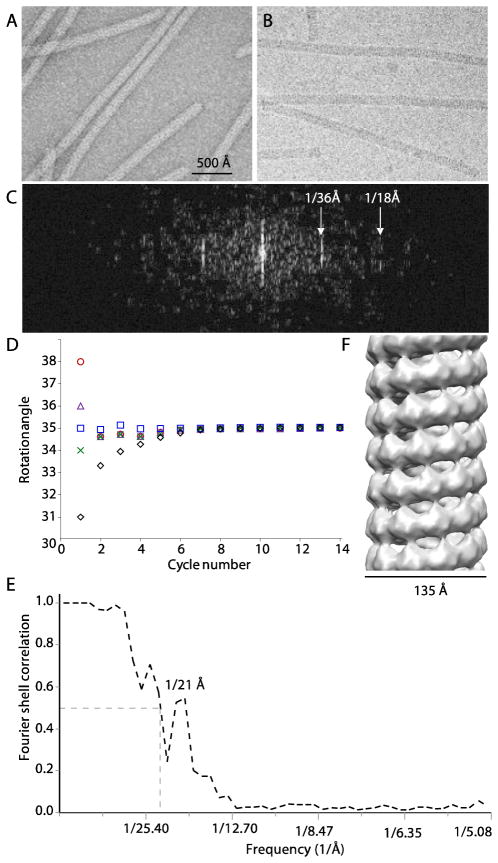
The flexibility of the Alphaflexiviridae members such as PapMV has been the main obstacle to orienting the viruses for high-resolution fiber diffraction study. To determine how individual CP molecules are arranged in PapMV, we carried out a preliminary cryo-EM study of the intact virus. The virus preparation was first examined by negative stain EM, revealing the expected filamentous morphology, about 14 nm wide and 530 nm long (Fig 4A). We then froze the purified viruses in vitreous ice and carried out cryo-EM data collection (Fig. 4B). The filamentous virus has a smooth surface. In the power spectrum of cryo-EM image of a straight segment of the virus, layer lines at the 1/36 Å and 1/18 Å reciprocal positions can be observed(Fig. 4C), clearly indicating the helical symmetry of the capsid proteins, and the 36 Å subunit repeat distance along the length of the virus. The vast majority of the viral particles are highly flexible, displaying no higher order layer lines beyond the first 1/36 Å layer line in their respective power spectrum, signifying that the reciprocal space based helical reconstruction algorithm is not suitable for dealing with the cryo-EM images of the PapMV filaments. We turned to the iterative helical real space reconstruction algorithm (IHRSR), a method shown to be effective for extracting structural information from less well ordered and flexible helical filaments. We previously designed an essentially the same real space single particle cryo-EM image reconstruction procedure for the flexible helical objects, and used the procedure to improve the microtubule 3D map to sub-nanometer resolution.
Fig 4. Cryo-EM of PapMV.
(A) Electron micrograph of negatively stained PapMV. (B) A selected area of a cryo-EM image ((A) and (B) are in same scale). (C) Power spectrum of a cryo-EM image from a straight section of PapMV, showing the helical symmetry of PMV. (D) The subunit rotational angles of PapMV converge to 35.2° after several cycles of refinement. (E) Fourier shell correlation of the cryo-EM 3D map. Note that the two dips of the FSC curve at 23 Å and 35 Å are caused by the uncompensated contrast transfer function effect due to the use of similar defocus values. (F) Surface rendering of the cryo-EM 3D map of PapMV.
We first manually selected 5616 PapMV segments based on the image contrast and the visibility of the first layer lines. Starting from a wide range of helical parameters, ranging from 31° to 38°, the helical symmetry search program converged to an inter-subunit rotation angle of 35.15°(Fig. 3D) and an axial rise per subunit of 3.6 Å. We note that the starting angular range (31° – 38°) does not limit the final angel to be within this range. In other words, if the true rotation angle were outside the range, say 30° or even smaller, the starting angle of 31° would have been sufficient to find that. The fact that such wide angular range we have used all converges to the same value signifies the accuracy of the determined angle. The 3D reconstruction had an estimated resolution of 21 Å (Fig. 4E). Further refinement against a larger dataset containing 36,643 segments, however, did not improve the resolution, indicating that the attainable resolution of PapMV 3D map may be limited by helical order or more likely by the cryo-EM image quality, rather than by the size of the dataset. The Fourier shell correlation curve contains two dips at frequencies 1/23-Å and 1/35-Å caused by the uncompensated contrast transfer function effect (Fig. 4E), owning to the use of similar under focus values (−2.5 to −3.0 μm) during image recording. Thus future cryo-EM work with a larger dataset recorded with broader focus value will likely increase the resolution. In our current low-resolution 3D reconstruction, the PapMV filament has an outer diameter of ~ 135 Å (Fig. 4F), similar to that of PVX and SMV. The helical symmetry of PapMV is approximately 41 subunits in 4 turns, about one subunit larger than the nearly 9-subunit per turn symmetry of PVX and SMV. This difference is perhaps not surprising because, although the diameter of these filamentous viruses appears to be similar, the size of PapMV CP (215 residues) is markedly smaller than that of PVX (237 residues) and SMV (347 residues), allowing one additional subunit of PMV within the similar circumferences. Furthermore, sequence alignment in ClustalW2 found only 3% sequence identity and 15% similarity among these viruses. Therefore, although all three viruses belong to the Alphaflexivirus family, their structures may have sufficiently diverged to accommodate altered axial interactions.
Docking with the PapMV CP Crystal Structure
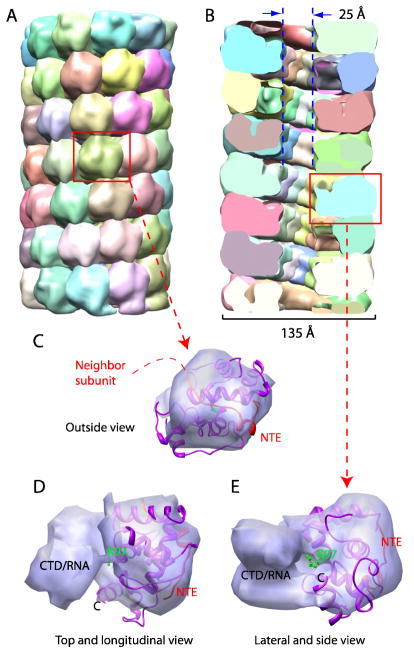
The low-resolution 3D EM map was subjected to semi-automatic density segmentation by Segger, as implemented in UCSF Chimera. Only default parameters were used during the process (Smoothing steps=3, step size = 1 voxel). The segmentation result is shown in Fig. 5 as both a surface view and as a centrally cut-open view with a vertical plane, with each density, presumably corresponding to an individual capsid protein subunit and its associated nucleic acids, colored differently (Fig. 5A and 5B). The cut-open view of the PapMV structure reveals a central low-density region, likely a channel about 25 Å in diameter (5B).
Fig 5. Density segmentation and docking of the cryo-EM map of PapMV.
(A) Surface rendered view of PapMV after automatic segmentation. Each presumptive protein subunit is shown in a different color. The display threshold is set to enclose the expected protein mass. (B) A vertical central section of the segmented cryo-EM map reveals the 25-Å diameter inner channel of PapMV. (C–E) Cryo-EM density of a presumptive single PapMV CP extracted from the segmented 3D map shown in outside surface view (C), in the longitudinal and top view (D), and in the lateral and side view (E). The docked crystal structure of PapMV CP with the N-terminal peptide of a neighbor is shown in magenta and red, respectively. The potential RNA-interacting Lys97(green)is shown in stick view.
The individual subunit density has an approximate dimension of 3.5 nm by 3.5 nm by 5 nm, longer in the radial direction (Fig. 5C–E). The crystal structure of PapMV CP together with the neighboring N-terminal peptide can be fitted as a single rigid body into the presumed subunit density, with the N-terminal peptide from the neighbor subunit facing outwards (Fig. 5C–E). The crystal structure fits well within the 3D map in all three perpendicular views, with a correlation coefficient of ~ 0.8 (Fig. 5C–E). In this docking, the N-terminal peptide from the neighbor subunit (NTE, red) is exposed to the filament surface (Fig. 5C–E). After docking, there is a smaller density region towards interior of the filament that is left unoccupied by the crystal structure. While its exact identity is currently unknown, we suggest that the unoccupied density contains the genomic RNA and the C-terminal fragment of CP that is proteolytically removed in our crystal structure. Consistent with this suggestion, residue Lys97 in the docked crystal structure, mutation of which was shown previously to affect the RNA binding activity of PapMV, faces the smaller interior density labeled as CTD/RNA (Fig. 5D and 5E). We should note that our tentative assignment of C-terminal region (40 residues) to the interior of PMV does not necessarily preclude the very C-terminus of the CP from being surface exposed; it requires only several residues to extend to the surface.
Because of the low resolution of the current cryo-EM map, and the small size of the crystal structure, we refrained from docking multiple crystal structures into the 3D map of the virus to deduce residues involved in lateral and longitudinal subunit-subunit interfaces. A fitting ambiguity of only a few degrees, quite likely at this low resolution, will result in different sets of interacting residues at the interface.
Conclusions and future directions
The crystal structure of the PapMV CP reported here represents, to our knowledge, the first of any flexible filamentous plant virus. The structure reveals a novel all helix fold of the PMV coat protein, suggests that CP may assemble in the filamentous virus by the flexible N-terminal loop via an open-ended loop swapping mechanism. We further suggest that the flexible loop mediated assembly may partially account for the intrinsically flexible morphology of this virus. If true, this mechanism will be fundamentally different from the assembly mechanism of TMV in which the long helix bundling interaction dominates the subunit interfaces, and likely confers the structural rigidity to the rod virus. Docking the crystal structure of the PMV CP core into the low-resolution cryo-EM map of the intact virus suggests that the N-terminal polymerizing loop is surface exposed, and that the C-terminal domain and the genomic nucleic acids are located to the interior surface of the hollow filamentous virus.
Although it is a significant step forward in revealing the structure and mechanism of PMV, our current work leaves several important questions: What is the structure and location of the C-terminal fragment of the PMV coat protein? How the genomic RNA is packaged within the filamentous virus? What are the longitudinal and lateral subunit-subunit interfaces? What are the structural features that eventually contribute to the mechanical flexibility of the virus? These questions await future structural investigation. The current crystal structure will guide us in designing non-aggregating CP constructs for future crystallographic studies that include the C-terminal peptide and that are capable of binding to nucleic acids. Admittedly, improving the resolution of the cryo-EM 3D map of the flexible filamentous virus will not be an easy task. Nevertheless, the small Fourier shell correlation peak at ~ 1/15-Å (Fig. 4E), well beyond the current 21 Å resolution, gives us confidence that a higher quality and larger dataset recorded in a broader under focus range will likely improve the 3D reconstruction. The structure of PapMV will eventually help improve our engineering of this vaccine platform and will be useful as a model to elucidate the key residues located at the surface of the nanoparticles that are involved in the interaction with the immune cells.
Materials and Methods
Expression and Purification of Recombinant PapMV CP(6-215) from E. coli
The recombinant PapMV CP(6-215) was cloned into the pET-3d expression vector, expressed in the strain BL21(DE3) RIL. The bacteria were grown at 37 °C in LB media containing 50 ug/ml of ampicillin. Protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) once OD600 of growing culture reaches 0.6. After addition of IPTG, the culture continued growing for 16 hours at 20 °C. Bacteria were harvested by centrifugation for 10 min for 8000 g. The pellet was then re-suspended in ice-cold lysis buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl) and the bacteria were lysed with microfluidics 4 times with the pressure of 80 psi. The lysate was centrifuged for 30 min at 32,000 g to remove cellular debris. The following steps of purification were performed with FPLC workstation. The lysate supernatant after centrifugation was applied onto 5 ml of Ni-NTA column at the flow rate of 2.5 ml/min. The loaded column was washed with 60 ml of washing buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl) containing increasing concentrations of imidazole (20 mM, 40 mM, 60 mM), respectively. Proteins then were eluted out in elution buffer (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 300 mM imidazole). Purified proteins were dialyzed against 10 mM Tris-HCl, pH 8.0, 50 mM NaCl at 4°C with 3 times dialysis buffer change. The purity of the proteins was determined by SDS-PAGE.
Limited Proteolysis and Mass Spectrometry
Purified recombinant PapMV CP proteins were subjected to trypsin proteolysis at amass ratio of 1:280 trypsinto protein at room temperature. The proteolysis was terminated by 1X complete protease inhibitor cocktail tablet (Roche) at different time points. Digested PapMV CP at different time points was then subjected to a 15% SDS-PAGE gel. Time points at which PapMV CP showed two clear fragment bands were considered good time points for limited proteolysis. These two bands were identified with mass spectrometry. The partially digested PapMV CP was then loaded on a column of Superdex 200 10/300GL(GE Healthcare) and washed out with buffer (10mM Tris-HCl, pH8.0, containing 50mM NaCl). The eluted PapMV CP was collected and concentrated to 5.0mg/ml in a concentrator with molecular weight cut-off of 10-kDa.
Crystallization of Limited Trypsin-Digested PapMV CP
The partially digested PapMV CP in 10 mM Tris-HCl, pH 8.0, 50 mM NaCl was set up in crystallization plates with the hanging drop method at 4 °C. The hanging droplet contains 2 μl of sample at 5.0 mg/ml and 1 μl of reservoir solution (100 mM Hepes, pH 7.6, 10% PEG 6000, 5% MPD). Crystals with approximate dimensions of 20 × 15 × 10 μm were observed after 13 days. A series of heavy atom derivatives were screened by native gel electrophoresis of purified PapMV CP and the results showed mercury acetate (Hg(Ac)2) caused a downward shift of the protein on the native gel, so Hg(Ac)2 was chosen for derivatizing the crystals. Crystal-containing hanging droplets were incubated with 2 mM mercury acetate at 4 °C for 12 hours for heavy metal derivatization. 24% glycerol (W/V) in the mother liquid was used as cryo-protectant for flash-freezing crystals.
Diffraction Data Collection and Crystal Structure Solution
All diffraction data were collected at 100 K at beamline X25 at the NSLS, Brookhaven National Laboratory. The mercury-derivatized crystal was used for anomalous data collection at peak position based on spectrum results. The native crystals diffracted up to 2.3Å but the dataset contained strong ice rings and spots; we therefore exclude dice diffractions by cutting off the dataset at 2.7Å. All datasets were indexed, integrated and processed with HKL2000. The heavy atom sites were searched by the SIR/AS method in the Shelx-HKL2MAP suite. Initial phases were improved by Phenix software suite. The model was manually built and expanded to full sequence coverage by using Coot. The structure was refined to Rwork of 0.17 and Rfree of 0.23, using Phenix and Refmac in CCP4. All crystal structure figures were drawn in PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC).
PapMV Production and Isolation
PapMV was purified from infected papaya leaves showing mosaic symptoms. Infected leaves (100 mg) were grounded in 100 mL 50 mM Tris/HCl pH 8.0 containing 10 mMEDTA in a commercial blender. The grounded leaves were filtered through the cheese-cloth and 1% of Triton X-100 was added to the filtrate and gently stirred for 10 min. Drop by drop, chloroform was added until one-quarter the volume of the filtrate. The solution was stirred for an additional 30 min at 4 °C and centrifuged for 20 min at 10 000 g to remove the precipitate. The supernatant was subjected to a high speed centrifugation (100 000 g) for 120 min in a Beckman 50.2 Ti rotor. The viral pellet was resuspended and subjected to another high speed centrifugation through a sucrose cushion (30% sucrose) at 100 000 g for 3.5 h. The final viral pellet was resuspended in 10 mL 50 mM Tris pH 8.0. If the colouration persisted, an additional clarification with chloroform was performed and the purified virus was collected by ultra centrifugation.
Cryo-EM
Sample grids for cryo-electron microscopy were prepared by applying 2.5 – 4.5 μl of 1 mg/ml papaya mosaic virus to a lacy carbon film substrate on copper grids, rendered hydrophilic via glow discharge. In an automated process carried out by a vitrobot, the excess liquid from the droplet was blotted away with filter paper, immediately followed by plunging of the sample grid into liquid ethane cooled by liquid nitrogen. Cryo-EM images were collected at 60,000X magnification using a Jeol 2010 (200 kV) transmission electron microscope equipped with a Gatan cryo-holder and using Kodak So-163 film. Film was developed under darkroom conditions and digitized using a Nikon scanner at 4000 dpi (14 bit) resolution; micrographs were then compressed to a final resolution of 2.54 Å/pixel. The defocus values of the micrographs were determined from the carbon film using the EMAN program suite, and were used to correct for the phase component of the contrast transfer function.
Image Processing and 3D Reconstruction
IHRSR was used for the three-dimensional reconstruction of virion images from cryo-electron micrographs. This method, based on single-particle reconstruction methods, has proven to be a highly effective method of reconstruction for flexible filamentous plant viruses too disordered over long distances to allow Fourier-Bessel methods to be reasonable. Segments of virions were selected with a 90% overlap between adjacent segments using the program BOXER, also from the EMAN suite. Of 36,643 segments selected from 76 micrographs, 5616 segments were carefully chosen, based on clearly visible and symmetric first layer line intensities in the power spectra, as well as compatibility with alignment, and used in subsequent IHRSR reconstructions using the SPIDER software package. Reconstructions starting from disparate rotation angles (31, 34, 35, 36, 38°) were performed with relaxed search boundaries. In other words, we used the shift search range that was larger than the subunit axial rise to ensure the identification of correct reference projections. The data converged after ~10 cycles (of 50 run) to the same value for all starting conditions. We noticed that the built-in resolution estimation routine in IHRSR was not able to produce a reliable FSC curve. Instead, we used the SPIDER program for this purpose. We split the dataset into two sets (even and odd numbered particles), calculated two 3D maps, and then used the SPIDER command “RF 3” to calculate the Fourier shell correlation curve. The resolution of the final reconstruction was 20.8 Å using a threshold value of 0.5. Although there is a second correlation peak that reaches above 0.5 threshold at ~ 15 Å, the resolution of the 3D reconstruction is ultimately limited by incomplete compensation of the zeros of the contrast transfer function due to the use of similar defocus values when recording EM images (−2.5 to −3 μm). We did not determine the handedness of PMV, instead, the previously shown right handedness of the related PVX and SMV is applied to our reconstruction. The low-resolution model provides a molecular envelope with which the orientation of crystal structure was derived via computer-based rigid body docking. UCSF Chimera was used for docking and for generating surface rendering. Surface rendering threshold was set to include the expected protein mass. Map segmentation was carried out automatically with Segger as implemented in Chimera v1.6.
Highlights.
No high-resolution structure was reported for any flexible filamentous plant virus.
We have determined the crystal structure of the papaya mosaic virus coat protein.
The structure reveals an amino terminal loop swapped asymmetrical dimer.
Docking crystal structure into cryo-EM map of the virus reveals its architecture.
Acknowledgments
We thank Edward Egelman for sharing his IHRSR program, William Rice and Andrew Pomfret for help with initial helical image processing. H.L. was supported by BNLLDRD 10-016 and NIHGM74985. D.L. was supported by the Canadian NSERC grant #227041. Diffraction data for this study were measured at beamline X25 of the National Synchrotron Light Source. Financial support comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources (P41RR012408) and the National Institute of General Medical Sciences (P41GM103473) of the National Institutes of Health
Footnotes
Accession number
The atomic coordinates and structural factor amplitudes for PapMV CP have been deposited in the Protein Data Bank with accession code 4DOX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009) Archives of virology. 2010;155(1):133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abouhaidar MG, Erickson JW. Molecular Plant Virology. CRC Press; Boca Raton, Florida: 1985. Structure and in vitro assembly of papaya mosaic virus; pp. 85–121. [Google Scholar]
- 3.Erickson JW, Bancroft JB, Horne RW. The assembly of papaya mosaic virus protein. Virology. 1976;72(2):514–517. doi: 10.1016/0042-6822(76)90180-x. [DOI] [PubMed] [Google Scholar]
- 4.Dolja VV, Boyko VP, Agranovsky AA, Koonin EV. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology. 1991;184(1):79–86. doi: 10.1016/0042-6822(91)90823-t. [DOI] [PubMed] [Google Scholar]
- 5.Verchot-Lubicz J, Ye CM, Bamunusinghe D. Molecular biology of potexviruses: recent advances. J Gen Virol. 2007;88(Pt 6):1643–1655. doi: 10.1099/vir.0.82667-0. [DOI] [PubMed] [Google Scholar]
- 6.Namba K, Pattanayek R, Stubbs G. Visualization of protein-nucleic acid interactions in a virus. Refined structure of intact tobacco mosaic virus at 2.9 A resolution by X-ray fiber diffraction. J Mol Biol. 1989;208(2):307–325. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Stubbs G. Structure determination of cucumber green mottle mosaic virus by X-ray fiber diffraction. Significance for the evolution of tobamoviruses. J Mol Biol. 1994;239(3):371–384. doi: 10.1006/jmbi.1994.1379. [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Culver JN, Stubbs G. Structure of ribgrass mosaic virus at 2.9 A resolution: evolution and taxonomy of tobamoviruses. J Mol Biol. 1997;269(5):769–779. doi: 10.1006/jmbi.1997.1048. [DOI] [PubMed] [Google Scholar]
- 9.Sachse C, et al. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J Mol Biol. 2007;371(3):812–835. doi: 10.1016/j.jmb.2007.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clare DK, Orlova EV. 4.6A Cryo-EM reconstruction of tobacco mosaic virus from images recorded at 300 keV on a 4k × 4k CCD camera. J Struct Biol. 2010;171(3):303–308. doi: 10.1016/j.jsb.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge P, Zhou ZH. Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci U S A. 2011;108(23):9637–9642. doi: 10.1073/pnas.1018104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richardson JF, Tollin P, Bancroft JB. The architecture of the potexviruses. Virology. 1981;112(1):34–39. doi: 10.1016/0042-6822(81)90609-7. [DOI] [PubMed] [Google Scholar]
- 13.Baratova LA, et al. The organization of potato virus X coat proteins in virus particles studied by tritium planigraphy and model building. Virology. 1992;188(1):175–180. doi: 10.1016/0042-6822(92)90747-d. [DOI] [PubMed] [Google Scholar]
- 14.Baratova LA, et al. The topography of the surface of potato virus X: tritium planigraphy and immunological analysis. J Gen Virol. 1992;73 (Pt 2):229–235. doi: 10.1099/0022-1317-73-2-229. [DOI] [PubMed] [Google Scholar]
- 15.Parker L, Kendall A, Stubbs G. Surface features of potato virus X from fiber diffraction. Virology. 2002;300(2):291–295. doi: 10.1006/viro.2002.1483. [DOI] [PubMed] [Google Scholar]
- 16.Tollin P, Bancroft JB, Richardson JF, Payne NC, Beveridge TJ. Diffraction studies of papaya mosaic virus. Virology. 1979;98(1):108–115. doi: 10.1016/0042-6822(79)90529-4. [DOI] [PubMed] [Google Scholar]
- 17.Shanmugam G, Polavarapu PL, Kendall A, Stubbs G. Structures of plant viruses from vibrational circular dichroism. J Gen Virol. 2005;86(Pt 8):2371–2377. doi: 10.1099/vir.0.81055-0. [DOI] [PubMed] [Google Scholar]
- 18.Kendall A, et al. Structure of flexible filamentous plant viruses. J Virol. 2008;82(19):9546–9554. doi: 10.1128/JVI.00895-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Todderud E, Stubbs G. Crystallization and preliminary X-ray analysis of papaya mosaic virus coat protein. J Mol Biol. 1993;234(3):885–887. doi: 10.1006/jmbi.1993.1634. [DOI] [PubMed] [Google Scholar]
- 20.Tremblay MH, et al. Effect of mutations K97A and E128A on RNA binding and self assembly of papaya mosaic potexvirus coat protein. Febs J. 2006;273(1):14–25. doi: 10.1111/j.1742-4658.2005.05033.x. [DOI] [PubMed] [Google Scholar]
- 21.Denis J, et al. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine. 2008;26(27–28):3395–3403. doi: 10.1016/j.vaccine.2008.04.052. [DOI] [PubMed] [Google Scholar]
- 22.Savard C, et al. Improvement of the trivalent inactivated flu vaccine using PapMV nanoparticles. PLoS One. 2011;6(6):e21522. doi: 10.1371/journal.pone.0021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denis J, et al. Immunogenicity of papaya mosaic virus-like particles fused to a hepatitis C virus epitope: evidence for the critical function of multimerization. Virology. 2007;363(1):59–68. doi: 10.1016/j.virol.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Leclerc D. Plant Viral Epitope Display Systems for Vaccine Development. Curr Top Microbiol Immunol. 2011 doi: 10.1007/82_2011_183. [DOI] [PubMed] [Google Scholar]
- 25.Lacasse P, Denis J, Lapointe R, Leclerc D, Lamarre A. Novel plant virus-based vaccine induces protective cytotoxic T-lymphocyte-mediated antiviral immunity through dendritic cell maturation. J Virol. 2008;82(2):785–794. doi: 10.1128/JVI.01811-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acosta-Ramirez E, et al. Translating innate response into long-lasting antibody response by the intrinsic antigen-adjuvant properties of papaya mosaic virus. Immunology. 2008;124(2):186–197. doi: 10.1111/j.1365-2567.2007.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laliberte Gagne ME, Lecours K, Gagne S, Leclerc D. The F13 residue is critical for interaction among the coat protein subunits of papaya mosaic virus. Febs J. 2008;275(7):1474–1484. doi: 10.1111/j.1742-4658.2008.06306.x. [DOI] [PubMed] [Google Scholar]
- 28.Lecours K, Tremblay MH, Gagne ME, Gagne SM, Leclerc D. Purification and biochemical characterization of a monomeric form of papaya mosaic potexvirus coat protein. Protein Expr Purif. 2006;47(1):273–280. doi: 10.1016/j.pep.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20(1):478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 30.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4(12):2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157(1):83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Li H, DeRosier DJ, Nicholson WV, Nogales E, Downing KH. Microtubule structure at 8 A resolution. Structure (Camb) 2002;10(10):1317–1328. doi: 10.1016/s0969-2126(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 33.Goujon M, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Web Server issue):W695–699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170(3):427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savard C, et al. Improvement of the PapMV nanoparticle adjuvant property through an increased of its avidity for the antigen [influenza NP] Vaccine. 2012 doi: 10.1016/j.vaccine.2012.01.085. [DOI] [PubMed] [Google Scholar]
- 37.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Academy Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 39.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- 40.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. The CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
- 43.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 44.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]