Abstract
Since aberrant wound healing and chronic inflammation can promote malignant transformation, we determined whether dietary bioactive fish oil (FO)-derived n-3 polyunsaturated fatty acids (n-3 PUFA) modulate stem cell kinetics in a colitis-wounding model. Lgr5-LacZ and Lgr5-EGFP-IRES-creERT2 mice were fed diets enriched with n-3 PUFA vs n-6 PUFA (control) and exposed to dextran sodium sulfate (DSS) for 5 days in order to induce crypt damage and colitis throughout the colon. Stem cell number, cell proliferation, apoptosis, expression of stem cell (Lgr5, Sox9, Bmi1, Hopx, mTert, Ascl2, and DCAMKL-1) and inflammation (STAT3) markers were quantified. DSS treatment resulted in the ablation of Lgr5+ stem cells in the distal colon, concurrent with the loss of distal crypt structure and proliferating cells. Lgr5, Ascl2 and Hopx mRNA expression levels were decreased in damaged colonic mucosa. Lgr5+ stem cells reappeared at day 5 of DSS recovery, with normal levels attained by day 6 of recovery. There was no effect of diet on the recovery of stem cells. FO fed animals exhibited higher levels of phospho-STAT3 at all time points, consistent with a higher wounding by DSS in FO feeding. n-3 PUFA-fed mice exhibited a reduction in stem cell associated factors, Ascl2, Axin2 and EphB3. These results indicate that rapidly cycling Lgr5+ stem cells residing at position 1 in the colon epithelium are highly susceptible to DSS-induced damage and that dietary cues can impact stem cell regulatory networks.
Keywords: Lgr5, dextran sodium sulfate, fish oil, Wnt signaling, colon
1. Introduction
“Adult” somatic stem cells of the colon are of particular interest because they sustain self-renewal and are target cells for cancer initiating mutations [1, 2]. Therefore, perturbations in stem cell dynamics are generally believed to represent the earliest step towards colon tumorigenesis. Exciting advances have been made in the identification of stem cells (approximately 6 per crypt), which replenish the intestinal epithelium every 3-5 days [3]. Although the exact identity of intestinal stem cells has proven controversial over the last 30 years, rapidly cycling crypt base columnar (CBC) Lgr5/GPR49 cells self-renew, actively divide, and give rise to all epithelial subtypes in the small intestine and colon [3-7]. Recent evidence indicates that intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5+ stem cells [8]. By crossing stem-cell-specific Lgr5-EGFP-IRES-creERT2 knockin mice to Apcflox/flox mice, it was unequivocally shown that crypt Lgr5+ stem cells are the cells-of-origin of intestinal cancer [4]. Collectively, these findings lend support to the cancer stem cell concept, i.e., that in some circumstances, normal intestinal stem/progenitor cells can initiate colon tumorigenesis and drive cancer progression towards metastasis [9-11]. Emerging evidence indicates that both quiescent +4 cells and CBCs have cooperative functional roles, exhibiting bi-directional lineage relationships in the small intestine and to some degree in the colon [8, 12-16]. Hence, it is now possible for the first time to visualize stem cells and examine their behavior in the context of tissue remodeling and cancer chemoprevention.
There is substantial experimental, epidemiological and clinical evidence indicating that fish oil-containing diets rich in n-3 polyunsaturated fatty acids (PUFA), e.g. docosahexaenoic acid (DHA, 22:6Δ4,7,10,13,16,19) and eicosapentaenoic acid (EPA, 20:5Δ5,8,11,14,17), are protective against colon tumorigenesis [17-25]. In a major recent finding, it was demonstrated that EPA reduced rectal polyp number and size in patients with familial adenomatous polyposis (FAP) [25]. Most impressive was the fact that fish oil derived n-3 PUFA suppressed FAP to a degree similar to the selective COX-2 inhibitor celecoxib. Collectively, these data indicate that n-3 PUFA hold promise as chemoprevention agents for FAP and sporadic colon cancer. In addition, we have recently demonstrated that n-3 PUFA impact genes which modulate the colon stem cell niche and tumor evolution, e.g., Wnt/TCF signature, Ephrin B1, BMP-4, and PGE2 [26, 27-29]. Despite the fact that aberrant wound healing/crypt regeneration can promote malignant transformation [27, 30,31], how the colonic stem cell population responds to environmental factors such as diet and chronic inflammation is not well understood. Therefore, we hypothesized that the chemoprotective effect of EPA and DHA may be attributed to a reduction in stem cell damage. Since little is known regarding homeostasis within the stem cell compartment in the large bowel, we examined the effect of dietary bioactive agents, i.e., EPA/DHA, on stem cell kinetics in a colitis-wounding model. These experiments are highly relevant because the effects of dietary cues in the context of DSS-induced damage on stem cell cytokinetics and regulatory networks in the large intestine have not been reported to date.
2. Materials and Methods
2.1 Animals and DSS treatment
All procedures involving animals followed guidelines approved by the Institutional Animal Care and Use Committee at Texas A&M University in accordance with EU Directive 2010/63/EU for animal experiments. Lgr5-EGFP-IRES-creERT2 and LacZ-Lgr5 mice, originally described by Barker et al [3, 5] (13-18 wk of age) were fed experimental diets for one wk prior to initiation of dextran sodium sulfate (DSS) treatment at 2.5% in drinking water, the only source of fluids during the treatment period. After 5 d of DSS treatment, plain water was provided during the DSS recovery period. Mice (n=3-5 per diet group and time point) were terminated on days 1-7 of DSS recovery, with EdU injected 2 h prior to termination in order to assess cell proliferation as previously described [32]. Untreated control mice were provided with tap water for the entire study period.
2.2 Experimental diets
Diets varied only in type of lipid. Experimental diets consisted of (by weight): 20% casein, 42% sucrose, 22% corn starch, 6% cellulose, 3.5% AIN-76A salt mix, 1% AIN-76A vitamin mix, 0.3% DL-methionine, 0.2% choline chloride, and 5% fat. The corn oil (CO) diet contained 5% corn oil (Dyets, Bethlehem, PA) and the vacuum deodorized Menhaden fish oil (FO) diet contained 4% fish oil (Omega Protein, Houston, TX) plus 1% corn oil. All diet ingredients except oils were obtained from Bio-Serv (Frenchtown, NJ). Three fish oil fed/DSS treated mice died during the study and were replaced. Control animals (no DSS treatment) were also fed either the CO or FO diets.
2.3 Tissue samples
Upon termination, the colon was removed, cut open, cleaned in PBS, placed lumen side up and cut in half longitudinally. Each half was Swiss rolled, half being fixed in 4% PFA in PBS for 4 h at 4°C, embedded in paraffin and used for immunohistochemistry assays. The other half was fixed in 1% formaldehyde/0.2% gluteraldehyde/0.02% NP-40 in PBS for 2 h at 4°C for LacZ staining. On a separate set of mice (n=5) receiving the same diet and treatment regimen, colons were removed, cut open and cleaned in PBS, the mucosal layer was scraped with a microscope slide and total RNA was isolated using Ambion mirVana kit followed by DNase treatment with Ambion DNAFree. RNA quality was assessed by Agilent Bioanalyzer and all samples had an RIN score greater than 9.0.
2.4 Immunohistochemistry
Cell proliferation was measured using the Click-IT EdU kit (Invitrogen, Carlsbad, CA) as per manufacturer's instructions on formalin fixed paraffin embedded (FFPE) sections. Apoptosis was assessed with the TACS 2 TdT TUNEL assay (Trevigen, Gaithersburg, MD) [33]. LacZ positive stem cells were detected with β-galactosidase staining as previously described [4]. Phospho-STAT3, Sox9 and DCAMKL1 were detected in FFPE sections after retrieval in 10 mM Tris base/1 mM EDTA (phospho-STAT3 and DCAMKL1) or 10 mM sodium citrate (Sox9) for 20 min in a sub-boiling bath. Primary antibodies were as follows: Phospho-STAT3 (Tyr705), Cell Signaling, 1:200 dilution; Sox9, Millipore, 1:1000 dilution; DCAMKL1, Abcam, 1:100 dilution. Prior to primary antibody incubation, Image-iT FX signal enhancer (Invitrogen) was used for phopho-STAT3 and Sox9 slides. Goat-anti-rabbit secondary antibody conjugated with Alexa-647 (Invitrogen) was used on all slides. For quantification of phospho-STAT3 expression, intensity of staining was measured using NIS Elements AR software on a Nikon Ti-E inverted microscope equipped with an X-cite 120 fluorescent microscopy illuminator using a 20X objective. Ten measurements were taken from each proximal or distal segment from each animal in order to determine average epithelial phospho-STAT3 expression.
2.5 Flow cytometry
Following tamoxifen induction, colonic crypts were isolated by the method of Sato et al [7] with minor modification. The intact colons were everted on a disposable mouse gavage needle (Instech Laboratories) and incubated with 20 mM EDTA in PBS at 37°C for 30 min. Following transfer to chilled Ca/Mg free HBSS, colons were vigorously vortexed to release crypts. The final crypt fraction was incubated with trypsin/DNase to produce a single cell suspension for flow cytometry of GFP positive cells using an Accuri C6 flow cytometer (BD Biosciences). In addition, for stem cell sorting, GFPhigh expressing stem cells and GPFnegative cells from the small intestine and colon were separately isolated using a BD FACSAria II cytometer/sorter (BD Biosciences).
2.6 Real Time PCR
Gene expression was quantified by TaqMan assay using an ABI 7900HT (Applied Biosystems) on control mice (no DSS treatment) and at 3 d of DSS recovery (n=5 per treatment and diet). Assays were tested for proportionality and efficiency prior to use and negative controls, omitting the reverse transcriptase enzyme, were performed for all genes. Gene expression was quantified for the following genes: Ascl2, Axin2, Bmi1, Eph3, Hopx, Lgr5, mTert, Rspo1, Sox9, Stat3 and 18S (for normalization). Because Rspo1 was nearly undetectable, quantification was repeated on a Bio-Rad QX-100 Droplet Digital PCR system using the same TaqMan assay.
2.7 RNA Sequencing
RNA (80 ng) was converted to cDNA using the NuGEN Ovation 3’-DGE kit. Three micrograms of cDNA was then fragmented (Covaris). The fragmented sample was quantified with the Quant-IT Picogreen kit (Invitrogen) and evaluated for proper fragmentation of 150 to 200 bp on an Agilent Bioanalyzer DNA 1000 chip. Subsequently, 200 ng sheared double stranded cDNA was used for the Encore Library System I kit (NuGEN) to create the Illumina libraries as per manufacturer's instructions. Libraries were quantified by Quant-IT and run on an Agilent DNA Chip 1000 to confirm appropriate size and absence of adapter dimers. Samples were sequenced on an Illumina GAII with single end, 75 bp reads.
2.8 Statistics
Data analyses were performed using either the exact Wilcoxon two-sample test or one-way ANOVA with Tukey's adjustment for multiple comparisons using SAS 9.2.
3. Results
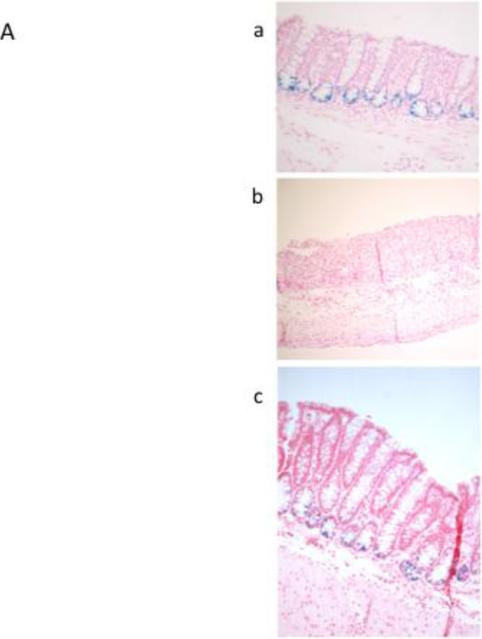
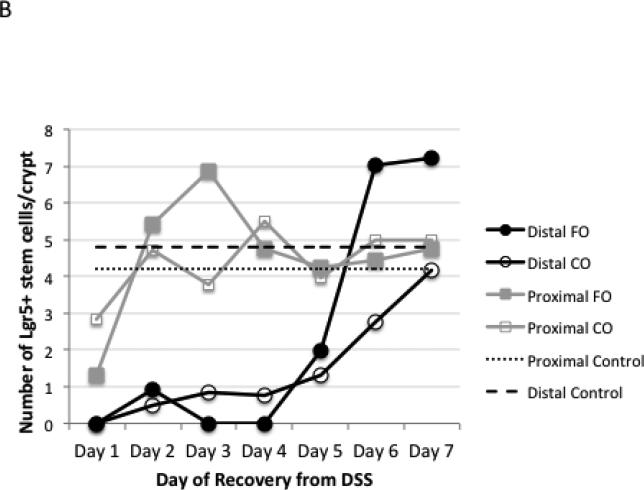
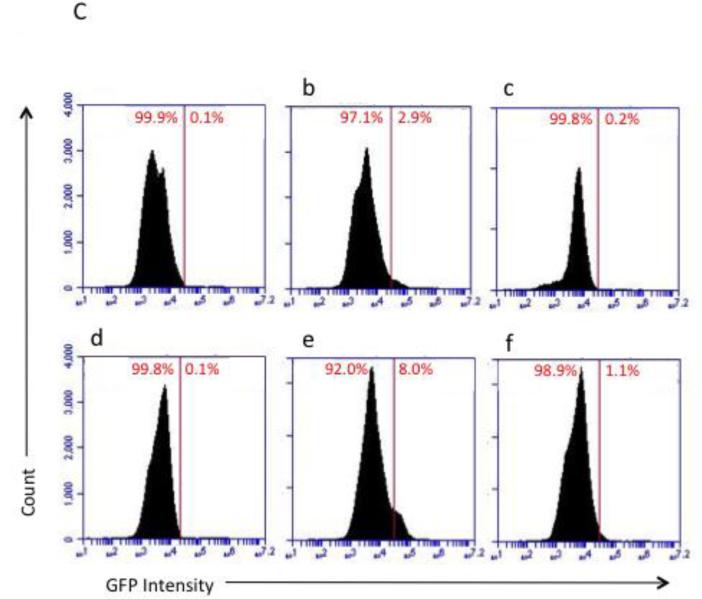
DSS treatment resulted in the ablation of LacZ positive-Lgr5+ stem cells in the distal colon (Figure 1A), concurrent with the extensive loss of distal crypt structure. In contrast, the proximal colon was protected from DSS damage, maintaining ~4-6 stem cells per crypt after the first day of DSS recovery, similar to untreated control animals (Figure 1A&B). Similar results were observed using flow cytometric analysis of Lgr5 EGFP+ colonic epithelial cells (Figure 1C). Stem cells in the distal colon, almost completely absent from days 1-4 of DSS recovery, reappeared at day 5, with higher than normal levels attained by days 6-7 of recovery. There was no difference in recovery of stem cells between the n-3 and n-6 PUFA groups (Figure 1B).
Figure 1.
Detection of Lgr5+ stem cells during recovery from DSS treatment. Animals were treated with DSS for 5 d to induce colitis and terminated at daily intervals during the “recovery” phase. A, representative photomicrographs depicting β-galactosidase staining of Lgr5+ stem cells in distal colon in (a) untreated control, (b) DSS treated, 2 d recovery, (c) DSS treated, 7 d recovery, x100-200. B, mean number of Lgr5+ stem cells per crypt during DSS recovery (at least 20 crypts each from n=3 mice per diet group and time point. However, during days 1-4 of recovery, many animals had no scoreable crypts in the distal colon.) There were no significant differences (P>0.05) between CO and FO fed mice. C, representative flow cytometric analysis of Lgr5 GFP+ colonic epithelial cells. a-c: distal colon; d-f: proximal colon. a and d are from wild type mice (no GFP) and were used for gating. b and e represent control animals displaying the expected levels of GFP+ cells, c and f are from DSS treated mice, d 1 recovery. FO, fish oil fed; CO, corn oil fed mice.
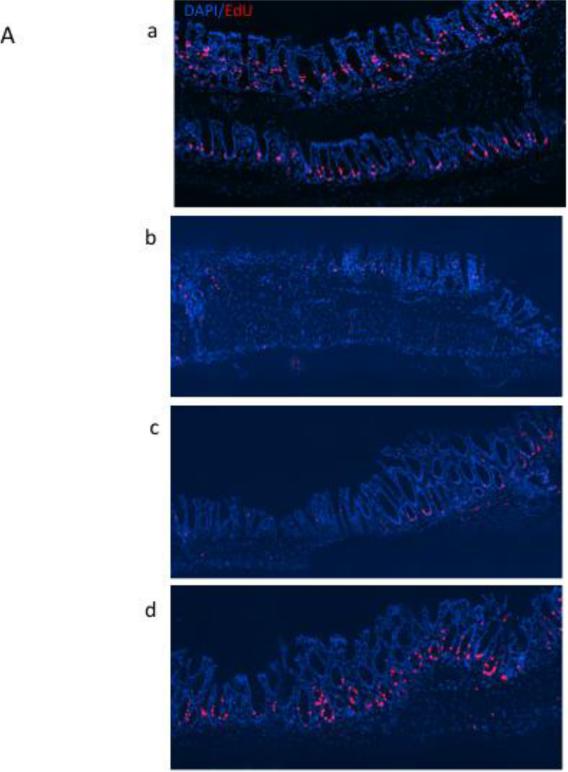
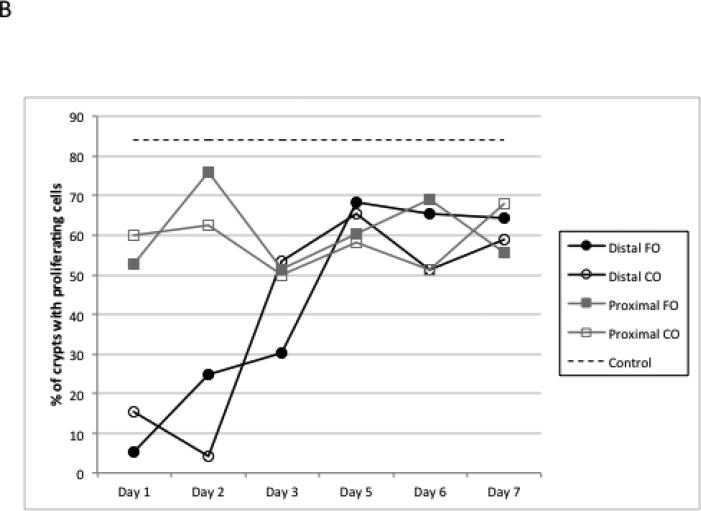
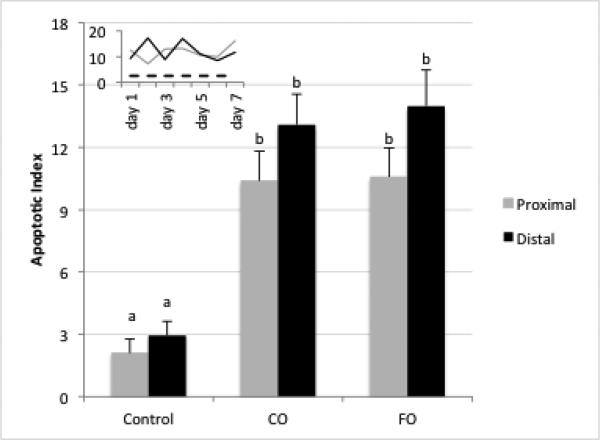
Since early alterations in cell proliferation and apoptosis occur following damage to the colonic epithelium [34], experiments were conducted to document the effects of DSS-induced wounding on crypt cell cytokinetics. The percentage of crypts with proliferating cells, i.e., crypts with EdU+ cells, remained relatively constant at 50-70% in the proximal colon region during all 7 days of recovery, compared to 85% of crypts containing proliferating cells in control animals. In contrast, in the distal colon, 5-20% of the crypt area contained proliferating cells during days 1-2 of recovery, increasing to 60-70% by day 5 (Figures 2A&B). These data indicate that, in general, the epithelial cell cycle has been arrested. In complementary analyses, apoptosis was measured using the dUTP nick end-labeling (TUNEL) assay. DSS exposure elevated the apoptotic index equally in the proximal and distal colon (Figure 3). Overall, diet had no effect on crypt cell cytokinetics.
Figure 2.
Detection of proliferating (EdU+) cells following DSS exposure. A, representative photomicrographs (×100) of distal colonic crypts stained for EdU (red) with all nuclei stained with DAPI (blue), (a) untreated control, (b) DSS treated, d 2 recovery, (c) DSS treated, d 3 recovery, (d) DSS treated d 7 recovery. B, mean percentage of crypts with proliferating cells during DSS recovery. Control represents non-DSS treated mice. There was no significant difference between CO and FO fed mice at any time point (p>0.05).
Figure 3.
Apoptotic index during recovery from DSS treatment. Data represent average apoptotic index within diet groups over the 7 d DSS recovery period (all days combined). At least 20 crypts each from n=3 mice per diet group and time point were scored for TUNEL positive (apoptotic) cells and expressed as apoptotic index (number of apoptotic cells per 100 crypts). Bars with different letters are statistically different (p<0.05). Inset: Apoptosis on individual days in proximal (grey) and distal (black) colon for both diets combined. Dotted line: untreated control mice.
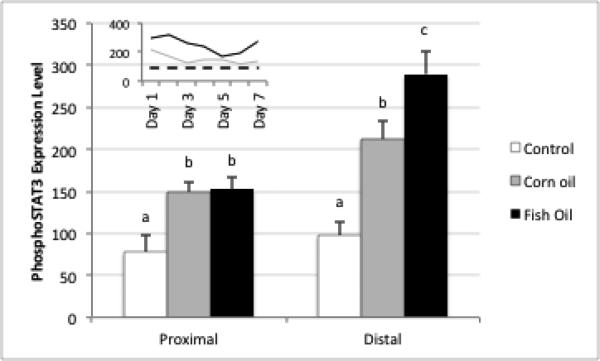
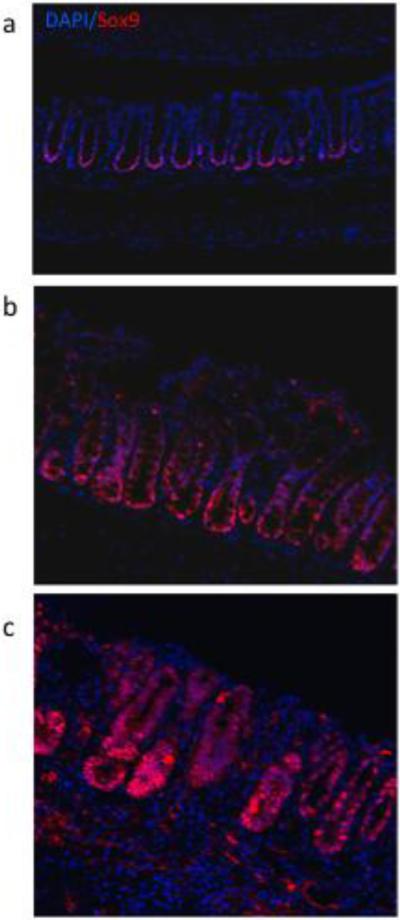
To better understand the impact of inflammation and dietary bioactive agents on the colonic epithelial progenitor niche during the regenerative response to colitis, we examined important regulators of intestinal homeostasis after induction of wounding injury. Phospho signal transducer and activator of transcription (pSTAT3) levels, a well-accepted marker of mucosal inflammation, wound healing and crypt cell survival [35, 36], were significantly elevated compared to untreated animals for days 1-5 of recovery, after which expression approached control levels (Figure 4). In general, STAT3 phosphorylation was localized primarily to damaged and regenerating crypts (Supplemental Figure 1). Fish oil fed animals exhibited higher levels of pSTAT3 compared to corn oil fed mice at all time points, consistent with a higher wounding by DSS in fish oil feeding [37] (Figure 4). Sox9, a marker of cell proliferation, self-renewal and repair in the intestine [38, 39], was strongly upregulated in recovering crypts, and was correlated to the regeneration of the damaged tissue and the reestablishment of Lgr5+ stem cells in the distal colon (Figure 5). For comparative purposes, an additional putative stem cell marker (DCAMKL1) was assessed by IHC (Supplemental Figure 2). In general, DCAMKL1 expression was dispersed throughout the crypt and was not associated with damaged and regenerating crypts.
Figure 4.
Detection of phospho-STAT3 (Y705) following DSS exposure. Mean fluorescence intensity of proximal versus distal colon during DSS recovery (n=3 mice per diet group, all time points combined). Inset: mean fluorescence intensity across time from pooled proximal and distal colon for corn oil (grey) and fish oil fed mice (black). Dotted line: untreated control mice. Bars with different superscripts are significantly different a p<0.05.
Figure 5.
Effect of DSS exposure on Sox9 crypt localization. Representative photomicrographs (×100) of Alexa-647 fluorescently labeled distal colonic crypt cells. (a) untreated control, (b) DSS treated, d 1 recovery, (c) DSS treated, d 3 recovery.
The continuous cell renewal of the intestinal epithelium is regulated by a number of homeostatic pathways, including Wnt signaling [6]. Therefore, initially in control animals (no DSS or n-3 PUFA exposure), we assessed the transcriptome profile of stem cells (GFP high population) as compared to GFP negative cells (non stem cell population) in the small intestine and colon using next generation sequencing (Table 1). Both small intestine and colon transcript GFP positive cells were enriched in genes associated with Wnt signaling (Lgr5, BMP4) [40], and stem cell renewal (EPHB3, HES1, SNAI2, SOX9) [38, 41-43]. Interestingly, the “master” intestinal transcription factor (CDX2) [44] was only enriched in colonic Lgr5 positive cells. We subsequently sought to determine the combined effect of wounding injury and an n-3 PUFA-enriched diet on the relative expression of Wnt signaling activators; R-spondin (Rspo-1); R-spondin receptor (Lgr5) [45]; Wnt signaling inhibitor, Axin2 [46] and intestinal stem cell associated factors, Achaete Scute-Like 2 (Ascl2), Ephrin type-B receptor 3 (EphB3) [6], mouse telomerase reverse transcriptase (mTERT) [47], and HOP homeobox (Hopx) [13]. At 3 days of recovery from DSS, a time when very few Lgr5+ stem cells are detectable (Figure 1B), Lgr5 mRNA was significantly (P<0.05) suppressed relative to control mice (no DSS treatment) (Table 2). Other stem cell markers, Ascl2 and Sox9, were similarly down regulated by DSS exposure. Interestingly, Axin2 was suppressed, consistent with an enhancement of Wnt signaling during the recovery phase. Consistent with the expression of pSTAT3 (Figure 4), STAT3 mRNA levels were also increased following DSS exposure.
Table 1.
Gene expression by RNA sequencing of colonic and small intestinal GFPHigh stem cells and non-stem cell (GFP negative) populations. Values are expressed as RPKM (reads per kilobase per million mapped reads). Cells were sorted from two pooled mice.
| Colon | |||
|---|---|---|---|
| Gene | Expression in stem cell population (GFP high) | Expression in non stem cell population (GFP negative) | Fold Change (GFP high/negative) |
| Adam9 | 18.665 | 5.991 | 3.116 |
| Ascl2 | 0 | 0.132 | 0 |
| Axin2 | 1.646 | 2.023 | 0.814 |
| Bmi1 | 0 | 1.783 | 0 |
| Bmp4 | 0.758 | 0.016 | 47.844 |
| Cdx2 | 44.720 | 7.527 | 5.941 |
| Dclk1 | 0 | 0.282 | 0 |
| Ephb3 | 21.991 | 0.622 | 35.344 |
| Foxn1 | 1.224 | 0.077 | 15.948 |
| Hes1 | 46.953 | 5.296 | 8.866 |
| Hopx | 0.317 | 2.477 | 0.128 |
| Hoxc8 | 1.230 | 0.024 | 51.831 |
| Igf1 | 0.028 | 0.004 | 7.974 |
| Lgr5 | 2.599 | 0.862 | 3.015 |
| Lgr6 | 0 | 0 | 0 |
| Lrig1 | 0.856 | 5.031 | 0.170 |
| Lrig2 | 0.020 | 2.548 | 0.008 |
| Lrig3 | 0.527 | 4.684 | 0.112 |
| Notch4 | 0.106 | 0.067 | 1.595 |
| Omlf4 | 0 | 0.041 | 0 |
| Prom1 | 6.541 | 40.642 | 0.161 |
| Rspo-1 | 0 | 0 | 0 |
| Runx3 | 0.042 | 0.007 | 5.980 |
| Smoc2 | 10.571 | 1.452 | 7.280 |
| Snai2 | 0.588 | 0.052 | 11.296 |
| Sox4 | 4.910 | 2.816 | 1.744 |
| Sox9 | 51.631 | 4.288 | 12.040 |
| Stat3 | 0.347 | 3.722 | 0.093 |
| Tert | 0.017 | 0.216 | 0.077 |
| Tgfbr2 | 4.761 | 2.454 | 1.940 |
| Small Intestine | |||
|---|---|---|---|
| Gene | Expression in stem cell population (GFP high) | Expression in non stem cell population (GFP negative) | Fold Change (GFP high/negative) |
| Adam9 | 1.448 | 2.858 | 0.506 |
| Ascl2 | 5.776 | 1.064 | 5.426 |
| Axin2 | 27.925 | 16.477 | 1.695 |
| Bmi1 | 2.333 | 3.054 | 0.764 |
| Bmp4 | 0.070 | 0 | 0 |
| Cdx2 | 7.731 | 8.247 | 0.938 |
| Dclk1 | 0.360 | 0.212 | 1.696 |
| Ephb3 | 13.987 | 2.026 | 6.904 |
| Foxn1 | 0.012 | 0.100 | 0.122 |
| Hes1 | 42.973 | 8.090 | 5.312 |
| Hopx | 9.810 | 9.202 | 1.066 |
| Hoxc8 | 0.053 | 0.024 | 2.198 |
| Igf1 | 0.004 | 0 | 0 |
| Lgr5 | 13.870 | 3.575 | 3.880 |
| Lgr6 | 0 | 0 | 0 |
| Lrig1 | 7.571 | 8.813 | 0.859 |
| Lrig2 | 0.710 | 2.207 | 0.321 |
| Lrig3 | 3.226 | 3.230 | 0.999 |
| Notch4 | 0.108 | 0.103 | 1.051 |
| Olfm4 | 647.212 | 678.583 | 0.954 |
| Prom1 | 36.511 | 29.596 | 1.234 |
| Rspo-1 | 0.020 | 0 | 0 |
| Runx3 | 0 | 0.057 | 0 |
| Smoc2 | 38.483 | 12.751 | 3.018 |
| Snai2 | 0.327 | 0.105 | 3.114 |
| Sox4 | 17.097 | 6.821 | 2.506 |
| Sox9 | 15.853 | 8.702 | 1.822 |
| Stat3 | 1.511 | 4.083 | 0.370 |
| Tert | 1.551 | 0.512 | 3.027 |
| Tgfbr2 | 1.441 | 1.219 | 1.182 |
Table 2.
mRNA expression profiles of stem cell mediators in colonic mucosa from control and DSS treated mice - 3 days of recovery.
| Gene | Diet | Expression Level (% of control) | pvalue vs control | pvalue CO vs FO |
|---|---|---|---|---|
| Ascl2 | CO | 61.3 | <0.001 | |
| FO | 24.1 | 0.002 | 0.016 | |
| Axin2 | CO | 53.1 | <0.001 | |
| FO | 28.8 | 0.002 | 0.016 | |
| EphB3 | CO | 65.8 | <0.001 | |
| FO | 43.7 | 0.002 | 0.032 | |
| STAT3 | CO | 168.8 | <0.001 | |
| FO | 161.3 | <0.001 | 0.111 | |
| Hopx | CO | 70.0 | 0.006 | |
| FO | 64.7 | 0.002 | 0.191 | |
| Lgr5 | CO | 43.8 | 0.008 | |
| FO | 24.1 | 0.002 | 0.191 | |
| Sox9 | CO | 59.9 | <0.001 | |
| FO | 50.8 | <0.001 | 0.310 | |
| Rspo-1 | CO | 114.6 | 0.550 | |
| FO | 128.0 | 0.413 | 0 342 | |
| mTERT | CO | 86.1 | 0.243 | |
| FO | 71.6 | 0.122 | 0 421 | |
| Bmi1 | CO | 127.3 | 0.222 | 0.548 |
| FO | 113.2 | 0.030 |
Real-time PCR was performed using Taqman probes (Applied Biosystems) and expression levels were normalized to ribosomal 18S. Values are means, n=5 mice per group. TaqMan gene expression assay IDs were as follows: Ascl2, Mm01268891_g1; Axin2, Mm00443610_m1; Bmi1, Mm03053308_g1; EphB3, Mm00802553_m1; Hopx, Mm00558630_m1; Lgr5, Mm00438890_m1; mTert, Mm00436931_m1; Rspo1, Mm00507077_m1; Sox9, Mm00448840_m1; Stat3, Mm01219775_m1; 18S, Hs99999901_s1.
Since dietary FO alters the levels of genes which modulate both intestinal inflammation [27, 48, 49] and the stem cell niche, e.g., Wnt/TCF signature, Ephrin B1, BMP4, and PGE2 [8, 27-29], it was noteworthy that n-3 PUFA-fed mice exhibited a significant (P<0.05) reduction of Ascl2, Axin2 and EphB3 expression (Table 2).
4. Discussion
Recent findings lend support to the concept that in some circumstances, inflammation-induced wound healing/intestinal crypt regeneration can perturb homeostasis within the stem cell compartment and promote malignant transformation [10, 27, 30, 31]. Therefore, it is important to know precisely how intestinal stem cell populations respond to environmental factors in the context of inflammation. Since little is known regarding the homeostatic mechanisms within the stem cell compartment in the large intestine, in this study we used the Lgr5-LacZ and Lgr5-EGFP-IRES-cre knockin mouse models to examine the adult colon stem cell response to DSS exposure and a putative therapeutic diet (fish oil).
Exposure to DSS represents a wounding model that induces mucosal injury and subsequent inflammation via the recruitment and activation of inflammatory cells and mediators, ultimately leading to the development of colitis [50]. As expected, acute DSS exposure ablated colonic crypts in the distal colon (Figure 1) [51]. The simultaneous abrupt reduction in crypt DNA synthesis (Figure 2) and enhanced deletion of damaged cells (Figure 3) represents a classical manifestation of damage to the colonic epithelium [34, 52]. Since colon stem cell kinetics have not been reported in the context of a colitis/wounding model, in the current study we also investigated how the DSS model impacts the colonic stem cell compartment. Rapidly cycling Lgr5+ stem cells were also completely eliminated in the distal colon and returned by day 5 of recovery. The elimination of stem cells and their subsequent recovery in response to acute DSS exposure was confirmed by (i) the disappearance of Lgr5-GPF and LacZ-positive cells and (ii) the absence of EdU at the base of crypts at positions 1-4. These data indicate that the rapidly cycling CBC stem cell (Lgr5+) compartment located at the base of the colonic crypt is highly susceptible to damage-induced intestinal injury. It is possible that Lgr5 gene expression is selectively turned off in response to damage, however, there is no evidence to date that this receptor is reversibly responsive to environmental insult.
Although it is not clear why the proximal colon is resistant to the effects of DSS, our findings extend a previous report indicating that following γ-irradiation, Lgr5 expressing cells are eliminated [53]. Interestingly, Lgr5+ cells in the small intestine are not susceptible to Doxorubicin-induced cell death [54]. With regard to the reestablishment of Lgr5+ cells, it is possible that a small number of rapidly cycling stem cells survive insult in the crypt base and repopulate the crypts. There is some evidence indicating that the small intestine contains a quiescent stem cell population at 4 cells above the base of the crypt (+4 position), e.g., Hopx, Bmi1, Lrig1, and mTert expressing cells [13, 15, 47, 55]. These slowly cycling stem cells are capable of interconverting with more rapidly cycling Lgr5 stem cells at the crypt base [13, 15]. Therefore, a putative intestinal model of stem cell renewal and crypt regeneration incorporates a bi-directional lineage relationship between active and quiescent stem cells in their niches. However, recent data imply that Bmi1, Hopx, mTert and Lrig1 cannot be used as markers for a quiescent reserve stem cell population [56]. This small intestinal model is further complicated by the fact that during regenerative responses to injury, stromal stem cells, including activated macrophages [57, 58], as well as bone marrow derived cells [59], transmit regenerative signals to neighboring colonic epithelial progenitors. Since many of the experiments focusing on stem cell identity and location have focused on the small intestine, additional work is needed in order to confirm these relationships in the colon.
Examination of the Lgr5 positive cell global transcriptome in the small intestine and colon revealed a significant enrichment in genes related to Wnt signaling and stemness (Table 1). These data are consistent with previous targeted qPCR analyses [60]. With respect to the DSS-induced colitis and crypt regeneration model, we showed that both Lgr5 and Hopx mRNA expression levels were decreased in damaged colonic mucosa (Table 2). Other Wnt targeted stem/progenitor cell markers, Ascl2 and Sox9, were also down regulated at the mRNA level by DSS exposure. This was unexpected given that Wnt signaling is activated during intestinal regeneration [6, 61], and may be attributed to the time point sampled (3 d DSS recovery). It was not surprising to see the inverse mRNA expression of Axin2 (suppression of Wnt signaling inhibitor) and Rspo-1 (enhancement of Wnt signaling activator) following DSS exposure. These two mediators reciprocally regulate Wnt signaling [45, 46].
To our knowledge, this is the first study to utilize Lgr5+ reporter mice to determine the impact of diet on intestinal stem cell survival and gene expression. Dietary fish oil, containing long chain n-3 PUFA, has been shown to attenuate intestinal inflammation [27, 37, 48, 49], and impact genes which modulate mediators of the colon stem cell niche, e.g., Wnt/TCF signature, Ephrin B1, BMP-4, and PGE2, in animal models of intestinal inflammation/carcinogenesis [26, 27-29]. Therefore, we hypothesized that the protective effect of n-3 PUFA may be partially attributed to a reduction in stem cell damage. To our surprise, there was no difference in the recovery of stem cells between the n-3 and n-6 PUFA groups (Figure 1B), although there was a modest elevation in the number of stem cells at recovery days 6 and 7 in FO fed mice. Interestingly, STAT3 activation (Figure 4) was highest in DSS exposed, FO-fed mice. This is noteworthy, because STAT3 mediates immune homeostasis in the gut by promoting mucosal wound healing and stem cell survival [35, 36]. The enhancement of STAT3 by n-3 PUFA is generally consistent with a higher wounding by DSS. Indeed, we have unambiguously demonstrated that FO feeding disrupts mucosa repair during the acute phase of DSS exposure in part due to the inhibition of NF-kB activity in the colonic mucosa [37].
FO feeding modulated the expression of genes known to regulate the stem cell niche, e.g., Ascl2, Axin2 and Eph3 (Table 2). Furthermore, we have recently demonstrated that dietary FO modulates a subset of non-coding microRNAs (miR 19b, 26b, 200c and 203) and their target genes (mRNAs) implicated in the regulation of the colon stem cell niche and tumor evolution [29]. Collectively, these results suggest that select dietary cues can impact stem cell regulatory networks. Additional research is needed in order to elucidate the biological relevance of the FO-induced changes in genes which define intestinal stem cells.
The present study has defined the impact of wounding/inflammation and dietary n-3 PUFA on the colonic epithelial progenitor niche during the regenerative response to colitis. Since transformation of adult stem cells is an extremely efficient route towards initiating intestinal cancer [4], including the transition from colitis to cancer [62], it is imperative that we understand how environmental factors modulate colonic stem cell survival and self-renewal in response to chronic inflammation and carcinogen exposure.
Supplementary Material
Highlights.
- Colon stem cell kinetics are altered in the colitis/wounding model
- Lgr5+ stem cells in the colon are highly susceptible to DSS-induced damage
- Diet modulates stem cell associated genes, Ascl2, Axin2 and Eph3
Acknowledgements
We thank Hans Clevers for insightful discussions and for the Lgr5 reporter mice.
Grants
Supported by Cancer Prevention and Research Institute of Texas grant RP100473 and National Institutes of Health grants CA168312 and CA129444. MSS was supported by the Texas A&M Whole Systems Genomics Initiative.
Abbreviations
- CBC
crypt base columnar cell
- CO
corn oil
- FO
fish oil
- DSS
dextran sodium sulfate
- PUFA
polyunsaturated fatty acids
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
No conflicts of interest are declared by the authors.
References
- 1.McDonald SA, Preston SI, Lovell MJ, Wright NA, Jankowski JA. Mechanisms of disease: from stem cells to colorectal cancer. Nature Clin. Prac. Gastroenterol. Hepatol. 2006;3:267–274. doi: 10.1038/ncpgasthep0473. [DOI] [PubMed] [Google Scholar]
- 2.Willis ND, Przyborski SA, Hutchinson CJ, Wilson RG. Colonic and colorectal cancer stem cells: progress in the search of putative biomarkers. J. Anat. 2008;213:59–65. doi: 10.1111/j.1469-7580.2008.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N, Clevers H. Tracking down the stem cells of the intestine: Strategies to identify adult stem cells. Gastroenterology. 2007;133:1755–1760. doi: 10.1053/j.gastro.2007.10.029. [DOI] [PubMed] [Google Scholar]
- 4.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–612. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, van Es HH, Kuipers J, Kujala P, van den Born M, Cozinjsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1008. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 6.Lin SA, Barker N. Gastrointestinal stem cells in self-renewal and cancer. J. Gastroenterol. 2011;46:1039–1055. doi: 10.1007/s00535-011-0424-8. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stems cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–266. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 8.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Gupta PB, Chaffer CI, Weinberg RA. Cancer stem cells: mirage or reality. Nature Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 10.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shackleton M. Normal stem cells and cancer stem cells: similar and different. Sem. Cancer Biol. 2010;20:85–92. doi: 10.1016/j.semcancer.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010 Jan 29;2327(5965):542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlone CL, Breault DT. Tales from the crypt: The expanding role of slow cycling intestinal stem cells. Cell Stem Cell. 2012;10:2–4. 2012. doi: 10.1016/j.stem.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, Guo Y, Shyr Y, Aronow BJ, Haigis KM, Franklin JL, Coffey RJ. The pan-ErbB negative regulator Lrig1 is an Intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensible. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anti M, Armelao F, Marra G, Percesepe A, Bartoli GM, Palozza P, Parrella P, Canetta C, Gentiloni N, De Vitis I, Gasbarrini G. Effects of Different doses of fish Oil on Rectal Cell Proliferation in Patients with Sporadic Colonic Adenomas. Gastroenterology. 1994;107:1709–1718. doi: 10.1016/0016-5085(94)90811-7. [DOI] [PubMed] [Google Scholar]
- 18.Anti M, Giancarlo M, Armelao F, Bartoli GM, Ficarelli R, Percesepe A, De Vitis I, Maria G, Sofo L, Rapaccini GL, Gentiloni N, Piccioni E, Miggiano G. Effect of -3 Fatty acids on Rectal Mucosal Cell Proliferation in Subjects at Risk for Colon Cancer. Gastroenterology. 1992;103:883–891. doi: 10.1016/0016-5085(92)90021-p. [DOI] [PubMed] [Google Scholar]
- 19.Bartram HP, Gostner A, Scheppach W, Reddy BS, Rao CV, Dusel G, Richter F, Richter A, Kasper H. Effects of Fish Oil on Rectal Cell Proliferation, Mucosal Fatty Acids, and Prostaglandin E2 Release in Healthy Subjects. Gastroenterology. 1993;105:1317–1322. doi: 10.1016/0016-5085(93)90135-y. [DOI] [PubMed] [Google Scholar]
- 20.Cheng J, Ogawa K, Kuriki K, Yokoyama Y, Kamiya T, Seno K, Okuyama H, Wang J, Luo C, Fujii T, Ickikawa H, Shirai T, Tokudome S. Increased intake of n-3 polyunsaturated fatty acids elevates the level of apoptosis in the normal sigmoid colon of patients polypectomized for adenomas/tumors. Cancer Lett. 2003;193:17–24. doi: 10.1016/s0304383502007176. [DOI] [PubMed] [Google Scholar]
- 21.Courtney ED, Matthews S, Finlayson C, Di Pierro D, Belluzzi A, Roda E, Kang JY, Leicester RJ. Eicosapentaenoic acid (EPA) rTheseces crypt cell proliferation and increases apoptosis in normal colonic mucosa in subjects with a history of colorectal adenomas. Int. J. Colorectal Dis. 2007;22:765–776. doi: 10.1007/s00384-006-0240-4. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez E, Chatenoud L, La Vecchia C, Negri E, Franceschi S. Fish consumption and cancer risk. Am. J. Clin. Nutr. 1999;70:85–90. doi: 10.1093/ajcn/70.1.85. [DOI] [PubMed] [Google Scholar]
- 23.Hall MN, Chavarro JE, Lee IM, Willett WC, Ma J. A 22-year prospective study of fish, n-3 fatty acid intake, and colorectal cancer risk in men. Cancer Epidemiol. Biomarkers Prev. 2008;17:1136–1143. doi: 10.1158/1055-9965.EPI-07-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavani A, Pelucchi C, Parpinel M, Negri E, Franeschi S, Levi F, La Vecchia C. n-3 polyunsaturated fatty acid intake and cancer risk in Italy and Switzerland. Int. J. Cancer. 2003;105:113–116. doi: 10.1002/ijc.11018. [DOI] [PubMed] [Google Scholar]
- 25.West NJ, Clark SK, Phillips RK, Hutchinson JM, Leicester RJ, Belluzzi A, Hull MA. Eicosapentaenoic acid reduces rectal polyp number and size in familial adenomatous polyposis. Gut. 2010;59:918–925. doi: 10.1136/gut.2009.200642. [DOI] [PubMed] [Google Scholar]
- 26.Vanamala J, Glagolenko A, Yang P, Carroll RJ, Murphy ME, Newman RA, Chapkin RS, Lupton JR. Dietary fish oil and pectin enhance colonocyte apoptosis in irradiated rats in part by suppressing protumorigenic signaling. Carcinogenesis. 2008;29:790–796. doi: 10.1093/carcin/bgm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia Q, Lupton JR, Smith R, Weeks BR, Callaway E, Davidson LA, Kim W, Fan YY, Yang P, Newman RA, Kang JX, McMurray DN, Chapkin RS. Reduced colitis-associated colon cancer in Fat-1 (n-3 fatty acid desaturase) transgenic mice. Cancer Res. 2008;68:3985–3991. doi: 10.1158/0008-5472.CAN-07-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kachroo P, Ivanov I, Davidson LA, Chowdhary BP, Lupton JR, Chapkin RS. Classification of diet-modulated gene signatures at the colon cancer initiation and progression stages. Digestive Diseases and Sciences. 2011;56:2595–2604. doi: 10.1007/s10620-011-1652-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah MS, Schwartz SL, Zhao C, Davidson LA, Zhou B, Lupton JR, Ivanov I, Chapkin RS. Integrated microRNA and mRNA expression profiling in a rat colon carcinogenesis model: Effect of a chemo-protective diet. Physiol. Genomics. 2011;43:640–654. doi: 10.1152/physiolgenomics.00213.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am. J. Physiol. 2004;287:G7–G17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- 31.Kasper M, Jaks V, Are A, Bergstrom A, Schwager A, Barker N, Toftgard R. Wounding enhances epidermal tumorigenesis by recruiting hair follicle keratinocytes. Proc. Natl. Acad. Sci. 2011;108:4099–4104. doi: 10.1073/pnas.1014489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma DWL, Finnell RH, Davidson LA, Callaway ES, Spiegelstein O, Piedrahita JA, Salbaum JM, Kappen C, Weeks B, James J, Bozinov D, Lupton JR, Chapkin RS. Folate transport gene inactivation in mice increases sensitivity to colon carcinogenesis. Cancer Research. 2005;65:887–897. [PMC free article] [PubMed] [Google Scholar]
- 33.Fan YY, Ran Q, Toyokuni S, Okazaki Y, Callaway ES, Lupton JR, Chapkin RS. Dietary fish oil promotes colonic apoptosis and mitochondrial proton leak in oxidatively stressed mice. Cancer Prevention Research. 2011;4:1267–1274. doi: 10.1158/1940-6207.CAPR-10-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirose Y, Yoshimi N, Makita H, Hara A, Tanaka T, Mori H. Early alterations of apoptosis and cell proliferation in azoxymethane-initiated rat colonic epithelium. Jpn. J. Cancer Res. 1996;87:575–582. doi: 10.1111/j.1349-7006.1996.tb00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews JR, Sansom OJ, Clarke AR. Absolute requirement for STAT3 function in small-intestine crypt stem cell survival. Cell Death & Differentiation. 2011;18:1934–1949. doi: 10.1038/cdd.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickert G, Neufert C, Leppkes M, Zheng Y, Wittkopf N, Wantjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, Ouyang W, Neurath MF, Becker C. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia Q, Ivanov I, Zlatev Z, Alaniz RC, Weeks BR, Callaway ES, Goldsby JS, Davidson LA, Fan YY, Zhou L, Lupton JR, McMurray DN, Chapkin RS. Dietary fish oil and curcumin combine to modulate colonic cytokinetics and gene expression in dextran sodium sulphate-treated mice. Br. J. Nutr. 2011;106:519–529. doi: 10.1017/S0007114511000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huch M, Clevers H. Sox9 marks adult organ progenitors. Nature Genetics. 2011;43:9–10. doi: 10.1038/ng0111-9. [DOI] [PubMed] [Google Scholar]
- 39.Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, Canamero M, Bujanda L, Schedl A, Cheah KS, Slotheim RI, Lothe RA, Lopez de Manain A, Briscoe J, Serrano M, Lovell-Badge R. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012;72:1301–1315. doi: 10.1158/0008-5472.CAN-11-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scoville DH, Sato T, He XC, Li L. Current View: Intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- 41.Kayahara T, Sawada M, Takaishi S, Fukui H, Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H, Chiba T. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003;535:131–135. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- 42.Holmberg J, Genander M, Halford MM, Anneren C, Sondell M, Chumley MJ, Silvany RE, Henkemeyer M, Frisen J. EphB receptors coodinate migration and proliferation in the intestinal stem cell niche. Cell. 2006;125:1151–1163. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 43.Cobaleda C, Perez-Caro M, Vicente-Duenas C, Sanchez-Garcia I. Function of the zinc-finger transcription factor SNAI2 in cancer development. Annu. Rev. Genet. 2007;41:41–61. doi: 10.1146/annurev.genet.41.110306.130146. [DOI] [PubMed] [Google Scholar]
- 44.Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell. 2010:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, Niehrs C. Lgr4 and Lgr5 are R-spondin receptors mediating Wnt/beta-catenin and Wn/PCP signaling. EMBO. J. 2011;12:1055–1061. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jho E, Zhang T, Domon C, Joo CKCK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol.Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl. Acad. Sci. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monk JM, Jia Q, Callaway E, Weeks B, Alaniz RC, McMurray DN, Chapkin RS. n-3 PUFA decrease Th17 cell accumulation in chronic dextran sodium sulfate (DSS)-induced colitis. J. Nutr. 2012;142:117–124. doi: 10.3945/jn.111.147058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monk JM, Kim W, Callaway E, He W, Weeks B, Alaniz RC, McMurray DN, Chapkin RS. Immunomodulatory action of dietary fish oil and targeted deletion of intestinal epithelial cell PPARδ in inflammation-induced colon carcinogenesis. American Journal of Physiology – GI Physiology Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G153–G167. doi: 10.1152/ajpgi.00315.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vowinkel T, Kalogeris TJ, Mori M, Krieglstein CF, Granger DN. Impact of dextran sulfate sodium load on the severity of inflammation in experimental colitis. Dig. Dis. Sci. 2004;49:556–564. doi: 10.1023/b:ddas.0000026298.72088.f7. [DOI] [PubMed] [Google Scholar]
- 51.Bancroft LK, Lupton JR, Davidson LA, Taddeo SS, Murphy ME, Carroll RJ, Chapkin RS. Dietary fish oil reduces oxidative DNA damage in rat colonocytes. Free Radical Biology & Medicine. 2003;35:149–159. doi: 10.1016/s0891-5849(03)00240-5. [DOI] [PubMed] [Google Scholar]
- 52.Wargovich MJ, Medline A, Bruce WR. Early histopathologic events to evolution of colon cancer in C57BL/6 and CF1 mice treated with 1,2-dimethylhydrazine. J. Natl. Cancer Inst. 1983;71:125–131. [PubMed] [Google Scholar]
- 53.Quyn AJ, Appleton PL, Carey FA, Steele RJ, Barker N, Clevers H, Ridgeway RA, Sansom OJ, Nathke IS. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6:175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 54.Dekaney CM, Gulati AS, Garrison AP, Helmrath MA, Henning SJ. Regeneration of intestinal stem/progenitor cells following doxorubicin treatment in mice. Am.J. Physiol. Gastro. Liver Physiol. 2009;297:G461–G470. doi: 10.1152/ajpgi.90446.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nature Genetics. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, Myant K, Versteeg R, Sansom OJ, van Es JH, Barker N, van Oudenaarden A, Mohammed S, Heck AJ, Clevers H. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012 Jun 12; doi: 10.1038/emboj.2012.166. doi: 10.1038/emboj.2012.166. [Epub ahead of print] PMID: 22692129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc. Natl. Acad. Sci. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stappenbeck TS, Miyoshi H. The role of stromal stem cells in tissue regeneration and wound repair. Science. 2009;324:1666–1669. doi: 10.1126/science.1172687. [DOI] [PubMed] [Google Scholar]
- 59.Verstappen J, Katsaros C, Torensma R, Von den Hoff JW. A functional model for adult stem cells in epithelial tissues. Wound Rep. Reg. 2009;17:296–305. doi: 10.1111/j.1524-475X.2009.00497.x. [DOI] [PubMed] [Google Scholar]
- 60.Van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker of stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 61.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda H, Sugihara H, Fujimoto K, Weissman II, Capecchi MR, Kuo CJ. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nature Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69:8208–8215. doi: 10.1158/0008-5472.CAN-09-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.