Summary
Pre-mRNA splicing is regulated by developmental and environmental cues, but little is known about how specific signals are transduced in mammalian cells to regulate this critical gene expression step. Here, we report massive reprogramming of alternative splicing in response to EGF signaling. By blocking individual branches in EGF signaling, we found that Akt activation plays a major role, while other branches, such as the JAK/STAT and ERK pathways, make minor contributions to EGF-induced splicing. Activated Akt next branches to SR protein-specific kinases, rather than mTOR, by inducing SRPK autophosphorylation that switches the splicing kinases from Hsp70- to Hsp90-containing complexes. This leads to enhanced SRPK nuclear translocation and SR protein phosphorylation. These findings reveal a major signal transduction pathway for regulated splicing and place SRPKs in a central position in the pathway, consistent with their reputed roles in a large number of human cancers.
Keywords: SR protein-specific kinases, Signal transduction, EGF signaling, SR protein phosphorylation, pre-mRNA splicing
Introduction
Alternative splicing is a prevalent phenomenon in mammalian cells (Nilsen and Graveley, 2010). Because the process is tightly coupled with transcription for co-transcriptional RNA processing as well as post-splicing steps for mRNA transport and stability control (Maniatis and Reed, 2002; Pandit et al., 2008; Moore and Proudfoot, 2009), it is widely anticipated that alternative splicing is subject to regulation by a variety of cellular signaling events. However, compared to numerous signal-induced gene expression events that are regulated at the transcriptional and translational levels, little is known about how specific signals are transduced to regulate alternative splicing in the nucleus (reviewed by Shin and Manley, 2004; Lynch, 2007).
The data collected, to date, suggest that many signaling molecules, particularly protein kinases and phosphatases, may directly modify and regulate activities of specific splicing regulators. One of the best examples is the modification of Sam68 in the MAP kinase pathway to regulate CD44 splicing (Konig et al., 1998; Weg-Remers et al., 2001). In another well-studied case, phorbol esters or cytokines activate Ras to regulate CD45 splicing during T cell development (Lynch and Weiss, 2000). In this pathway, GSK3 phosphorylates the splicing regulator PSF to promote its interaction with TRAP150 in resting T cells; upon T cell activation, GSK3 is decreased, which leads to PSF release from the inhibitory complex with TRAP150, allowing PSF to bind and repress CD45 exon 4 in mature T cells (Heyd and Lynch, 2010; reviewed by Heyd and Lynch, 2011).
The Akt pathway appears to modulate the function of the SR family of splicing factors and regulators that act on exonic splicing enhancers (Liu et al., 2003; Shultz et al., 2010). Activated Akt has been further implicated in directly acting on SR proteins (Blaustein et al., 2005; Jiang et al., 2009), or indirectly relaying its signal to the nucleus through SR protein-specific kinases, such as SRPK2 (Jang et al., 2009) or Clk/Sty (Jiang et al., 2009). Interestingly, GSK3 appears to act both upstream and downstream of Akt (Lu et al., 2011) and is able to phosphorylate SR proteins once they are primed by other SR protein kinases (Hernandez et al., 2004). While these studies have introduced potential players, systematic analysis has been lacking in connecting upstream signal transducers to downstream effectors to regulate the splicing program in the nucleus.
We have been focusing on the SRPK family of kinases in regulated splicing, which are highly specific for the SR family of splicing factors (Lin and Fu, 2007). Mammalian genomes encode two such kinases with SRPK1 being ubiquitously expressed in most cell types and tissues and SRPK2 being relatively restricted in neurons (Wang et al., 1998). Interestingly, while SRPK1 and SRPK2 share similar enzymatic activities towards SR proteins, they each associate with distinct complexes in the spliceosome (Mathew et al., 2008; Hegele et al., 2011). Most SRPK molecules are localized in the cytoplasm until the cell is simulated by a signal (Ding et al., 2006; Jang et al., 2009). We recently showed that this is because SRPKs are anchored by molecular chaperones in the cytoplasm, a common mechanism for restricting signal transducers in some specific cellular compartments, and that a stress signal is able to trigger SRPK nuclear translocation to regulate the phosphorylation state of SR proteins and alternative splicing (Zhong et al., 2009). Therefore, SRPKs appear to fulfill the classic definition of signal transducers for regulated splicing in mammalian cells.
In the present work, we systematically dissected EGF-induced alternative splicing. By monitoring global response to EGF signaling at the level of alternative splicing, we found that SRPKs are the central transducers of EGF signaling, while all other previously established branches in the EGF pathway play relatively minor roles, suggesting that the Akt-SRPK-SR axis constitutes a major branch in transducing EGF signaling to regulate the splicing program in the nucleus. Interestingly, unlike classic signal transduction pathways, we found that activated Akt binds and stimulates SRPK1 autophosphorylation to trigger a series of switches in its interaction with molecular chaperones, which leads to nuclear translocation of the splicing kinase and hyper-phosphorylation of SR proteins. These findings, coupled with altered expression of SRPK1 in diverse human cancers (Hayes et al., 2007; Krishnakumer et al., 2008) and its direct contribution to renal failure and development of Wilms’ tumors (Amin et al., 2011), place the signal branch involving Akt, SRPKs and SR proteins in a strategic position for growth control in metazoans.
Results
EGF regulates pre-mRNA splicing via activated Akt and SRPKs
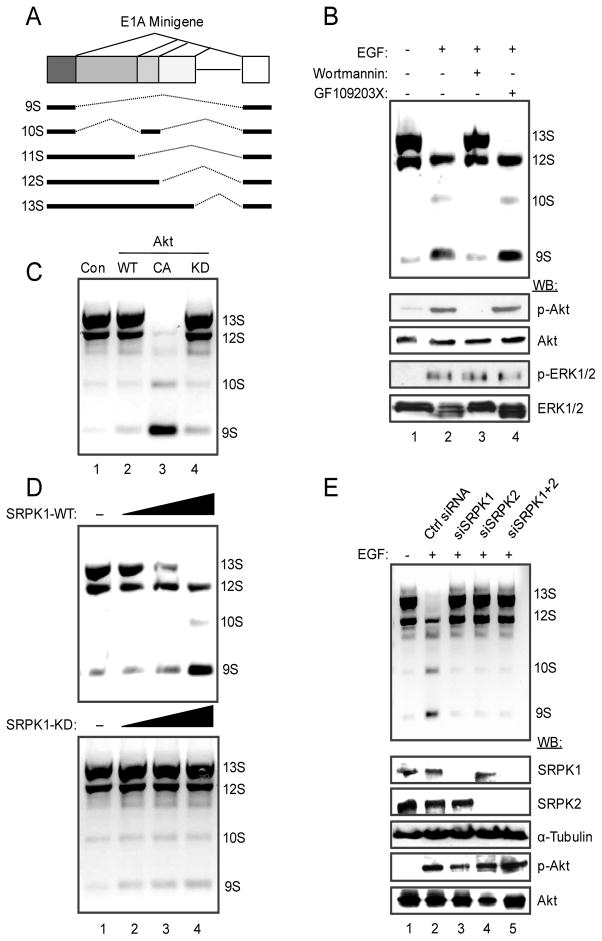
Prior studies have demonstrated a key role of the PI3 kinase pathway in regulated splicing based on analysis of splicing reporters or a limited number of endogenous genes (Liu et al., 2003; Blaustein et al., 2005; Jiang et al., 2009). This system thus serves as a good model for mechanistic dissection of the signaling cascade that leads to regulated splicing in the nucleus. Using an E1A splicing reporter (Fig. 1A), we found that EGF induced a dramatic switch in splice-site selection towards the production of 9S and 10S E1A mRNA isoforms in transfected HEK293T (Fig. 1B) and HeLa cells (Fig. S1A). This effect depends on PI3K activation because the PI3K inhibitor Wortmannin prevented the switch, while a PKC inhibitor showed no effect (Fig. 1B and Fig. S1A). As expected, Akt is activated in response to EGF treatment (Fig. 1B), and a constitutively active (CA) Akt, but not the kinase dead (KD) mutant, mimicked the EGF effect (Fig. 1C and Fig. S1B). These results demonstrate a critical role of Akt in EGF-induced alternative splicing, thus establishing a cellular system to dissect the pathway(s) involved in transducing EGF signaling to regulate the splicing program in the nucleus.
Figure 1.
EGF regulates E1A splicing via activated Akt and SRPKs. (A) Structure of the E1A minigene. The common three isoforms (13S, 12S, and 9S) are products of three alternative 5′ splice sites in competition for the same downstream 3′ splice site. Additional splicing events produce the 11S and 10S isoforms, which are much less abundant compared to the three common isoforms. (B) E1A splicing in response to EGF treatment. EGF signaling induced the switch from the proximal 5′ splice sites (for 13S and to some extent 12S) to the distal 5′ splice site (for 9S). The switch could be blocked by the PI3K inhibitor Wortmannin, but not by the PKC inhibitor GF109203X. The effects of EGF and Wortmannin on Akt activation were determined by Western blotting on the lower panels. The ERK/MAPK pathway was unaffected in HEK293T cells by either inhibitor. (C) Induced E1A splicing switch by a constitutively activated (CA) Akt, but not by a kinase dead (KD) Akt. (D) Overexpression of SRPK1, but not the kinase dead mutant, induced a similar switch in E1A splicing to favor the production of the 9S isoform. (E) Requirement for SRPKs in EGF-induced splicing. The effect of EGF on E1A splicing was blocked by siRNA-mediated knockdown of either SRPK1 or SRPK2 or both (upper panel). The knockdown was efficient and specific as determined by Western blotting (lower panel). Control siRNA had no effect and Akt was still active after SRPK knockdown in EGF-treated cells.
Since SRPKs appear to occupy a strategic position in the cell to relay external signals to the nucleus, we determined whether SRPKs were involved in EGF-induced E1A splicing. We found that overexpression of either SRPK1 or SRPK2 in HEK293T cells caused a similar switch in E1A splicing whereas the kinase dead mutants had no effect (Fig. 1D and Fig. S1C). To determine if SRPKs are essential for transducing EGF signaling to regulate E1A splicing, we performed siRNA knockdown of SRPK1, SRPK2 or both in EGF-treated HEK293T cells, finding that these treatments abolished EGF-induced splicing despite full activation of Akt (Fig. 1E). While these results demonstrate the essential role of SRPKs in EGF-induced splicing, we were surprised by the nearly complete effect when either kinase was inactivated by RNAi since SRPK1 and SRPK2 are thought to have redundant kinase activities on SR proteins (Wang et al., 1998). These findings suggest that the two kinases may be coordinately regulated by some common mechanisms, such as sequestration by heat shock proteins as shown previously (Zhong et al., 2009). As a result, reduction of one kinase may induce the shielding of the other, and consequently, the ability to transduce EGF signaling may depend on the overall level of both kinases in the cell.
SRPKs mediate the global response to EGF in regulated splicing
To initially determine the position of SRPKs in the EGF pathway, we tested EGF and SRPK-induced E1A splicing in the presence of specific inhibitors against some key components in the EGF pathway. We observed that EGF-induced splicing could be blocked by Wortmannin, an inhibitor to PI3K, but not by Rapamycin, an inhibitor to mTOR (Fig. S1D). Similarly, Wortmannin, but not Rapamycin, prevented induction of E1A splicing in SRPK1 or SRPK2 overexpressed cells (Fig. S1E–F). These data suggest that SRPKs act below PI3K, but above mTOR in the EGF pathway.
We next addressed how EGF signaling might elicit a global effect on alternative splicing of endogenous genes and the role of SRPKs in such response. For this purpose, we coupled the oligonucleotide-mediated RNA Annealing, Selection, and Ligation assay (Yeakley et al., 2002) with high throughput sequencing (referred to as RALS-seq as diagrammed in Fig. S2A) to target 3726 alternative splicing events that are conserved between human and mice. Although this technology focuses on annotated targets, as opposed to completely unbiased profiling of alternative splicing by RNA-seq, we found that RASL-seq is robust in detecting quantitative differences in mRNA isoform expression. We detected 954 alternative splicing events that expressed both isoforms in HEK293T cells (by requiring a minimal of 5 counts for each isoform in a given event). This allowed us to compute isoform ratio change for each alternative splicing event under two biological conditions (Fig. S2B, see Du et al., 2010). The splicing ratio changes deduced by RASL-seq were highly consistent with the RT-PCR results (Fig. S2C).
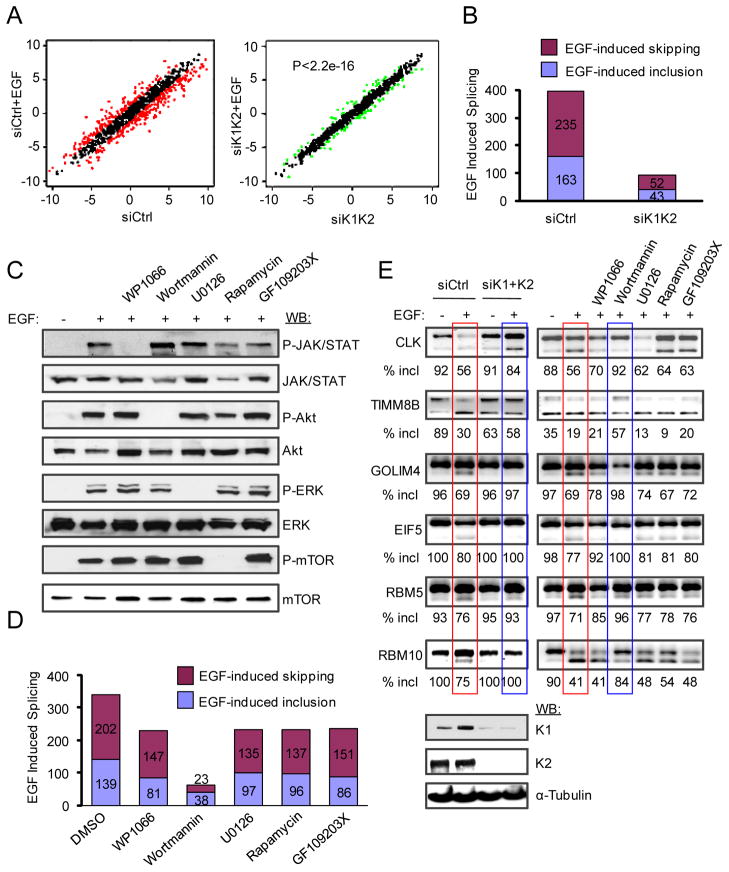
Based on analysis of biological triplicates, we found that EGF-induced splicing switches with the ratio change ≥ 2 are highly statistically significant with p-value <0.05. Based on this cut-off, we scored 398 splicing events (41.7% out of the total 954 detectable alternative splicing events) that were responsive to the EGF treatment, indicating that EGF triggered dramatic splicing reprogramming in the cell (Fig. 2A–B). Significantly, inactivation of SRPKs by RNAi diminished EGF-induced splicing response (p<2.2e-16 based on two-sided Kolmogorov-Smirnov (KS) test in comparison between EGF-induced splicing in control siRNA- and siSRPK1/K2-treated cells) (Fig. 2A–B). These results suggest that SRPKs are largely responsible for transducing EGF signaling to regulate alternative splicing in the cell.
Figure 2.
SRPKs are the major branch in the EGF pathway for global regulation of alternative splicing. (A) EGF induced widespread splicing changes from a global scale, which was largely diminished in SRPK1/2 knockdown cells (p<2.2e-16 according to KS-test). Red dots in the left panel represent splicing ratio changes ≥ 2. Although some changes were detectable in response to double knockdown of SRPK1 and SRPK2 (green dots in the right panel), the magnitude of ratio changes in those cases was much lower. (B) EGF-induced alterative splicing before and after knocking down SRPK1/2. (C) EGF activated multiple signaling branches, including the JAK/STAT, PI3K/Akt, ERK/MAPK and mTOR pathways, each of which could be blocked by a specific inhibitor. (D) Wortmannin effectively blocked EGF-induced splicing, while inhibition of all other pathways had much less effects (see the degree of individual responses in Table S1). (E) A selective panel of splicing events induced by EGF was examined by RT-PCR under different treatment conditions. SRPK knockdown and Wortmannin treatment showed a similar effect in each case, while other inhibitors had minor, if any, effects. The efficiency of SRPK knockdown was shown at bottom.
SRPKs are the major branch in the EGF pathway for global regulation of alternative splicing
Because EGF is known to activate multiple signal transduction pathways, we used a panel of specific kinase inhibitors to block each of the major pathways, including WP1066 against the JAK/STAT pathway, Wortmannin against the PI3K/Akt pathway, U0126 against the ERK pathway, and Rapamycin against the mTOR pathway. GF109203X is an inhibitor of PKC, which was included as a negative control. Each of these inhibitors was able to block the respective pathway (Fig. 2C). Wortmannin was the most potent inhibitor to EGF-induced alternative splicing among all inhibitors tested based on the significance of the global impact determined by KS test (Wortmannin: p<3.386e-14; Rapamycin: p=0.6087; WP1066: p=0.2201; U0126: p=0.4928; GF109203X: p=0.2089) (Fig. S2D and Table S1) or simply on the reduction of EGF-responsive alternative splicing events in the presence of individual kinase inhibitors (Fig. 2D). This is further illustrated on a panel of specific EGF-induced splicing events validated by RT-PCR, showing that siRNAs against SRPK1/K2 blocked EGF-induced splicing, and in comparison, only Wortmannin showed similar effects to SRPK RNAi (Fig. 2E).
Relatively to the PI3K/Akt pathway, inhibition of other signaling branches in the EGF pathway each reduced a subset of EGF-induced splicing events below the cut-off and blocking the PKC pathway had a similar effect (Fig. 2D and Fig. S2E). Out of these suppressed alternative splicing events (113 by WP1066, 109 by U0126, 108 by Rapamycin, and 104 by GF109203X), 49 were commonly affected by all 4 inhibitors, and we further noted that most of these events were affected to a much less degree compared to Wortmannin treatment (Table S1). These observations indicate some general effects on cell fitness by these kinase inhibitors, although each may have some specific, but limited contribution to the overall EGF-induced splicing program. We conclude from these analyses that the PI3K/Akt branch transduces the EGF signal through SRPKs, rather than the well-established downstream effector mTOR, to induce large-scale splicing responses in the nucleus.
Activated Akt acts as a regulatory factor in inducing SRPK1 autophosphorylation
Having established a key role of SRPKs in transducing EGF signaling to regulate splicing in the nucleus, we next wished to determine how EGF regulates the activity of SRPKs. A previous study showed that activated Akt can directly phosphorylate SRPK2, thereby inducing catastrophic neuronal death by forcing differentiated neurons to re-enter mitosis (Jang et al., 2009). This study mapped the Akt phosphorylation site to Thr491 in the HDRSRT motif located in the spacer region of SRPK2, which only loosely fits the Akt consensus RXRXXS/T. We scanned the amino acid sequence of SRPK1 and found no such Akt consensus sequences in the kinase, raising the question of whether SRPK1 is a direct substrate for Akt. We therefore took a proteomics approach to first determine whether SRPK1 could be phosphorylated in response to EGF treatment or overexpression of a constitutively activated Akt. Analysis of phosphopeptides by mass spectrometry from immunoprecipitated SRPK1 revealed multiple phosphorylation sites in SRPK1 that could be induced by EGF or activated Akt (Fig. 3A and S3A). This observation suggests that SRPK1 may be extensively phosphorylated in vivo, despite the fact that recombinant SRPKs expressed from bacteria are highly active in phosphorylating SR proteins in vitro (Nolen et al., 2001; Aubol et al., 2003; Ngo et al., 2005), indicating that some of these phosphorylation events may regulate other aspects of the kinase function in the cell.
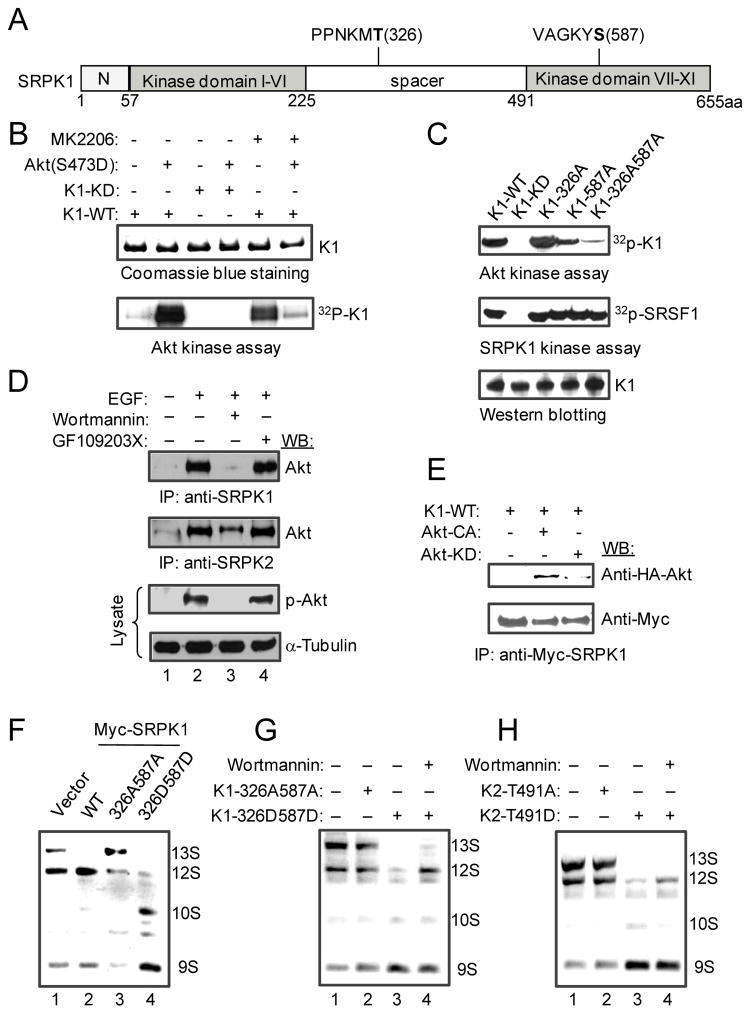
Figure 3.
Activated Akt triggers SRPK1 autophosphorylation. (A) Schematic presentation of SRPK1 domains with two candidate Akt-mediated phosphorylation sites highlighted in bold, which were deduced from in vivo and in vitro phosphorylation experiments analyzed by mass spectrometry (Fig. S3A). (B) In vitro kinase assay. Purified activated Akt phosphorylated WT SRPK1, which could be blocked by Akt inhibitor MK2206 (1 μM). Activated Akt was unable to phosphorylate the kinase dead SRPK1 mutant while MK2206 alone could induce SRPK1 autophosphorylation. (C) Akt-mediated phosphorylation of SRPK1 at T326 and S587. Double Alanine mutations on these two sites abolished Akt-mediated phosphorylation, but had no effect on the kinase activity of SRPK1 on the SR protein SRSF1. (D) Interaction of both SRPK1 and SRPK2 with Akt only after Akt activation by EGF. (E) Co-IP experiments further showed the interaction of SRPK1 only with activated Akt. (F) Akt-mediated phosphorylation in SRPK1 is necessary and sufficient to induce splicing switch on the E1A reporter. The 326A587A mutant appears to act in a dominant negative fashion whereas the phospho-mimicking 326D587D mutant was more potent than WT SRPK1 in inducing the splicing switch. (G and H) The phospho-mimicking SRPK mutants bypassed the requirement for Akt signaling in regulated splicing. In both cases, the phospho-mimicking mutants (K1-326D587D or K2-T491D) could induce E1A splicing in the presence of Wortmannin.
To determine which of the in vivo mapped sites might be direct targets for activated Akt, we used highly purified Akt to phosphorylate bacterially expressed SRPK1. We found that activated Akt could indeed induce SRPK1 phosphorylation in vitro, which could be blocked by the Akt-specific inhibitor MK2206 (Fig. 3B). Unexpectedly, we noted that the SRPK1 kinase dead mutant lost the ability to be phosphorylated by Akt, even though it could compete with WT SRPK1 for binding (Velazques-Dones et al., 2005) and phosphorylating an SR protein (Fig. S3B). Also surprising was the observation that MK2206 alone was able to induce SRPK1 autophosphorylation in the absence of Akt, although it could efficiently suppress SRPK1 phosphorylation by Akt (Fig. 3B). This may be related to the fact that MK2206 is a non-ATP competitive allosteric inhibitor of Akt (Hirai et al., 2010), which may occupy a regulatory pocket on SRPK1 to induce its autophosphorylation. These observations strongly suggest that SRPK1 is regulated by an Akt-dependent allosteric mechanism.
Akt induces two critical autophosphorylation events in SRPK1
The ability of activated Akt to induce SRPK1 autophosphorylation permitted us to determine the in vitro phosphorylation sites by mass spectrometry and compare them to those in EGF-treated cells. This led to the identification of two autophosphorylation sites (Ser33 and Ser309) and two additional sites that were induced only in the presence of active Akt, one (Thr326) located in the spacer domain and the other (Ser587) in the C-terminal region of SRPK1 (Fig. 3A). Further consistent with the possibility that these phosphorylation events result from autophosphorylation is the observation that a fragment of SRPK1 containing T326 could not be phosphorylated with purified Akt (data not shown). Importantly, T326 matches the in vivo site in EGF-treated cells. However, phosphorylation at S587 escaped the detection in our in vivo experiments, which might be due to two lysines (K) that flank this site, making it difficult to detect after extensive trypsin digestion of the limited amount of immunoprecipitated SRPK1. In any case, these in vivo and in vitro mapping studies indicate that T326 and S587 might be the major sites that were induced by activated Akt. We therefore mutated both sites to Alanine, either individually or in combination, finding that only the double mutation abolished Akt-induced SRPK1 autophosphorylation in vitro, even though the mutation had no effect on the kinase activity of SRPK1 towards an SR protein (Fig. 3C).
Akt-induced SRPK phosphorylation appears to require binding of activated Akt to the splicing kinases because only activated Akt could efficiently co-immunoprecipitate with SRPK1 and SRPK2, which could be blocked by Wortmannin (Fig. 3D). We further confirmed binding of only activated Akt to SRPK1 by co-IP (Fig. 3E). This is consistent with the observation that even highly purified constitutively active Akt from a commercial source appears to contain both Akt and SR kinase activities. We further tested this possibility by using a well-characterized Akt substrate GSK3β to suppress the authentic Akt activity towards another Akt substrate H2B. We found that, while GSK3β was able to suppress H2B phosphorylation, it enhanced the associated kinase activity towards the SR protein SRSF1 (Fig. S3C), which is consistent with the reported effect of GSK3β in phosphorylating primed SR proteins (Hernandez, et al., 2004). Conversely, a synthetic SRPK substrate containing 16 Ser/Arg repeats (SR16) was able to suppress the kinase activity towards SRSF1 (Fig. S3C). These data provide a plausible explanation to a previous observation that immunopurified Akt could phosphorylate SR proteins, which led to the suggestion that SR proteins might be direct substrates for activated Akt (Blaustein et al., 2005). The evidence presented here strongly indicates that this SR protein kinase activity is due to the association of SRPKs with purified Akt.
Akt-induced SRPK phosphorylation relays EGF signaling to the nucleus
The evidence presented above indicates that, while SRPK1 may be phosphorylated on multiple sites in response to EGF signaling, two such sites appear to be directly induced by activated Akt. To determine the biological significance of such Akt-induced phosphorylation events, we asked whether phosphorylation at T326 and S587 is critical for SRPK1-dependent splicing activity. We therefore mutated both sites to either Alanine (A) or Aspartic Acid (D), the latter mimicking Akt-induced phosphorylation on SRPK1, and tested both 326A587A and 326D587D mutants in E1A splicing. We found that, while the 326A587A mutant lost the ability to trigger switch in E1A splicing, the 326D587D mutant was more potent than WT SRPK1 in inducing E1A splicing (Fig. 3F). Importantly, we found that the phospho-mimicking mutants of SRPK1 and SRPK2 rendered both kinases insensitive to Wortmannin inhibition, suggesting that the mutations bypassed the requirement for Akt activation in inducing alternative splicing (Fig. 3G–H). Therefore, although it remains to be seen whether multiple other phosphorylation events on SRPK1 have a biological function, two of these sites induced by activated Akt appear to be necessary and sufficient to transmit EGF signaling to the nucleus to regulate alternative splicing.
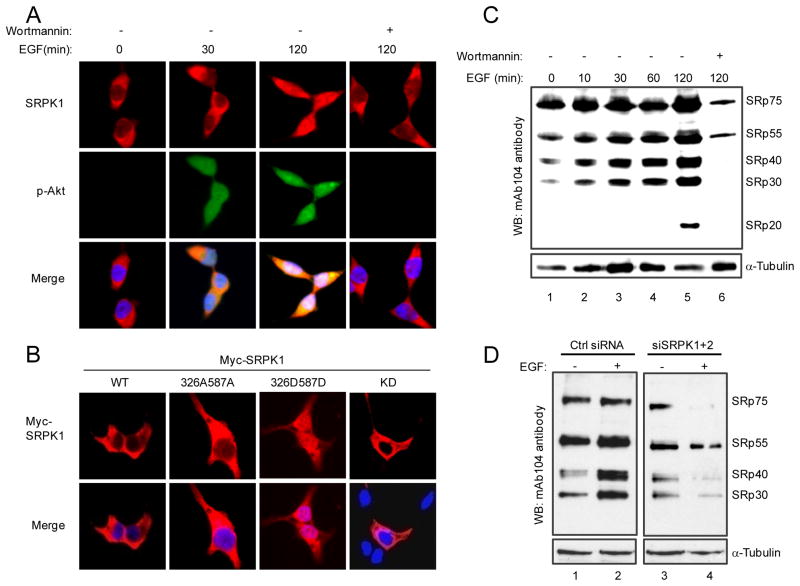
To further understand how activated Akt relays EGF signaling through SRPKs to the nucleus and in light of our previous observation that SRPKs could be induced to relocate from the cytoplasm to the nucleus in response to a stress signal (Zhong et al., 2009), we asked whether EGF signaling and Akt-mediated phosphorylation might trigger nuclear translocation of SRPKs. Indeed, we found that EGF treatment induced nuclear translocation of both SRPK1 and SRPK2, which could be blocked by Wortmannin (Fig. 4A and S4A). Again, a constitutively activated Akt was able to trigger a similar response (Fig. S4B), and as expected, none of the inhibitors against other branches in the EGF pathway showed detectable effect on blocking SRPK1 nuclear translocation (Fig. S4C). Consistently, while the phosphorylation-defective mutant SRPK1 (326A587A) was restricted to the cytoplasm, the Akt phosphorylation-mimicking mutant (326D587D) was constitutively localized in the nucleus in the absence of EGF treatment (Fig. 4B). EGF treatment showed little effect on the constitutive localization of these SRPK1 mutants in the cell (Fig. S4D). Together, these findings established signal-induced SRPK nuclear translocation under physiological conditions and demonstrated that activated Akt is both necessary and sufficient for this EGF-induced event.
Figure 4.
Induction of SRPK nuclear translocation and dynamic regulation of SR protein phosphorylation in response to EGF signaling. (A) EGF induced SRPK1 translocation to the nucleus and Wortmannin blocked the translocation. (B) Phospho-mimicking SRPK1 entered the nucleus in the absence of EGF signaling. (C) EGF induced SR protein phosphorylation whereas blockage of EGF signaling by Wortmannin triggered extensive dephosphorylation of SR proteins. (D) SRPKs are responsible for EGF-induced SR protein phosphorylation. In the absence of EGF treatment, SRPK knockdown had a detectable but modest effect on the steady state of SR protein phosphorylation (compare lane 1 with lane 3). EGF induced dramatic reduction of SR protein phosphorylation in SRPK knockdown cells (compare lane 2 with lane 4). These data suggest increased competition by SR protein phosphatases when SR protein kinases were reduced by RNAi in EGF-treated cells.
In agreement with induced translocation of SRPKs to the nucleus, SR proteins became hyperphosphorylated, as detected by a pan-phospho-SR antibody (mAb104), which could be effectively blocked by Wortmannin (Fig. 4C). SRPKs appear to be responsible for such EGF-induced increase in the steady-state phosphorylation of SR proteins because RNAi-mediated knockdown of SRPK1/K2 abolished this effect (Fig. 4D). As expected, the Alanine mutant of SRPK1 lost the effect in enhancing SR proteins phosphorylation while the Aspartic Acid mutant of the kinase potently induced SR protein phosphorylation, similar to the WT kinase, in transfected HEK293T cells (Fig. S4E). Interestingly, both Wortmannin treatment and SRPK siRNA accelerated SR protein dephosphorylation, indicating that the steady-state level of SR protein phosphorylation is dynamically regulated by both kinase and phosphatase systems, as previously noticed (Shi and Manley, 2007). Together, these results connect a series of causal events downstream of EGF signaling from Akt activation to SRPK nuclear translocation to SR protein hyperphosphorylation, leading to regulated splicing in the nucleus.
SRPKs are subject to multi-layer control before and after activation by Akt
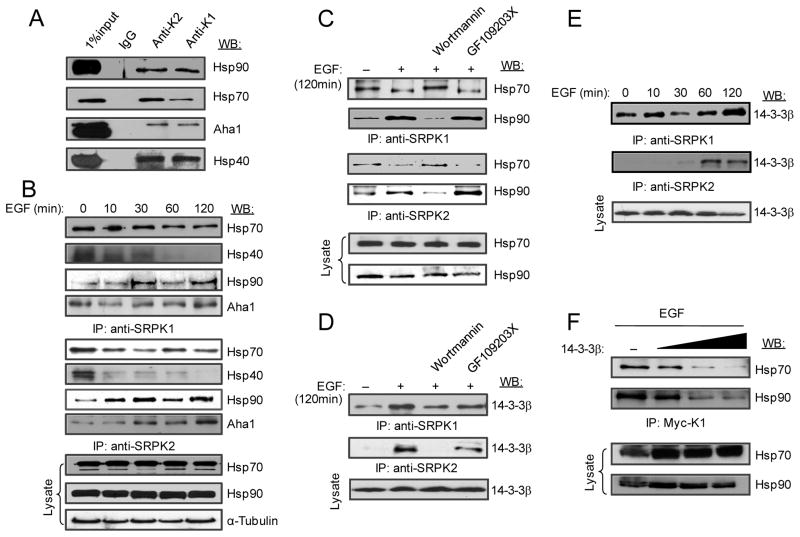
To further understand the mechanism for phosphorylation-induced nuclear translocation of SRPKs, we examined dynamic interactions of SRPKs with their molecular chaperones, which we previously showed to be responsible for anchoring the splicing kinases in the cytoplasm (Zhong et al., 2009). We first confirmed that both SRPK1 and SRPK2 are associated with Hsp70 and Hsp90 as well as their respective co-chaperones Hsp40 and Aha1 in HEK293T cells (Fig. 5A). To determine how EGF might modulate such interactions, we preformed a time-course co-immunoprecipitation experiment. We found that the association of Hsp70 and its co-chaperone Hsp40 with SRPK1 and SRPK2 was progressively reduced (Fig. 5B). We noted that the association of Hsp70 with both kinases was less sensitive than Hsp40 to EGF treatment, likely due to multiple members of the Hsp40 family expressed in the cell, thus providing redundant functions in mediating Hsp70 binding. In contrast, EGF signaling progressively induced the association of Hsp90 and its co-chaperone Aha1 with both kinases (Fig. 5B). Furthermore, the reduced association with Hsp70 and enhanced binding with Hsp90 were sensitive to Wortmannin, but not the PKC inhibitor GF109203X (Fig. 5C). These data indicate that EGF signaling induces a cascade of switches in the interaction of SRPKs with their molecular chaperones.
Figure 5.
EGF signaling modulates the interaction of SRPKs with heat shock and 14-3-3 proteins. (A) Co-immunoprecipitation of SRPK1 and SRPK2 with Hsp70 and Hsp90 along with their co-chaperones in the presence of serum. (B) EGF-induced release of Hsp70 and its co-chaperone Hsp40 from SRPKs while enhanced the association of the splicing kinases with Hsp90 and its co-chaperone Aha1. (C) The differential interaction of SRPKs with heat shock proteins required the activation of the PI3K pathway. (D-F) EGF induced the interaction of SRPKs with 14-3-3β. Both SRPKs showed similar association in proliferating HEK293T cells (D). Interestingly, SRPK2 seems to be more inducible than SRPK1 in association with 14-3-3β in response to EGF signaling (E). Increased 14-3-3β was able to progressively suppress the interaction of SRPKs with both Hsp70 and Hsp90 in EGF-treated cells (F).
An additional layer of SRPK sequestration in the cytoplasm is likely provided by the 14-3-3 family of proteins, particularly 14-3-3β, as previously demonstrated on SRPK2 (Jang et al., 2009). Indeed, we found that, like SRPK2, SRPK1 was also associated with 14-3-3β, which could be blocked by Wortmannin, but not the PKC inhibitor GF109203X (Fig. 5D), and the interaction with 14-3-3β was progressively enhanced in response to EGF signaling (Fig. 5E). Conversely, in EGF-treated cells, 14-3-3β overexpression effectively blocked the interaction of SRPK1 with both Hsp70 and Hsp90 (Fig. 5F). Together, these data suggest that SRPKs are tightly regulated by heat shock complexes and by 14-3-3 family members during the course of EGF signaling. These results explain why SRPKs are not fully relocated to the nucleus in EGF-induced cells (Fig. 4A and Fig. S4A). This tight control of SRPK nuclear translocation is likely biologically important because our early studies showed that constitutive localization of the kinases in the nucleus caused a severe cell lethal phenotype in both yeast and mammalian cells (Siebel et al., 1999; Ding et al., 2006). 14-3-3 proteins might thus function to prevent excessive localization of SRPKs even under strong stimulation conditions, which may cause toxic effects in the nucleus.
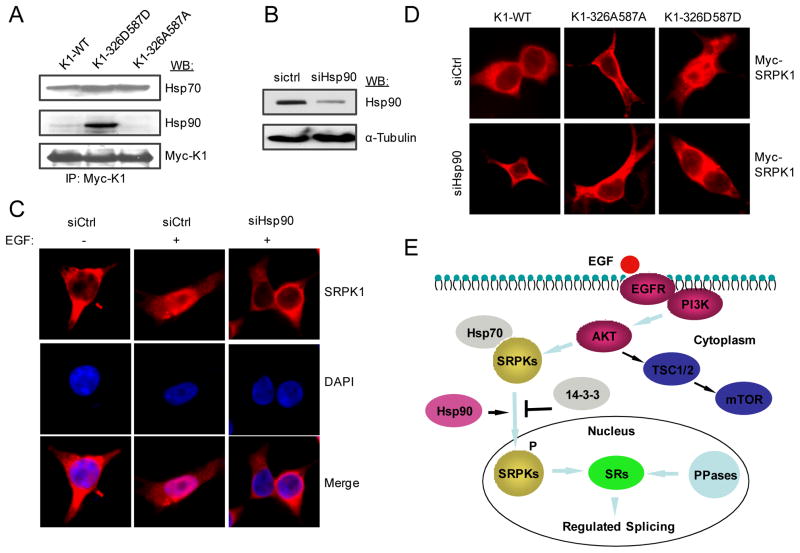
It is curious that Hsp90 became increasingly associated with SRPKs in response to EGF signaling, which was coincident with the kinetics of nuclear translocation of the kinases (Fig. 4A and Fig. S4A). As Hsp90 has been implicated in facilitating nuclear translocation of many cellular factors, such as p53 (Zylicz et al., 2001) and the nuclear receptor GRβ (Zhang et al., 2006), we asked whether the interaction of SRPK1 with Hsp90 in EGF-treated cells plays a critical role in SRPK1 nuclear translocation. We first showed that the phospho-mimicking mutant of SRPK1 caused increased association with Hsp90 (Fig. 6A). RNAi-mediated knockdown of Hsp90 (Fig. 6B) effectively blocked EGF-induced nuclear translocation of SRPK1 (Fig. 6C) as well as nuclear translocation of the phospho-mimicking mutant SRPK1-326D587D (Fig. 6D). These data strongly support a key role of Hsp90 in facilitating nuclear translocation of SRPK1 in response to EGF signaling.
Figure 6.
A critical role of Hsp90 in facilitating SRPK nuclear translocation in response to EGF signaling. (A) The phospho-mimicking mutant K1-326D587D gained elevated association with Hsp90 compared to WT SRPK1 and the double Alanine mutant K1-326A587A. (B) RNAi-mediated knockdown of Hsp90. (C) Hsp90 knockdown blocked EGF-induced SRPK1 nuclear translocation. (D) The phospho-mimicking SRPK1 mutant entered the nucleus in the absence of EGF signaling (upper panel), but in an Hsp90-dependent fashion (lower panel). (E) Summary of EGF signaling through the Akt-SRPK-SR axis to regulate alternative splicing in the nucleus. Hsp70 is responsible for inhibiting nuclear import of the splicing kinases, Hsp90 knocks off Hsp70 to facilitate nuclear import, and 14-3-3 functions to prevent excessive accumulation of the splicing kinases in the nucleus.
Discussion
The data presented here reveal a major signal transduction pathway for regulated splicing in mammalian cells. As depicted in Fig. 6E, EGF treatment activates the PI3K and then Akt. Although EGF is well known to activate multiple other signaling branches, including the ERK and JAK/STAT pathways (Jorrissen et al., 2003), our data indicate that activated Akt plays a dominant role in transducing EGF signaling to the nucleus for regulated splicing. Although mTOR is a major downstream effector in the Akt pathway (Inoki et al., 2005), we found that activated mTOR has a limited contribution to EGF-induced alternative splicing events. Instead, EGF signaling branched from activated Akt to SRPKs to regulate most of EGF-induced alternative splicing events. Therefore, SRPKs represent an important branch of the EGF signal transduction pathway for regulated splicing in the nucleus.
Previous work has placed SR proteins in growth factor-induced splicing pathway (Liu et al., 2003). However, it has been suggested that activated Akt may directly act on SR proteins (Blaustein et al., 2005) and/or relay through the Clk family of kinases that are constitutively localized in the nucleus (Jiang et al., 2009). Our current data suggest that the ability of immunopurified Akt to phosphorylate SR proteins is likely due to associated SRPKs. With respect to the Clk family of kinases, it is interesting to note that the SRPK and Clk families of kinases can act in a synergistic fashion to control the phosphorylation state of SR proteins and alternative splicing in mammalian cells (Ngo et al., 2005; Cho et al., 2011). This synergy may be further enhanced by other kinases, such as GSK3, either directly through additional phosphorylation events on SR proteins (Hernandez et al., 2004) or via GSK3-dependent feedback regulation of the Akt pathway (Lu et al., 2011). Therefore, it is entirely possible that multiple kinases are involved in EGF-induced alternative splicing.
The data presented in the current work strongly support that Akt activates SRPKs in EGF-treated cells by employing an unusual allosteric mechanism. Instead of directly transferring phosphates to its targets, like in most signal transduction cascades, we found that activated Akt binds and induces SRPK1 autophosphorylation because Akt-mediated phosphorylation depends on the kinase activity of SRPK1 and an allosteric kinase inhibitor could also induce SRPK1 autophosphorylation. This explains why Akt is able to trigger SRPK1 phosphorylation in the absence of any consensus motif in SRPK1. It also explains a previous observation that the kinase activity of SRPK1 is required for its nuclear import (Ding et al., 2006). Although published data suggest that Akt can directly transfer phosphates to SRPK2 (Jang et al., 2009), it remains to be determined whether activated Akt could also induce other phosphorylation events on SRPK2 through the autophosphorylation mechanism.
Since SR proteins are efficiently phosphorylated by bacterially expressed SRPKs, attainment of an active kinase conformation is not dependent on Akt. Instead, Akt-mediated phosphorylation appears to induce a series of re-arrangements with molecular chaperones and other regulatory factors to regulate the cellular distribution of the splicing kinases. Although the signal-dependent interaction of SRPK1 with molecular chaperones has been established in our previous studies (Zhong et al., 2009), we have now further extended the work by showing that the Hsp70-containing complexes are responsible for anchoring the splicing kinases in the cytoplasm, whereas the Hsp90-containing complexes actually facilitate SRPK translocation to the nucleus. This cascade of events is reminiscent of the regulatory p53 nuclear import pathway where the Hsp70/Hsp90-containing complex first assists p53 folding; subsequently, correctly folded p53 is imported to the nucleus in a Hsp90 dependent manner (King et al., 2001; Song et al., 2001; Zylicz et al., 2001).
The recognition of SRPKs as key signal transducers in mammalian cells paves the way to understand the role of this important family of kinases in a variety of human diseases, particularly cancer. Multiple components in the Akt pathways have been shown to function as oncogenes or tumor suppressors (Inoki et al., 2005; Carracedo and Pandolfi, 2008). Our current findings add a dimension in understanding diverse disease phenotypes from the prospective of regulated splicing, because dysregulation of RNA splicing has been attributed to many different kinds of human diseases (Cooper et al., 2009). Potential roles of SRPKs in cancer are underscored by the observed overexpression of SRPK1 in adult T-cell leukemia (Hishizawa et al., 2005) as well as in several types of solid tumors, such as colon, pancreatic, and breast carcinomas (Hayes et al., 2006; 2007; Plasencia et al., 2006). A more recent study demonstrated that SRPK1 is transcriptionally repressed by WT1, a well-known tumor suppressor, and overexpression of SRPK1 directly contributes to angiogenesis through induced VEGF alternative splicing that causes renal failure and Wilms’ tumors (Amin et al., 2011). Interestingly, SRPK1 down regulation has also been linked to tumorigenesis in male germ cell tumors (Schenk et al., 2004) and late stage retinoblastoma (Krishnakumar et al., 2008). These observations suggest that altered SRPK expression in either direction may contribute to tumorigenesis in different biological contexts. By placing SRPKs in a central position in the Akt pathway, we can now begin to dissect key molecular events from Akt activation to regulated splicing in understanding the etiology and progression of human cancers.
Experimental Procedures
Reagents
EGF, Wortmannin, GF109203X, WP1066, U0126, GSK3β, H2B, Rapamycin, anti-α-Tubulin, and anti-Myc were from Sigma. Anti-phospho-Akt, anti-JAK2, and anti-phospho-JAK2 were from Cell Signaling. Antibodies against Akt, HSP70, HSP40, 14-3-3β, p-ERK1/2, ERK1/2, HA, and IgG were from Santa Cruz. MK2206 was from Selleck Chemicals. Antibodies against HSP90, SRPK1 and SRPK2 were from BD Pharmingen. Anti-Aha1 was a gift from the laboratory of William E. Balch. The mAb104 hybridoma was from ATCC and 1:4 dilution of culture supernatant was used for Western blotting.
Protein A/G Sepharose was from GE Healthcare. Ni-resin and On-column DNase kit were from Qiagen. Lipofectamine 2000, trizol, SuperScript III First-Strand Synthesis System and RNase-free DNase I were from Invitrogen. SiSRPK1 and siSRPK2 were from Dharmacon. SiHsp90 was from Bioneer. Activated Akt1 was from Millipore. Applied Biosystem AmpliTaq® Gold kit was from Applied Biosystems.
Cell culture, transfection, and drug treatment
Cells were cultured in Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum. Transient transfections were performed using Lipofectamine 2000. Cells were first starved for 12 hrs and pretreated with various pharmaceutical inhibitors (20 nM Wortmannin, 10 μM GF109203X, 100 nM Rapamycin, 10 μM U0126 or 3 μM WP1066) for 30 min, followed by EGF treatment (100 ng/ml) from various time points as indicated in the text.
Minigene analysis of regulated splicing
Cells were cotransfected with the E1A minigene plasmid along with different plasmids. Total RNA was extracted and reverse-transcribed to cDNA and then was used for PCR amplification as previously described (Zhong et al., 2009; Yang et al., 1994).
RASL-Seq analysis of alternative splicing
A pool of oligonucleotides was designed to detect 3726 alternative splicing events. The RASL reaction (Fig. S2A) was carried out as previously described (Li et al., 2012).
Two-sided Kolmogorov-Smirnov statistics
Two-sided Kolmogorov-Smirnov statistics (in the R package, http://cran.r-project.org/) was used to determine the significance of SRPK knockdown and various kinase inhibitors in blocking EGF-induced alterative splicing (Conover, W. J. 1971).
Expression of recombinant proteins and in vitro phosphorylation assay
Different SRPK1 mutants were individually cloned into pRSET-A and expressed as His-tagged proteins in BL-21 DE3. After incubated with IPTG at 20°C for 12 hrs, cells were harvested and disrupted in a lysis buffer (20 mM MES-pH6.5, 150 mM NaCl, 20% glycerol, 1 mM PMSF, protease inhibitor, 1 mg/ml lysozyme, RNAse A (1500 U), and DNase I (1000 U)) by sonication. The supernatant was loaded onto the Nickel resin and was washed three times. Individual His fusion proteins were eluted with the lysis buffer containing 300 mM imidazole. Purified proteins were dialyzed and stored in the kinase reaction buffer (20 mM Tris-pH 7.5 and 10 mM MgCl2).
The kinase assay was carried out in a 25 μl kinase reaction containing 25 μM ATP and 2.5 μCi of [γ-32P]-ATP and incubated for 20 min at 30 °C. Reactions were terminated by boiling in 10 μl 2× SDS sample buffer for 5 min followed by SDS-PAGE and autoradiography.
Co-immunoprecipitation analysis
Cells were lysed in 1 ml of lysis buffer (10 mM Tris-pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.5% Triton-100, 1× phosphate, protease inhibitors), and was added to 40 μl slurry (1:1 ratio) of protein A/G Sepharose pre-bound with various antibodies. After incubation for 5 hrs at 4°C with rocking, antigen/antibody beads were collected. After elution in 30 μL SDS loading buffer and separation by SDS-PAGE, the samples were transferred to nitrocellulose membrane. The membrane was blocked in buffer TBST plus 5% nonfat milk and then incubated with individual primary antibodies. After extensive rinse with TBST, the blot was incubated with appropriate HRP-conjugated secondary antibody and analyzed by ECL.
Phosphopeptide generation and LC-MS/MS analysis
Protein samples were prepared as described previously (McCormack et al. 1997). After prepared protein samples were digested with trypsin (trypsin:protein ratio=1:50) overnight at 37°C, the peptides were extracted and desalted using Aspire RP30 desalting columns (Thermo Scientific). Then, trypsin-digested peptides were analyzed by high pressure liquid chromatography (HPLC) coupled with tandem mass spectroscopy (LC-MS/MS) using nano-spray ionization. Finally, the collected data were analyzed using MASCOT® (Matrix Sciences) and Protein Pilot 4.0 (ABSCIEX) for peptide identifications.
Fluorescence Microscope Analysis
Cells were grown onto coverslips and were fixed with 3.7% paraformaldehyde in PBS for 10 min. The cells were then permeabilized with 0.2% Triton X-100 for 10 min and blocked with 1% fetal bovine serum in PBS for 20 min. After washing with PBS three times, the coverslips were loaded with primary antibodies, incubated for 1 hr, washed with PBS three times, and treated with fluorescence-conjugated secondary antibodies with 1hr. Washed coverslips were mounted in a solution containing 4,6-diamidino-2-phenylindole (DAPI) and images captured on a Zeiss Axiophot microscope.
Supplementary Material
Highlights.
SR kinases are a major branch in the EGF pathway to regulate alternative splicing.
Activated Akt induces SRPK1autophosphorylation.
Activated SRPK1 translocates to the nucleus in an Hsp90-dependent manner.
Induced SR protein hyper-phosphorylation leads to widespread changes in splicing.
Acknowledgments
The authors are grateful to W. Balch for the anti-Aha1 antibody and to colleagues in the Fu lab, particularly Xiangyang Zhong, and Pingping Wang, for stimulating discussion and Jie Huang for contribution to statistic analysis during the course of investigation. This work was supported by NIH grants (GM52872 and HG004659) to X-D.F. R.M.P. was supported by NIH under the Ruth L. Kirchstein National Research Service Award (GM090484). J.A.A. was supported by an NIH grant (GM67969). M.G.R. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession number
The RASL-seq data for profiling alternative splicing in response to EGF or individual kinase inhibitors are available at the Gene Expressing Omnibus under the accession number XXX.
Supplemental information includes four figures and a table summarizing the RASL-seq results under different experimental conductions, which can be found with this article online at XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amin EM, Oltean S, Hua J, Gammons MV, Hamdollah-Zadeh M, Welsh GI, Cheung MK, Ni L, Kase S, Rennel ES, Symonds KE, Nowak DG, Royer-Pokora B, Saleem MA, Hagiwara M, Schumacher VA, Harper SJ, Hinton DR, Bates DO, Ladomery MR. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell. 2011;20:768–780. doi: 10.1016/j.ccr.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc Natl Acad Sci USA. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Tanos T, Muñoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Cáceres JF, Coso OA, Srebrow A. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by Akt. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Pandolfi PP. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 2008;27:5527–5541. doi: 10.1038/onc.2008.247. [DOI] [PubMed] [Google Scholar]
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembley. Proc Natl Acad Sci USA. 2011;108:8233–8238. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover WJ. Practical Nonparametric Statistics. New York: John Wiley & Sons; 1971. Statistical analysis was performed using “R”; pp. 309–314. ( http://cran.r-project.org/), a freely available open-source software package. [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JH, Zhong XY, Hagopian JC, Cruz MM, Ghosh G, Feramisco J, Adams JA, Fu XD. Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol Biol Cell. 2006;17:876–885. doi: 10.1091/mbc.E05-10-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle D, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares M., Jr Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struc Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes GM, Carrigan PE, Beck AM, Miller LJ. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res. 2006;66:3819–3827. doi: 10.1158/0008-5472.CAN-05-4065. [DOI] [PubMed] [Google Scholar]
- Hayes GM, Carrigan PE, Miller LJ. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67:2072–2080. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- Hegele A, Kamburow A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Luhrmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell. 2011;45:567–580. doi: 10.1016/j.molcel.2011.12.034. [DOI] [PubMed] [Google Scholar]
- Hernandez F, Perez M, Lucas JJ, Mata AM, Bhat R, Avila J. Glycogen synthase kinase-3 plays a cruical role in Tau exon 10 splicing and intranuclear distribution of SC35. J Biol Chem. 2004;279:3801–3806. doi: 10.1074/jbc.M311512200. [DOI] [PubMed] [Google Scholar]
- Heyd F, Lynch KW. Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing. Mol Cell. 2010;40:126–137. doi: 10.1016/j.molcel.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyd F, Lynch KW. DEGRADE, MOVE, REGROUP: signaling control of splicing proteins. Trend Biomed Sci. 2011;36:397–404. doi: 10.1016/j.tibs.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Sootome H, Nakatsuru Y, Miyama K, Taguchi S, Tsujioka K, Ueno Y, Hatch H, Majumder PK, Pan BS, Kotani H. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Ther. 2010;9:1956–1967. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- Hishizawa M, Imada K, Sakai T, Ueda M, Hori T, Uchiyama T. Serological identification of adult T-cell leukaemia-associated antigens. Br J Haematol. 2005;130:382–390. doi: 10.1111/j.1365-2141.2005.05619.x. [DOI] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Jang SW, Liu X, Fu H, Rees H, Yepes M, Levey A, Ye K. Interaction of Akt-phosphorylated SRPK2 with 14-3-3 mediates cell cycle and cell death in neurons. J Biol Chem. 2009;284:24512–24525. doi: 10.1074/jbc.M109.026237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M, Cooper DR. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology. 2009;150:2087–2097. doi: 10.1210/en.2008-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorrissen RN, Walker F, Pouliot N, Garrett TPJ, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signaling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- King FW, Wawrzynow A, Höhfeld J, Zylicz M. Co-chaperones Bag-1, Hop and Hsp40 regulate Hsc70 and Hsp90 interactions with wild-type or mutant p53. EMBO J. 2001;20:6297–6305. doi: 10.1093/emboj/20.22.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König H, Ponta H, Herrlich P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998;17:2904–2913. doi: 10.1093/emboj/17.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar S, Mohan A, Kandalam M, Ramkumar HL, Venkatesan N, Das RR. SRPK1: a cisplatin sensitive protein expressed in retinoblastoma. Pediatr Blood Cancer. 2008;50:402–406. doi: 10.1002/pbc.21088. [DOI] [PubMed] [Google Scholar]
- Li H, Qiu J, Fu X-D. RASL-seq for massive parallel and quantitative analysis of gene expression. Curr Protocol Mol Biol. 2012;Chapter 4(unit 4.13) doi: 10.1002/0471142727.mb0413s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Adv Exp Med Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Liu X, Mayeda A, Tao M, Zheng ZM. Exonic splicing enhancer-dependent selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site can be rescued in a cell lacking splicing factor ASF/SF2 through activation of the phosphatidylinositol 3-kinase/Akt pathway. J Virol. 2003;77:2105–2115. doi: 10.1128/JVI.77.3.2105-2115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Muller M, Smith D, Dutta B, Komurov K, Iadevaia S, Ruths D, Tseng JT, Yu S, Yu Q, Nakhleh L, Balazsi G, Donnelly J, Schurdak M, Morgan-Lappe S, Fesik S, Ram PT, Mills GB. Kinome siRNA-phosphoproteomic screen identifies networks regulating Akt signaling. Oncogene. 2011;30:4567–4577. doi: 10.1038/onc.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]
- Lynch KW, Weiss A. A model system for activation-induced alternative splicing of CD45 pre-mRNA in T cells implicates protein kinase C and Ras. Mol Cell Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- McCormack AL, Schieltz DM, Goode B, Yang S, Barnes G, Drubin D, Yates JR 3rd. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B, Yun CY, Wong CF, McCammon JA, Fu XD, Ghosh G. The structure of Sky1p reveals a novel mechanism for constitutive activity. Nat Struct Biol. 2001;8:176–183. doi: 10.1038/84178. [DOI] [PubMed] [Google Scholar]
- Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–265. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasencia C, Martínez-Balibrea E, Martinez-Cardús A, Quinn DI, Abad A, Neamati N. Expression analysis of genes involved in oxaliplatin response and development of oxaliplatin-resistant HT29 colon cancer cells. Int J Oncol. 2006;29:225–235. doi: 10.3892/ijo.29.1.225. [DOI] [PubMed] [Google Scholar]
- Schenk PW, Stoop H, Bokemeyer C, Mayer F, Stoter G, Oosterhuis JW, Wiemer E, Looijenga LH, Nooter K. Resistance to platinum-containing chemotherapy in testicular germ cell tumors is associated with downregulation of the protein kinase SRPK1. Neoplasia. 2004;6:297–301. doi: 10.1593/neo.03406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Shin C, Manley JL. Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol. 2004;5:727–738. doi: 10.1038/nrm1467. [DOI] [PubMed] [Google Scholar]
- Shultz JC, Goehe RW, Wijesinghe DS, Murudkar C, Hawkins AJ, Shay JW, Minna JD, Chalfant CE. Alternative splicing of caspase 9 is modulated by the phosphoinositide 3-kinase/Akt pathway via phosphorylation of SRp30a. Cancer Res. 2010;70:9185–9196. doi: 10.1158/0008-5472.CAN-10-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Feng L, Guthrie C, Fu XD. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc Natl Acad Sci USA. 1999;96:5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Takeda M, Morimoto RI. Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat Cell Biol. 2001;3:276–282. doi: 10.1038/35060068. [DOI] [PubMed] [Google Scholar]
- Velazques-Dones A, Hagopian J, Aubol BE, Ma CT, Zhong XY, Zhou H, Ghosh G, Fu XD, Adams JA. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J Biol Chem. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weg-Remers S, Ponta H, Herrlich P, König H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2001;20:4194–4203. doi: 10.1093/emboj/20.15.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bani MR, Lu SJ, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Fan JB, Doucet D, Luo L, Wickham E, Ye Z, Chee MS, Fu XD. Profiling alternative splicing on fiber-optic arrays. Nat Biotechnol. 2002;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- Zhang X, Clark AF, Yorio T. Heat shock protein 90 is an essential molecular chaperone for nuclear transport of glucocorticoid receptor β. Investigative Ophthalmology Visual Sci. 2006;47:700–708. doi: 10.1167/iovs.05-0697. [DOI] [PubMed] [Google Scholar]
- Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009;23:482–495. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylicz M, King FW, Wawrzynow A. Hsp70 interactions with the p53 tumour suppressor protein. EMBO J. 2001;20:4634–4638. doi: 10.1093/emboj/20.17.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.