Abstract
Purpose
The β3 subunit of the γ-aminobutyric acid type A receptors (GABAA–Rs) is an essential component of GABAA–Rs in fetal, perinatal and adult mammalian brain. Various transcripts of the β3 subunit gene (GABRB3) produce various proteins with different N-termini. Rare variants in this N-terminus (exon 1A and exon 2) of GABRB3 protein segregate in affected family members of two multigeneration-multiplex families with remitting childhood absence epilepsy (rCAE), suggesting GABRB3 is a major Mendelian epilepsy gene for rare families with CAE. Thus the N-terminus of GABRB3 could be important for GABRB3 regulation in development and its alteration could produce rCAE. Here we determine if SNPs within the 1148 bp region upstream from exon1a influence the expression of GABRB3.
Methods
We studied luciferase reporter expression for promoter activity, 1148 bp upstream from exon 1A, using human embryonic kidney 293 cells. We generated constructs of the promoter region and compared different SNP haplotypes in 48 patients with rCAE. Then, we compared frequencies of rs20317, located in the core promoter region and rs4906902 located in the enhancer region between 48 patients with rCAE and more than 500 healthy controls matched for ethnicity and ancestral origin.
Key Findings
Highest luciferase expression occurred 230 bp upstream of exon 1A. The construct which excluded this region lost luciferase activity. Thus, this region contains the core promoter of exon1A. Allele C but not allele G (rs20317) significantly increased luciferase expression activity. Allele C creates binding motifs for cMYB and EGR-3. Longer constructs overlapping this region have a binding motif for REST (RE1-silencing transcription factor), a critical epigenetic modulator for neuronal genes. REST represses expression of neuronal genes in non-neuronal tissues, resulting in reduced luciferase expression activity. Even in the suppressed condition, the longer construct enhanced luciferase expression activity of the shorter construct, which excluded the distal end containing rs4906902. However allele frequencies of rs20317 and rs4906902 were not significantly associated with 48 rCAE patients in comparison to > 500 controls matched for ethnicity and ancestral origin.
Significance
Common SNPs in the promoter region increase luciferase expression activity. An epigenetic modulator, REST specifically alters expression of GABRB3 exon 1A transcripts, suggesting epigenetic regulation by REST dominantly controls the expression of GABRB3 variant2 transcript in early life GABAA signaling. Abnormal epigenetic regulation could be involved in absence seizures.
Keywords: luciferase activity, rs20317, rs4906902, epigenetic modulator, REST, Childhood absence epilepsy
Introduction
The GABAA receptor β3 gene (GABRB3) product emerges early in embryonic and neonatal brain (Laurie et al., 1992; Ben-Ari et al.,1997; Herlenius & Lagercrantz, 2004). Reduced GABRB3 expression is involved in three neurological developmental disorders that have seizures: Angelman syndrome, autism spectrum disorder, and Rett syndrome (Samaco et al,. 2005; Hogart et al., 2007). In our cohort of remitting childhood absence epilepsy (rCAE) patients, three variants (P11S, S15F, G32R) in the N-terminus of GABRB3, i.e., in an alternative signal sequence (exon 1A), and in exon 2, segregate with affected members of multigeneration families with rCAE, suggesting GABRB3 is a major Mendelian epilepsy gene in rare families with rCAE (Tanaka et al., 2008). Replication of epilepsy in mice lacking the β3 subunit of GABAA receptor further suggests a causative role for GABRB3 in epilepsy (DeLorey et al., 1998). To clarify the influence of SNPs within the 5′ upstream region on the expression of alternative transcripts of GABRB3 exon 1A, we studied luciferase reporter assays within the 1148 bp region upstream from exon 1A in human embryonic kidney 293 (HEK) cells, and examined the difference between haplotypes or alleles in SNPs found in rCAE patients. To determine if SNPs on the promoter region also affect as minor susceptibility genes in rCAE patients at large, we compared allele frequencies between our cohort of 48 rCAE patients and 568 controls matched for ethnicity and ancestral origin.
Materials and Methods
Sample collection and Family material: We studied 48 persons with rCAE, as defined by our inclusion and exclusion criteria and the 1989 and 2010/ILAE clarification of Epilepsies (Supplement; Table 1). Additionally we analyzed 568 healthy controls, matched by ethnicity and ancestral origin, obtained from Mexico and Honduras.
-
Detection of Polymorphisms in the 5′ region of GABRB3:
DNA was extracted from each blood sample with the QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, CA). We amplified two overlapped proximal regions in the 5′ region of GABRB3 by polymerase chain reaction (PCR) using two pair of primers (5′tgtgtccattagacaaaagtctgc/acctacggccacaggtttta3′, 5′cagaacacaaaaacgagcttgatg/ccgacggtgcctgcagaa3′), and the AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA) or Fast Taq polymerase (Roche, Indianapolis, IN). Each PCR product of probands was screened by direct sequencing with the use of ABI 3700 capillary automated-sequencing system (Applied Biosystems, Foster City, CA). SNP genotyping for healthy ethnically matched controls was performed by restriction endonuclease digestion or direct sequencing.
-
Reporter constructs:
Four 5′ primers and one common 3′ primer were designed to amplify four regions from −1148 bp to −18 bp in the 5′ region of exon 1A by PCR of human genomic DNA (Fig 1). Additionally one construct was designed to contain from −1148 bp to −206 bp upstream. The products were subcloned into a TA-vector (Invitrogen, Carlsbad, CA) and cleaved with suitable restriction enzymes (KpnI/XhoI), gel-purified, and subcloned into promoterless pGL3 Enhancer vectors (Promega, Madison, WI).
-
Cell transfection and luciferase assay
HEK 293 cells were maintained in Minimum Essential Medium (MEM) Alpha Medium (Invitrogen, Carlsbad, CA) and transfected with Lipofectamine Reagent (Invitrogen, Carlsbad, CA) on 24 well plates. GABRB3 promoter pGL3 enhancer vector constructs (1 μg), 0.5 μg pGL3 control vector, 0.5 μg pGL3-promoterless enhancer vector: each was cotransfected with 2 ng phRG-TK vector containing the Renilla luciferase gene (Promega, Madison, WI) to correct for transfection efficiency. Twenty-four to thirty hours after transfection, cells were harvested and luciferase expression activities were assessed using the Stop & Glo kit (Promega, Madison, WI). Firefly luciferase activity was normalized to Renilla luciferase activity, and each activity was indicated relative to the activity of the promoterless enhancer vector.
-
In silico analysis
Alignment of mouse and human 5′ regions were performed with ClustalW (1.82). Putative transcription factor binding sites in the −1148 bp upstream flanking region of exon 1A were predicted with MatInspector Release professional 8.01 (http://www.genomatix.de) and Transcription Element Search System (TESS) (www.cbil.upenn.edu/tess/). Putative transcription factor binding motifs in the conserved region were searched by rVista 20 (http://rvista.dcode.org/) and comparison with the prediction of mice genome with MatInspector. The REST/NRSF binding site was confirmed by Quantitative Enrichment of Sequence Tags (QuEST 2.4) (Valouev et al., 2008) (http://mendel.stanford.edu/SidowLab/downloads/quest/indexhtml) and ENCODE Transcription Factor Binding Sites by ChIP-seq in Regulation Tracks in UCSC Genome Browser on Human Mar 2006 (NCBI36/hg18) Assembly (http://genomeucscedu/). Prediction for CpG islands was performed by MethPrimer v11 beta (http://www.urogene.org/methprimer/index1.html).
-
Statistics
Allele frequencies were compared with healthy ethnically matched controls using Fisher exact test. Differences in luciferase activity were compared using a t-test, and p values less than p<0.05 were considered significant.
Table 1.
Allele frequency and genotype frequency for dbSNP rs20317 and rs4906902
| rs20317 | N | C/C | G/C | G/G | Allele C | Allele G | The two-tailed P |
| CAE | 48 | 0.437 | 0.396 | 0.167 | 0.635 | 0.365 | 0.1955 |
| Control | 505 | 0.323 | 0.476 | 0.201 | 0.562 | 0.438 | - |
| rs4906902 | N | C/C | T/C | T/T | Allele T | Allele C | The two-tailed P |
| CAE | 48 | 0.205 | 0.438 | 0.357 | 0.594 | 0.406 | 0.1667 |
| Control | 521 | 0.241 | 0.478 | 0.281 | 0.52 | 0.48 | - |
Each N of healthy controls corresponds to N of obtained PCR products.
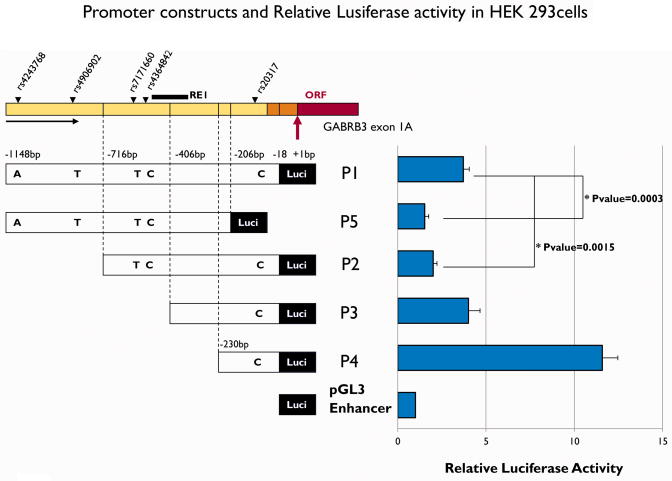
Fig 1. Promoter constructs and relative luciferase activity in HEK 293 cells.
Five deletion constructs of 5′ region are employed in the analyses. They were generated as described under Methods. Data are presented as average-fold difference of luciferase activity versus control vector (pGL3 enhancer in bottom of Fig 2) ± SE of six independent experiments. Luciferase activity in P4 is 11-fold higher compared to controls. All constructs containing RE1 (P1, P2, P3, P5) have decreased activities.
Results
Promoter Activities
Five constructs were generated for deletion analysis in the 5′ region of GABRB3 exon1A. Construct P1 is upstream from −18 bp to −1148 bp; construct P2 is −18 to −715 bp; construct P3 is −18 to −405 bp; and construct P4 is −18 to −230 bp. Construct P5 is from −206 bp to −1148 bp and excludes the −205 bp upstream flanking region of the translational start codon from construct P1 (see Figs 1, 3). Each construct was cloned into pGL3 promoterless enhancer vector and the five were compared for stimulation of luciferase expression in HEK 293 cells (Fig 1& 2). Each value was normalized for Renella luciferase activity. The construct P4 shows 11 times higher activity than a control vector in which no construct was inserted into pGL3 enhancer vector. In contrast, constructs longer than P4 and containing P4 (P1, P2, P3) had activities less than P4 but more than a control vector. Activity of the construct P3 showed 35% of P4, the construct P2 showed 17 % of P4, but the construct P1 showed a little greater than P2, 32 % of P4. The construct P5 was 13% of P4. The difference in luciferase activities between constructs P1 and P5 was statistically significant, (P=0.0003). The difference between constructs P1 and P2 was also statistically significant, (P=0.0015). These results suggest that the 205 bp upstream region from the transcriptional initiation site contains the core promoter region and the last 432 bp distal end has an enhancing effect (see Fig 1).
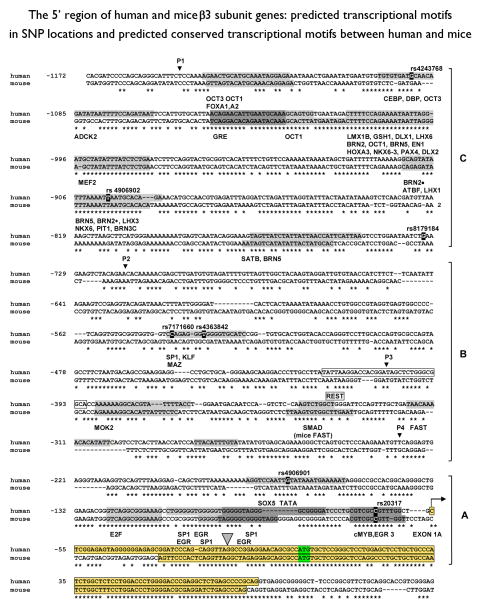
Figure 3.
The 5′ region of human and mice β3 subunit genes: Predicted transcriptional motifs in SNP locations and predicted conserved transcriptional motifs between human and mice
The nucleotide sequences of human and mice β3 subunit genes were aligned by inserting horizontal dashed lines (- - - -) to maximize homology, and conserved nucleotides are indicated by asterisks. Each exon 1A is surrounded by a yellow colored rectangle (
 ) and each translational start codon is shadowed by green. The arrow (
) and each translational start codon is shadowed by green. The arrow (
 ) shows the transcriptional start site of exon 1A.
) shows the transcriptional start site of exon 1A.
Location of SNPs are painted white against a black background (
 ) and each accession number is shown above each nucleotide. The black triangles (▼) represent the border of each segment for the deletion study. The gray larger triangle (
) and each accession number is shown above each nucleotide. The black triangles (▼) represent the border of each segment for the deletion study. The gray larger triangle (
 ) represents the common end nucleotide for each segment of the deletion study except P5. The region which was predicted to contain transcriptional motifs associated with neuronal cells and several ubiquitous major motifs are shown by shadowed letters (
) represents the common end nucleotide for each segment of the deletion study except P5. The region which was predicted to contain transcriptional motifs associated with neuronal cells and several ubiquitous major motifs are shown by shadowed letters (

 ), and each annotation is presented under each motif. The REST binding motif (RE1) is also surrounded by a rectangle. Common predicted matrixes between human and mice have over 0.89 of matrix similarity in MatInspector version 8.01.
), and each annotation is presented under each motif. The REST binding motif (RE1) is also surrounded by a rectangle. Common predicted matrixes between human and mice have over 0.89 of matrix similarity in MatInspector version 8.01.
In region A, SNP rs20317 is located in the potential core promoter region, which is over 75 % identical between mouse chromosome 7 and human chromosome 15. Three specificity protein 1 (SP1) binding sites which overlap three EGR motifs and one E2F motif are predicted in the region from −124 bp to −77 bp upstream from the translational start codon in region A. These three sorts of motifs are also predicted in the mouse genome. GpC island clustering is found in the flanking region −140 bp upstream from the start codon ATG.
The matrix of the TATA box lies in the region of SNP rs4906901, but three SP1 binding sites reside downstream of this motif. The allele G of rs4906901 makes SOX 5 binding motif but the allele T does not. SOX5 has been shown the involvement in neurogenesis, neuronal differentiation and neocortical neuron diversity (Lai et al., 2008).
In region B, FAST1 and SMAD protein plays a critical role in early embryonic development as well as REST (Silvestri, 2008).
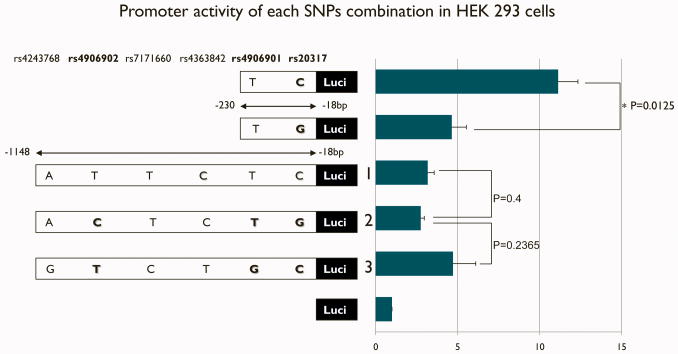
Figure 2. Promoter activity of each SNP combination in HEK 293 cells.
Graph shows luciferase activity in HEK 293 cells for each construct with the SNP haplotype and the length of the construct in the description on the left. Data are presented as average-fold difference of luciferase activity versus negative control (pGL3 enhancer) vector ± SE of three independent experiments. Luciferase activity of the short construct with the C allele of db SNP ID rs20317 was significantly higher than the G allele; however, all longer constructs have no significant difference in activity.
Haplotype and promoter activity
We observed 7 SNPs in 48 rCAE probands in the 5′ region. No novel mutations were found in this region (Figure 3). Haplotypes (Fig 2) were cloned into pGL3 Enhancer vector and compared for transfection in HEK 293 cells. The luciferase promoter activity of the C allele of SNP (rs23017) in the construct P4 were significantly higher than the luciferase promoter activity of allele G (t=4.3135, P =0.0125) (see Fig 2). The difference in DNA sequence of these P4 constructs is solely this allele. Therefore, the transcription factor binding motif which contains this SNP is suggested to play an important part in the regulation of the GABRB3 promoter in HEK cells. Transcription factor analysis using MatInspector and TESS predicted a match of cMYB and EGR 3 for allele C. However, the relative luciferase activities of P1 constructs containing different alleles in different combinations including the same alleles in the P4 region, were not significantly different from each other (t=0.933, p=0.409). The relative luciferase activities were suppressed in equal amounts (see Fig 2). In silico analysis revealed that the repressor element 1 (RE1) was predicted to at the position, −418 to −398 bp upstream region. This region was coincident with the same result reported by the genome-wide study by Valouev et al (Valouev et al., 2008) and ENCODE data (The ENCODE Project Consortium, 2007) in UCSC Genome Browser on Human Mar 2006 (NCBI36/hg18) Assembly (see Fig 3).
Statistical analyses
Allele frequencies for rs20317 and rs4906902 in the 48 probands with rCAE were not statistically significant in comparison with 504 and 521 healthy ethnically matched controls respectively (P=0.1955 for rs20317, P= 0.1667 for rs4906902 Table 1).
Discussion
GABRB3 has at least five transcription start sites, potentially creating various GABRB3 protein isoforms. GABRB3 transcript variant 2 (exon1A) and transcript variant 1(exon1), shorter transcripts started from intron3 (transcript variant 3 and variant 4) and a longer transcript (CR749803.1) from the intron of the contiguous gene GABRA5 have all been identified. All these transcripts have common exons (exon 4~exon 9). The longest transcript has common exons 2 ~ 9. Distinct N-termini of these GABRB3 protein isoforms suggest differing roles in brain development (Kirkness & Fraser, 1993). Our study therefore targeted the 1148 bp 5′-upstream region of exon1A because of its importance in immature brain.
Our studies showed: 1) highest luciferase reporter acitivities resulted from the P4 construct, 230 bp upstream of exon 1A. In contrast, luciferase reporter assays showed almost background level in the construct P5, deleted −205 bp upstream of the translational start codon from construct P1 (Fig. 1). These results suggested that the P4 region contains the core promoter of exon 1A. 2) Longer constructs (P1, P2, P3) which specifically contain REST/NRSF binding sites suppressed luciferase activities (Fig. 1). 3) The longer construct P1 enhanced the luciferase activity more than the shorter construct P2, which lacks the 432 bp distal end of the construct P1, containing alleles of db SNP; rs 4906902 (Fig, 1). These results also showed the 432 bp distal end of the construct P1 has an enhancing effect. 4) The allele G (db SNP ID: rs20317) in the potential core promoter region, showed significantly decreased luciferase activity in HEK cells compared to allele C (Fig. 2). 5) However, allele frequencies in rs 20317 and rs 4906902 were not statistically and significantly associated with rCAE patients when compared with healthy controls, suggesting they are not minor susceptibility alleles in rCAE patients at large. A larger cohort of rCAE patients will need to be studied to verify this suggestion.
Here, we discuss characteristics of the promoter region of exon1A in GABRB3 and the interpretations of our results. Fig 3 illustrates in detail the 5′ region of human and mouse beta3 subunit genes and their predicted transcriptional motifs.
Region B is not highly conserved; however, it contains the important DNA sequence element, RE-1, which is where the transcriptional repressor binds. This repressor is called “neuron-restrictive silencer factor (NSRF), and is also known as [the repressor element 1 (RE1)-silencing transcription factor (REST)]”. RE1 is a well-specified site regulating a large network of neuronal genes epigenetically (Chen et al., 1998; Ballas & Mandel, 2005). The same REST binding site was also found in the human Jurkat cell line (Valouev et al., 2008) and in various cells including HEK cells in the ENCODE project (The ENCODE Project Consortium, 2007). It is reasonable to suggest from our results that REST binds RE-1 and is probably responsible for suppressing promoter activities in all our constructs except P4.
Clustering of neuron-related transcription factors of the POU family, e.g., BRN2, BRN5, OCT1, PIT1, OCT3, and BRN3 including DLX1 and DLX2 transcription factors in region C is speculated by in silico analysis to also potentially play a role in enhancing promoter activity. Indeed, our construct with this region significantly improved luciferase reporter expression activity (t=4.3127, p=0.0015) (Fig 1). Although these results were actually produced in HEK 293 cells and promoter activity was still suppressed, the larger construct P1 did produce greater expression than the P2 construct lacking the distal end. Urak et al. (2006) similarly studied luciferase promoter activities for the construct which contains the SNP rs4906902 in NT2 cells (human neuronal-like cell) where actually rs4906901 and rs20317 lie. In results of Urak et al. (2006), the activity of the construct with T allele of rs4906902 which combine with G allele of rs490690 and C allele of rs20317 (Combination 3, T-G-C in Fig2) was significantly higher than the construct which combined C allele of rs490602, T allele of rs490601, and G allele of rs20317 (Combination 2, C-T-G in Fig2) in NT2 cells (p< 0.0001 (Urak et al., 2006)). Interestingly, allele T (db SNP ID: rs4906902) in the region that had an enhancing effect, have various predicted neuronal-related motifs, including two BRN2 motifs (Table 2). On the other hand, allele C of rs4906902 does not match transcription factors in table 2 except one of two predicted BRN2 motifs. It matches the matrix of GFI1 which is the nuclear zinc finger protein that works as a transcriptional repressor (see table 2). The study of Urak et al. (2006) also showed differences of the binding condition of BRN2 between each alleles at rs 4906902 using gel shift assay of nucler proteins extracts in NT2cell (human neuronal-like cell). Even though SNPs in the promoter region are functional, we could not replicate association between rs4096092 and CAE as reported by Urak et al. Our results used HEK cells and showed no significantly different relative luciferase expression between constructs that contained SNPs rs4906902. Instead, we observed promoter activities which were suppressed in HEK cells, probably due to REST (Fig 2).
Table 2.
Change of predicted transcriptional motifs associated with neuronal cell by common sequence SNPs in 5′ region of GABRB3
| SNPID | rs4243768 | rs4906902 | rs7171660 | rs4906901 | rs20317 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs4363842 | ||||||||||
|
| ||||||||||
| Allele | G | A | T | C | C | T | G | T | G | C |
| T | T | |||||||||
|
| ||||||||||
| Predicted transcriptional matrix | CEBP+ DBP+ OCT3+ |
SIX1+ |
ATBF+ LHX1+ BRN5+ BRN2(1)+ BRN2(2) LHX3+ NKX6+ PIT1+ BRN3+ |
GFI1+ BRN2(2) |
SP1, MAZ+ | SOX5+ TATA |
TATA | CpG+ | EGR+ cMYB+ |
|
|
| ||||||||||
| T | C | |||||||||
| C | C | |||||||||
|
| ||||||||||
| SP1 | ||||||||||
Matrixes were selected by matrix similarity over 0.83, +means the additional motif which is produced by the sequence with only one allele difference. CpG means CpG islands. Allele G for rs20317 makes an additional CpG island. Matrixes with bald letters are conserved in mice genome (NT_0394247). Alleles in painted cells make the combination which produced a significant high promoter activity in Urak et al.’s report (Urak et al., 2006). The C allele at rs20317 contains cMYB and ERG3 motifs; in contrast, the G allele does not contain either motifs but gains a CpG island.
REST is a zinc finger protein, that contains a DNA-binding domain and two repressor domains: one at the N-terminal and the other at the C-terminal of the protein. The DNA-binding domain of REST binds to a canonical DNA sequence, the RE-1. Two repressor domains recruit various corepressor elements including MeCP2 and CoREST and cause chromatin remodeling context-dependently to inhibit gene transcription. (Majumder, 2006). But also REST can be changed from a repressor to an activator by the nuclear-localized small modulatory double-strand RNA coding the RE1/NRSE sequence (Kuwabara et al., 2004). REST prevents extraneural expression of neuronal genes in nonneural tissue but also regulates neuronal development by repressing the neuronal differentiation of neuronal subtypes for proper tissue differentiation during embryonic development in mammalian brain (Ballas & Mandel, 2005; Jones & Meech, 1999; Sun et al., 2005; Abrajano et al., 2009). As a corollary to our results, we suggest that REST is involved in the developmental regulation and maintenance of GABRB3 in neuronal cells at embryonic, fetal, and perinatal periods when GABRB3 exon 1A expression is most abundant in mammalian brain (Laurie et al., 1992; Kirkness & Fraser, 1993). GABRB3 produces a main component of GABAA-R in embryonic, fetal, and perinatal brain. GABAAR in these early stages depolarizes neural progenitor cells and promotes neurogenesis (Laurie et al., 1992; Ben-Ari et al., 1997; Herlenius & Lagercrantz, 2004). Thus REST plays a significant role on early brain development.
In conclusion, common SNPs on functional promoter regions of GABRB3 variant 2 which has N-terminal exon1A, affects promoter activities by changing transcription binding motifs. An epigenetic modulator, REST regulates expression of GABRB3 variant 2. Epigenetic regulation of GABRB3 expression could clarify genetic mechanisms underlying pathogenesis of CAE and other neurological developmental disorders with absence seizures, such as Angelman syndrome, autism spectrum disorder, and Rett syndrome.
Supplementary Material
Acknowledgments
We thank all CAE patients and their families for their cooperation, GENESS site neurologists and staff for their help, Ziwei Chen for helpful advice, the J Huang laboratory, and Computational Sciences Group, UCLA Pharmacology Dept, for their cooperation. This work was supported by National Institutes of Health Grant NS35985 to RWO; by National Institutes of Health NINDS Grant NS055057 to AVDE, and a Veterans Administration Merit Review Grant to AVDE.
Footnotes
Disclosure
The authors declare that they have no conflicts of interest. We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
References
- Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS One. 2009 doi: 10.1371/journal.pone.0007936. published on line: 7-Dec-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y, Khazipov R, Leinekugel X, Caillard O, Gaiarsa JL. GABAA, NMDA and AMPA receptors: a developmentally regulated ‘ménage à trois’. Trends Neurosci. 1997;20 :523–529. doi: 10.1016/s0166-2236(97)01147-8. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- DeLorey TM, Handforth A, Homanics GE, Anagnostaras SG, Homanics GE, Minassian BA, Asatourian A, Fanselow MS, Delgado-Escueta A, Ellison GD, Olsen RW. Mice lacking the β3 subunit of the GABAA receptor have the epilepsy phenotype and many of the behavioral characteristics of Angelman syndrome. J Neurosci. 1998;18:8505–14. doi: 10.1523/JNEUROSCI.18-20-08505.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlenius E, Lagercrantz H. Development of neurotransmitter systems during critical periods. Exp Neurol. 2004;(Supp1):S8–S21. doi: 10.1016/j.expneurol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Hogart A, Nagarajan RP, Patzel KA, Yasui DH, Lasalle JM. 15q11-13 GABAA receptor genes are normally biallelically expressed in brain yet are subject to epigenetic dysregulation in autism-spectrum disorders. Hum Mol Genet. 2007;16:691–703. doi: 10.1093/hmg/ddm014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FS, Meech R. Knockout of REST/NRSF shows that the protein is a potentrepressor of neuronally expressed genes in non-neural tissues. Bioessays. 1999;21 :372–376. doi: 10.1002/(SICI)1521-1878(199905)21:5<372::AID-BIES3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Fraser CM. A strong promoter element is located between alternativeexons of a gene encoding the human gamma-aminobutyric acid-type A receptor beta 3 subunit (GABRB3) J Biol Chem. 1993;268:4420–4428. [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Wisden W, Seeburg PH. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain III Embryonic and postnatal development. J Neurosci. 1992;12 :4151–4172. doi: 10.1523/JNEUROSCI.12-11-04151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5:1929–35. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- Samaco RC, Hogart A, LaSalle JM. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum Mol Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri C, Narimatsu M, von Both I, Liu Y, Tan NB, Izzi L, McCaffery P, Wrana JL, Attisano L. Genome-wide identification of Smad/Foxh1 targets reveals a role for Foxh1 in retinoic acid regulation and forebrain development. Dev Cell. 2008;14:411–423. doi: 10.1016/j.devcel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Sun YM, Greenway DJ, Johnson R, Street M, Belyaev ND, Deuchars J, Bee T, Wilde S, Buckley NJ. Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol Biol Cell. 2005;16:5630–5638. doi: 10.1091/mbc.E05-07-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Olsen RW, Medina MT, Schwartz E, Alonso ME, Duron RM, Castro-Ortega R, Martinez-Juarez IE, Pascual-Castroviejo I, Machado-Salas J, Silva R, Bailey JN, Bai D, Ochoa A, Jara-Prado A, Pineda G, Macdonald RL, Delgado-Escueta AV. Hyperglycosylation and reduced GABA currents of mutated GABRB3 polypeptide in remitting childhood absence epilepsy. Am J Hum Genet. 2008;82:1249–1261. doi: 10.1016/j.ajhg.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urak L, Feucht M, Fathi N, Hornik K, Fuchs KA. GABRB3 promoter haplotype associated with childhood absence epilepsy impairs transcriptional activity. Hum Mol Genet. 2006;15:2533–2541. doi: 10.1093/hmg/ddl174. [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.