Abstract
The definitive demonstration of a role for a recently acquired gene is a difficult task, requiring exhaustive genetic investigations and functional analysis. The situation is indeed much more complicated when facing multicopy gene families, because most or portions of the gene are conserved among the hundred copies of the family. This is the case for the ERVWE1 locus of the human endogenous retrovirus W family (HERV-W), which encodes an envelope glycoprotein (syncytin) likely involved in trophoblast differentiation. Here we describe, in 155 individuals, the positional conservation of this locus and the preservation of the envelope ORF. Sequencing of the critical elements of the ERVWE1 provirus showed a striking conservation among the 48 alleles of 24 individuals, including the LTR elements involved in the transcriptional machinery, the splice sites involved in the maturation of subgenomic Env mRNA, and the Env ORF. The functionality and tissue specificity of the 5′ LTR were demonstrated, as well as the fusogenic activity of the envelope polymorphic variants. Such functions were also shown to be preserved in the orthologous loci isolated from chimpanzee, gorilla, orangutan, and gibbon. This functional preservation among humans and during evolution strongly argued for the involvement of this recently acquired retroviral envelope glycoprotein in hominoid placental physiology.
We molecularly characterized the human endogenous retrovirus W family (HERV-W) family (1) by screening a placental cDNA library with a polymerase (pol) probe derived from a retroviral sequence named multiple-sclerosis associated retrovirus (MSRV) (2) isolated from biological samples from multiple sclerosis patients. Surprisingly, this multicopy family contained a unique proviral locus, located on chromosome 7, flanked by two intact LTRs, and which retained a 538-aa envelope ORF that exhibited all of the characteristic features of the precursor polypeptide of classical retroviral envelope proteins (1, 3). The presence of inactivating mutations in the gag and pol genes of this provirus, termed ERVWE1 (OMIM 604659), led us to propose that Env function has been selectively preserved (1).
Although HERV-W mRNA expression was detected in various physiological (1, 4) and pathological (2, 5, 6) contexts, HERV-W transcript pattern observed in placenta correlates with a LTR5′-U3 driven transcription coupled with a specific splicing strategy compatible with the ERVWE1 locus (1). Env expression was evidenced in placenta (3) at the syncytiotrophoblast layer (7, 8) covering the chorionic villi, bathed by maternal blood in the intervillous spaces, which is the site of nutrient exchanges and of the synthesis of hormones required for fetal growth and development (9, 10). We and others recently demonstrated that the product of the HERV-W env gene is a highly fusogenic membrane glycoprotein (4, 7), inducing the formation of syncytia upon interaction with the type D mammalian retroviruses receptor (7). The in vitro fusogenic activity of Env HERV-W and its in vivo localization suggested that this glycoprotein may have a critical role in human placental development steps involving the syncytiotrophoblast layer. Such a hypothesis was supported by the observations that an anti-Env-W polyclonal antibody was able to inhibit heterologous fusion between a BeWo cell line and COS reporter cells (4) and that anti-ERVWE1 antisens oligonucleotides were able to inhibit primary human trophoblast cell fusion and differentiation (8).
Nevertheless, as previously highlighted (7, 8, 11), there is a huge gap between the demonstration of the fusogenic function and a definitive proven physiological role. The ERVWE1 locus is a young element, as the HERV-W ancestor active element entered the primate lineage 25–40 million years ago (12). Our working hypothesis is that this element has been selected during primate evolution to play a role in placentation. However, it is also possible that this locus is a still-active pseudogene in the process of extinction, and that its activity in the syncytiotrophoblast formation is simply a remnant of its original function for the propagation of retroviruses, but that has no real physiological role. To rule out this latter hypothesis, we have designed a genetic and functional analysis of the ERVWE1 locus. We show in this article that the ERVWE1 locus is functionally preserved in the human population and in the identified orthologous locus of chimpanzee, gorilla, orangutan, and gibbon.
Materials and Methods
DNA Samples. The human rodent monochromosomal National Institute of General Medical Sciences somatic hybrid mapping panel 2 was obtained from the Coriell Institute (Camden, NJ). Stored human DNA from 180 individuals were used, including 59 Caucasians, three Asians, two Africans, five Metis, and one Ashkenazi. Thirty samples were derived from multiple sclerosis patients, including 22 familial forms (13). Ninety-three individuals were male, and 87 individuals female, as determined by PCR amplification of a segment of the X–Y homologous gene amelogenin (14). Animal DNAs from three chimpanzees (Pan troglodytes), three gorillas (Gorilla gorilla), two orangutans (Pongo pygmaeus), three gibbons (Hylobates pileatus) (one DNA per species from Quantum Biotechnologies Inc, Canada), six rhesus monkeys (Macaca mulatta), and one guinea baboon (Papio papio) were used.
Long-Distance (LD) PCR. The ERVWE1 locus was amplified by using sense primer U6198R, 5′-CAAAACGCCTGGAGATACAGCAATTATC-3′ and antisense primer L6186R, 5′-GCACCCTCATGGTTGTGTTACTTGG-3′ as illustrated in Fig. 1. DNA quality was checked by amplifying a 16.2-kb fragment from the mitochondrial genome and a 13.5-kb fragment from the β-globin gene cluster essentially as described (15). Detailed protocols are included in Appendix, which is published as supporting information on the PNAS web site.
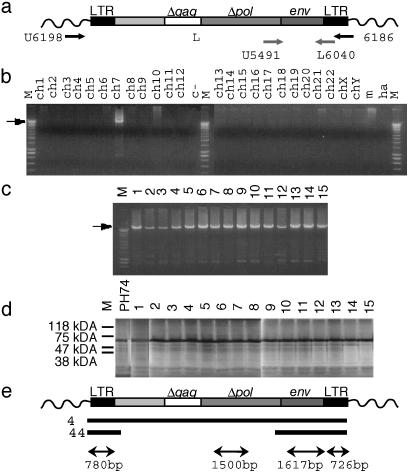
Fig. 1.
Conservation of the ERVWE1 locus in the human population. (a) Schematic representation of the human ERVWE1 locus including flanking nonretroviral sequences (wavy line), LTRs (black boxes), 2-kb nonretroviral insert (light gray box), Δgag (white box), Δpol (gray box) interrupted ORFs, env ORF (dark gray box), and positioning of the locus-specific amplification primers (black arrows) U6198 and L6186 and env ORF amplification primers (gray arrows) U5491 and L6040. (b) Evaluation of the U6198-L6186 primer pair on DNA extracted from human–rodent monochromosomal cell lines. M, molecular mass; ch1-chY, human chromosomes; c-, negative control; m, mouse DNA; ha, hamster DNA; the arrow indicates the 10-kb amplification product. (c) Amplification of the ERVWE1 locus from 15 human individuals. (d) SDS/PAGE analysis of the in vitro transcription–translation products of the env ORF from the same 15 individuals. PH74, env ORF containing placental cDNA clone (1). (e) Global sequencing strategy of the ERVWE1 locus. Two alleles of two individuals were entirely sequenced (4), and two alleles of 22 individuals (44) were partially sequenced to characterize transcriptional and encoding elements. The double arrows indicated the four regions used to identify polymorphic sites.
Protein Truncation Test. The conservation of env ORF was evaluated by using the in vitro transcription–translation procedure as described (3). Briefly, 0.02 μl of the LD-PCR amplification product mixture was amplified by nested-PCR with env-specific primer pair U5491-L 6040. Four microliters of nonpurified PCR product were transcribed and translated for 120 min at 30°C in a TNT-T7-coupled reticulocyte lysate system (Promega) as recommended by the manufacturer and analyzed by SDS/PAGE. A detailed protocol is included in Appendix.
Sequence Analysis. The LD-PCR product was purified and cloned in pCR-XL-TOPO plasmid by using TOPO XL PCR cloning kit (Invitrogen). One cloned fragment was sequenced, and direct sequencing of nested PCR products derived from the 10-kb LD-PCR product were simultaneously performed, to identify both alleles and true polymorphic sites. A detailed protocol is included in Appendix. Alignments were performed with clustalw (16), and phylogenetic trees were computed with phylo_win (17).
Cell Lines. The b30 BeWo cell line was grown in F-12K medium (Life Technologies) containing 10% FCS and fungizone. Other cell lines were as follows: LC5, human lung fibroblasts; HeLa, human epithelioid carcinoma cells (ATCC CCL2); TELCeB6 rhabdomyosarcoma cells (7); XC, rat sarcoma cells (ATCC CCL-165); and XC-RDR cells, XC cells expressing the RDR receptor (8). LC5, HeLa, TELCeB6, XC, and XC-RDR cells were cultured in DMEM (Life Technologies) (without sodium pyruvate for LC5) supplemented with 10% FCS (5% FCS for TELCeB6) and fungizone.
LTR Luciferase Assays. LTRs were PCR amplified and cloned into pGL3-basic plasmid (Promega). HERVWE1[5] and HERVWE1[3] clones corresponded to nucleotide -35/+309 and -25/+311 of the human ERVWE1 5′ and 3′ LTRs, respectively. HERV-WHS14[3] and HERV-W8[3] (plasmid pBL-W8, ref. 18) clones corresponded to nucleotides -34/+375 and -16/+325 of 3′ LTRs derived from a provirus located on chromosome 14 (GenBank accession number AL365295) and from a mRNA expressed in a breast cancer cell line, respectively. pGL3-basic and parental pBL-basic plasmids (control) exhibited similar background activity in all tested cell lines (data not shown). ERVWE1 5′ and 3′ ape cloned sequences corresponded exactly to their human counterparts. A total of 0.7 μg of firefly luciferase LTR-ERV-W plasmid and 0.035 μg of Renilla luciferase pRL-TK plasmid (standardization) were transiently transfected in choriocarcinoma b30 BeWo and fibroblastic LC5 cell lines by using Lipofectamine Plus (Life Technologies) method in 12-well plate format. Forskoline (50 μM) was added after 12 h to induce BeWo differentiation. All experiments were performed in triplicate. Promoter activities were calculated as [firefly/Renilla] × 100 luciferase activities by using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions.
Envelope Fusion Assays. The mutations corresponding to the four observed human polymorphic variants were introduced into the phCMV-env plasmid (7) by using a QuikChange mutagenesis kit (Stratagene) and verified by sequencing. This produced five expression vectors named according to the residues localized in each polymorphic site, i.e., VRSS, VRnS, VqnS, aRSS, and VqSf. Similar ape ERVWE1 env-containing fragments were cloned into the phCMV vector. The envelopes fusion efficiencies were determined as described (7). The overall experiment was repeated five times, and differences between envelopes fusion properties were assessed by using the Kruskal–Wallis test (Stat-View II, Abacus Concepts, Berkeley, CA). Receptor-dependent fusion activity was determined by using XC and XC-RDR cell lines as described (7). Detailed protocols are included in Appendix.
Results
Positional Conservation of the ERVWE1 Locus Containing a Preserved env ORF. Primers designed to demonstrate the positional conservation of the ERVWE1 locus (Fig. 1a) were evaluated in LD-PCR using human-rodent monochromosomal DNA. A 10-kb band can be detected only on chromosome 7 as expected (Fig. 1b). The same protocol was applied to 180 individuals, and 155 individuals were found positive as partially illustrated in Fig. 1c. The 25 ERVWE1-negative DNA were also LD-PCR-negative by using the 16.2-kb mitochondrial genome and the 13.5-kb human β-globin gene cluster control sequences, showing that those DNA preparations were not suitable for LD-PCR purpose (data not shown). Thus, the ERVWE1 locus is present in 100% of the 155 analyzable individuals. The conservation of the envelope ORF in all 155 ERVWE1 LD-PCR products was evaluated by an in vitro transcription–translation assay subsequent to a specific env gene nested PCR step (Fig. 1a). A full-length ORF was observed in all 155 individuals, as partially illustrated in Fig. 1d. Because the 5′ primer contained the initiation codon, the presence of the env natural ATG was confirmed by direct sequencing of the env nested-PCR product of the 155 individuals. The sequence was remarkably conserved, as not a single mutation was observed within a -20/+20 interval surrounding the ATG codon (AACTAAAATCATAAATCCCCATGGCCCTCCCTTATCATATTTT), indeed in a relatively favorable context, although slightly different from the known GCC(A/G)CCATGG favorable context for translation (19).
Global Sequence Analysis of the ERVWE1 Locus. The sequencing and analysis strategy is summarized in Fig. 1e. The 10-kb PCR-derived sequences obtained from two individuals presented exactly the same structure as the previously identified env-containing HERV-W provirus (1) contained in human bacterial artificial chromosome clone RG083M05 from 7q21 to 7q22 (GenBank accession no AC007566), i.e., LTR–2kb insert–gag–pol–env–LTR, gag and pol genes containing many stop codons. The comparative analysis of the 48 sequences obtained from all 24 individuals have shown the presence of few polymorphic sites exhibiting an heterogeneous distribution along the locus (see below). The global mean divergence between the four full-length proviruses was ≈0.05%. The local mean divergence calculated with the 48 partial sequences ranged from 0.005% to 0.2% for the 5′ LTRs and the 3′ LTRs, respectively, and was 0.04% for the env ORF, similar to the overall provirus.
Transcriptional Elements Are Functionally Preserved. The 5′ LTR was particularly conserved as a single mutation appeared in the U5 region in one of the 48 sequences (Fig. 7, which is published as supporting information on the PNAS web site), i.e., 47 sequences were strictly identical (e.g., isolate 132 allele A). Conversely, nine different alleles were observed for the 48 sequenced 3′ LTR, with variable frequencies: 18 of 48 (e.g., isolate 37B), 14 (e.g., isolate 24A), 6 (e.g., isolate 22B), 3 (e.g., isolates 83A and 37A), and 1 (isolates 132A, 71A, 80B, 132B). Most of these polymorphic sites relied within the U5 subdomain of the 3′ LTR.
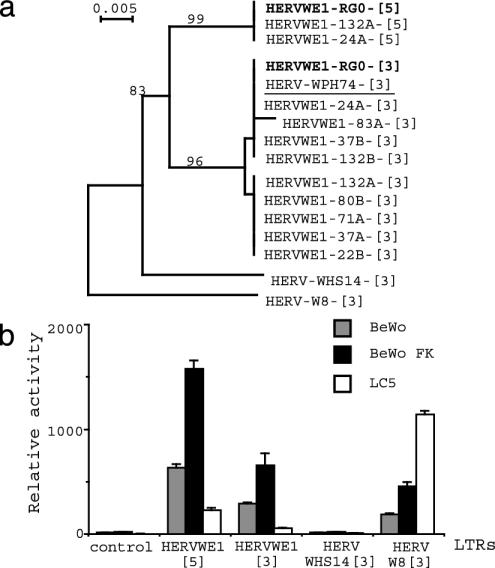
The alignment of the two ERVWE1 5′ LTRs with the nine ERVWE1 3′ LTRs, the 3′ LTR of env containing clone PH74 derived from a placental cDNA library (HERV-WPH74), the 3′ LTR of an mRNA expressed in a breast cancer cell line (HERV-W8), and another HERV-W LTR locus from chromosome 14 (HERV-WHS14) highlighted some specific features of the ERVWE1 locus (Fig. 7). Thus, all ERVWE1 5′ LTRs are distinct from 3′ LTRs. Although all LTRs contained a CAAT box and a TATA box, nine sites discriminate between 5′ and 3′ U3 region. The phylogenic analysis of HERV-W U3 regions clearly shown at least four different groups consisting of 5′ LTRs of ERVWE1, 3′ LTRs of ERVWE1 (two subgroups characterized by a G or an A in position 151), then the HERV-WHS14 element and finally the HERV-W8 more distant element (Fig. 2a). A functional analysis of all four U3 elements revealed that the HERVWE1 5′ LTR was the most active element in BeWo cells, then came the HERVWE1 3′ LTR, the HERV-W8 LTR which exhibited a 4-fold lower activity, and the HERV-WHS14 sequence which was almost not active (Fig. 2c). Intriguingly, although the levels of expression were different, the observed increase caused by forskolin addition (an inducer of BeWo differentiation into syncytiotrophoblast-like fused cells) was similar for all three active promoters, 2.5, 2.3, and 2.5 for HERVWE1 5′ LTR, HERVWE1 3′ LTR, and HERV-W8 LTR, respectively. Conversely, the level of expression of HERVWE1 5′ LTR was five times lower than the HERV-W8 LTR in LC5 fibroblastic cells. These data supported a specific tropism of HERVWE1 5′ LTR for placenta cells as suggested (1).
Fig. 2.
Analysis of the ERVWE1 LTRs in the human population. (a) Phylogenetic analysis of the U3 region of 5′ ([5]) and 3′ ([3]) HERV-W LTRs derived from 24 amplified ERVWE1 loci (HERVWE1, 11 sequences), one bacterial artificial chromosome clone containing the ERVWE1 locus (bold letters), the PH74 placental cDNA clone containing a full-length env ORF (underlined), and two other HERV-W LTRs (HERV-WHS14-[3], HERV-W8-[3]). Neighbor-joining trees were based on 244 nucleotides, and all-gap removal was used. Bootstrap values for 500 replicated trees are indicated. (b) Relative activities of ERVWE1 5′ (HERVWE1 [5]) and 3′ (HERVWE1 [3]) LTRs, and other HERV-W 3′ LTRs (HERV WHS14 [3], HERV W8 [3]) in choriocarcinoma BeWo b30 cell line in absence (gray bar) or presence (black bar) of forskolin and in lung fibroblastic LC5 cell line (white bar). Control, basic vector without LTR.
Elements located within the R region required for the cleavage/polyadenylation pathway were conserved for all LTRs, including the ATTAAA polyadenylation signal, the upstream T-rich region and the CA dinucleotide poly(A) site located 12-bp downstream from the ATTAAA signal. Nevertheless, two mutations, a single base deletion and a dinucleotide insertion, discriminated between 5′ and 3′ HERVWE1 R regions (Fig. 7). The U5 region contained 23 sites and two deletions that discriminated between 5′ and 3′ LTR of the locus. Elements involved in the posttranscriptional steps were also found conserved, such as the previously described major splice donor site 1 and acceptor site 2 involved in producing the env spliced mRNA (1).
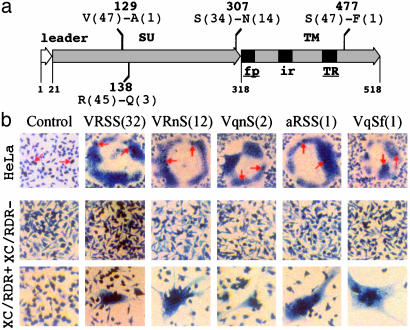
Env Polymorphic Variants Are Functionally Preserved. Within the env ORF, five mutations were detected including one synonymous and four nonsynonymous mutations (Fig. 3a). This corresponded to five envelope polymorphic variants named according to the residus present in the four polymorphic positions, i.e., VRSS, VRnS, VqnS, aRSS, VqSf, representing 67%, 25%, 4%, 2%, and 2% of the sequenced population. The two major alleles VRSS (Fig. 8, which is published as supporting information on the PNAS web site) and VRnS were present in all of the individuals of the sequenced population. The VRSS predominant form was identical to the one we previously identified in a placental cDNA library (1), and the VRnS second major form was identical to the one previously dubbed syncytin (4). Twelve of 24 individuals were homozygous, 11 and 1 individuals exhibiting the VRSS and VRnS sequences, respectively. The major heterozygous genotype VRSS-VRnS was found in eight individuals and four minor heterozygote forms were found, VqnS-VRSS, VqnS-VRNS, aRSS-VRSS, and VqSf-VRNS. No skewed allele distribution was observed with respect to sex or health state (multiple sclerosis). All five polymorphic variants expressed in the phCMV plasmid were found to induce fusion in an heterotypic cell-cell fusion assay (Fig. 3b). More precisely, no significant difference was found in the number of syncytia per well determined for each mutant in five independent experiments based on the same format assay (P value = 0.837). Moreover, the observed fusogenic property was strictly dependent on the recognition of the RDR receptor (Fig. 3b) as described (7).
Fig. 3.
Polymorphism and functional analysis of the ERVWE1 envelope in the human population. (a) Schematic drawing of the Env glycoprotein indicating the location of the polymorphic sites and the nature and frequency of the amino acids. SU, surface protein; TM, transmembrane protein; fp, fusion peptide; ir, immunosuppressive region; TR, carboxy-transmembrane region. (b) Formation of syncytia by ERVWE1 envelopes. TELCeB6 cells expressing a nuclear β-galactosidase were transfected with plasmids expressing the five polymorphic forms of ERVWE1 env ORF, VRSS, VRnS, VqnS, aRSS, and VqSf (less represented amino acids are shown in lowercase; allele frequency is shown in parentheses) or one env gene in antisense orientation (Control). Transfected cells were overlaid with HeLa indicator cells. Cocultures were stained with 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) substrate to visualize the nuclei of the producer cells (arrows) and then with May–Grünwald and Giemsa solutions. Receptor-mediated fusion was analyzed by cocultivating the env transfected TELCeB6 cells with XC rat cells lacking (XC/RDR-) or expressing (XC/RDR+) the human RDR receptor.
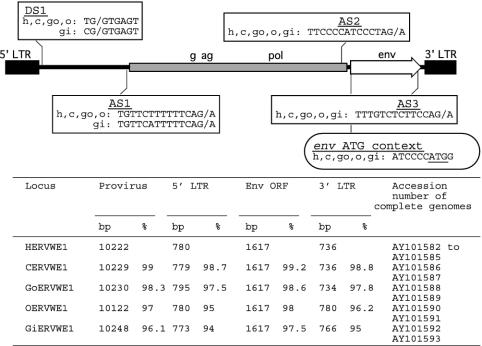
Functional Preservation of the ERVWE1 Locus in Nonhuman Hominoids. The positional amplification and sequencing strategy for apes was identical to the one described above for human. A 10-kb fragment was obtained for all apes from chimpanzee to gibbon, but no signal was detected for one baboon and six rhesus monkeys, although the 16.2-kb positive control from the mitochondrial genome of these two Old World monkeys was detectable. In vitro transcription–translation after a specific env nested-PCR step showed conservation of the full-length ORF for all tested animals, three chimpanzees, three gorillas, two orangutans, and three gibbons (data not shown). Both alleles of the orthologous ERVWE1 loci were sequenced for one animal from each ape species. The main features of all four proviruses are summarized in Fig. 4. They presented exactly the same structure than the human ERVWE1 provirus, i.e., LTR–2-kb insert–gag–pol–env–LTR and, as in human, gag and pol regions contained numerous stop codons and frameshifts. Fifty-two gap events were detected in the whole locus of the five ERVWE1 orthologs. Interestingly, none of these gaps occurred within the env ORF, whose length was strictly constant. The even distribution of the gaps along the other regions of the locus suggested that there was a strong selective pressure to maintain the env reading frame intact. Moreover, the env ATG context remained identical to the one observed in all human samples. Splice sites previously identified (1) were also remarkably conserved, notably the DS1 and AS2 donor and acceptor sites involved in env mRNA processing (Fig. 4).
Fig. 4.
Comparison of the main features of the human ERVWE1 locus (h, HERVWE1) with the chimpanzee (c, CERVWE1), gorilla (go, GoERVWE1), orangutan (o, OERVWE1), and gibbon (gi, GiERVWE1) orthologous loci. Sequences of the major donor (DS1) and acceptor (AS1, AS2, and AS3) splice sites and the env initiation codon context are indicated in boxes on the schematic representation of the locus. The length and the percentage of similarity (ape/human) of the full-length provirus, 5′ and 3′ LTRs, and the env ORF are expressed at the nucleic acid level.
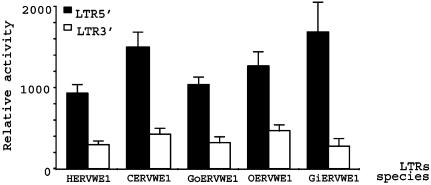
The alignment of U3 domains of 5′ and 3′ LTRs revealed that both LTRs are distinct within each species (Fig. 9, which is published as supporting information on the PNAS web site). All LTRs contained a CAAT box and a TATA box, identical or slightly different from the human motifs, e.g., the TATA box of the 5′ LTRs of both alleles of the gibbon was AATAAA as compared to the human sequence. The sites that were shown to discriminate between 5′ and 3′ human U3 did not systematically discriminate for each species, with the exception of the 3′ last base of U3, which was an A vs. a G in 5′ and 3′ LTRs, respectively. Nevertheless, the functional analysis of all U3 elements revealed that the ape ERVWE1 5′ LTRs were always more active in BeWo cells than the ERVWE1 3′ LTRs, as observed for the human ERVWE1 locus (Fig. 5). Within the R region, the T-rich region, the polyadenylation signal, and the CA dinucleotide poly(A) site were conserved for all LTRs; two mutations located within the first 30 bases or R, which discriminated between the 5′ and 3′ human LTR, also specified the 3′ R region in apes (data not shown).
Fig. 5.
Analysis of the ERVWE1 LTRs in hominoids. Shown are relative activities of human (HERVWE1), chimpanzee (CERVWE1), gorilla (GoERVWE1), orangutan (OERVWE1), and gibbon (GiERVWE1) 5′ (black bars) and 3′ (white bars) LTRs in BeWo b30 cell line. GenBank accession numbers AY101584, AY101586, AY101589, AY101590, and AY101593.
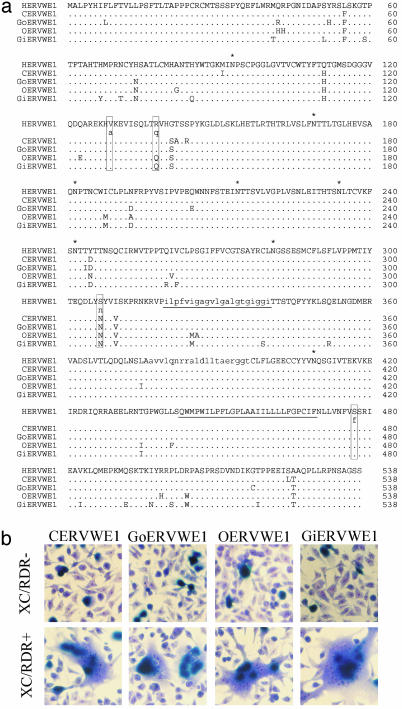
Sequencing of the four ape env ORFs showed that all these animals were homozygous for this protein. The similarities observed by pairwise alignment of Env proteins between human and chimpanzee, gorilla, orangutan, and gibbon were 98.0%, 97.2%, 95.7%, and 94.6%, respectively. Alignments of human and ape envelopes showed a high conservation of critical domains essential for classical retroviral envelope expression and function (Fig. 6a). The topology of the tree obtained by the phylogenetic analysis of the Env amino acid sequences was identical to the typical higher ape species tree (data not shown), suggesting that this locus has evolved as a classical Mendelian gene. Nevertheless, the analysis of the ratio of synonymous over nonsynonymous substitution rate did not reveal any obvious selective pressure on the protein sequence (Ka/Ks = 0.8 over the whole phylogenetic tree). Functionally, the four ape envelopes were found to present similar fusion activities in the heterologous cell assay described above (P value = 0.900). Because the orthologous ASCT-2 receptors have not yet been identified in apes, the receptor-dependent fusion process of ape envelopes was evaluated with the human receptor. The observed fusogenic property was strictly dependent on the recognition of the hASCT-2 receptor for all ape envelopes (Fig. 6b).
Fig. 6.
Analysis of the ERVWE1 envelope in hominoids. (a) Alignments of the env translated region. The furine cleavage site (gray letters), the hydrophobic fusion peptide (lowercase letters, underlined), the immunosuppressive region (lowercase letters), the carboxy-transmembrane region (underlined), and potential glycosylation sites (asterisks) are indicated on the human sequence. Human polymorphic sites are boxed, and the less represented amino acid are indicated in lowercase. (b) Receptor-mediated fusion analyzed as described in Fig. 3.
Discussion
Comparative analysis of the ERVWE1 locus for 24 individuals showed a pattern of sequence variation that was variable along the proviral locus. The 5′ LTR exhibited an unusually low polymorphism (one variable site every 18.0 kb) as compared to the one in 0.47 kb and one in 0.31 kb variability described for noncoding sequences and repeated sequences (20), respectively, suggesting that there has been a selective sweep of this region. Conversely, the variability of the 3′ LTR (one in 0.5 kb) was typical of repeated sequences (20). The observed variability of the env ORF (one variable site every 2.2 kb) fell within the same range as the one in 1.08–2.00 kb variability described for human coding sequences (20), highlighting that the behavior of this gene of retroviral origin is similar to any essential cellular gene, as opposed to infectious retroviruses or more generally RNA-based organism (21).
The identification of the ERVWE1 locus exclusively in hominoids may reflect two situations. First, the ERVWE1 locus had retrotransposed in the hominoid lineage after the split of hominoids from the Old World monkeys ≈15–20 million years after the germ line insertion of the ancestral virus in the Catarrhini lineage (12). Second, this locus was already present in the ancestor of the Catarrhini lineage and was fixed in the hominoid lineage, but has degenerated in the Old World monkey lineage. Further analyses will be required to choose between these two hypotheses. Nevertheless, the structure conservation of both LTRs and the sequence conservation of the 5′ and 3′ proviral-cellular junctions, for all hominoid species, suggested that the ERVWE1 transposition process depended on a functional and specific HERV-W reverse transcriptase and not a LINE-mediated retrotransposition process as observed for other HERV-W elements (22, 23).
Although HERV-W expression was detected in autoimmune diseases (2, 5) and schizophrenia (6), no direct link between mRNA expression and (re)activation of a specific HERV-W locus (or a subset of loci) was demonstrated to date (24, 25). Conversely, the ERVWE1 locus is the unique HERV-W locus for which a tissue specificity was proposed (1, 4). Interestingly, the U3 region of the 5′ HERVWE1 LTR was extremely well conserved in the human population, and comparison with the other HERV-W LTRs tested showed that it contained some specific signatures. The presence of such a combination of sites was apparently correlated with the HERVWE1 promoter efficiency and tropism, as the 5′ HERVWE1 LTR was the most active promoter in BeWo cells. Nevertheless, among the nine sites that were shown to discriminate between 5′ and 3′ HERVWE1 U3 regions, only a single one was conserved during the whole hominoid evolution. In addition, because cloned sequences contained some residues upstream from U3 and a downstream R sequence providing the natural cap site, further analysis will be required to identify the determinants involved in the observed tropism. Nevertheless, it is noticeable that all active ERVWE1 LTRs lacked a Sp1 site identified in a subset of inactive HERV-W LTRs and absent from an other subset of active HERV-W LTRs (18).
All five human envelope polymorphic variants and four ape orthologous envelopes were found to be functional in an in vitro fusogenic assay, a process dependent on the recognition of the hASCT-2/RDR receptor. This was also true for the VRnS human mutant, which has been previously proposed to be involved in a receptor-independent fusion process (4), in contradiction with the apparent selectivity of trophoblast fusion, as highlighted elsewhere (11). The similar recognition of the human ASCT-2 receptor by all hominoid ERVWE1 envelopes suggested that the ape ASCT-2 receptors were closely related to the hASCT-2. Indeed, it would be interesting to isolate such hASCT-2 orthologous receptors with the potential of resolving the conflict between the coevolution of the receptor with the endogenous envelope and the introduction of hypervariability to avoid putative pathogenic retroviral infection (26, 27). In addition, the sequencing of the 48 HERVWE1 envelopes confirmed that the MSRV envelope (GenBank accession no. AF331500), which has been proposed to trigger a superantigen effect (28) involved in multiple sclerosis, was not encoded by the ERVWE1 locus. First, Env HERVWE1 presented an 81% identity with Env MSRV, and second, a 4-aa gap that was shown to be a unique signature of Env HERVWE1 among all HERV-W envelope sequences (data not shown) is absent from Env MSRV.
The presence of inactivating mutations in the gag and pol genes of the ERVWE1 provirus initially led us to propose that Env function has been selectively preserved (1). The demonstrations that the Env HERV-W glycoprotein was (i) fusogenic in vitro (4, 7), (ii) localized at the syncytiotrophoblast layer of the placenta (7, 8), and (iii) directly involved in primary human trophoblast cell fusion and differentiation (8) supported the hypothesis of a role in placentation. The functional preservation of the transcriptional and posttranscriptional regulatory elements and of the fusogenic property of the Env glycoprotein of the ERVWE1 locus in the human population and in hominoids demonstrated that this recently acquired gene has become a bona fide gene. The corollary of the implication of this envelope in the formation of the continuous multinucleate syncytiotrophoblast is that a deregulation of Env expression could be associated with pathologies ranging from implantation to delivery. Interestingly, Env was poorly expressed in pathologies associated with hypoxia (29), hypoxia-impairing cytotrophoblast fusion, and differentiation in vitro (30). Because the majority of the Eutherians do not contain the young ERVWE1 locus, it will be challenging to ascertain whether this retroviral acquisition represents an additional adaptive or substitutive mechanism for placental morphogenesis (31). The recent finding of an in vitro fusogenic HERVFRD envelope conserved during primate evolution (32) raises questions about the apparent redundancy of recently acquired genes sharing common function(s). This may thereby contribute to our understanding of the extreme diversity of the placental structures observed in nature (33).
Supplementary Material
Acknowledgments
We thank B. Rudkin for critical reading of the manuscript. We thank B. Mougin, S. Mazoyer, A. Blancher, J. Brosius, C. Coiffier, and G. Hunsmann for providing us with human and ape DNAs. We thank A. Ponge and A. Chedal for technical assistance and O. Sémonin at ESGS Cybergène for help in sequencing. We thank J. Strauss III for providing the b30 BeWo cell line and C. Leib Mösch for gift of the LC5 cell line and of the pBL-W8 plasmid. S.P. is supported by a doctoral fellowship from the Ministère de la Recherche et de la Technologie, and B.B. is supported by a doctoral fellowship from bioMérieux and Centre National de la Recherche Scientifique.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HERV, human endogenous retrovirus; LD-PCR, long-distance PCR.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos.: human full-length ERVWE1 locus (4 entries), AY101582–AY101585; human 5′ and 3′ partial ERVWE1 locus (88 entries), AF520477–AF520564; and chimpanzee, gorilla, orangutan, and gibbon full-length ERVWE1 locus (8 entries), AY101586–AY101593].
References
- 1.Blond, J. L., Beseme, F., Duret, L., Bouton, O., Bedin, F., Perron, H., Mandrand, B. & Mallet, F. (1999) J. Virol. 73, 1175-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perron, H., Garson, J. A., Bedin, F., Beseme, F., Paranhos-Baccala, G., Komurian-Pradel, F., Mallet, F., Tuke, P. W., Voisset, C., Blond, J. L., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voisset, C., Bouton, O., Bedin, F., Duret, L., Mandrand, B., Mallet, F. & Paranhos-Baccala, G. (2000) AIDS Res. Hum. Retroviruses 16, 731-740. [DOI] [PubMed] [Google Scholar]
- 4.Mi, S., Lee, X., Li, X., Veldman, G. M., Finnerty, H., Racie, L., LaVallie, E., Tang, X. Y., Edouard, P., Howes, S., et al. (2000) Nature 403, 785-789. [DOI] [PubMed] [Google Scholar]
- 5.Komurian-Pradel, F., Paranhos-Baccala, G., Bedin, F., Ounanian-Paraz, A., Sodoyer, M., Ott, C., Rajoharison, A., Garcia, E., Mallet, F., Mandrand, B., et al. (1999) Virology 260, 1-9. [DOI] [PubMed] [Google Scholar]
- 6.Karlsson, H., Bachmann, S., Schroder, J., McArthur, J., Torrey, E. F. & Yolken, R. H. (2001) Proc. Natl. Acad. Sci. USA 98, 4634-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blond, J. L., Lavillette, D., Cheynet, V., Bouton, O., Oriol, G., Chapel-Fernandes, S., Mandrand, B., Mallet, F. & Cosset, F. L. (2000) J. Virol. 74, 3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frendo, J. L., Olivier, D., Cheynet, V., Blond, J. L., Bouton, O., Vidaud, M., Rabreau, M., Evain-Brion, D. & Mallet, F. (2003) Mol. Cell. Biol. 23, 3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sibley, C. P. (1999) in Encyclopedia of Reproduction, eds. Knobil, E. & Neill, J. D. (Academic, New York), Vol. 3, pp. 881-888. [Google Scholar]
- 10.Hofmann, G. E. (1999) in Encyclopedia of Reproduction, eds. Knobil, E. & Neill, J. D. (Academic, New York), Vol. 2, pp. 670-680. [Google Scholar]
- 11.Stoye, J. P. & Coffin, J. M. (2000) Nature 403, 715-717. [DOI] [PubMed] [Google Scholar]
- 12.Voisset, C., Blancher, A., Perron, H., Mandrand, B., Mallet, F. & Paranhos-Baccala, G. (1999) AIDS Res. Hum. Retroviruses 15, 1529-1533. [DOI] [PubMed] [Google Scholar]
- 13.Lucotte, G. L. & The French MS Consortium (2002) Genet. Couns. 13, 133-138. [PubMed] [Google Scholar]
- 14.Sullivan, K. M., Mannucci, A., Kimpton, C. P. & Gill, P. (1993) BioTechniques 15, 636-631. [PubMed] [Google Scholar]
- 15.Van Houten, B., Cheng, S. & Chen, Y. (2000) Mutat. Res. 460, 81-94. [DOI] [PubMed] [Google Scholar]
- 16.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galtier, N., Gouy, M. & Gautier, C. (1996) Comput. Appl. Biosci. 12, 543-548. [DOI] [PubMed] [Google Scholar]
- 18.Schön, U., Seifarth, W., Baust, C., Hohenadl, C., Erfle, V. & Leib-Mosch, C. (2001) Virology 279, 280-291. [DOI] [PubMed] [Google Scholar]
- 19.Kozak, M. (2002) Gene 299, 1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nickerson, D. A., Taylor, S. L., Weiss, K. M., Clark, A. G., Hutchinson, R. G., Stengard, J., Salomaa, V., Vartiainen, E., Boerwinkle, E. & Sing, C. F. (1998) Nat. Genet. 19, 233-240. [DOI] [PubMed] [Google Scholar]
- 21.Drake, J. W., Charlesworth, B., Charlesworth, D. & Crow, J. F. (1998) Genetics 148, 1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costas, J. (2002) Mol. Biol. Evol. 19, 526-533. [DOI] [PubMed] [Google Scholar]
- 23.Pavlicek, A., Paces, J., Elleder, D. & Hejnar, J. (2002) Genome Res. 12, 391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujinami, R. S. & Libbey, J. E. (1999) Trends Microbiol. 7, 263-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston, J. B., Silva, C., Holden, J., Warren, K. G., Clark, A. W. & Power, C. (2001) Ann. Neurol. 50, 434-442. [DOI] [PubMed] [Google Scholar]
- 26.Lavillette, D., Marin, M., Ruggieri, A., Mallet, F., Cosset, F. L. & Kabat, D. (2002) J. Virol. 76, 6442-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marin, M., Lavillette, D., Kelly, S. M. & Kabat, D. (2003) J. Virol. 77, 2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perron, H., Jouvin-Marche, E., Michel, M., Ounanian-Paraz, A., Camelo, S., Dumon, A., Jolivet-Reynaud, C., Marcel, F., Souillet, Y., Borel, E., et al. (2001) Virology 287, 321-332. [DOI] [PubMed] [Google Scholar]
- 29.Knerr, I., Beinder, E. & Rascher, W. (2002) Am. J. Obstet. Gynecol. 186, 210-213. [DOI] [PubMed] [Google Scholar]
- 30.Alsat, E., Wyplosz, P., Malassine, A., Guibourdenche, J., Porquet, D., Nessmann, C. & Evain-Brion, D. (1996) J. Cell Physiol. 168, 346-353. [DOI] [PubMed] [Google Scholar]
- 31.Harris, J. R. (1998) BioEssays 20, 307-316. [DOI] [PubMed] [Google Scholar]
- 32.Blaise, S., Parseval, N. N., Benit, L. & Heidmann, T. (2003) Proc. Natl. Acad. Sci. USA 100, 13013-13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross, J. C., Baczyk, D., Dobric, N., Hemberger, M., Hughes, M., Simmons, D. G., Yamamoto, H. & Kingdom, J. C. (2003) Placenta 24, 123-130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.