Abstract
Normal cell growth consists of two distinct phases, quiescence and proliferation. Quiescence, or G0, is a reversible growth arrest in which cells retain the ability to reenter the proliferative cycle (G1, S, G2, and M). Although not actively dividing, quiescent cells are metabolically active and quiescence is actively maintained. Our results from microRNA PCR arrays and Taqman PCR assays showed a significant decrease (4-fold) in miR-302 levels during quiescence compared to proliferating normal human fibroblasts, suggesting that miR-302 could regulate cellular proliferation. Results from a Q-RT-PCR and dual luciferase-3'-UTR reporter assays identified ARID4a (AT-Rich Interacting Domain 4a, also known as RBP1) and CCL5 (C-C motif Ligand 5) as targets for miR-302. Ionizing radiation decreased miR-302 levels, which was associated with an increase in its target mRNA levels, ARID4a and CCL5. Such an inverse correlation was also observed in cells treated with hydrogen peroxide as well as SOD2 overexpressing cells. Overexpression of miR-302 suppresses ARID4a and CCL5 mRNA levels, and increased the percentage of S-phase cells. These results identified miR-302 as an ROS-sensitive regulator of ARID4a and CCL5 mRNAs as well as demonstrate a regulatory role of miR-302 during quiescence and proliferation.
Keywords: ARID4a, CCL5, cell cycle, quiescence, microRNAs, miR-302, SOD2
Introduction
Normal cell proliferation has two distinct phases: quiescence (G0) and proliferation (G1, S, G2, and M). Quiescence is defined as a reversible growth arrest, allowing for cells to reenter and exit from the proliferative phase if the necessary conditions are met. Reversible growth arrest is important for preservation of proliferative-competent normal cells that are required for biological processes such as wound healing and regeneration [1–3]. Quiescent cells are also known to exhibit age-dependent loss in their proliferative capacity. We and others have shown that quiescent fibroblasts cultured for a long duration are unable to reenter the proliferative cycle [4, 5].
Previous results have demonstrated that the abundance of the cellular reactive oxygen species (ROS) varies during the cell cycle, coordinating cellular metabolism to the cell cycle regulatory processes [6–8]. Cellular ROS levels are the result of the production of pro-oxidants and their removal by the antioxidant network. An imbalance between the production of pro-oxidants and their removal results in oxidative stress. Recent results have demonstrated that the cellular ROS levels may also regulate quiescence [2, 5]. SOD2 expression increases in quiescence, and fibroblasts lacking the SOD2 gene are unable to exit the cell cycle to enter quiescence [2]. Ionizing radiation is well known to induce oxidative stress and transient quiescence [9–16]. However, the molecular mechanisms mediating ROS-sensitive regulation of quiescence and proliferation are not completely understood.
MicroRNAs (miRs) are small RNAs involved in mRNA stability and translational regulation of genes [17–19]. miRs function to repress translation or induce degradation of target mRNAs by binding to target sites in the 3'-untranslated region (UTR). miRs have been shown to affect cell cycle traversal with either oncogenic or tumor suppressor functions [20, 21]. Multiple miRs have been reported to induce cell cycle arrest in the G1 phase by repressing target genes (e.g. miR-16, -34a, -192, -215 and the let-7 family) involved in cell cycle traversal [22–25]. Reversible and irreversible growth states may be defined by their own distinct set of miRs, some of which are shared amongst different types of growth arrest [25].
miR-302 is a family of 8 miRs (mir-302a, mir-302a*, miR-302b, mir-302b*, miR-302c, mir-302c*, miR-302d, and miR-367). Previous results have shown transcription factors Oct4 and Sox2 regulate miR-302 expression [26]. miR-302 has also been shown to regulate stem cell related genes, including NR2F2, as well as cell cycle genes, including CDK 4/6 and Cyclin D1 [26–28]. We used bioinformatics to identify two miR-302 targets, ARID4a and CCL5. ARID4a (AT-Rich Interacting Domain, also known as RBP1, RB Binding Protein 1) is known to bind to the retinoblastoma protein and prevent transcription of E2F related genes, causing cell cycle arrest in G1 [29]. CCL5 (C-C motif Ligand 5, also known as RANTES) is expressed and secreted in many cell types, including growth arrested fibroblasts [30, 31].
The present study was designed to identify miRs that are differentially expressed in quiescent and proliferating cells. Our results showed miR-302 levels significantly decreased in quiescent and irradiated cells, which was associated with a significant increase in its target mRNA levels, ARID4a and CCL5. Results from dual luciferase-3'-UTR reporter assays further support miR-302 regulating ARID4a and CCL5 mRNA levels. Additional results show miR-302 as an ROS-sensitive regulator of ARID4a and CCL5 mRNAs as well as demonstrate a regulatory role of miR-302 during quiescence and proliferation.
Materials and Methods
Cell Culture and Transfections
AG01522 normal human fibroblasts (NHFs) were obtained from Coriell cell repository and cultured following our previously published protocol [2]. MDA-MB-231 human mammary adenocarcinoma cells were cultured in RPMI media and supplemented with 10% FBS and PenStrep [32]. FaDu human oral squamous carcinoma cells were purchased from ATCC and cultured in DMEM media supplemented with 10% FBS, PenStrep, and Gentamycin. Irradiations were performed using a cesium-137 source set at a dose rate of 0.83 Gy/min. miR-302a mimic and negative control mimic were purchased from GenePharma (Shanghai, China). miR-302 expression plasmid and negative control plasmid were purchased from OriGene (Rockville, MD). Transfections were performed using either Lipofectamine 2000 (Invitrogen) or Arrest-In (Thermo Scientific) transfection reagents according to manufacturer supplied protocols. Replication deficient adenovirus containing CMV promoter driven human SOD2 cDNA (AdSOD2) or negative control cDNA (AdBglII) were obtained from the University of Iowa DNA-vector core. Cells were infected following our previously published protocol [2, 5]. Hydroxytyrosol (Cayman) was diluted in complete media immediately prior to application to cells.
Dual Luciferase Reporter Assay
Sequences encompassing miR-302 target sequence in ARID4a (NM_002892.3) and CCL5 (NM_002985.2) 3'-UTRs were cloned into the multiple cloning site of the psiCheck-2 dual luciferase reporter vector (Promega). The primers used to amplify the miR-302 target sites were: luc-ARID4a-1 containing the first miR-302a target site (amplicon size 229 bp, nt: 4229–4457): forward primer, 5'-TTTTCTCGAGAAGCAGCCTGCCATATTTGT-3', and reverse primer, 5'- AAAAGCGGCCGCCCCCTACTCACCAAGTGTTCA-3'; luc-ARID4a-2 containing the second miR-302a site (amplicon size, 220 bp, nt: 4334–4553): forward primer, 5'-TTTTCTCGAGGCTTTCAGGTGTTACAGAAATCG-3', reverse primer, 5'-AAAAGCGGCCGCTGGGCACACTGCTGAACATA-3'; luc-ARID4a-1,2 containing both miR-302a target sites was amplified using the forward primer from luc-ARID4a-1 and the reverse primer of luc-ARID4a-2 (amplicon size, 325 bp, nt: 4229–4553); luc-CCL5 containing the only miR-302 target site forward primer (amplicon size, 414 bp, nt: 344–757), 5'-TTTTCTCGAGGGATGGAGAGTCCTTGAACC-3', reverse primer, 5'-AAAAGCGGCCGCCAGGCTGGAGTGCAGT-3'. NHFs or MB231 cells were seeded on a 24-well plate and transfected the following day with 100 ng of luciferase reporter plasmid and 100 nM miR mimic oligonucleotides using Lipofectamine 2000 or Arrest-In according to manufacturer instructions. Seventy-two hours after transfection, measurements of firefly luciferase and renilla luciferase were performed stepwise using the Dual-Glo luciferase assay system (Promega) and a luminometer (Infinite m200, Tecan). Fold change was calculated following our previously published protocol [33].
RNA extraction and real-time quantitative PCR
Total RNA was isolated with Trizol reagent according to the manufacturer's protocol (Invitrogen) and cDNAs were synthesized using the cDNA Archive Kit (Applied Biosystems). MicroRNAs were reverse transcribed into cDNAs with a TaqMan MicroRNA reverse transcription kit using microRNA-specific primers (Applied Biosystems). The quantitation of mRNA or microRNA (miR) levels was performed by real-time PCR (StepOnePlus; Applied Biosystems) using Sybr green gene expression assays and TaqMan MicroRNA assays, respectively. Quantitative RT-PCR data were normalized to the RNA 18S or small RNA RNU44. Primers used for Sybr green gene expression assays were: ARID4a, Forward primer: 5'-AATATCCCCGCACATCAAAGATGGAG-3', Reverse primer: 5'-TGGTTGCAACTTCAGACTTCAAAGACA-3', amplicon size: 200 bp, nt: 3683–3882; CCL5, Forward primer: 5'- GCAGCCCTCGCTGTCATCCT -3', Reverse primer: 5'- AAGACGACTGCTGGGTTGGAGC -3', amplicon size: 176 bp, nt: 84–238; SOD2 (NM_000636.3), Forward primer: 5'-GGCCTACGTGAACAACCTGAA-3', Reverse primer: 5'-CTGTAACATCTCCCTTGGCCA-3', amplicon size: 70 bp, nt: 322–372; and 18S, Forward primer: 5'-CCTTGGATGTGGTAGCCGTTT-3', Reverse primer: 5'-AACTTTCGATGGTAGTCGCCG-3', amplicon size: 104 bp.
Antioxidant Activity Assay
SOD2 activity was determined by indirect competitive inhibition assay [34].
Western Blot
Cells were collected by scraping in phosphate buffer and lysed using sonication. Proteins were separated on a 12.5% SDS-PAGE gel and blotted on to nitrocellulose membrane. The blots were incubated with antibody to SOD2 (1:2000). Anti-rabbit secondary antibody (1:10000) was conjugated to HRP and visualized with Western Blot ECL Plus reagent on a Typhoon FLA 7000 (GE Healthcare). For ARID4a immunoblotting, proteins were separated on a 4–15% gradient gel (Bio-Rad) and transferred on to nitrocellulose membrane. The blots were incubated with ARID4a antibody (1:200) (Santa Cruz Biotechnology) followed by incubation with anti-goat secondary antibody (1:3000). Beta-actin was used for loading control.
ELISA Assay
CCL5 protein abundance was determined using the Quantikine Human CCL5/RANTES Immunoassay (R&D Systems). Media was collected from cultures and stored at −80° C until time of analysis. Standards were prepared according to manufacturer directions and samples were used undiluted. Optical density was determined using the Infinite m200 plate reader (Tecan) at 450 nm with wavelength correction at 540 nm. Data Analysis Software (Enzo Life Sciences) was used to generate a four parameter logistic curve-fit and analysis.
Confocal Microscopy
Monolayer cultures of control and miR-302 overexpressing cells were incubated with bromodeoxyuridine (BrdU) for 1 h followed by fixation with 4% para-formaldehyde. Cells were incubated with anti-BrdU primary antibody, 1:200 dilution overnight, and Alexa 568 conjugated goat anti-mouse secondary antibody or anti-GFP primary antibody, 1:200 dilution overnight, and Alexa 488 conjugated goat anti-rabbit secondary antibody. Cells were counter stained with DAPI for nuclear staining. ImageJ software with the ITCN (Image-based Tool for Counting Nuclei) plug-in was used to calculate the percentage of BrdU positive cells.
Statistics
Statistical analysis was done using the one and two-way analysis of variance with LSD significant difference test; student t test was used for experiments with less than three groups. Homogeneity of variance was assumed with 95% confidence interval level. Results from at least n ≥ with p < 0.05 were considered significant. All statistical analyses were done using SPSS version 17 and/or Microsoft Excel.
Results
miR-302 levels decreased in growth arrested cells
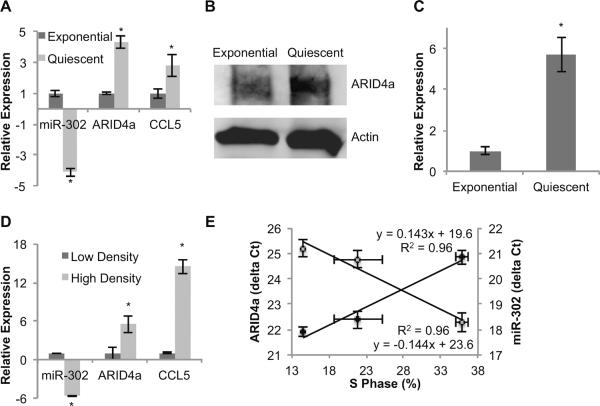
To determine if the abundance of specific miRs varies between proliferating and quiescent normal human fibroblasts (NHFs) contact inhibited monolayers were continued in cultures for 7 d (early quiescence) and 60 d (late quiescence) with media change in 3 d intervals. Exponential cultures were collected at 24 h after re-plating of NHFs from 2 d contact inhibited cultures. Flow cytometry measurements of DNA content showed more than 90% G0/G1-cells in early quiescence and more than 95% in late quiescent NHFs. Exponential cultures exhibited 29% G0/G1-cells. Total RNA was isolated and enriched for small RNAs. In vitro reverse transcribed miRs were used to hybridize miFinder Sybr Green based PCR arrays (SABiosciences). The PCR array contains 88 of the most common and well-studied miRs. Of the 88 miRs profiled, 4 up-regulated and 16 down-regulated miRs were common in both early and late quiescence compared to exponential cultures (data not shown). miR-302 exhibited the largest decrease among the down-regulated miRs. Cell growth state specific variations in miR-302 levels were further verified by performing an Applied Biosystems Taqman Assay. miR-302 levels decreased 4-fold during quiescence compared to exponential cultures of NHFs (Figure 2A).
Figure 2. miR-302 expression is repressed in quiescence compared to proliferating cells.
Normal human fibroblasts were induced to quiescence by contact inhibition. Exponential cultures were collected 24 h after replating of contact inhibited quiescent cultures. (A) RNA samples from quiescent and proliferating fibroblasts were isolated and analyzed for miR-302 abundance as well as mRNA levels of ARID4a and CCL5. Fold change was calculated relative to RNA levels in exponential cultures. Asterisks represent statistical significance relative to exponential cultures. (B) Protein samples from quiescent and proliferating cells were isolated and immunoblotting for ARID4a was performed. Actin levels were used for comparison. (C) An ELISA assay was used to measure CCL5 in media collected from exponential and quiescent fibroblasts. Asterisks represent statistical significance relative to exponential cultures. (D) RNA samples from low and high density monolayer cultures of FaDu cells were assayed for miR-302, ARID4a, and CCL5. Asterisks represent statistical significance relative to low density cultures. (E) An inverse correlation between miR-302 and ARID4a was obtained when the delta Ct of ARID4a (closed circles) and miR-302 (open circles) were plotted relative to the percentage of S-phase. n = 3, error bars represent standard deviation.
Potential targets for miR-302 were found using GOmir, which integrates results from 4 different prediction algorithms (TargetScan, RNAhybrid, miRanda, and PicTar-4way) or TargetScan alone [35]. In the search for potential targets we limited our scope to those genes involved in cell cycle control, proliferation, or response to oxidative stress. We found that both ARID4a and CCL5 are putative targets of miR-302 and involved in cell proliferation. ARID4a contained 2 highly conserved target sites for miR-302 (Figure 1A and 1B) and CCL5 contained 1 target site for miR-302 (Figure 1C).
Figure 1. Conserved binding sites of miR-302 in the 3'-UTR of ARID4a and CCL5 mRNAs.
(A and B) Nucleotide pairings of two conserved binding sites for miR-302 in the 3'-UTR of ARID4a (NM_002892); (C) one poorly conserved binding site for miR-302 in the 3'-UTR of CCL5 (NM_002985). Aligned sequences show the conserved seed sequences for miR-302 in the 3'-UTRs of six mammalian species. The seed sequence used in the prediction is underlined (TargetScanHuman 6.0, [35]).
ARID4a mRNA levels showed an approximately 1:1 inverse correlation with miR-302 in quiescence relative to exponential cells; ARID4a mRNA levels increased 4-fold while miR-302 levels decreased 4-fold (Figure 2A). CCL5 mRNA levels showed 2.7-fold increase in quiescence compared to exponential cultures (Figure 2A). The changes in mRNA levels were associated with corresponding changes in ARID4a and CCL5 protein levels. ARID4a protein levels increased approximately 2-fold in quiescence relative to exponential cultures (Figure 2B). CCL5 protein levels increased approximately 6-fold in quiescent cells (Figure 2C). To determine the generality of the observed phenomenon quiescence was induced in FaDu human head and neck cancer cells by culturing cells to low and high cell density and the above measurements were repeated. Flow cytometry measurements of DNA content showed 75% G0/G1 cells in high cell density cultures compared to 35% G0/G1 cells in sparsely populated cultures. Consistent with results presented in Figure 2A, an inverse correlation between miR-302 and its target mRNAs was also observed in FaDu cells (Figure 2D). miR-302 levels decreased approximately 6-fold in high cell density cultures compared to sparsely populated cultures (Figure 2D). Likewise, mRNA levels of miR-302 targets, ARID4a and CCL5, increased 6- and 14-fold, respectively. Growth state specific 1:1 inverse correlation between miR-302 and ARID4a levels was clearly evident from the delta Ct plots of gene expression relative to the percentage of S-phase (Figure 2E).
miR-302 regulates ARID4a and CCL5 mRNA levels
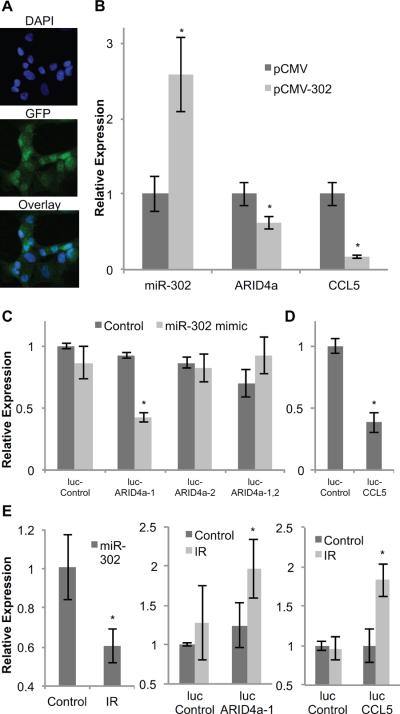
To further determine if miR-302 regulates ARID4a and CCL5 mRNA levels NHFs were transfected with plasmid DNAs containing miR-302 (pCMV-302) and ARID4a and CCL5 mRNA levels were measured at 72 h after transfection. NHFs transfected with pCMV-302 showed 2.6-fold increase in miR-302 levels compared to pCMV transfected cells (Figure 3B). Overexpression of miR-302 was associated with a significant decrease in ARID4a (38%) and CCL5 (84%) mRNA levels (Figure 3B).
Figure 3. miR-302 regulates ARID4a and CCL5 mRNA levels through target sequence in the 3'-UTRs.
(A) Fibroblasts were transfected with CMV-promoter driven miR-302 expression vector (pCMV-302) or a control vector (pCMV). Microscopy images of nuclei (DAPI) and GFP expression were used to assess transfection efficiency. (B) Expression of miR-302, ARID4a, and CCL5 in pCMV-302 transfected fibroblasts is shown relative to pCMV transfected cells. Asterisks represent statistical significance relative to pCMV transfected cells. (C) Dual-luciferase reporter assay: fibroblasts were co-transfected with miR-302 mimic and reporter plasmid DNAs carrying either luc-ARID4a-1, luc-ARID4a-2, luc-ARID4a-1,2, or luc-control. Reporter activity was measured at 48 h post-transfection and fold-change calculated following our previously published method [33]. Asterisks represent statistical significance relative to psiCheck-2 transfected cells. (D) MB-231 cells were co-transfected with pCMV-302 and luc-CCL5 or pCMV and luc-control plasmid DNAs. Luciferase expression was measured at 48 h post-transfection and fold-change calculated [33]. Asterisk represents statistical significance relative to pCMV and luc-control co-transfected cells. (E) miR-302 levels in 8 Gy irradiated fibroblasts were measured at 48 h after irradiation (left panel). Radiation induced reporter activity of constructs carrying miR-302 target sequence of ARID4a and CCL5 are shown in middle and right panels, respectively. Asterisks represent statistical significance relative to un-irradiated controls. n = 3, error bars represent standard deviation.
A dual-luciferase reporter assay was used to determine if the miR-302 target sequence in the 3'-UTRs of ARID4a and CCL5 mRNAs regulate their mRNA levels. miR-302 target sequences that are present in the 3'-UTRs of ARID4a and CCL5 mRNAs (Figure 1) were engineered into psiCheck-2 reporter vector. NHFs were co-transfected with reporter plasmids and miR-302 mimic. Luciferase activity was measured at 48 h after transfection using the dual-luciferase assay system. Luciferase activity decreased more than 60% (Figure 3C) in cells transfected with psiCheck-2 plasmid DNAs carrying the proximal miR-302 target sequence of ARID4a (luc-ARID4a-1). However, there were no significant changes in luciferase activity in cells carrying the distal miR-302 target sequence of ARID4a (luc-ARID4a-2) as well as cells carrying both target sites (luc-ARID4a-1, 2). Bioinformatics identified only one miR-302 target site in the 3'-UTR of CCL5 mRNA (Figure 1C). Luciferase activity in MB-231 cells cotransfected with pCMV-302 and luc-CCL5 reporter plasmid DNAs decreased 60% compared to pCMV and luc-control transfected cells (Figure 3D). The interaction between miR-302 and its target sequence in 3'-UTRs of ARID4a and CCL5 was also observed in radiation induced growth arrested NHFs (Figure 3E). NHFs were transfected with plasmid DNAs carrying luc-ARID4a-1 and luc-CCL5 constructs and irradiated with 8 Gy. miR-302 levels and reporter activity were measured at 48 h after irradiation. Radiation decreased miR-302 levels approximately 40%. It is interesting to note that radiation induced decrease in miR-302 was associated with a 2-fold increase in luciferase reporter activity in cells transfected with plasmid DNAs carrying luc-ARID4a-1 and luc-CCL5 reporter constructs (Figure 3E). These results demonstrate in vivo interactions between miR-302 and its target sequences in ARID4a and CCL5 mRNAs. Because radiation is well known to generate ROS, these results also suggest that the interaction between miR-302 and its target sequences in ARID4a and CCL5 could be sensitive to changes in cellular ROS levels.
ROS sensitivity of miR-302 regulation of ARID4a and CCL5 mRNA levels
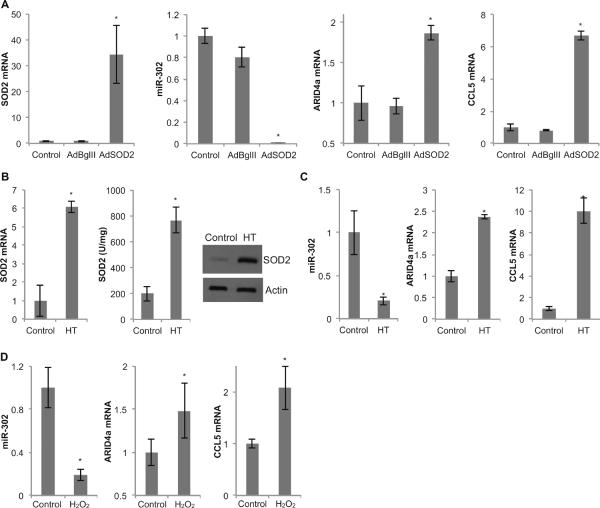
Manganese superoxide dismutase (SOD2) is an antioxidant enzyme that is well known to regulate cellular ROS levels by converting superoxide to hydrogen peroxide. To determine the ROS sensitivity of miR-302 regulation of ARID4a and CCL5 mRNA levels, SOD2 was overexpressed in NHFs by adenoviral gene transfer method [5]. Interestingly, overexpression of SOD2 nearly abolished miR-302 levels (Figure 4A). SOD2 induced decrease in miR-302 levels inversely correlated with a corresponding increase in ARID4a and CCL5 mRNA levels (Figure 4A). These results derived from genetic manipulations of SOD2 expression were also evident from pharmacological manipulations of SOD2 expression. We have recently shown that hydroxytyrosol (active ingredient of olives) undergoes semiquinone-quinone redox cycling generating superoxide, which was associated with a significant increase in SOD2 expression [36]. To determine if hydroxytyrosol induced SOD2 expression influences the inverse correlation between miR-302 and ARID4a and CCL5 mRNA levels, NHFs were incubated in culture medium containing 200 μM hydroxytyrosol for 15 d in quiescence, with regular change in media every 3 d. Similar to our earlier observations, hydroxytyrosol increased SOD2 expression at the mRNA and protein levels as well as activity (Figure 4B). Hydroxytyrosol induced increase in SOD2 activity was associated with more than 80% decrease in miR-302 levels (Figure 4C). The decrease in miR-302 levels correlated with a corresponding increase in ARID4a and CCL5 mRNA levels (Figure 4C). It is interesting to note that while SOD2 overexpression decreased miR-302 levels, cells with lower levels of SOD2 activity showed an increase in miR-302 levels. SOD2 activity was manipulated in FaDu cells by harvesting cells with a higher and lower percentage of S-phase. Cells with a higher percentage of S-phase showed lower SOD2 activity (10 U/mg), which was associated with an increase in miR-302 levels and a corresponding decrease in its target mRNAs, ARID4a and CCL5 (Figure 2D and 2E). miR-302 levels was significantly lower in FaDu cells with higher SOD2 activity (50 U/ml; lower percentage of S-phase), which was consistent with a significant increase in its target mRNAs, ARID4a and CCL5 (Figure 2D and 2E).
Figure 4. miR-302 as an ROS-sensitive regulator of ARID4a and CCL5 mRNAs.
(A) Fibroblasts were infected with 30 MOI of adenovirus carrying either the SOD2 gene (AdSOD2) or a control vector (AdBglII). miR-302 abundance as well as mRNA levels of SOD2, ARID4a and CCL5 were analyzed in control, AdBglII and AdSOD2 infected fibroblasts. Asterisks represent statistical significance relative to un-infected controls. (B) SOD2 mRNA, activity, and protein levels were measured in control and 15 d 200 μM hydroxytyrosol (HT)-fed fibroblasts. (C) Fibroblasts were treated with 200 μM HT for 15 d and analyzed for miR-302 abundance as well as mRNA levels of ARID4a and CCL5. (D) Fibroblasts were treated with 200 μM H2O2 and analyzed for miR-302 and its target mRNAs ARID4a and CCL5. Asterisks represent statistical significance relative to respective controls. n = 3, error bars represent standard deviation.
The ROS sensitivity of miR-302 regulation of ARID4a and CCL5 mRNA levels was also evident from hydrogen peroxide treated NHFs (Figure 4D). Hydrogen peroxide treatments resulted in a 5-fold down-regulation of miR-302, which was associated with approximately 1.5 and 2-fold increase in its target mRNAs, ARID4a and CCL5 (Figure 4D). These results demonstrate ROS sensitivity of miR-302 regulation of ARID4a and CCL5 mRNA levels.
miR-302 overexpression facilitates cellular proliferation
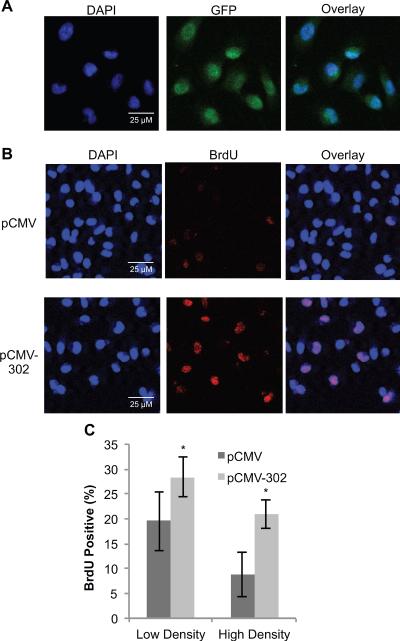
To determine if miR-302 promotes cellular proliferation, MB231 cells were transfected with 250 ng of pCMV or pCMV-302 plasmid DNAs. Monolayer cultures were incubated with BrdU for 1 h at 48 h post-transfection. Confocal microscopy was used to measure transfection efficiency (GFP-positive cells) and proliferative index (BrdU-positive cells). Transfection efficiency was calculated to be approximately 90% in pCMV and pCMV-302 transfected cells (Figure 5A). The percentage of BrdU-positive cells (S-phase) was calculated to be 19% in pCMV transfected cells compared to 29% in pCMV-302 transfected cells (Figure 5B and 5C). Comparable results were obtained from experiments using high cell density cultures (Figure 5C). The percentage of S-phase was approximately 8% in pCMV transfected high cell density cultures. Overexpression of miR-302 in these high cell density cultures increased the percentage of S-phase to 21% (Figure 5C). These results suggest miR-302 regulates transitions between quiescence and proliferation presumably by ROS-sensitive regulation of ARID4a and CCL5 mRNA levels.
Figure 5. miR-302 overexpression increases the percentage of BrdU-positive cells.
MB-231 cells were transfected with pCMV or pCMV-302. (A) Transfected monolayers were immunostained for GFP followed by counterstaining with DAPI. Representative microscopy images of nuclei (DAPI), GFP, and overlay images are shown. Transfection efficiency was calculated by counting the percentage of GFP-positive cells. (B) Representative microscopy images of nuclei (DAPI) and BrdU-positive cells in pCMV (top) and pCMV-302 (bottom) transfected cells. (C) ImageJ software was used to calculate the percentage of BrdU-positive cells in pCMV and pCMV-302 transfected low and high cell density cultures. Five hundred cells were counted in low cell density cultures, and 2000 cells were counted in high cell density cultures, over a 4mm × 4mm field. n = 3, error bars represent standard deviation.
Discussion
Reversible growth arrest, quiescence, is important for preservation of proliferative-competent cells that are required for biological processes such as wound healing and regeneration [1–3]. Results from microRNA expression array showed that the abundance of the majority of the 88 most commonly studied miRs are suppressed in quiescent compared to exponential cultures (data not shown). Among the down-regulated miRs, miR-302 was found to be significantly suppressed in quiescent cultures of normal human fibroblasts (Figure 2A). Quiescence related changes in miR-302 levels were also observed in density arrested monolayer cultures of cancer cells as well as in radiation induced transient cell cycle arrest (Figures 2D and 3E). Our observation of growth-state related changes in the abundance of specific miRs is consistent with an earlier study by Maes et al. [25] where the authors reported growth-state specific changes in the abundance of individual miRs in WI-38 human lung fibroblasts. Although the authors did not pursue studies on miR-302, it is interesting to note that miR-302 levels were lower in quiescent WI-38 fibroblasts. A regulatory role of miR-302 during quiescence and proliferation was also evident from results shown in Figure 5. The percentage of BrdU-positive (S-phase) cells increased significantly in miR-302 overexpressing MB231 cells compared to controls. These results indicate that miR-302 may regulate transitions between quiescence and proliferation by influencing the expression of specific miR-302 target genes.
We used bioinformatics to search for miR-302 target genes and restricted our search to cell cycle and oxidative stress related genes. These efforts combined with results obtained from quantitative RT-PCR and dual luciferase reporter assays show ARID4a and CCL5 as newly identified miR-302 targets whose expression inversely correlated with the abundance of miR-302 during quiescence and proliferation (Figure 2). ARID4a is a cell cycle regulatory gene that also responds to oxidative stress. ARID4a contains an LXCXE domain through which it binds the pocket region of the retinoblastoma (RB) protein. In addition, ARID4a co-localizes with E2F and histone deacetylases (HDAC1, HDAC3, and HDAC6) to discrete regions within the nuclei of quiescent fibroblasts [37]. Cells stably overexpressing ARID4a formed significantly fewer colonies than controls [38], suggesting that ARID4a is a negative regulator of proliferation. This hypothesis is supported by our observations of increased ARID4a expression in contact-inhibited quiescent fibroblasts and density arrested cancer cells as well as during radiation induced transient cell cycle arrest (Figures 2 and 3). Earlier reports identified cell cycle regulatory proteins cyclin D1, cyclin D2 and CDK2 as targets for miR-302 [26, 28]. The authors utilized a tetracycline inducible miR-302 expression vector and a luciferase target vector similar to the methods presented here [28]. The authors demonstrated decreased expression of cyclin D1, cyclin D2, and CDK2 in the presence of doxycycline compared to without [28]. Our results identified ARID4a as an additional cell cycle target for miR-302. These previous reports and results presented here (Figure 5) suggest that miR-302 may have a major role regulating proliferation and quiescence presumably by regulating ARID4a dependent modifications of the chromatin.
We have identified an additional new target of miR-302, CCL5. CCL5 is a chemokine that functions through binding to its receptor, CCR5 that belongs to the G protein-coupled receptor family [39]. A recent study reports CXCR4, another chemokine receptor for CCL5, as a target for miR-302 [40]. CCL5 expression is believed to be sensitive to changes in cellular ROS levels. Murine mesangial cells incubated with xanthine oxidase and hypoxanthine increased CCL5 expression [41], suggesting that superoxide-signaling activates CCL5 expression. Overexpression of CCL5 also increased extracellular hydrogen peroxide levels in human eosinophils [42]. Furthermore, a role of mitochondrial ROS has been suggested in regulating CCL5 expression [43]. Results presented here show CCL5 expression is higher in quiescent and irradiated cells, conditions that are well known to affect cellular ROS levels. These results suggest ROS sensitivity of miR-302 regulation of CCL5 expression.
The identification of ARID4a and CCL5 as new targets of miR-302 is also evident from results presented in Figure 3. Overexpression of miR-302 was associated with decreased levels of ARID4a and CCL5 mRNA levels (Figure 3A and 3B). Results from dual-luciferase reporter assay identified miR-302 target sequence in ARID4a and CCL5 3'-UTRs (Figure 3C and 3D). Reporter activity decreased in cells transfected with plasmid DNAs carrying the proximal miR-302 target sequence of ARID4a 3'-UTR, while cells transfected with plasmid DNAs carrying the distal miR-302 target site as well as cells transfected with plasmid DNAs carrying both target sites of miR-302 did not show any significant change in reporter activity. These results indicate the complexity of 3'-UTR mediated regulation of ARID4a expression, and suggest that additional regulatory pathways (e.g. AU-rich elements) may also be involved in the post-transcriptional regulation of ARID4a expression.
We have shown previously that both quiescence and radiation induced growth arrest are associated with changes in cellular ROS levels [2, 5, 10]. These previously published results and results presented in Figure 4 suggest ROS-sensitive miR-302 regulation of ARID4a and CCL5 mRNAs. miR-302 levels decreased significantly in both genetic and pharmacological based overexpression of SOD2 (Figure 4A–4C). Because SOD2 converts superoxide to hydrogen peroxide, overexpression of SOD2 is anticipated to increase hydrogen peroxide levels, and an increase in hydrogen peroxide may lead to the ROS sensitivity of miR-302 regulation of ARID4a and CCL5 mRNA levels. In fact, this is what we observed. miR-302 levels decreased in hydrogen peroxide treated cells, and a decrease in miR-302 levels was associated with an increase in ARID4a and CCL5 mRNA levels (Figure 4D).
In summary, our results identified ARID4a and CCL5 as additional cell cycle targets of miR-302 along with earlier reports of Oct4/Sox2, cyclin D1, cyclin D2, and CDK2 [26–28]. Because SOD2 regulates cellular ROS levels and it is localized to the mitochondrial matrix, our results also suggest that the ROS sensitivity of miR-302 regulation of ARID4a and CCL5 mRNAs could be of mitochondrial origin.
Highlights
miR-302 levels decrease in quiescence and ionizing radiation induced growth arrest.
miR-302 is an ROS-sensitive regulator of ARID4a and CCL5 mRNAs.
Overexpression of miR-302 increased the percentage of S-phase cells.
Acknowledgements
We thank Jian Shao for his expertise and assistance with the microscopy methods, and the Holden Comprehensive Cancer Center, Central Microscopy Research Facility, Flow Cytometry, and Radiation and Free Radical Research Core facilities. Funding from NIH R01 CA 111365 supported this work.
Abbreviations
- ARID4a
AT-Rich Interacting Domain 4a
- CCL5
C-C motif Ligand 5
- HT
Hydroxytyrosol
- IR
ionizing radiation
- miR
microRNA
- NHFs
normal human fibroblasts
- ROS
reactive oxygen species
- SOD2
manganese superoxide dismutase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest The authors declare no conflict of interest.
References
- [1].Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sarsour EH, Venkataraman S, Kalen AL, Oberley LW, Goswami PC. Manganese superoxide dismutase activity regulates transitions between quiescent and proliferative growth. Aging Cell. 2008;7:405–417. doi: 10.1111/j.1474-9726.2008.00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dubay DA, Franz MG. Acute wound healing: the biology of acute wound failure. Surg Clin North Am. 2003;83:463–481. doi: 10.1016/S0039-6109(02)00196-2. [DOI] [PubMed] [Google Scholar]
- [4].Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sarsour EH, Agarwal M, Pandita TK, Oberley LW, Goswami PC. Manganese superoxide dismutase protects the proliferative capacity of confluent normal human fibroblasts. J Biol Chem. 2005;280:18033–18041. doi: 10.1074/jbc.M501939200. [DOI] [PubMed] [Google Scholar]
- [6].Menon SG, Sarsour EH, Spitz DR, Higashikubo R, Sturm M, Zhang H, Goswami PC. Redox regulation of the G1 to S phase transition in the mouse embryo fibroblast cell cycle. Cancer Res. 2003;63:2109–2117. [PubMed] [Google Scholar]
- [7].Menon SG, Goswami PC. A redox cycle within the cell cycle: ring in the old with the new. Oncogene. 2007;26:1101–1109. doi: 10.1038/sj.onc.1209895. [DOI] [PubMed] [Google Scholar]
- [8].Goswami PC, Sheren J, Albee LD, Parsian A, Sim JE, Ridnour LA, Higashikubo R, Gius D, Hunt CR, Spitz DR. Cell cycle-coupled variation in topoisomerase IIalpha mRNA is regulated by the 3'-untranslated region. Possible role of redox-sensitive protein binding in mRNA accumulation. J Biol Chem. 2000;275:38384–38392. doi: 10.1074/jbc.M005298200. [DOI] [PubMed] [Google Scholar]
- [9].Cmielova J, Havelek R, Jiroutova A, Kohlerova R, Seifrtova M, Muthna D, Vavrova J, Rezacova M. DNA damage caused by ionizing radiation in embryonic diploid fibroblasts WI-38 induces both apoptosis and senescence. Physiol Res. 2011;60:667–677. doi: 10.33549/physiolres.932083. [DOI] [PubMed] [Google Scholar]
- [10].Du C, Gao Z, Venkatesha VA, Kalen AL, Chaudhuri L, Spitz DR, Cullen JJ, Oberley LW, Goswami PC. Mitochondrial ROS and radiation induced transformation in mouse embryonic fibroblasts. Cancer Biol Ther. 2009;8:1962–1971. doi: 10.4161/cbt.8.20.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fisher CJ, Goswami PC. Mitochondria-targeted antioxidant enzyme activity regulates radioresistance in human pancreatic cancer cells. Cancer Biol Ther. 2008;7:1271–1279. doi: 10.4161/cbt.7.8.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gao Z, Sarsour EH, Kalen AL, Li L, Kumar MG, Goswami PC. Late ROS accumulation and radiosensitivity in SOD1-overexpressing human glioma cells. Free Radic Biol Med. 2008;45:1501–1509. doi: 10.1016/j.freeradbiomed.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kalen AL, Sarsour EH, Venkataraman S, Goswami PC. Mn-superoxide dismutase overexpression enhances G2 accumulation and radioresistance in human oral squamous carcinoma cells. Antioxid Redox Signal. 2006;8:1273–1281. doi: 10.1089/ars.2006.8.1273. [DOI] [PubMed] [Google Scholar]
- [14].Papadopoulou A, Kletsas D. Human lung fibroblasts prematurely senescent after exposure to ionizing radiation enhance the growth of malignant lung epithelial cells in vitro and in vivo. Int J Oncol. 2011;39:989–999. doi: 10.3892/ijo.2011.1132. [DOI] [PubMed] [Google Scholar]
- [15].Robles SJ, Adami GR. Agents that cause DNA double strand breaks lead to p16INK4a enrichment and the premature senescence of normal fibroblasts. Oncogene. 1998;16:1113–1123. doi: 10.1038/sj.onc.1201862. [DOI] [PubMed] [Google Scholar]
- [16].Wang Y, Schulte BA, LaRue AC, Ogawa M, Zhou D. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 2006;107:358–366. doi: 10.1182/blood-2005-04-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- [18].Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- [19].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [20].Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- [21].Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- [22].He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- [24].Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- [25].Maes OC, Sarojini H, Wang E. Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J Cell Physiol. 2009;221:109–119. doi: 10.1002/jcp.21834. [DOI] [PubMed] [Google Scholar]
- [26].Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rosa A, Brivanlou AH. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lin SL, Chang DC, Ying SY, Leu D, Wu DT. MicroRNA miR-302 inhibits the tumorigenecity of human pluripotent stem cells by coordinate suppression of the CDK2 and CDK4/6 cell cycle pathways. Cancer Res. 2010;70:9473–9482. doi: 10.1158/0008-5472.CAN-10-2746. [DOI] [PubMed] [Google Scholar]
- [29].Lai A, Lee JM, Yang WM, DeCaprio JA, Kaelin WG, Jr., Seto E, Branton PE. RBP1 recruits both histone deacetylase-dependent and - independent repression activities to retinoblastoma family proteins. Mol Cell Biol. 1999;19:6632–6641. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Eyman D, Damodarasamy M, Plymate SR, Reed MJ. CCL5 secreted by senescent aged fibroblasts induces proliferation of prostate epithelial cells and expression of genes that modulate angiogenesis. J Cell Physiol. 2009;220:376–381. doi: 10.1002/jcp.21776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Begley LA, Kasina S, MacDonald J, Macoska JA. The inflammatory microenvironment of the aging prostate facilitates cellular proliferation and hypertrophy. Cytokine. 2008;43:194–199. doi: 10.1016/j.cyto.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Menon SG, Coleman MC, Walsh SA, Spitz DR, Goswami PC. Differential susceptibility of nonmalignant human breast epithelial cells and breast cancer cells to thiol antioxidant-induced G(1)-delay. Antioxid Redox Signal. 2005;7:711–718. doi: 10.1089/ars.2005.7.711. [DOI] [PubMed] [Google Scholar]
- [33].Chaudhuri L, Nicholson AM, Kalen AL, Goswami PC. Preferential selection of MnSOD transcripts in proliferating normal and cancer cells. Oncogene. 2012;31:1207–1216. doi: 10.1038/onc.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Spitz DR, Oberley LW. An assay for superoxide dismutase activity in mammalian tissue homogenates. Anal Biochem. 1989;179:8–18. doi: 10.1016/0003-2697(89)90192-9. [DOI] [PubMed] [Google Scholar]
- [35].Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- [36].Sarsour EH, Kumar MG, Kalen AL, Goswami M, Buettner GR, Goswami PC. MnSOD activity regulates hydroxytyrosol-induced extension of chronological lifespan. Age (Dordr) 2011 doi: 10.1007/s11357-011-9223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge MC, Tsai SC, Seto E, Zhang Y, Kuzmichev A, Lane WS, Reinberg D, Harlow E, Branton PE. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol. 2001;21:2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lai A, Marcellus RC, Corbeil HB, Branton PE. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene. 1999;18:2091–2100. doi: 10.1038/sj.onc.1202520. [DOI] [PubMed] [Google Scholar]
- [39].Balistreri CR, Caruso C, Grimaldi MP, Listi F, Vasto S, Orlando V, Campagna AM, Lio D, Candore G. CCR5 receptor: biologic and genetic implications in age-related diseases. Ann N Y Acad Sci. 2007;1100:162–172. doi: 10.1196/annals.1395.014. [DOI] [PubMed] [Google Scholar]
- [40].Fareh M, Turchi L, Virolle V, Debruyne D, Almairac F, de-la-Forest Divonne S, Paquis P, Preynat-Seauve O, Krause KH, Chneiweiss H, Virolle T. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012;19:232–244. doi: 10.1038/cdd.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Satriano JA, Banas B, Luckow B, Nelson P, Schlondorff DO. Regulation of RANTES and ICAM-1 expression in murine mesangial cells. J Am Soc Nephrol. 1997;8:596–603. doi: 10.1681/ASN.V84596. [DOI] [PubMed] [Google Scholar]
- [42].Kapp A, Zeck-Kapp G, Czech W, Schopf E. The chemokine RANTES is more than a chemoattractant: characterization of its effect on human eosinophil oxidative metabolism and morphology in comparison with IL-5 and GM-CSF. J Invest Dermatol. 1994;102:906–914. doi: 10.1111/1523-1747.ep12383399. [DOI] [PubMed] [Google Scholar]
- [43].Kim JM, Kim JS, Lee JY, Kim YJ, Youn HJ, Kim IY, Chee YJ, Oh YK, Kim N, Jung HC, Song IS. Vacuolating cytotoxin in Helicobacter pylori water-soluble proteins upregulates chemokine expression in human eosinophils via Ca2+ influx, mitochondrial reactive oxygen intermediates, and NF-kappaB activation. Infect Immun. 2007;75:3373–3381. doi: 10.1128/IAI.01940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]