Abstract
Alpha-synuclein has been reported to be present in the nucleus and levels enhanced by oxidative stress. Herein, we sought to investigate the mechanistic role of nuclear alpha-synuclein. We found that alpha-synuclein nuclear localization coincided with enhanced chromatin binding both in an in vitro and a corresponding in vivo brain oxidative stress model previously characterized by our laboratory as well as in PD brain tissues. Genome-wide chromatin immunoprecipitation (ChIP)-on-chip analysis of alpha-synuclein:promoter binding in response to oxidative stress in vitro revealed that binding occurs at several promoters belonging to a range of functional categories including transcriptional regulation. Interestingly, given the important role of mitochondrial dysfunction in PD, this included binding to the promoter for the master mitochondrial transcription activator, PGC1alpha in vitro, in vivo, and in human brain tissue with age and PD. To test the possible mechanistic impact of alpha-synuclein PGC1alpha promotor binding, we assessed PGC1alpha promoter activity, mRNA, and protein levels and expression of candidate PGC1alpha-target genes in our in vitro model. All were found to be reduced in conjunction with increased levels of aberrant mitochondrial morphology and impaired mitochondrial function. Exogenous PGC1alpha expression was found to attenuate alpha-synuclein-mediated mitochondrial dysfunction and subsequent neurotoxicity in vitro. Our data suggests that nuclear alpha-synuclein localization under conditions of oxidative stress may impact on mitochondrial function in part via the protein’s capacity to act as a transcriptional modulator of PGC1alpha. This represents a novel role for alpha-synuclein as it relates to mitochondrial dysfunction in PD.
Keywords: Parkinson’s disease (PD), 3-synuclein, PGC1-3, histone, chromatin, mitochondria, oxidative stress
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder that impacts approximately 1:100 people in the United States over the age of 65 [1]. PD is accompanied by hallmark formation of intraneuronal inclusions termed “Lewy bodies” which contain alpha-synuclein as their major protein component. Interest in alpha-synuclein was initially sparked when rare autosomal dominant familial disease forms were linked first to point mutations in the gene and then to wildtype gene duplication and triplication events which increase wildtype protein levels; interest was further heightened when the protein was revealed to be a major component of Lewy bodies in sporadic PD [2–5]. Recent gene-wide association studies have identified several polymorphisms of the alpha-synuclein gene as susceptibility factors for the idiopathic form of the disorder [6, 7]. Post-mortem studies suggest that temporal patterns of alpha-synuclein-containing Lewy body accumulation within various brain regions track with disease progression [8–11]. It has become increasingly clear that obtaining a fuller mechanistic ‘picture’ of how this protein contributes to PD-associated neuropathology would not only improve our understanding of the disease itself, but also potentially uncover novel therapeutic targets for its treatment.
Nuclear α-synuclein localization has recently been demonstrated in a variety of experimental systems and nuclear levels have been reported to increase under conditions of oxidative stress both in vitro and in vivo [1]. The function of nuclear alpha-synuclein is unknown [12] but it has been reported to co-localize with histones in conjunction with reduced levels of histone acetylation [13, 14]. Alpha-synuclein expression has also been shown to affect the expression of genes involved in various cellular or neuronal functions including transcription [15, 16]. Given known affects of alterations in histone acetylation on transcription, this suggests that nuclear alpha-synuclein may contribute to neurotoxicity in part via its ability to impact on this epigenetic event [17–21]. In this report, we demonstrate select binding of alpha-synuclein to promoters including PGC1-alpha which may contribute to mitochondrial affects associated with alpha-synuclein via its transcriptional modulation of this master regulator of mitochondrial gene expression.
Materials and Methods
All chemicals were obtained from Sigma unless otherwise noted.
Cell culture and transfection
Stable doxycycline (dox)-inducible MAO-B PC12 cell lines used for these studies were previously described [22]. Cells were maintained in DMEM containing 10% FBS, 5% horse serum, 1% streptomycin–penicillin, and 200 mg/ml of G418. Cells differentiated via 50 ng/ml nerve growth factor (NGF, 2 d) were transfected with either wildtype (WT) or mutant (A53T) alpha-synuclein cDNAs (plasmids, gift of Dr C. Ross; John Hopkins) using Lipofectamine 2000 reagent (Invitrogen). To assure equivalent transfection efficiency, cells were collected 4 hrs after transfection, mixed and re-plated for subsequent experiments. 32 hr following transfection, oxidative stress was induced via dox addition (40 μg/ml, 16 hrs). Untransfected non-induced cells were used as negative controls.
MAO-B transgenic primary cultures and brain tissue
Primary cortical cultures were prepared as previously described [26]. Briefly primary mixed cultures were prepared from the midbrain of 14-day-old mice embryos (n=5 per condition) from MAO-B transgenics and WT controls. Tissue was digested in Neurobasal medium containing 30 U/ml papain and 20 μg/ml DNase at 37°C for 30 min and mechanically triturated. Dissociated cells were centrifuged at 500 × g, resuspended in growth medium (Neurobasal medium supplemented with 10% FBS, 2 mM glutamate, B25 supplement without antioxidants, 50 U/ml penicillin, 50 U/ml streptomycin and 50 ng/ml GDNF [24]), and plated on poly-d-lysine-coated 8 well chamber slides (BD-Biocoat) at a density of 105 cells per ml. Mixed cultures were grown at 37°C for 3–5 days before induction with 40 μg/ml doxycycline for 12 hours as per [25]. Cells isolated from MAO-B transgenic mice [26] were either treated with dox or left untreated (24 hrs). Cortical tissues were isolated from inducible transgenic MAO-B lines fed dox versus vehicle for 3 weeks for further analyses as previously described [26].
Human brain sample collection
Post-mortem tissues isolated from late-onset sporadic PD patients with mild-to-moderate neuronal loss versus age-matched controls (n = 3; average postmortem period, 7.25 +/− 5 hr; average age, 69.7 +/− 9 years) were provided by Dr. Carole Miller, the University of Southern California brain bank. All PD cases were diagnosed clinically and neuropathologically confirmed whereas controls had no clinical or neuropathological signs of PD or dementia.
Cellular fractionation
Cellular fractionation was performed as previously described [23]. Briefly, cells were homogenized in L1 buffer (10 mM Hepes/0.1 mM EGTA/10 mM KCl/1.5 mM Mg2Cl), centrifuged for 5 minutes at 5000 x g, and the pellet (P1) and supernatant (S) retained. Following 10% glycerol addition, S1 was retained as the cytoplasmic fraction. The P1 pellet was resuspended in L1 buffer, passed through a 16 gauge needle, centrifuged at 5000 x g, 5 minutes, the pellet (P2) resuspended in L2 buffer (10 mM Hepes/0.1 mM EGTA/400 mM NaCl/1.5 mM Mg2Cl) at 4°C for 60 minutes, centrifuged at 14,000 x g, 30 min, and the supernatant retained as the nuclear fraction.
Immunoblotting
For western blots, 25 μg protein samples were separated on 4–12% bisacrylamide gels prior to transfer to polyvinylidine difluoride membranes. Membranes were blocked with 5% powdered milk solution in 0.3% Triton/phosphate-buffered saline solution before incubation with either 1:500 alpha-synuclein (BD Transduction, San Jose CA), 1:250 PGC1alpha (Abcam, Cambridge, UK), or 1:1000 histone H3 or acetylated H3 (Abcam, Cambridge, UK) antibodies. Beta-actin (1:5000, Sigma) antibody was used as loading control and Parp (1:1000, Cell Signaling, Danvers, MA) and beta-tubulin (1:1000, Sigma) antibodies to assess cytoplasmic and nuclear subfraction purity. Protein bands were detected via chemiluminescence substrate for horseradish peroxidase (Amersham Biosciences), resulting bands scanned, and densitometry measured by NIH image J.
Chromatin immunoprecipitation
Chromatin was prepared from cells or tissues using the MAGnify chromatin immunoprecipitation system (Invitrogen, Carlsbad, CA). 200 ug of chromatin was immunoprecipitated using alpha-synuclein, H3, acetyl H3, or PGC1alpha Abs and de-crosslinked by heating at 65°C, 3 hours prior western blot analysis. For DNA isolation for ChIP-chip promoter tile arrays, the MAGnify DNA isolation protocol was followed per the manufacturer’s instructions.
Nuclear co-localization of alpha-synuclein via confocal immunocytochemistry
Cells or tissues were fixed with 4% paraformaldehyde (PFA) for 15 min, blocked with 10% donkey serum for 30 min, then subjected to primary antibodies (1:500 H3, Cell Signaling; 1:1700 alpha-synuclein, BD Transduction) overnight, 4° C. Samples were incubated with Alexa-conjugated secondary antibodies (1:1000) for 60 min in the dark, rt. DAPI staining was performed as a nuclear localization marker. Images were captured on a Zeiss LSM 510 confocal microscope.
Biochemical histone acetylation assay
Histone acetylation was assayed biochemically in vitro via the histone acetyltransferase assay (Active Motif kit, Carlsbad, CA). The assay was carried out using 0.1 ug of highly purified nuclear protein fractions obtained as described above incubated in 17 ul HAT assay buffer containing 50 mm Tris-HCl, pH 8.0, 10% (v/v) glycerol, 1 mm dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.1 mm EDTA, pH 8.0, and 10 mm sodium butyrate. 0.5 mM of acetyl-co-A was added followed by 50 uM of H3 peptide, and the reaction incubated for 30 min, rt. The reaction was terminated by 50 ul stop solution followed by addition of 100 ul developing solution. Samples were incubated for 15 min in the dark, rt. Fluorescence was read at an excitation wavelength of 360–390 nm and emission wavelength of 450–470 nm.
High resolution ChIP-chip analysis of alpha-synuclein promoter binding sites
DNA samples were purified from wildtype alpha-synuclein ChIP from de-crosslinked chromatin samples [27]; DNA quantitation was performed and 4 μg of amplified DNA simultaneously labeled with Cy3 and controls with Cy5 to eliminate inter-array variations. DNA was hybridized to Nimblegen’s high-density 3X720K rat promoter array RN34 (720,000 50–75 mer probes per array, median probe spacing, 100 bp, 3 per slide) containing 4280 base pairs upstream and 1070 base pairs downstream of transcription start sites using high stringency conditions (http://www.vmrf.org/researchcenters/gene-chip/chromatin-immunoprecipitation.pdf). Data was extracted by Nimblegen as part of their chromatin immunoprecipitation custom array service and displayed using SignalMap software. For GO ontologies, the Database for Annotation, Visualization and Integrated Discovery (DAVID) was used. MEME suite tools were used for MEME-CHIP analysis.
ChIP qPCR
DNA samples were purified from alpha-synuclein ChIP samples as above. Samples were PCR-amplified using primers designed against regions of the PGC1-α gene using conventional qPCR; GAPDH was included as a normalization control [27]: promoter/5′ UTR: forward, tgaggtctaattgagactggctc; reverse, gctaaggtgacggtgctgtc. Intron 5: forward, gagtgagatagggtgacactcaag; reverse, ctatgagatgactaaggcccaag. PCR consisted of 50 cycles of 95°C, 30 sec, 65° C, 30 sec, and 72°, 25 sec, for human and mouse samples. The following primer sequences are used: forward, gctgcacaggagaagggaggct; reverse, ccagctcctgaatgacgccagtc. PCR cycling conditions consisted of 35 cycles of 95° C, 30 sec, 61° C, 30 sec, and 72°, 25 sec
PGC1alpha promoter luciferase activity assay
PC12 MAO-B cells transfected with WT or A53T-expressing plasmids were co-transfected with PGC-1alpha promoter-luciferase reporter plasmid (Addgene) and pGL3 as control (Promega). Luciferase activity was measured +/− dox using the Luciferase assay system kit (Promega) on a Victor Perkin-Elmer luminescence plate reader.
Quantitative real-time RT-PCR (qRT-PCR)
RNA was isolated using Trizol reagent (Invitrogen) and purified using the RNeasy Mini Kit (QIAGEN). RNA quality and concentration was determined by measuring absorbance at 260/280 nm and RNA integrity confirmed by agarose gel electrophoresis. cDNA was synthesized using purified RNA (~5 μg), GO script reverse transcriptase (Promega), and random hexamers. Gene expression was determined via qRT-PCR using the Sybr Green system in a Light cycler 480 sequence detection system (Roche Applied Science). The expression of PPARGC1A, Nrf1, Cox5b, Cyc1, and Atp51a were measured using the following primer sequences: PPARGC1A; forward 5′-gcactcagaaccatgcaaaccaca-3′ and reverse5′-gcactcagaaccatgcaaaccaca-3′; NRF1 forward 5′ttcccactcacccattgatggaca-3′ and reverse 5′-tgggcttctatggtagccatgtgt-3′ COX5b, forward 5′-gctgcatctgtgaagaggacaact -3′ and reverse 5′-agccagtgcgttggctagtcttta-3′; CYC1, forward 5′-gttgttgatgatgggcttgctgct-3′ and reverse 5′-ttcctggtctgctgagagcttgtt-3′; and ATP5a1, forward 5′-tgacgacttatccaagcaggctgt-3′ and reverse 5′-actggtaaggcagtcaaagagcca-3′. At least three technical replicates were performed for each gene. Each qRT-PCR well (20 μl total volume) contained 10 μl Sybr Green SSO fast master mix, 0.4 μl of each primer giving a final concentration of 400 nM, 8.2 μl of molecular-grade H2O, and 1.0 μl of a 50 ng/μl dilution of the stock template. The cycling conditions consisted of 45 cycles of 95° C, 30 sec, 62° C, 30 sec, 72°, 25 sec. Relative expression between samples was calculated based on the threshold cycle value following normalization to GAPDH [27].
Live mitochondria imaging via LSM510 confocal microscopy
40,000 cells were loaded per chamber on 8 chamber glass slides on the day of the experiment. Cell media was replaced with imaging buffer (3.5 mM KCl, 120 mM NaCl, 1.3 mM CaCl2, 0.4 mM KH2PO4, 1.2 mM Na2SO4, 2 mM Mgcl2, 25 mM d-glucose, and 20 mM TES, pH 7.4), 1 μM tetraphenyl boron, and 50 μM Mitotracker green (Molecular Probes) followed by incubation for 40 min at 37° C. Fluorescence was monitored in a Zeiss LSM510 confocal microscope with a Plan-Apochromat 60×/0.95 oil objective using the ‘Multi Time Lapse’ module of the Zeiss LSM software. To quantify mitochondrial length, Image Analyst software MKII was used. The green channel of MitoTracker Green stained cells was inverted to show mitochondria-specific fluorescence as black pixels and thresholded to optimally resolve individual mitochondria. Macro traces the mitochondrial outlines using “analyze particles.” The mean area/perimeter ratio was employed as an index of mitochondrial interconnectivity with inverse circularity used as a measure of mitochondrial length in microns.
Electron microscopy
Cells were cultured on Thermanox coverslips before fixing for 30 min in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer. Coverslips were rinsed in ice-cold 0.1 M sodium cacodylate buffer and post-fixed in 1% osmium tetroxide and 0.8% potassium ferrocyanide in 0.1 M sodium cacodylate for 60 min. They were then rinsed and stained with 2% uranyl acetate for 30 min on ice. Cells were dehydrated using a series of ice-cold ethanol dilutions followed by 100% ethanol and infiltrated with propylene oxide followed by Epon-812. Coverslips were divided into 8 sections, cells embedded in epon-filled beem capsules and allowed to polymerize at 60°C for 72 h. Capsules were sectioned using a MT-7000 ultramicrotome at 60 nm and imaged on a Phillips Tecnai 12 transmission electron microscope. To quantify number of swollen mitochondria scoring was performed on stereo investigator software using 8 different fields.
Spare respiratory capacity measurement via XF24 Seahorse microrespirometry
40,000 cells were seeded into 20 wells of XF24 V7 Seahorse microplates (Seahorse Bioscience, North Billerica, MA) and analyzed via the Seahorse XF24 extracellular flux analyzer per the manufacturer’s instructions. DMEM medium was replaced with 500 μL of “Seahorse medium” as previously described [28]. Plates were incubated at 37 °C for 25 min to allow temperature and O2 equilibration and then loaded into the XF24 where samples were further incubated for 12 min before the baseline was recorded. Experiments consist of a total of 9 cycles of 30 sec mixing, 150 sec pause, and 3 min measurement unless otherwise stated.
Mitochondrial complex I activity
Complex I activity was assayed in isolated mitochondrial fractions as rotenone-sensitive NADH dehydrogenase activity as previously described [29]. Values were normalized/protein using BioRad reagent.
Cell viability via 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay following PGC1alpha transfection
Cells seeded into 12 well plates were either co-transfected with PGC1alpha cDNA-expressing plasmid and WT or A53T-expressing plasmids for 24 hrs or pre-transfected with PGC1alpha plasmid for 12 hrs followed by transfection of WT or A53T plasmids for 24 hrs. Four hrs following the final transfection, 10 × 103 cells were re-plated in 96 well plates and 28 hr later, dox administered for a period of 16 hours. Cells were then exposed to 5 mg/ml MTT for 4 h at 37 °C in the dark. Supernatant was removed, 100 μL of dimethyl sulfoxide added, and OD measured at 570 nm. Values from each treatment were calculated as a percentage relative to untreated control.
Statistics
One-way ANOVA followed by Newman-Keuls Multiple comparison was used to compare differences between groups.
Results
Oxidatively-induced increases in nuclear alpha-synuclein levels
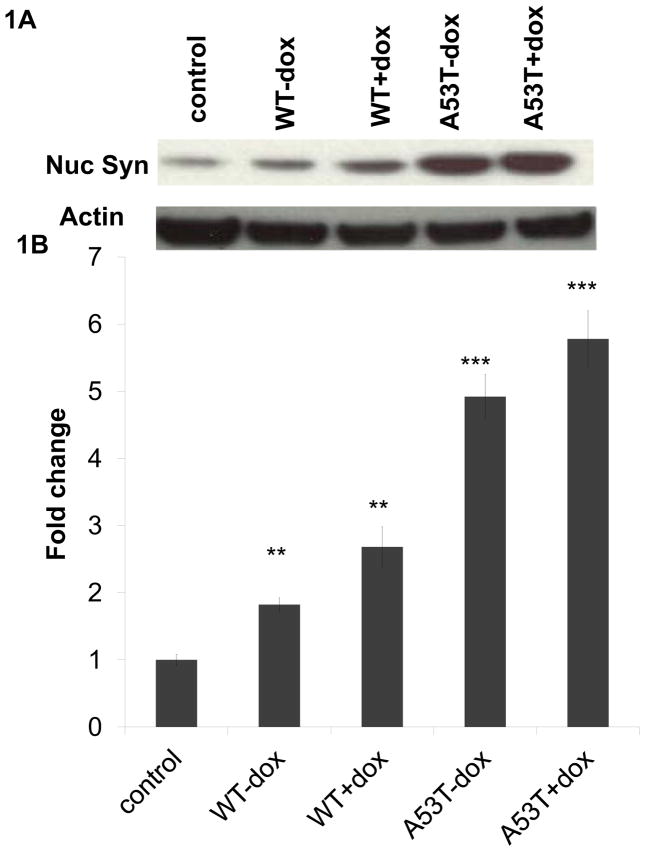
In order to verify the impact of oxidative stress on nuclear alpha-synuclein levels, we initially utilized an in vitro dopaminergic cell model system previously described by our laboratory in which oxidative stress can be induced by doxycycline (dox)-mediated increases in expression of the enzyme monoamine oxidase B (MAO-B), mimicking changes which occur with aging and in PD [22]. Data from both this model as well as comparable induction of MAO-B expression in vivo [30, 31] demonstrate that neurodegenerative affects are a direct consequence of increased oxidative stress. Cells were transfected with equivalent levels of either wildtype (WT) or familial A53T mutant alpha-synuclein, resulting in expression levels of approximately 2-fold over normal cellular levels, mimicking gene duplication events in human patients (Fig. S1A). In agreement with several previous studies [1, 13, 14], we found that increasing levels of oxidative stress resulted in significant increases in levels of alpha-synuclein (A53T mutant > WT) in isolated nuclear fractions (Fig. 1A,B). Purity of subcellular fractions analyzed were verified by western blot analyses (Fig. S1B).
Fig 1. Nuclear 3-synuclein levels are increased by oxidative stress in vitro.
(A) Representative alpha-synuclein western blot of nuclear fractions (Syn nuc) from wildtype (WT) and A53T alpha-synuclein-transfected cells +/− dox induction versus untransfected control; actin was used as a normalization control. (B) Densitometric analyses of nuclear alpha-synuclein levels normalized to actin and reported as fold change (n= 4). Values are expressed as mean ± SD; **p<0.01 & ***p<0.001 versus control.
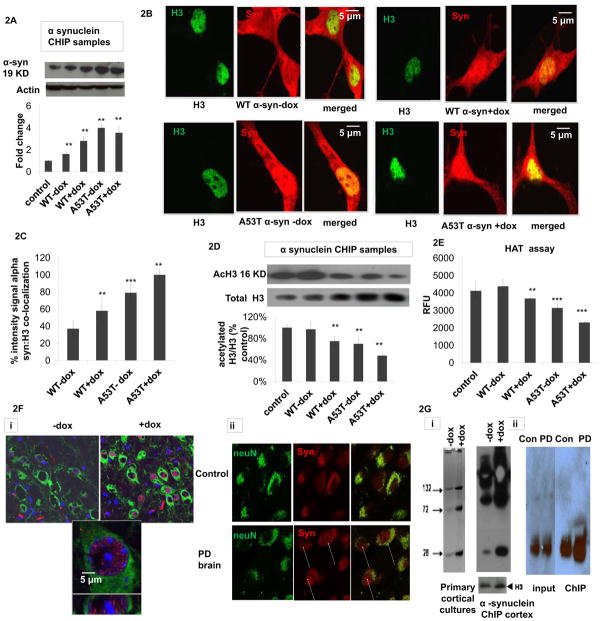
Increased oxidative stress in a corresponding inducible MAO-B transgenic mouse model [26] resulted in increased levels of endogenous nuclear synuclein (Fig. 2F). In order to assess whether endogenous alpha-synuclein was also present in the nuclei of neurons in affected human disease tissues, fluorescent immunocytochemistry was also performed on post-mortem tissues isolated from late-onset sporadic PD patients with mild-to-moderate neuronal loss versus age-matched controls (n = 3). Alpha-synuclein was found in both the cytoplasm and nuclei of PD neurons while mainly in the cytoplasm of age-matched control neurons (FIG. 2F).
Fig 2. Oxidatively-induced increase in nuclear 3-synuclein coincides with increased chromatin binding in vitro and in vivo and in Parkinsonian neurons versus age-matched controls.
(A) Representative alpha-synuclein western blot of isolated alpha-synuclein ChIP samples from untransfected (control), wildtype (WT) and A53T alpha-synuclein (α-syn) transfected cells +/− dox (actin was used as loading control) and corresponding densitometric analysis (n=4). Values are expressed as mean ± SD; **p<0.01 versus control. (B) Representative fluorescent confocal images of dual alpha-synuclein (red, BD Transduction) versus histone (H3, Cell Signaling) immunocytochemistry (merged, yellow) of WT and A53T 3-synuclein PC12 +/− dox induction (−/+Dox). (C) Co-localization as quantitated by yellow versus green nuclei in 5 fields per chamber (n=2) using NIH imageJ free software. Values are expressed as mean ± SD; **p<0.01 versus WT-dox. (D) Representative acetylated H3 (acH3) and total H3 western blots of isolated nuclear α-synuclein ChIP samples from untransfected controls, wildtype (WT) and A53T alpha-synuclein (α-syn) transfected cells +/− dox and corresponding densitometric analysis of acH3 normalized to total H3 (n=3). Values are expressed as mean ± SD; **p<0.01 versus WT-dox group. (E) Biochemical histone actyl transferase (HAT) activity reported as relative fluorescence units (RFU) in nuclear fractions from untreated controls, WT or A53T α-synuclein cells +/−dox; values are expressed as mean ± SD; **p<0.01 & ***p<0.001 versus WT-dox. (F) Co-alpha-synuclein (red) and neuronal (NeuN, green) immunostaining in (i) mouse brain tissues −/+ induction of oxidative stress via doxycycline (-dox versus +dox) versus nuclear (blue) DAPI staining; lower panel is a higher magnification of the upper dox-treated panel and (ii) in brain tissues from PD patients versus age-matched controls (yellow, merged). Arrows denote presence of alpha-synuclein in the nuclei of PD neurons. Results shown are representative of those observed in 10 separate fields. (G) Alpha-synuclein ChIP was performed on (i) samples from either primary cortical cultures isolated from MAO-B transgenic mice (left panel) either treated with dox (D) or left untreated (-dox, 24 hrs) or on brain tissues (right panel) isolated from inducible transgenic MAO-B lines fed dox versus vehicle (+dox and -dox, 3 weeks) using the alpha-synuclein Ab Syn 205 (Cell Signaling) per the Abcam Protein A ChIP kit instructions. Immunoblots were probed with Ab Syn 205 and H3 antibody as a normalization control. Samples were run on a 10% Bis-Tris NuPAGE gel (Invitrogen), blotted and probed with either an alternate alpha-synuclein antibody (BD Biociences) or H3 antibody (Abcam). Bands represent synuclein multimers of higher MW; P>0.05,+dox versus -dox. (ii) ChIP was performed on chromatin IP’d from affected SN PD tissues versus age-matched controls; samples were subsequently analyzed by alpha-synuclein ChIP-western. IP = immunoprecipitation, I = 100% input; p>0.05, PD versus Con.
Increased nuclear alpha-synuclein levels correspond with increased chromatin binding
Alpha-synuclein ChIP western analyses revealed that increased nuclear protein levels correlated with increased chromatin binding (Fig. 2A, A53T>WT) in conjunction with increased immunocytochemical co-localization with histones in alpha-synuclein-expressing cells (Fig. 2B,C). Histone acetylation levels in alpha-synuclein-bound chromatin (alpha-synuclein ChIP samples) were found to be decreased in conjunction with elevations in nuclear alpha-synuclein:chromatin binding compared to untreated WT (Fig. 2D); efficiency of alpha-synuclein IP was confirmed by western blot versus 10% input control normalized for total chromatin (H3) levels (data not shown). Loss of histone acetylation in the context of alpha-synuclein binding was verified by biochemical histone acetylation activity assay (Fig. 2E). These data suggest that increases in levels of nuclear synuclein under conditions of oxidative stress in vitro (A53T>WT) results in increased chromatin binding of the protein and prevention of histone acetylation at these binding sites.
Chromatin immunoprecipitation (ChIP) analyses using alpha-synuclein antibody in primary cortical neurons and in brain tissues isolated from induced versus uninduced MAO-B mice also revealed increases physical interaction of the protein with chromatin in the presence of increased oxidative stress (Fig 2G). Alpha-synuclein westerns were also performed on ChIP samples isolated from PD brains versus age-matched controls; although appreciable levels of alpha-synuclein:chromatin binding exist even in the age-match control samples (likely as a consequence of normal age-related increases in both oxidative stress and DNA damage), levels were found to be elevated in diseased tissue samples.
Genome-wide mapping of alpha-synuclein:chromatin binding reveals select binding to a subset of promoters
In order to assess whether alpha-synuclein:chromatin binding is a selective event, we used alpha-synuclein antibody to perform a genome-wide ChIP-on-chip experiments to identify whether binding of wildtype alpha-synuclein to specific promoter sequences was enhanced in the presence of oxidative stress in vitro; untransfected cells were used as control. Out of 8,483 promoters analyzed, 3,089 sites of significant alpha-synuclein binding were preliminarily identified (data not shown). To gain mechanistic insight into alpha-synuclein binding to select promoter sequences, we used MEME suite tools to analyse our ChIP-chip data. MEME-ChIP analysis [32, 33] of 600 randomly chosen central 100 bp core promoter sequences from the identified alpha-synuclein promoter binding regions identified a consensus sequence, CCCCTTCC. GOMO (gene ontology for motifs) analysis revealed that this consensus sequence is associated with transcription factor activity, in particular negative regulation of transcription from RNA polymerase II-associated promoters. GO enrichment analysis of the CHiP-chip data revealed a significant enrichment in 14 separate GO categories including several previously identified to be impacted by elevations in alpha-synuclein expression (Table 1). Of particular interest, amongst the 15 PPAR pathway gene sequences identified displaying significant alpha-synuclein binding particularly under conditions of oxidative stress included the PGC1-alpha promoter. PGC1alpha is a master mitochondrial transcriptional co-activator involved in activation of several genes involved in mitochondrial biogenesis and respiration and has recently been identified by genome-wide meta-analysis as a potential therapeutic target for early intervention in PD [34].
Alpha-synuclein binds to the promoter region of the master transcriptional regulator PGC1alpha and correlates with repression of PGC1alpha function in vitro
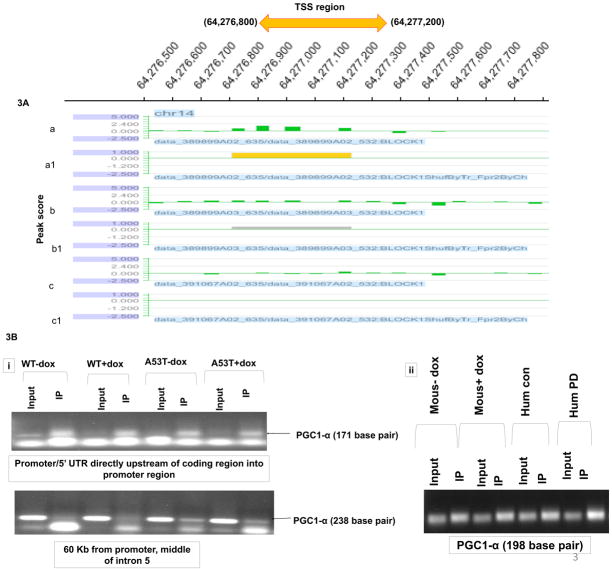
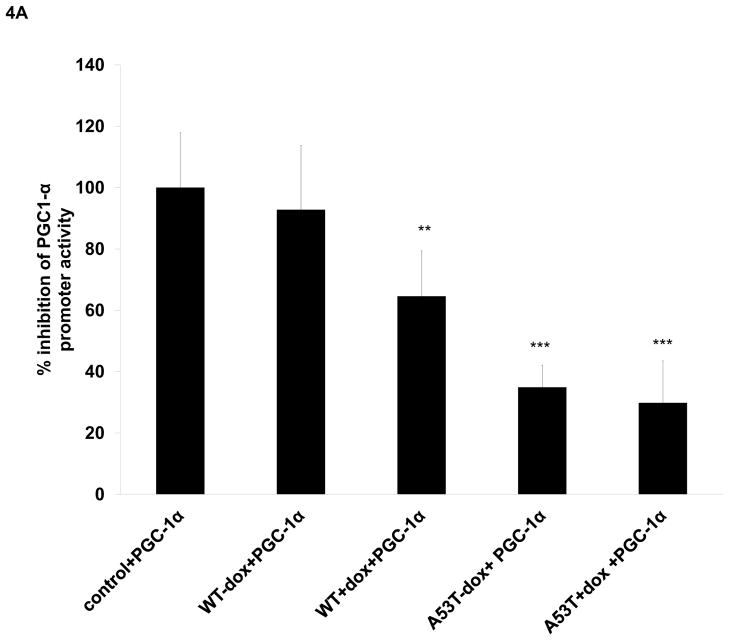
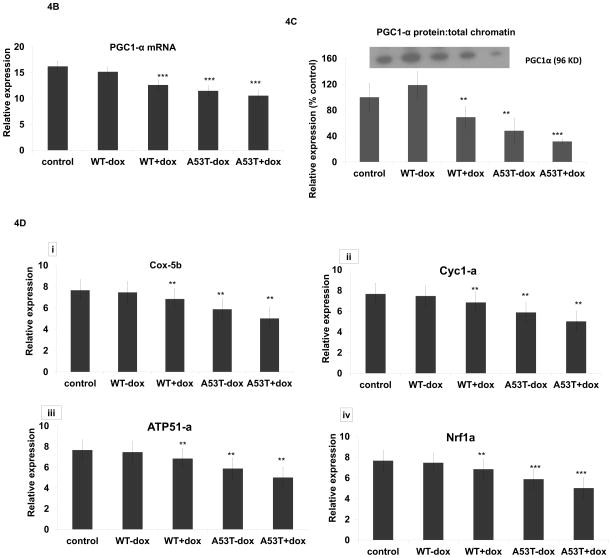
In preliminary in vitro ChIP-on-chip experiments, hybridization of wildtype alpha-synuclein protein to PGC1alpha promoter sequences appeared elevated even in the absence of oxidative stress in comparison to untransfected controls (Fig. 3A, although basal elevations unlike induced did not reach statistical significance). We subsequently confirmed the presence of WT (and A53T) alpha-synuclein:PGC1alpha promoter binding by subsequent qPCR analysis of DNA isolated from in vitro alpha-synuclein ChIP samples (Fig. 3B). We further confirmed alpha-synuclein binding to the PGC1alpha promoter not only in vivo in our mouse model in the context of increased oxidative stress, but also in the SN of PD brain and age-matched controls (Fig 3B). Binding of WT alpha-synuclein to PGC1alpha promoter sequences in the presence of oxidative stress was found in vitro to coincide with a 35% loss in PGC1alpha promoter activity while A53T resulted up to 60% loss in the absence of dox and a 70% loss in its presence (Fig. 4A). Lack of affects with WT alpha-expression alone may be due to the short time of transfection or timing of measurements. This was found to coincide with significant decreases in PGC1-3 mRNA levels (Fig. 4B), PGC1alpha protein:chromatin binding (Fig. 4C) in all conditions other than WT-dox versus control, and with reduced expression of several candidate PGC1alpha-dependent mitochondrial genes in comparison to uninduced WT (Fig. 4D).
Fig 3. Alpha-synuclein binds to PGC1alpha promoter sequences in vitro and in vivo and in PD brain tissues.
(A) Cross-linked and sonicated chromatin samples was prepared from WT transfected cells +/− dox; untransfected cells were used as controls. Total input and chipped DNA were used to prepare amplicons by LM-PCR (using genomeplex whole genome amplification kit WGA2 from sigma)which were hybridized to high-density HG18_RefSeq_promoter arrays. (a) WT + dox, (b) WT-dox, and (c) untreated control hybridization signals over PGC1alpha promoter region on chromosome 14; (a′) “yellow peak” indicative of significant binding (false discovery ratio-fdr<0.2), (b′) grey peak indicative of likely binding (fdr.>0.2), (c′) indicates no detectable binding. (B) Selective binding of CHIP DNA isolated from (i) WT and A53T +/− dox and (ii) MAO-B transgenics +/− dox and PD patients versus age-matched controls amplified with primers specific to the PGC1alpha promoter/5′UTR region. Primers specific for intron 5 of the PGC1alpha gene from in vitro samples as used as a negative binding control; arrows indicate positions of expected PGC1alpha-specific bands and primer dimers. PCR of GAPDH was used as a DNA input control (not shown).
Fig 4. Alpha synuclein overexpression results in reduced PGC1alpha promoter activity, reduced expression of PGC1alpha, and downstream transcriptional effects.
(A) Co-transfection of a PGC1alpha promoter-reporter or control pGL3 vector to normalize the promoter reporter activity along with WT and A53T alpha synuclein with and without MAOB induction greatly inhibited promoter activity in cells expressing mutant A53T alpha synuclein; values are expressed as mean ± SD; ***p<0.001 versus control cell expressing WT alpha synuclein also show loss in PGC1alpha promoter activity, significant reduction of *p<0.05 versus control was observed only after MAOB induction compared to cells that were transfected only with PGC1alpha promoter-reporter. (B) Relative PGC1alpha mRNA levels as analyzed via RT-PCR of RNA isolated from untreated control, WT and A53T +/− dox. N=3, *** p<0.001 in all groups vs control. (C) PGC1alpha western blot analysis of ChIP samples isolated from untreated controls, WT and A53T +/− dox; **p<0.01, ***p<0.001 vs control (D) Relative mRNA levels of representative PGC1alpha-dependent genes (Nrf-1, Atp51a, Cox5b, and Cyc1) as analyzed via RT-PCR of RNA isolated from untreated controls, WT and A53T +/− dox. N=3, **p<0.01,*** p<0.001 in all groups vs WT-dox.
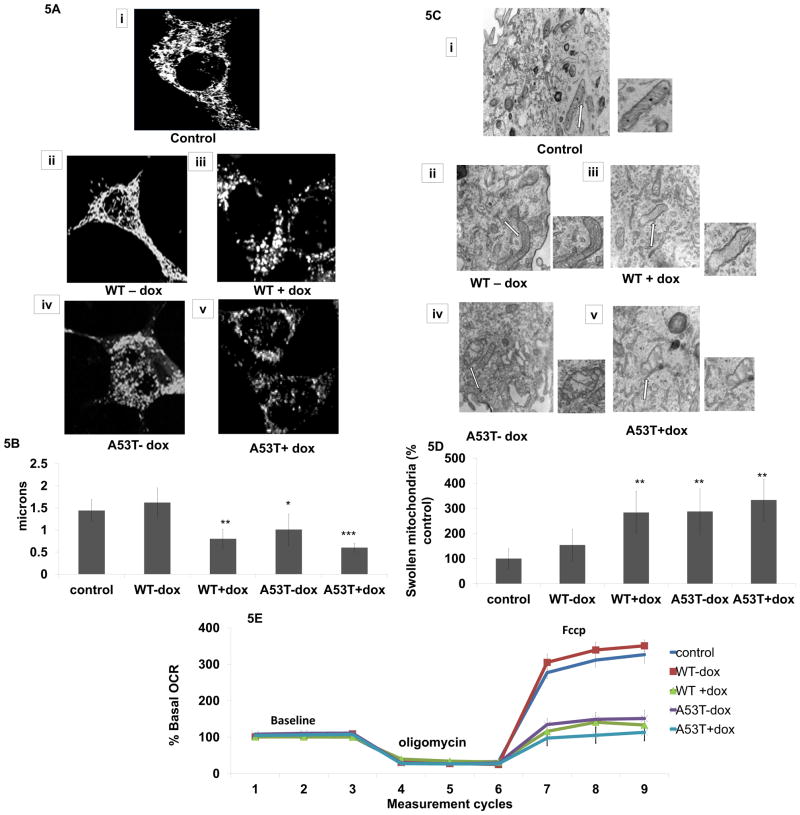
PGC1alpha expression has previously been shown to reduce the presence of mitochondrial anomolies and to protect against alpha-synuclein-mediated biochemical defects in vivo [35]. In order to explore whether reduced PGC1alpha activity as a consequence of alpha-synuclein PGC1alpha promotor binding corresponded with either alterations in mitochondrial morphology or function, we performed a series of experiments to assess both in our in vitro model. We observed that alpha-synuclein expression resulted in decreased mitochondrial length in the presence of oxidative stress and/or mutant protein (Fig. 5A,B) which coincided with significant increased numbers of swollen mitochondria containing diffuse cristae (Fig. 5C,D). As a measure of mitochondrial function, we assessed oxygen consumption rates (Fig. 5E). While neither basal respiration nor ATP synthesis (rate following the ATP synthase inhibitor, oligomycin) were impacted, maximal respiratory capacity was significantly affected versus untreated WT (Fig. 5E).
Fig 5. Impact of α-synuclein on mitochondrial morphology and function in vitro.
(A) Representative images of live mitochondrial imaging in untreated control, WT and A53T +/− dox. Z-stacks of six planes at 1 mm steps were taken at 0.14 mm/pixel resolution. Images were high pass filtered, binarized, and skeletonized using Analyst MKII (Image Analyst Software, Novato, CA). (B) Quantitative morphological analysis of mean mitochondrial length; values are expressed as mean ± SD; *p<0.05, **p<0.01, ***p<0.001 versus control. (C) Representative electron microscopy images from untreated control, WT and A53T +/− dox. Note presence of swollen mitochondria with loss of cristae in A53T cells alone and in both cell types + dox. (D) scoring of swollen mitochondria performed using Stereo Investigator; values are expressed as mean ± SD of % control; **p<0.01 versus control. (E) Relative oxygen consumption rates measured in untreated controls (blue diamond), WT-dox (red square), WT+dox (green triangle), A53T-dox (purple X), and A53T+dox (aqua X). Experiments consist of 9 cycles of a 30 sec mix, a 150 sec pause, and a 3 min measurement unless otherwise noted. All experiments were run at 37°C.
Overexpression of PGC1alpha protects against alpha-synuclein-mediated loss in mitochondrial complex I activity and subsequent cell loss in vitro
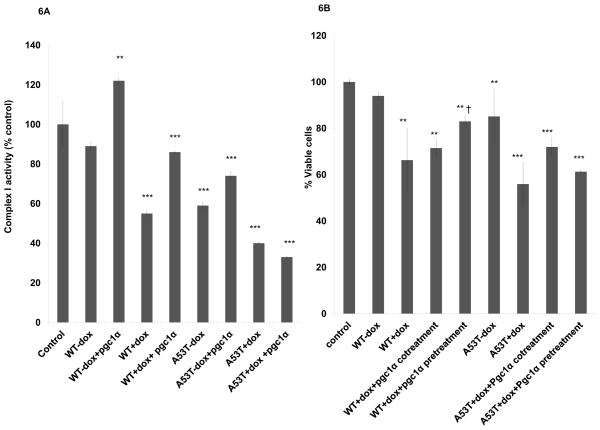
We had previously demonstrated in our inducible in vitro MAO-B cell lines that complex I activity is not only instrumental in determining overall rates of metabolic respiration due to its relatively low spare capacity, but that its levels are reduced in the presence of oxidative stress [36]. This was found to correspond with age-related nigrostriatal neurodegeneration and decreased locomotor function in vivo and affects were attenuated by inhibition of oxidative stress [26]. Reduced mitochondrial complex I activity is associated with PD [37]. In vitro complex I activity levels were found to undergo similar reductions to those previously reported in PD patient tissues [38, 39]. PGC1-alpha expression resulted in partial restoration in complex I activity in all conditions but mutant A53T expression in presence of dox (Fig 6A). Transfection of PGC1alpha in vitro also reduced WT and A53T alpha-synuclein-mediated in vitro cell death in all conditions but when transfected prior to dox treatment in A53T transfectants (Fig. 6B).
Fig 6. Constitutive PGC1-3 overexpression in vitro protects against 3-synuclein-mediated reductions in complex I activity and losses in cell viability.
(A) Mitochondrial complex I activity reported as % untransfected controls in untreated controls, WT and A53T +/− dox, +/− PGC1 alpha. Values are expressed as mean ± SD**p<0.01 & ***p<0.001 compared to control. (B) Cell viability as analyzed in MTT assay reported as % cell viability versus WT syn in WT and A53T transfected cells in the absence or presence of constitutive PGC1alpha cDNA pre- or co-transfection. (looks like a repeat so I deleted it)***p<0.001 versus uninduced WT and A53T, †p<0.05 versus WT dox, §p<0.001 versus A53T dox.
Discussion
Several previous studies have demonstrated that increased nuclear alpha-synuclein under conditions of oxidative stress results in its co-localization with histones coupled with reduced histone acetylation levels, although neither the mechanism nor the functional significance of this phenomenon was investigated. Using previously characterized in vitro and in vivo oxidative stress models [36], we report that this corresponds with the protein’s physical interaction with chromatin. Increased chromatin binding was also observed in conjunction with elevated nuclear alpha-synuclein levels in post-mortem PD brains versus age-matched controls. A subsequent genome-wide ChIP-on-chip promotor tile array in our in vitro cell model demonstrated specific binding of wildtype alpha-synuclein to a subset of the promoter sequences including the master mitochondrial transcription factor PGC1alpha. This was of special interest to us given that loss of PGC1alpha expression results in increased susceptibility to subsequent neurodegeneration and increased expression to block morphological mitochondrial dysfunction and dopaminergic cell loss associated with alpha-synuclein and oxidative stress-mediated events (3, 14, 35–37, 42, 47). PGC1alpha expression has recently been reported to be compromised in the sporadic PD brain in conjunction with a large genome-wide meta-expression analysis [34, 34]. Binding of alpha-synuclein to the PGC1alpha promoter was confirmed both in vitro and in vivo by subsequent qPCR analysis. Binding of alpha-synuclein to the PGC1alpha promoter was also found to be present in brain tissues from PD patients and older age-matched controls. Given that the aging brain itself is prone to increased oxidative stress, it would be of interest how binding varies in normal older versus young brain samples.
In terms of functional affects, the degree of alpha-synuclein:chromatin binding following oxidative induction in the various in vitro conditions was found to correlate with reduced PGC1-alpha promoter activity, mRNA, and protein levels and decreased expression of PGC1-alpha target genes (Fig. 4). The degree of chromatin binding also corresponded with reduced histone acetylation levels, a possible mechanism for PGC1alpha repression. In support of this, histone deacetylase (HDAC) inhibitors have been reported to up-regulate PGC1alpha transcription [1]. Alpha-synuclein has also recently been reported to impact on DNA methylation, another epigenetic modification known to impact on gene expression [40].
Live cell imaging and EM analyses in various in vitro cellular conditions demonstrate that repression of PGC1alpha generally corresponded with an increase in morphological mitochondrial anomolies. Alterations in mitochondrial morphology were found in turn to correspond with mitochondrial dysfunction including alterations in mitochondrial complex I activity and spare metabolic capacity, factors that may have a major impact on neuronal survival particularly under stress conditions. One exception is in the case of increased A53T alpha-synuclein expression in the presence of oxidative stress versus its absence; these were not found to be statistically different in terms of either the percentage of swollen mitochondria or uncoupled respiration (although both showed a trend in this direction). These data may imply that A53T alone has reached a maximal threshold in terms of its affects on these parameters which are not then exacerbated in the presence of increased oxidative stress. It may also be confounded by other potential non-PGC1alpha promoter binding-related affects of A53T on mitochondrial function, for example alpha-synuclein’s ability to physically interact with mitochondria as previously described by our laboratory [41]. Constitutive overexpression of PGC1alpha in the presence of dox induction in our in vitro paradigm was found to attenuate reduced complex I activity in all conditions but in dox-induced A53T-expressing MAO-B cells, the most severe stress condition (Fig 6A). We speculate that this may be due to the inability of this level of PGC1alpha expression to overcome repression of non-PGC1alpha-binding dependent affects on mitochondrial function. Co-PGC1alpha expression was found to be capable of protecting against cell loss in all conditions. Interestingly, pre-treatment was found to have statistically significant affects in all conditions other than dox-treated A53T-transfected cells (although there is a trend in this direction). We speculate that pre-transfection of exogenous PGC1alpha may allow interact particularly of the A53T protein in the cytoplasm, resulting in transcriptional inhibition of PGC1alpha prior to oxidative translocation of A53T to the nucleus that may confound the affects of exogenous PGC1alpha expression. This is currently under investigation in the laboratory.
Conclusion
Loss of mitochondrial homeostasis is implicated in the pathogenesis of PD [43–45]. We report here direct binding of nuclear alpha synuclein to the PGC1alpha promoter both in vitro and in vivo, providing a novel mechanistic link between oxidative stress, nuclear alpha-synuclein localization, and mitochondrial function. These data suggest that nuclear alpha-synuclein may impact on mitochondrial function in part via its capacity to act as a transcriptional modulator of PGC1alpha. Given the recent identification of PGC1alpha as a therapeutic PD target [35, 46–48], this has important implications for our understanding of the mechanistic role of alpha-synuclein as it relates to PD. PARIS, a protein whose levels are elevated in the brains of PD parkin mutation patients, has also been reported to act as a transcriptional repressor of PGC1alpha, suggesting that PGC1alpha repression may be a shared mechanism in familial and sporadic PD [49].
Table l. GO gene categories significantly enriched for in vitro WT alpha synuclein: promoter binding.
The threshold of EASE score, a modified Fisher exact p value, for gene-enrichment analysis; p values of ≤_0.05 are considered strongly enriched in the annotation categories.
| Go category | 3-syn Target genes | pvalue |
|---|---|---|
| Regulation of transcription | 21 | 3.2E-2 |
| Neuron development | 54 | 1.7E-3 |
| Regulation of inflammatory response | 17 | 6.2E-3 |
| Cellular homeostasis | 65 | 4.1E-3 |
| Histone modification | 18 | 5.7E-2 |
| Regulation of transcription from RNA polymerase II promoter | 97 | 5.2E-6 |
| PPAR signaling pathway | 15 | 1.7E-2 |
| Methylation | 16 | 1.6E-2 |
| Regulation of behavior | 13 | 6.1E-3 |
| Central nervous system neuron differentiation | 11 | 5.7E-3 |
| Cell death | 30 | 3.8E-3 |
| Learning or memory | 52 | 1.5E-2 |
| Regulation of neurogenesis | 23 | 1.4E-2 |
highlights.
Oxidatively-induced nuclear 3-synuclein binds PGC1-alpha promoter regions.
Alpha-synuclein appears to act as a transcriptional modulator of PGC1-alpha.
Findings link PD, oxidative stress, alpha-synuclein, and mitochondrial dysfunction.
Acknowledgments
These studies were funded by R01 NS045615 (JKA), Allison Foundation and a grant from the National Parkinson’s Foundation (JKM). We are thankful to Drs. Brand and Lunyak and their laboratories for discussions related to this manuscript and to the Buck Institute Morphology Core (Danielle Crippen and Cathy Vitelle) for their help with imaging studies.
Abbreviations used are
- A53T
mutant alpha synuclein
- ChIP
chromatin immunoprecipitation
- dox
doxycycline
- -dox
absence of doxycycline
- +dox
doxycycline addition
- IP
immunoprecipitation
- I
100% input
- Nuc
nuclear
- syn
synuclein
- TSS
transcription start site
- WT
wildtype alpha-synuclein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Monti B, Gatta V, Piretti F, Raffaelli SS, Virgili M, Contestabile A. Valproic acid is neuroprotective in the rotenone rat model of Parkinson’s disease: involvement of alpha-synuclein. Neurotox Res. 2010;17:130–41. doi: 10.1007/s12640-009-9090-5. [DOI] [PubMed] [Google Scholar]
- 2.Keri S, Moustafa AA, Myers CE, Benedek G, Gluck MA. {alpha}-Synuclein gene duplication impairs reward learning. Proc Natl Acad Sci U S A. 2010;107:15992–4. doi: 10.1073/pnas.1006068107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C, Liu L, Zhang L, Peng Y, Zhou F. Redox reactions of the alpha-synuclein-Cu(2+) complex and their effects on neuronal cell viability. Biochemistry. 2010;49:8134–42. doi: 10.1021/bi1010909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurz A, Double KL, Lastres-Becker I, Tozzi A, Tantucci M, Bockhart V, et al. A53T-alpha-synuclein overexpression impairs dopamine signaling and striatal synaptic plasticity in old mice. PLoS One. 2010;5:e11464. doi: 10.1371/journal.pone.0011464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wills J, Jones J, Haggerty T, Duka V, Joyce JN, Sidhu A. Elevated tauopathy and alpha-synuclein pathology in postmortem Parkinson’s disease brains with and without dementia. Exp Neurol. 2010;225:210–8. doi: 10.1016/j.expneurol.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myhre R, Klungland H, Farrer MJ, Aasly JO. Genetic association study of synphilin-1 in idiopathic Parkinson’s disease. BMC Med Genet. 2008;9:19. doi: 10.1186/1471-2350-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–7. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 9.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al Chalabi A, Durr A, Wood NW, Parkinson MH, Camuzat A, Hulot JS, et al. Genetic variants of the alpha-synuclein gene SNCA are associated with multiple system atrophy. PLoS One. 2009;4:e7114. doi: 10.1371/journal.pone.0007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholz SW, Houlden H, Schulte C, Sharma M, Li A, Berg D, et al. SNCA variants are associated with increased risk for multiple system atrophy. Ann Neurol. 2009;65:610–4. doi: 10.1002/ana.21685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu S, Li X, Liu G, Han J, Zhang C, Li Y, et al. Extensive nuclear localization of alpha-synuclein in normal rat brain neurons revealed by a novel monoclonal antibody. Neuroscience. 2007;145:539–55. doi: 10.1016/j.neuroscience.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 13.Goers J, Manning-Bog AB, McCormack AL, Millett IS, Doniach S, Di Monte DA, et al. Nuclear localization of alpha-synuclein and its interaction with histones. Biochemistry. 2003;42:8465–71. doi: 10.1021/bi0341152. [DOI] [PubMed] [Google Scholar]
- 14.Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15:3012–23. doi: 10.1093/hmg/ddl243. [DOI] [PubMed] [Google Scholar]
- 15.Crews L, Mizuno H, Desplats P, Rockenstein E, Adame A, Patrick C, et al. Alpha-synuclein alters Notch-1 expression and neurogenesis in mouse embryonic stem cells and in the hippocampus of transgenic mice. J Neurosci. 2008;28:4250–60. doi: 10.1523/JNEUROSCI.0066-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winner B, Rockenstein E, Lie DC, Aigner R, Mante M, Bogdahn U, et al. Mutant alpha-synuclein exacerbates age-related decrease of neurogenesis. Neurobiol Aging. 2008;29:913–25. doi: 10.1016/j.neurobiolaging.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 18.Voutsinas GE, Stavrou EF, Karousos G, Dasoula A, Papachatzopoulou A, Syrrou M, et al. Allelic imbalance of expression and epigenetic regulation within the alpha-synuclein wild-type and p.Ala53Thr alleles in Parkinson disease. Hum Mutat. 2010;31:685–91. doi: 10.1002/humu.21248. [DOI] [PubMed] [Google Scholar]
- 19.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317:516–9. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 20.Duce JA, Smith DP, Blake RE, Crouch PJ, Li QX, Masters CL, et al. Linker histone H1 binds to disease associated amyloid-like fibrils. J Mol Biol. 2006;361:493–505. doi: 10.1016/j.jmb.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 21.Leng Y, Chuang DM. Endogenous alpha-synuclein is induced by valproic acid through histone deacetylase inhibition and participates in neuroprotection against glutamate-induced excitotoxicity. J Neurosci. 2006;26:7502–12. doi: 10.1523/JNEUROSCI.0096-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar MJ, Nicholls DG, Andersen JK. Oxidative alpha-ketoglutarate dehydrogenase inhibition via subtle elevations in monoamine oxidase B levels results in loss of spare respiratory capacity: implications for Parkinson’s disease. J Biol Chem. 2003;278:46432–9. doi: 10.1074/jbc.M306378200. [DOI] [PubMed] [Google Scholar]
- 23.Siddiqui A, Lacroix T, Stasko MR, Scott-McKean JJ, Costa AC, Gardiner KJ. Molecular responses of the Ts65Dn and Ts1Cje mouse models of Down syndrome to MK-801. Genes Brain Behav. 2008;7:810–20. doi: 10.1111/j.1601-183X.2008.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith DW, Friedmann T. Discrepant effects of culture conditions on survival and function of dopaminergic neurons. Neuroreport. 2004;15:1025–8. doi: 10.1097/00001756-200404290-00018. [DOI] [PubMed] [Google Scholar]
- 25.Kumar MJ, Nicholls DG, Andersen JK. Oxidative alpha-ketoglutarate dehydrogenase inhibition via subtle elevations in monoamine oxidase B levels results in loss of spare respiratory capacity: implications for Parkinson’s disease. J Biol Chem. 2003;278:46432–9. doi: 10.1074/jbc.M306378200. [DOI] [PubMed] [Google Scholar]
- 26.Mallajosyula JK, Kaur D, Chinta SJ, Rajagopalan S, Rane A, Nicholls DG, et al. MAO-B elevation in mouse brain astrocytes results in Parkinson’s pathology. PLoS One. 2008;3:e1616. doi: 10.1371/journal.pone.0001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thuraisingam T, Xu YZ, Moisan J, Lachance C, Garnon J, Di MS, et al. Distinct role of MAPKAPK-2 in the regulation of TNF gene expression by Toll-like receptor 7 and 9 ligands. Mol Immunol. 2007;44:3482–91. doi: 10.1016/j.molimm.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Gerencser AA, Neilson A, Choi SW, Edman U, Yadava N, Oh RJ, et al. Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem. 2009;81:6868–78. doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 30.Mallajosyula JK, Kaur D, Chinta SJ, Rajagopalan S, Rane A, Nicholls DG, et al. MAO-B elevation in mouse brain astrocytes results in Parkinson’s pathology. PLoS One. 2008;3:e1616. doi: 10.1371/journal.pone.0001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siddiqui A, Mallajosyula JK, Rane A, Andersen JK. Ability to delay neuropathological events associated with astrocytic MAO-B increase in a Parkinsonian mouse model: implications for early intervention on disease progression. Neurobiol Dis. 2010;40:444–8. doi: 10.1016/j.nbd.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–7. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey TL, Boden M, Whitington T, Machanick P. The value of position-specific priors in motif discovery using MEME. BMC Bioinformatics. 2010;11:179. doi: 10.1186/1471-2105-11-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med. 2010;2:52ra73. doi: 10.1126/scitranslmed.3001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wareski P, Vaarmann A, Choubey V, Safiulina D, Liiv J, Kuum M, et al. PGC-1{alpha} and PGC-1{beta} regulate mitochondrial density in neurons. J Biol Chem. 2009;284:21379–85. doi: 10.1074/jbc.M109.018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallajosyula JK, Chinta SJ, Rajagopalan S, Nicholls DG, Andersen JK. Metabolic control analysis in a cellular model of elevated MAO-B: relevance to Parkinson’s disease. Neurotox Res. 2009;16:186–93. doi: 10.1007/s12640-009-9032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenner P, Olanow CW. Understanding cell death in Parkinson’s disease. Ann Neurol. 1998;44:S72–S84. doi: 10.1002/ana.410440712. [DOI] [PubMed] [Google Scholar]
- 38.Schapira AH, Cooper JM, Dexter D, Jenner P, Clark JB, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 39.Davey GP, Peuchen S, Clark JB. Energy thresholds in brain mitochondria, Potential involvement in neurodegeneration. J Biol Chem. 1998;273:12753–7. doi: 10.1074/jbc.273.21.12753. [DOI] [PubMed] [Google Scholar]
- 40.Desplats P, Spencer B, Coffee E, Patel P, Michael S, Patrick C, et al. Alpha-synuclein sequesters Dnmt1 from the nucleus: a novel mechanism for epigenetic alterations in Lewy body diseases. J Biol Chem. 2011;286:9031–7. doi: 10.1074/jbc.C110.212589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chinta SJ, Mallajosyula JK, Rane A, Andersen JK. Mitochondrial alpha-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett. 2010;486:235–9. doi: 10.1016/j.neulet.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Danielson SR, Held JM, Schilling B, Oo M, Gibson BW, Andersen JK. Preferentially increased nitration of alpha-synuclein at tyrosine-39 in a cellular oxidative model of Parkinson’s disease. Anal Chem. 2009;81:7823–8. doi: 10.1021/ac901176t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dagda RK, Cherra SJ, III, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–55. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005;12:1613–21. doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- 45.Yu WH, Dorado B, Figueroa HY, Wang L, Planel E, Cookson MR, et al. Metabolic activity determines efficacy of macroautophagic clearance of pathological oligomeric alpha-synuclein. Am J Pathol. 2009;175:736–47. doi: 10.2353/ajpath.2009.080928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spiegelman BM. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Found Symp. 2007;287:60–3. [PubMed] [Google Scholar]
- 47.St Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 48.Ebrahim AS, Ko LW, Yen SH. Reduced expression of peroxisome-proliferator activated receptor gamma coactivator-1alpha enhances alpha-synuclein oligomerization and down regulates AKT/GSK3beta signaling pathway in human neuronal cells that inducibly express alpha-synuclein. Neurosci Lett. 2010;473:120–5. doi: 10.1016/j.neulet.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, et al. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]