SUMMARY
In the model organism Escherichia coli, the coupling protein CheW, which bridges the chemoreceptors and histidine kinase CheA, is essential for chemotaxis. Unlike the situation in E. coli, Borrelia burgdorferi, the causative agent of Lyme disease, has three cheW homologues (cheW1, cheW2, and cheW3). Here, a comprehensive approach is utilized to investigate the roles of the three cheWs in chemotaxis of B. burgdorferi. First, genetic studies indicated that both the cheW1 and cheW3 genes are essential for chemotaxis, as the mutants had altered swimming behaviors and were non-chemotactic. Second, immunofluorescence and cryo-electron tomography studies suggested that both CheW1 and CheW3 are involved in the assembly of chemoreceptor arrays at the cell poles. In contrast to cheW1 and cheW3, cheW2 is dispensable for chemotaxis and assembly of the chemoreceptor arrays. Finally, immunoprecipitation studies demonstrated that the three CheWs interact with different CheAs: CheW1 and CheW3 interact with CheA2 whereas CheW2 binds to CheA1. Collectively, our results indicate that CheW1 and CheW3 are incorporated into one chemosensory pathway that is essential for B. burgdorferi chemotaxis. Although many bacteria have more than one homologue of CheW, to our knowledge, this report provides the first experimental evidence that two CheW proteins co-exist in one chemosensory pathway and that both are essential for chemotaxis.
Keywords: Lyme disease, Borrelia burgdorferi, Chemotaxis, Receptor-kinase coupling protein CheW
INTRODUCTION
Chemotaxis allows motile bacteria to swim towards a favorable environment or away from one that is toxic. The signaling transduction system controlling bacterial chemotaxis has been extensively studied in two model organisms, Escherichia coli and Salmonella enterica [for recent reviews, see (Wadhams and Armitage, 2004;Sourjik and Armitage, 2010;Hazelbauer et al., 2008)]. The core structural unit in the chemotaxis signaling pathway consists of a ternary complex of chemoreceptors (often referred to as methyl-accepting chemotaxis proteins, MCPs), a histidine autokinase CheA, and a coupling protein CheW (Gegner et al., 1992;Liu and Parkinson, 1989). CheW is a single-domain cytoplasmic protein (Griswold and Dahlquist, 2002).
MCPs sense various environmental signals, which control the activity of CheA. Activated CheA (CheAP) transfers its phosphoryl group to CheY, a response regulator that controls the rotational direction of flagellar motors. The phosphorylated CheY (CheY-P) diffuses from the core complex to the flagellar motors, where it binds motor-switch complex proteins to promote a switch in the rotational direction from counterclockwise (CCW) to clockwise (CW). CCW rotation results in smooth swimming (also referred to as run), and CW rotation leads to tumbling. Cells responding to a positive response (binding of an attractant to MCPs) lengthen the intervals between tumbling events and hence have longer runs that allow the bacteria to swim preferentially toward higher concentrations of attractants (Sourjik and Armitage, 2010;Porter et al., 2011). In the enteric bacteria, there are single homologues of cheA, cheW and cheY, and null mutations in any of these genes cause cells to run constantly and to become deficient in chemotaxis (Parkinson, 1977;Parkinson and Houts, 1982).
Borrelia burgdorferi, the causative agent of Lyme disease (Burgdorfer et al., 1982), is highly motile and shows chemotactic responses to several attractants produced by the hosts (Charon and Goldstein, 2002; Bakker et al., 2007;Shih et al., 2002). Our recent study shows that chemotaxis is involved in the pathogenicity of B. burgdorferi (Sze et al., 2012). Chemotaxis in B. burgdorferi differs from that of E. coli and S. enterica in several important respects [for recent reviews, see (Charon and Goldstein, 2002; Charon et al., 2012)]. B. burgdorferi cells are relatively long (10 to 20 μm in length) and thin (0.3 μm in diameter), and two flat ribbons of periplasmic flagella (PFs) arise in the subpolar region at each cell end (Charon et al., 2009;Liu et al., 2009). Motility is powered by the coordinated rotation of the PFs. This architecture requires that the swimming behavior of spirochetes is very different from that of the peritrichously flagellated enteric bacteria (Dombrowski et al., 2009;Yang et al., 2011;Harman et al., 2012;Goldstein et al., 1994;Li et al., 2002;Motaleb et al., 2011b;Motaleb et al., 2005). B. burgdorferi has three swimming modes: run, flex, and reversal. A run occurs when the bundle of PFs at the anterior end rotates CCW and that at the posterior end rotates CW. A reversal happens when both bundles change their rotational direction nearly simultaneously. A flex represents a non-translational mode when the two bundles of PFs rotate in the same direction (both CCW or both CW).
During a chemotaxis response, the spirochetes must coordinate the rotation of the motors at the two ends of cells (i.e., repressing the time spent in flexing and reversing, and increasing the time spent in running). A long-standing question about the spirochete chemotaxis is how the cells achieve this coordination (Li et al., 2002;Charon and Goldstein, 2002;Charon et al., 2012). In the spirochetes, the motors at the two ends of the cells are located at a considerable distance from one another (at least 10 μm), and the MCPs form clusters that are in close proximity to the motors (Xu et al., 2011;Briegel et al., 2009;Charon et al., 2009;Liu et al., 2009). It would seem too slow to transmit signals from one end of the cell to the other simply by diffusion of CheY-P (Motaleb et al., 2011b;Sarkar et al., 2010;Porter et al., 2011).
Unlike E. coli and S. enterica, B. burgdorferi contains more than one homologue of cheA, cheW, and cheY: two cheAs (cheA1 and cheA2), three cheWs (cheW1, cheW2 and cheW3), and three cheYs (cheY1, cheY2 and cheY3) (Fraser et al., 1997;Charon and Goldstein, 2002). Many of these genes reside within two gene clusters: the flaA operon (flaA-cheA2-cheW3-cheX-cheY3) and the cheW2 operon (cheW2-bb0566-cheA1-cheB2-bb0569-cheY2) (Ge and Charon, 1997;Li et al., 2002). We have recently identified several genes that are essential for the chemotaxis of B. burgdorferi, including cheA2, cheY3, and cheX (an analogue of cheZ from E. coli). The cheA2 and cheY3 mutants fail to reverse and constantly run, whereas the cheX mutant constantly flexes. None of these mutants is able to carry out chemotaxis (Motaleb et al., 2011b;Motaleb et al., 2005;Li et al., 2002;Bakker et al., 2007;Sze et al., 2012).
In contrast to the flaA operon, the genes studied to date in the cheW2 operon are not required for the chemotaxis of B. burgdorferi, e.g., the cheA1 and cheY2 mutants have a chemotaxis phenotype that is similar to wild type (Li et al., 2002;Motaleb et al., 2011b). It has been speculated that B. burgdorferi may possess two chemotaxis pathways that function in different hosts during the infection cycle (Li et al., 2002;Charon and Goldstein, 2002;Sze et al., 2012). For example, the chemotaxis genes (cheA2-cheW3-cheX-cheY3) in the flaA operon may form a pathway that executes chemotaxis in mammalian hosts, whereas the genes in the cheW2 operon (cheW2-cheA1-cheY2) may constitute a pathway that controls chemotaxis in the tick vector. In E. coli, CheW interacts with both MCPs and CheA and plays a pivotal role in chemotaxis and formation of the MCP-CheW-CheA ternary complexes (Gegner et al., 1992;Liu and Parkinson, 1989;Vu et al., 2012;Boukhvalova et al., 2002b). Thus, elucidating the roles of the three CheWs of B. burgdorferi in chemotaxis will help us determine whether this organism has two different chemotaxis pathways.
In this report, the three cheW genes of B. burgdorferi were separately inactivated by allelic exchange mutagenesis, and their roles in chemotaxis and chemoreceptor assembly were investigated by an approach consisting of computer-based bacterial tracking analysis, swim plate and capillary assays, immunofluorescence assay (IFA), and cryo-electron tomography (cryo-ET). Furthermore, the interactions between the two CheAs and three CheWs were studied by co-immunoprecipitation (co-IP). The results support the idea that B. burgdorferi has two different chemosensory pathways: CheW1/CheW3-CheA2-CheY3, which form a pathway that is essential for chemotaxis under the tested in vitro conditions; CheW2-CheA1-CheY2 and/or CheY1, whichform another pathway that is either required for chemotaxis under other conditions or is involved in a different signaling pathway.
RESULTS
Conservation of functionally important residues in CheW1, CheW2, and CheW3
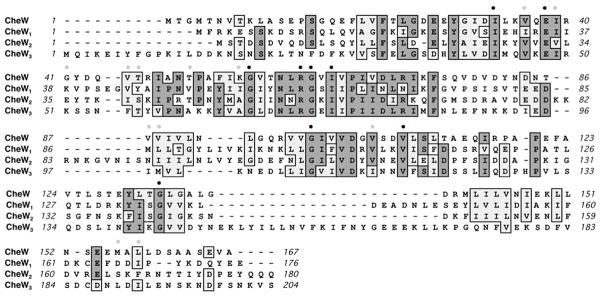
Among the three cheW genes, cheW2 (bb0565) is the first gene in the cheW2 operon, cheW3 (bb0670) is the third gene in the flaA operon, and cheW1 (bb0312) is located in a gene cluster where no other putative chemotaxis or motility genes are evident (Fraser et al., 1997;Charon and Goldstein, 2002;Li et al., 2002). CheW1 consists of 176 amino acids (aa) with a predicated molecular weight (MW) of 20 kDa. CheW2 is 180 aa in length with a predicted MW of 21 kDa. CheW3 contains 466 aa, and its predicted MW is 53 kDa. A Blast search showed that the N-terminus of CheW3 is a conserved CheW domain (aa 26 to 165) and that its C-terminus (aa 196 to 466) contains a CheR-like domain (Figure S1) (Djordjevic and Stock, 1997;Djordjevic and Stock, 1998;Shiomi et al., 2002). The E. coli CheA contains a CheW-like domain, P5, which mediates the interaction between CheA and CheW (Bilwes et al., 1999;Park et al., 2006). Sequence alignment showed that the three CheW proteins also share certain similarity to the P5 domains from the CheA proteins of E. coli and B. burgdorferi (Figure S2).
The function of CheW has been extensively studied in E. coli, and the key residues involved in the CheW/MCP and CheW/CheA interactions have been identified (Boukhvalova et al., 2002a; Boukhvalova et al., 2002b;Liu and Parkinson, 1989;Liu and Parkinson, 1991;Cardozo et al., 2010;Vu et al., 2012). B. burgdorferi CheWs share 28% (CheW1), 28% (CheW2), and 30% (the CheW domain in CheW3) sequence identity with E. coli CheW (CheWEc). Sequence alignment disclosed that the majority of the residues essential for the function of CheWEc are conserved among the three CheWs (Figure 1), including I33, E38, G57, R62, G63, G99, V108, and G133. A few residue variations were also observed (e.g., V36/I in CheW1, V88/M and V105/I in CheW3; see Figure 1). These similarities suggest that all three CheWs may function like CheWEc.
Figure 1. Sequence comparison between E. coli CheW and the three CheWs of B. burgdorferi.
The numbers show the positions of residues in E. coli CheW and B. burgdorferi CheW1, CheW2, and the CheW domain of CheW3. Dots represent functionally important residues identified in E. coli CheW (Liu and Parkinson, 1989;Boukhvalova et al., 2002a;Boukhvalova et al., 2002b;Alexandre and Zhulin, 2003). The black dots represent residues conserved in all four CheWs, and grey dots represent residues that are different in one or more of the three CheWs of B. burgdorferi. The boxes represent conserved residues of CheWs. The alignments were performed using the program MacVector 10.6.
CheW1 and CheW3 have more structural similarities with CheW
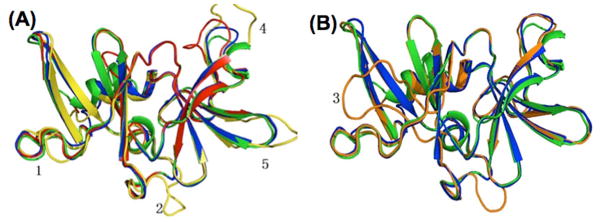
The structure of T. maritima CheW (designated as CheWTm) has been determined by nuclear magnetic resonance (NMR) (Griswold and Dahlquist, 2002;Park et al., 2006), and CheWEc and CheWTm appear to share a very similar 3D structure (Li et al., 2007). To reveal the structural features of CheW1, CheW2, and CheW3, homology modeling analysis was conducted using CheWTm as a structure template. Like CheWTm, all three CheW proteins are predicted to contain two β-sheet domains (domain 1 and domain 2), and each domain consists of a five-stranded β-barrel (Figure 2). In addition, five highly variable regions (HVR) were identified (Figure 2). Structural alignment revealed that the root-mean-square deviations (RMSD) of backbone atoms between CheWTm (blue) and CheW1 (yellow), CheW2 (orange), or the N-terminal CheW domain of CheW3 (red) were 0.566 Å, 1.617 Å, and 0.347 Å, respectively. In contrast to CheW1 and CheW3, CheW2 had a long loop inserted near the N-terminus of β strand 6 in domain 2 (Figure 2B), within the binding interface predicted for CheA (Griswold and Dahlquist, 2002;Park et al., 2006). These structural features suggest that CheW1 and CheW3 are more structurally similar to CheWEc and CheWTm than is CheW2.
Figure 2. Homology modeling of B. burgdorferi CheWs.
(A) Structure alignment of CheW1 (yellow), CheW3 (red), E. coli CheW (green), and T. maritima CheW (blue). (B) Structure alignment of CheW2 (orange), E. coli CheW, and T. maritima CheW. The N-terminal regions ahead of β strand were removed for better visualization. T. maritima CheW (Griswold and Dahlquist, 2002)(Protein Data Bank ID: 1K0S) was selected as the basis for structural modeling using the program Modeller 9v7 (Sali and Blundell, 1993). All structures were analyzed and visualized in PyMol. The numbers represent the highly variable regions (HVR) identified.
Immunoblot analysis of cheW mutants and their cognate complemented strains
As a coupling protein, CheW interacts with both MCPs and CheA. In E. coli, CheW plays a critical role in chemotaxis; a cheW null mutant constantly runs and is deficient in chemotaxis (Parkinson, 1977;Liu and Parkinson, 1989;Liu and Parkinson, 1991). To investigate the roles of CheW1, CheW2, and CheW3 in chemotaxis, the genes encoding these three proteins were inactivated by allelic exchange mutagenesis (described in Materials and Methods). A PCR analysis showed that the individual cheW genes were targeted by the antibiotic resistant makers as expected (Figure S3).
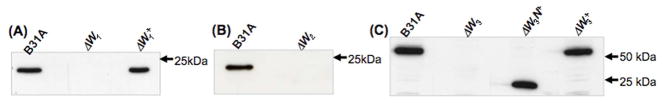
A single clone representing each mutation (ΔW1, ΔW2, and ΔW3, which represent the cheW1, cheW2, and cheW3 mutants, respectively) was selected for further characterizations. Immunoblot analyses using anti-CheW antisera (designated asαCheW1, αCheW2, and αCheW3) showed that CheW1, CheW2, and CheW3 were all detected in the wild-type strain B31A but not in the corresponding mutant clones (Figure 3). Among these three mutants, as ΔW1 and ΔW3 had altered chemosensory behaviors, these two mutants were complemented using the vectors CheW1/pBSV2G and CheW3/pBSV2G, which were constructed as described in the Materials and Methods. Immunoblot analyses showed that the complementation of cheW1 (ΔW1+) and cheW3 (ΔW3+) by the corresponding wild-type genes restored the synthesis of CheW1 (Figure 3A) and CheW3 (Figure 3C).
Figure 3. Immunoblot analysis of the three cheW mutants and their complemented strains.
(A) Immunoblot analysis of the cheW1 mutant (ΔW1) and its complemented strain (ΔW1+) using αCheW1. (B) Immunoblot analysis of the cheW2 mutant (ΔW2) using αCheW2. (C) Immunoblot analysis of the cheW3 mutant (ΔW3), its complemented strain (ΔW3+), and the mutant complemented with the N-terminal CheW domain (aa 1–210) of CheW3 (ΔW3N+) using αCheW3. The predicted molecular weights of CheW1, CheW2, CheW3, and the N-terminal CheW domain of CheW3 are approximately 20 kDa, 21 kDa, 53 kDa, and 24 kDa, respectively.
The cheW1 and cheW3 mutants are defective in chemotaxis
Chemotaxis in the ΔW1, ΔW2, and ΔW3 mutants was characterized using swim plate and capillary assays. In the swim plate assay, the ΔW2 mutant formed similar-sized colonies as the B31A strain (Figure 4B). However, the ΔW1 and ΔW3 mutants formed considerably smaller rings that were similar to that of a ΔflaB strain (Figure 4A & C), a previously documented non-motile mutant (Motaleb et al., 2000). Thus, cheW1 and cheW3, but not cheW2, are critical for chemotaxis under the tested conditions. Consistent with the results of swim plate assay, the capillary assay demonstrated that ΔW1 and ΔW3 do not respond to GlcNAc as an attractant (Figure 4D & F), whereas the ΔW2 mutantshowed the same response to GlcNAc as the wild-type strain (Figure 4E). The cognate complemented strains, ΔW1+ and ΔW3+, exhibited spreading on the swim plates and chemotactic responses to GlcNAc at wild-type levels (Figure 4A, C, D, and F). Collectively, these results indicate that cheW1 and cheW3 are required for B. burgdorferi chemotaxis, whereas cheW2 is dispensable for chemotaxis.
Figure 4. B. burgdorferi cheW1 and cheW3 mutants are non-chemotactic.
Swim plate (A) and capillary (D) assays of the cheW1 mutant (ΔW1) and its complemented strain (ΔW1+). Swim plate (B) and capillary (E) assays of the cheW2 mutant (ΔW2). Swim plate (C) and capillary (F) assays of the cheW3 mutant (ΔW3) and its complemented strains (ΔW3+ and ΔW3N+). The swim plate and capillary assays were carried out as previously described (Motaleb et al., 2000;Li et al., 2002;Bakker et al., 2007). For the swim-plate assay, ΔflaB, a previously constructed non-motile mutant (Motaleb et al., 2000), was used as a control to determine the size of non-spreading colonies on the plates. For the capillary assay, N-acetyl-D-glucosamine (GlcNAc) was used as an attractant. Results are expressed as the means ± SEM from five plates or capillary tubes. * represents a P value < 0.01.
The cheW1 and cheW3 mutants show an altered swimming behavior
Non-chemotactic mutants often show altered swimming behaviors, e.g., the cheA2 and cheY3 mutants of B. burgdorferi fail to reverse and constantly run (Motaleb et al., 2011b;Li et al., 2002). The tracking analysis using a computer-assisted cell tracker coupled with video microscopy disclosed that the ΔW2 mutant had swimming behavior indistinguishable from (Video 2, Table 1) the wild type (Video 1), whereas the ΔW1 and ΔW3 mutants had altered swimming behaviors. The ΔW3 mutant failed to reverse and constantly ran in one direction (Video 3, Table 1), like the cheA2 and cheY3 mutants of B. burgdorferi. The behavior of the ΔW1 mutant is mixed (Video 4): approximately half of the cells (21 out of 50) failed to reverse and swam exclusively in one direction. The remainder of the cells (29 out of 50) reversed, but at a lower reversal frequency (9 reversals/min) compared to the wild type (23 reversals/min). A similar pattern was observed in a reconstructed ΔW1 mutant, suggesting that the observed mixed phenotype is stochastic and not caused by genetic heterogeneity. The complemented mutants (ΔW3+and ΔW1+) had a similar swimming behavior as the wild type (Video 3A, Video 4A, and Table 1). All three cheW mutants had similar swimming velocities as the wild type (Table 1), ranging from 9 to12 μm/sec. Thus, none of the cheW mutations causes a decrease in the propulsive force generated by the flagella.
Table 1.
Effects of CheWs on swimming behaviors of B. burgdorferi.
| Strains | Mean velocity (μm/sec) ± SEMa | Mean number of reversals/min ± SEMa |
|---|---|---|
| B31 | 10.0 ± 0.8 | 23.3 ± 4.2 |
| ΔW1 | 12.3 ± 1.5 | 9.0 ± 6.0 b |
| ΔW2 | 10.0 ± 1.4 | 20.0 ± 3.2 |
| ΔW3 | 11.0 ± 0.8 | 0.0c |
| ΔW1+ | 9.8 ± 2.2 | 25.0 ± 2.6 |
| ΔW3+ | 9.7 ± 1.8 | 26.0 ± 1.9 |
| ΔW3N+ | 9.0 ± 1.7 | 24.0 ± 2.1 |
Standard errors of the means were calculated from data obtained from at least 30 individual tracked cells of each strain.
Approximately one half of the cells ran in one direction and did not reverse; the other half of the cells did reverse and the mean reversal frequency was calculated from this group of the cells.
Cells ran in one direction and did not reverse.
The CheR-like domain in CheW3 is not required for chemotaxis
CheW3 possesses a CheR-like domain at its C-terminus (Figure S1). In E. coli, CheR functions as a methyltransferase that is involved in chemoreceptor adaptation (Djordjevic and Stock, 1997;Djordjevic and Stock, 1998;Porter et al., 2011). Searching large sets of CheW homologues from microbial genome databases revealed that only CheWs from some spirochete species have a similar domain composition as CheW3, including CheW1 (TP_0364) of Treponema pallidum and CheW1 (TDE_1492)of Treponema denticola (Fraser et al., 1998;Seshadri et al., 2004). To determine whether the CheR-like domain is required for normal chemotaxis, the ΔW3 mutantwas complemented with a plasmid producing only the N-terminal CheW domain of CheW3 (aa 1–210). Immunoblotting using αCheW3 showed that the expression of the N-terminal CheW domain was restored in the complemented clone (ΔW3N+) (Figure 3C). The swim plate (Figure 4C), capillary (Figure 4F), and tracking (Table 1) assays demonstrated that chemotaxis in the ΔW3N+ strain was indistinguishable from that of the wild-type and ΔW3+ strains, indicating that deletion of the CheR-like domain does not affect the chemotactic function of CheW3 under the conditions tested.
Loss of CheW1 or CheW3 affects chemoreceptor assembly at the cell poles
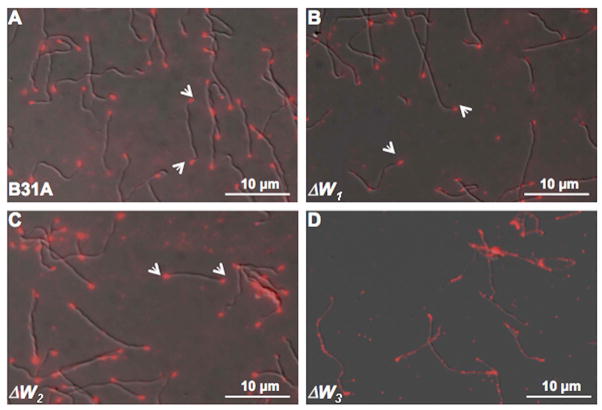
In E. coli, CheW is essential for the assembly of chemoreceptor arrays at the cell poles (Studdert and Parkinson, 2005;Maddock and Shapiro, 1993;Sourjik and Berg, 2000). Our previous studies showed that B. burgdorferi MCPs also form arrays at the cell poles (Xu et al., 2011). To determine whether the B. burgdorferi cheW mutants are defective in chemoreceptor assembly, the cellular location of the MCPs in the three mutants was determined by IFA using an antibody targeted specifically against B. burgdorferi MCP3 (Xu et al., 2011). As expected, bright fluorescent loci were observed at both cell poles in wild-type cells (Figure 5A). A similar pattern was observed in ΔW2 cells (Figure 5C), but not in ΔW3 cells, in which the fluorescence was diffused (Figure 5D). Although fluorescent loci were still evident at the poles of ΔW1 mutant cells, the fluorescence signals were considerably reduced, and even absent in many cells (Figure 5B). The IFA results suggest that CheW1 and CheW3, but not CheW2, are involved in the assembly and localization of the chemoreceptor arrays.
Figure 5. Localization of B. burgdorferi chemoreceptor arrays using IFA.
The wild-type (A), the ΔW1 (B), ΔW2 (C), and ΔW3 (D) mutant cells were fixed with methanol, stained with anti-MCP3 antibody, and counterstained with anti-rat Texas red antibody as previously described (Li et al., 2010;Xu et al., 2011). The micrographs were taken under DIC light microcopy or fluorescence microscopy with a tetramethylrhodamine isothiocyanate (TRITC) emission filter, and the resultant images were merged. Arrows point to the location of the chemoreceptor arrays within cells.
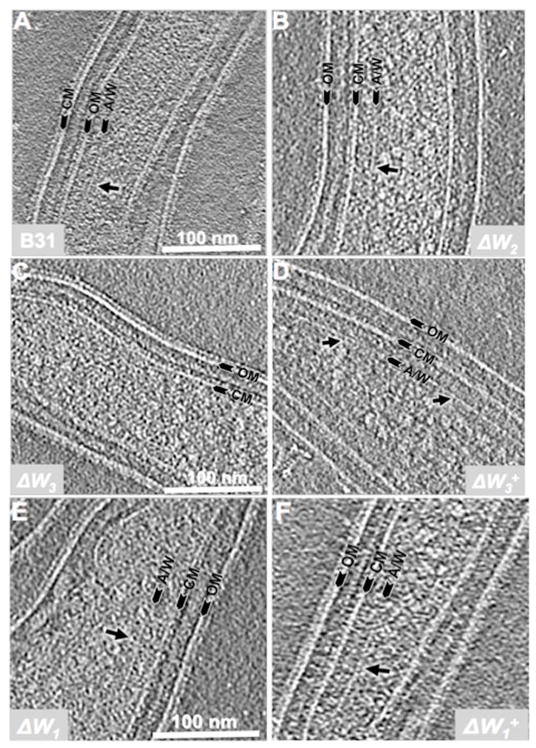
Cryo-ET was conducted to determine the cellular locations and ultrastructures of the chemoreceptor arrays in the three cheW mutants more precisely. Chemoreceptor arrays could be readily recognized as prominent ‘basal plate’-like structures (Zhang et al., 2004;Briegel et al., 2009;Briegel et al., 2012;Xu et al., 2011;Liu et al., 2012) at the poles of wild-type (Figure 6A) and ΔW2 cells (Figure 6B). The arrays had an average length of 159 ± 86 nm (n=19 cells, Table 2). No chemoreceptor arrays were observed in any of the ΔW3 cells examined (0 out of 25 cells, Figure 6C). However, the arrays could be readily detected in its complemented strain ΔW3+ (12 out of 30 cells, Figure 6D). With the ΔW1 mutant, the arrays were still evident in a small portion of the cells (4 out 31 cells, Figure 6E), but their sizes were substantially reduced (average length of 75 ± 7 nm, n=4 cells) compared to those of the wild type or the complemented ΔW1+strain (Figure 6F, 152 ± 58 nm, n = 10 cells). The cryo-ET results are consistent with the IFA data and thus further confirm that both CheW1 and CheW3 are involvedin assembly of the chemoreceptor arrays, whereas CheW2 is not.
Figure 6. Detection of B. burgdorferi chemoreceptor arrays by cryo-ET.
The cryo-ET analysis was carried out as previously described (Xu et al., 2011). Six strains were included: (A) B31A, (B) ΔW2, (C) & (D) ΔW3 and its complemented strain ΔW3+, and (E) & (F) ΔW1 and its complemented strain ΔW1+. Arrows point to chemoreceptor arrays. OM: outer membrane; CM: cytoplasmic membrane; A/W: the basal plate composed of CheA and CheW.
Table 2.
Impact of CheWs on B. burgdorferi chemoreceptor assembly.
| Strains | Total cells | Positive cells | Chemoreceptor array length (nm) |
|---|---|---|---|
| B31A | 30 | 19 | 159 ± 86 |
| ΔW1 | 31 | 4 | 75 ± 7 |
| ΔW2 | 30 | 12 | 130 ± 30 |
| ΔW3 | 25 | 0 | NA |
| ΔW1+ | 20 | 10 | 152 ± 58 |
| ΔW3+ | 30 | 12 | 155 ± 77 |
CheW1 and CheW3 interact with CheA2, whereas CheW2 binds CheA1
In E. coli, the ternary complex of MCP-CheW-CheA is the core structural unit in the signaling pathway of chemotaxis (Wadhams and Armitage, 2004;Hazelbauer et al., 2008). B. burgdorferi has two CheA homologues, CheA1 and CheA2. Identifying the interactions between the two CheAs and the three CheWs will help us understand the complexity of chemotaxis signaling pathways in B. burgdorferi. Co-IP experiments were carried out to reveal the interactions between the two CheAs and the three CheWs. For the co-IP assays, either CheA1 antibody (αCheA1) or CheA2 antibody (αCheA2) was first co-incubated with whole cell lysates of the B31A wild type and a previously constructed double cheA1cheA2 mutant (designated as ΔA1A2 and used as a negative control) (Li et al., 2002). The co-precipitated products were then probed with αCheW1, αCheW2, or αCheW3, respectively. As shown in Figure 7, CheW1 (Figure 7A) and CheW3 (Figure 7C) were detected in the samples precipitated by αCheA2 (left panel, Figure 7) but not by αCheA1 (right panel, Figure 7), whereas CheW2 was detected in the samples precipitated by αCheA1 (right panel, Figure 7B) but not by αCheA2 (left panel, Figure 7B), suggesting that both CheW1 and CheW3 interact with CheA2, whereas CheW2 binds CheA1. To confirm that CheW1 and CheW3 interact with CheA2, αCheW1 and αCheW3 were used in the co-IP assays, and the co-IP samples were probed with αCheA2. As expected, CheA2 was detected in the co-precipitated products from the wild type but not from the ΔW1 and ΔW3 mutants (Figure 7D). Collectively, the results of the co-IP assays show that CheW1 and CheW3 interact with CheA2 but not with CheA1, whereas CheW2 interacts with CheA1 but not with CheA2.
Figure 7. Detecting the interactions between two CheAs and three CheWs of B. burgdorferi by co-IP.
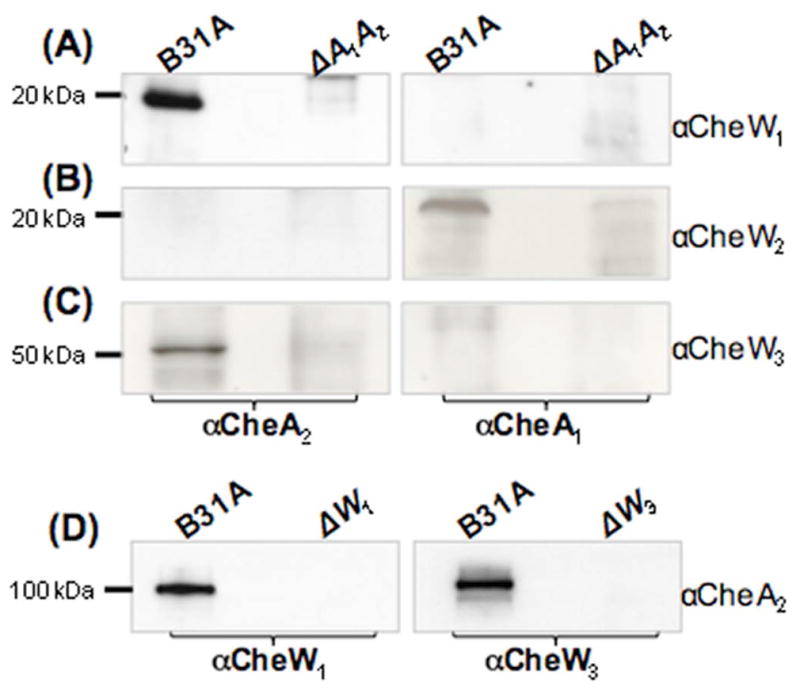
Pull down of the CheWs using αCheA1 (right panel) or αCheA2 (left panel). Precipitated proteins were probed with αCheW1 (A), αCheW2 (B), or αCheW3 (C). A previously constructed cheA1A2 double-deletion mutant (ΔA1A2) of B. burgdorferi (Li et al., 2002) was used as a negative control for the co-IP. (D) Pull down of CheA2 using αCheW1 (left panel) and αCheW3 (right panel). Precipitated proteins were probed with CheA2. Extracts from the ΔW1 or ΔW3 mutants were used as negative controls for the co-IP.
DISCUSSION
As a coupling protein, CheWEc has four known activities: binding to CheA, binding to MCPs, promoting formation of MCP-CheW-CheA ternary complexes and chemoreceptor arrays, and enabling MCPs to modulate CheA autokinase activity (Gegner et al., 1992;Cardozo et al., 2010;Liu and Parkinson, 1989). In this report, a comprehensive approach has been applied to investigate the roles of the products of the three cheW genes in B. burgdorferi. The results indicate that CheW1 and CheW3 play a similar role as the CheW of E. coli, because the ΔW1 and ΔW3 mutants showed an altered swimming behavior (Table 1 and Videos 3 & 4) and failed to respond to attractant stimuli (Figure 4D & F). Also, the IFA and cryo-ET studies showed that these two mutants are unable to assemble intact chemoreceptor arrays at the cell poles of B. burgdorferi (Figures 5 & 6). In contrast to ΔW1 and ΔW3, the ΔW2 mutant behaved like the wild type with respect to chemotactic response to attractants (Figure 4E), swimming behavior (Table 1), and chemoreceptor assembly (Figure 5C & Figure 6B). Collectively, these results indicate that CheW1 and CheW3 are essential for the chemotaxis of B. burgdorferi, whereas CheW2 is dispensable for the chemotaxis under the tested in vitro conditions. Consistent with this proposition, the homology modeling analysis predicts that CheW2 shares the least structural similarity to CheWEc and CheWTm (Figure 2). It is noteworthy to point out that CheW2 has a long loop insertion near the binding interface of CheW and CheA (Figure 2B). This insertion may disrupt the local environment of the CheA binding surface and consequently prevent CheW2 from interacting effectively with CheA2, a histidine kinase that is essential for chemotaxis of B. burgdorferi (Li et al., 2002;Sze et al., 2012).
IFA and cryo-ET assays demonstrate that CheW3 plays a more important role than CheW1 in the assembly of chemoreceptor arrays at the cell poles of B. burgdorferi. The IFA results showed that the polar-localized chemoreceptor arrays were completely disrupted in ΔW3 cells(Figure 5D & Figure S5), nor did cryo-ET analyses find any array-like structures in the mutant (Figure 6C, Table 2). The observed phenotype of the ΔW3 mutant is very similar to that of an E. coli cheW mutant (Zhang et al., 2004; Sourjik and Berg, 2000;Maddock and Shapiro, 1993). Unlike the situation in ΔW3, IFA still detected weak polar localized signals in ΔW1 cells (Figure 5B), and arrays could still be observed by cryo-ET in a small portion of the ΔW1 cells (Figure 6E). The average length of the chemoreceptor arrays observed in the ΔW1 cells was approximately two fold less than those in the wild type and its complemented strain, ΔW1+ (Table 2). Recent cryo-ET studies of E. coli MCPs show that the basal plates of the arrays consist primarily of CheA and CheW (Briegel et al., 2009;Briegel et al., 2012;Liu et al., 2012). Thus, it is conceivable that CheW1 contributes to the stability of the basal plates but is not essential for their formation. Approximately one half of the ΔW1 cells swim smoothly, and the other half still reverse but with a lower frequency than wild type (Video 4 and Table 1). The observed heterogeneous phenotype of the ΔW1 mutant is not due to genetic heterogeneity because when the mutation was recloned, the same mixed phenotype of the original ΔW1 mutant was observed. Moreover, genetic complementation totally restored the wild-type phenotype (Video 4A and Table 1).
In E. coli, CheW tethers CheA to the MCPs and affects the activity of CheA, which in turn controls the level of CheY-P. The inactivation of cheW completely blocks production of CheY-P. Thus, the flagellar motors are locked in CCW rotation, and a cheW mutant constantly runs (Gegner et al., 1992;Wadhams and Armitage, 2004;Hazelbauer et al., 2008). Our previous studies show that, of the two CheAs and three CheYs of B. burgdorferi, only CheA2 and CheY3 are involved in chemotaxis (Li et al., 2002; Motaleb et al., 2011b). The cheA2 and cheY3 mutants are smooth swimming and non-chemotactic, suggesting that CheY3-P directly controls the rotation of flagellar motors. Because the chemoreceptor arrays in the ΔW1 cells are only partially disrupted (Figure 5B and Figure 6E), it is possible that CheW1 plays an auxiliary role in coupling CheA2 to the MCPs.The decrease in CheY3-P associated with the reduced coupling of CheA2 results in a baseline concentration that spans the threshold required to elicit reversals.
The results presented here raise the possibility that B. burgdorferi may have two different chemosensory pathways (Li et al., 2002;Motaleb et al., 2011b;Charon and Goldstein, 2002). Among the multiple homologues of cheA, cheW, and cheY, only cheA2, cheY3, cheW1, and cheW3 are essential for chemotaxis in vitro. Other than cheW1, all of the essential che genes are located within the flaA operon (cheA2, cheW3, cheX, and cheY3), whereas most of the che genes that are dispensable for chemotaxis reside within anoperon that contains cheA1, cheY2, and cheW2 (Charon and Goldstein, 2002;Ge and Charon, 1997;Li et al., 2002;Fraser et al., 1997). Co-IP assays demonstrated that CheW1 and CheW3 interact with CheA2, whereas CheW2 binds CheA1. Thus, we favor the idea that B. burgdorferi has two chemosensory pathways: CheW1/CheW3-CheA2-CheY3 form the pathway that is essential for chemotaxis under the conditions usually used in vitro, and CheW2-CheA1-CheY2 and/or CheY1 form another pathway that may be used only under other conditions that have yet to be duplicated in the laboratory.
Why might B. burgdorferi have two chemosensory pathways? In nature, B. burgdorferi is maintained via an enzootic cycle comprising both mammalian hosts and an Ixodes tick vector [for recent reviews, see (Radolf et al., 2012;Samuels, 2011;Rosa et al., 2005;Steere et al., 2004)]. The enzootic cycle begins with the feeding by an uninfected tick on an infected vertebrate. After the feeding, the spirochetes remain in the tick gut throughout the molting process. At the time that the infected tick takes a blood meal on a mammal, the spirochetes begin to multiply and migrate from the tick gut to the salivary glands, from which they are transmitted to a new host, thereby completing the enzootic cycle. To adapt to different hosts and complete its enzootic cycle, B. burgdorferi may need one chemosensory pathway, perhaps represented by CheW1/CheW3-CheA2-CheY3, for chemotaxis in mammalian hosts. The pathway involving CheW2-CheA1-CheY2 and/or CheY1 may be activated in the tick vector and/or during the transmission from tick to mammal. Our recent study of the role of cheA2 in the enzootic cycle of B. burgdorferi (Sze et al., 2012) is consistent with the proposal that the CheW1/CheW3-CheA2-CheY3 pathway is important in the mammalian host. Inactivation of cheA2 decreased the ability of B. burgdorferi to establish infection in mice, but not in ticks. The true function of the CheW2-CheA1-CheY2 and/or CheY1 pathway remains obscure. It could be involved in chemotaxis in the tick vector, perhaps in migration of the spirochetes to the salivary glands, or it may function in a signal transduction pathway that regulates gene expression of B. burgdorferi.
The incorporation of two coupling proteins (CheW1 and CheW3) into one chemosensory pathway is different from the situation in other bacteria that have more than one homologue of CheW, such as Vibrio cholera and Rhodobacter sphaeroides [for recent review, see (Porter et al., 2011;Butler and Camilli, 2005;Alexander et al., 2010;Rao et al., 2008)]. In these organisms, either only one CheW homologue functions as a key coupling protein essential for chemotaxis, or CheW homologues are functionally redundant. For instance, V. cholera has three CheW homologues, but only CheW-1 is required for chemotaxis (Butler et al., 2006). Among the four CheW homologues of R. sphaeroides, CheW2 is essential for chemotaxis and chemoreceptor clustering; and deletions of other three cheWs either have no impact on chemotaxis or only conditionally affect chemotactic responses and chemoreceptor localization (Martin et al., 2001;Hamblin et al., 1997a;Hamblin et al., 1997b). It is intriguing to think that the requirement for two CheW proteins in B. burgdorferi may have to do with the extra task of coordinating flagellar reversals at the two ends of an elongated cell body.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
High-passage, avirulent Borrelia burgdorferi sensu stricto strain B31A (wild type) (Bono et al., 2000) and its derivative mutants were grown in BSK-II liquid medium or on semi-solid agar plates at 34°Cin a humidified incubator in the presence of 3.4% CO2, as previously documented (Li et al., 2002). The E. coli strains were grown in LB medium at 37°C with appropriate antibiotics.
Construction of cheW mutants
The cheW1, cheW2 and cheW3 genes were inactivated by allelic exchange mutagenesis as illustrated in Figure S6. To construct the vector for inactivation of cheW1 (gene locus bb0312; gene length, 531 bp), a 120 bp HindIII fragment was deleted and replaced by a kanamycin-resistance cassette (aphI) (Elias et al., 2003). To construct the vector for inactivation of cheW2 (bb0565; gene length, 543 bp), the aphI cassette was directly inserted into an EcoRV restriction cut site within the gene. To construct the vector for inactivation of cheW3 (bb0670; gene length, 1,401 bp), the entire open reading frame (orf) was deleted and replaced with a promoterless streptomycin resistance marker (aadA1), as recently described (Frank et al., 2003;Motaleb et al., 2011a). The resultant constructs were designated as W1::aphI, W2::aphI, and W3::aadA1 (Figure S6), respectively. The PCR primers for constructing these vectors are listed in Table S1. To knock out the cheW genes, these vectors were first linearized and then separately transformed into B31A competent cells via electroporation as previously reported (Samuels, 1995). Transformants were selected on BSK-II agar plates containing 350 μg/ml kanamycin (for W1::aphI and W2::aphI) or 50 μg/ml streptomycin (for W3::aadA1).
Constructing genetic complementation vectors
To construct the vector for the complementation of the cheW3 mutant, the entire cheW3 gene and its native promoter (Pami) (Yang and Li, 2009) were first amplified by PCR with two pairs of primers (P17/P18 for Pami; P19/P20 for cheW3). The resultant PCR products were then fused together via PCR using primers P17/P20. The resultant PamicheW3 fragment was first cloned into the pGEM®-T Easy vector (Promega, Madison, WI) and then subcloned into pBSV2G, a shuttle vector of B. burgdorferi that contains a gentamicin-resistance cassette (aacC1) (Elias et al., 2003;Rosa et al., 2005). The final construct was named CheW3/pBSV2G (Figure S6). A similar strategy was used to construct a vector for complementation of the cheW1 mutant (CheW1/pBSV2G) and the vector for complementation of the cheW3 mutant (CheW3N+/pBSV2G) with the N-terminal domain of CheW3 (1–210 amino acids). The PCR primers for constructing the complementation vectors are listed in Table S1.
Generation of polyclonal antisera against CheW1, CheW2 or CheW3
The entire orfs (without the translation initiation ATG/GTG codon) of cheW1, cheW2 and cheW3 were amplified by PCR (the primers are listed in Table S1). The obtained PCR products were first cloned into the pGEM®-T Easy vector (Promega), and then subcloned into the pQE30 expression vector (Qiagen, Valencia, CA), which encodes an amino-terminal histidine tag. The expression of these three genes was induced using 1 mM isopropyl-β-D-thiogalactoside (IPTG). The recombinant proteins were purified by a nickel agarose column and concentrated in 10 kDa molecular weight cut off Amicon Ultra centrifugal concentrators (Millipore, Billerica, MA). Rats (for rCheW1 and rCheW2) and rabbits (for rCheW3) were immunized with 1 to 5 mg of purified recombinant proteins during a one-month period using standard methods. The obtained polyclonal antisera were further purified using affinity chromatography with the AminoLink Plus Immobilization Kit (Thermo Scientific, Rockford, IL) and eluted as recommended by the manufacturer.
Bacterial motion tracking analysis, swim plate, and capillary assays
The swimming velocity of B. burgdorferi cells was measured using a computer-based motion tracking system. Swim plate assays were carried out using 0.35% agarose with BSK-IImedium diluted 1:10 with Dulbecco’s phosphate-buffered saline (DPBS, pH 7.5) without divalent cations, as previously documented (Motaleb et al., 2000;Li et al., 2002). The plates were incubated for 3–4 days at 34°C in the presence of 3.4% CO2. Diameters of the swim rings that appeared on the plates were measured and recorded in millimeters (mm). A previously constructed non-motile flaB− mutant (ΔflaB) (Motaleb et al., 2000) was used as a negative control to determine the initial inoculum size. Capillary assays were carried out as previously documented with minor modifications (Li et al., 2002;Bakker et al., 2007). Briefly, B. burgdorferi cells were grown to late-logarithmic-phase (~5–7 × 107 cells/ml) and harvested by low-speed centrifugations (1,800 × g). The harvested cells were then resuspended in the motility buffer (Bakker et al., 2007). Capillary tubes filled with either the attractant (0.1 M N-acetyl-glucosamine [GlcNAc] dissolved in the motility buffer) or only motility buffer (negative control) were sealed and inserted into microcentrifuge tubes containing 200 μl of resuspended cells (7 × 108 cells/ml). After 2 hrs incubation at 34°C in a humidified chamber, the solutions were expelled from the capillary tubes, and the spirochete cells were enumerated using Petroff-Hausser counting chambers under a dark-field microscope. A positive chemotactic response was defined as at least twice as many cells entering the attractant-filled tubes as the buffer-filled tubes. For the swim plate, motion tracking, and capillary assays, results are expressed as means ± standard errors of the means (SEM). The significance of the difference between different strains was evaluated with an unpaired Student t test (P value < 0.01).
Electrophoresis and immunoblot analyses
Sodium-dodecyl-sulfate polyacrylamide-gel electrophoresis (SDS-PAGE) and immunoblotting using the enhanced chemiluminescent detectionsystem were carried out as described before (Li et al., 2010;Sze et al., 2011). B. burgdorferi cells were grown at 34°C and harvested at early stationary phase (approximately108 cells/ml). The whole cell lysates were prepared by washingcells once in PBS buffer (phosphate-buffered saline, pH 7.5) and then boiling for 5 min in Laemmlisample buffer. The same amount of cell lysates (~10–20 μg) were separated on SDS-PAGE gels and transferred to PVDF membrane (Bio-Rad Laboratories, Hercules, CA). The immunoblots were probed with specific antibodies against various proteins (CheA1, CheA2, CheW1, CheW2, and CheW3) and developed using horseradish peroxidase-coupled secondary antibody with an ECL luminol assay.
Co-IP assay
The co-IP assay was carried out as previously described (Motaleb et al., 2004). Briefly, 200 ml of the late-logarithmic-phase (~5–7 × 107 cells/ml) B. burgdorferi cultures were harvested by centrifugation and washed twice with PBS buffer containing 5 mM MgCl2. The resultant cell pellets were resuspended in TSEA buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.05% sodium azide, pH 7.5) containing Nonidet P-40 (1%, v/v) and phenylmethylsulfonyl fluoride (50 μg/ml) and then incubated at 37°Cfor 1 hr. After the incubation, the obtained samples were centrifuged (1,600 × g for 30 min, 25°C). The resultant cell pellets were resuspended in the PBS buffer and French pressed followed by centrifugation (15,000 × g for 30 min, 25°C). Approximately 200 μl of the obtained supernatants were incubated with 50 μl of the polyclonal anti-CheAs (αCheA1 and αCheA2) or anti-CheWs (αCheW1, αCheW2, and αCheW3) for 1 hr at 25°C in the presence of 1% bovine serum albumin (BSA). After the incubation, 50 μl of protein A (Calbiochem-Behring Corporation, La Jolla, CA) was added to each sample and further incubated at 25°C for 1 hr. The immunoprecipitates and controls were centrifuged at 1,600 × g at 25°C and washed three times with 1 ml of TSEA buffer containing 0.05% Tween-20. The final pellets were suspended in 100 μl of electrophoresis sample buffer, boiled for 5 min, and briefly centrifuged. For the immunoblots, 10 μl of the supernatants was applied to each lane of SDS-PAGE gels as described above.
IFA and cryo-ET
IFA and cryo-ET assays were carried out to determine the cellular locations of MCPs in B31A and the three cheW mutants as previously described (Xu et al., 2011). For the IFA, αMCP3, a specific antibody against B. burgdorferi MCP3, was used. For the cryo-ET analysis, freshly prepared B. burgdorferi cultures were deposited onto a glow-discharged holey carbon EM grid, blotted, and rapidly frozen in liquid ethane. The frozen-hydrated specimens were imaged at −170°C using a Polara G2 electron microscope (FEI Company, Hillsboro, Oregon) equipped with a field emission gun and a 4K × 4K CCD camera (TVIPS; GMBH, Germany). The microscope was operated at 300 kV with a magnification of 31,000×. Low-dose single-axis tilt series were collected from each bacterium at −6 μm defocus with a cumulative dose of ~100 e−/Å2 distributed over 65 images with an angular increment of 2°, covering a range from −64° to +64°. The tilt series images were aligned and reconstructed using the IMOD software package (Kremer et al., 1996). In total, cryo tomograms of B31A (30 cells), a cheW1 mutant (31 cells) and its complemented strain (20 cells), a cheW2 mutant (30 cells), a cheW3 mutant (25 cells) and its complemented strain (30 cells) were reconstructed and visualized using IMOD (Kremer et al., 1996).
Homology model construction of CheW1, CheW2, and CheW3
The NMR structure of T. maritima CheW (Protein Data Bank ID: 1K0S) (Griswold and Dahlquist, 2002) was selected as a template for the homology modeling analysis of CheW1, CheW2, and N-terminus CheW like domain of CheW3. Pairwise sequence alignment of CheW homologues was conducted using Clustal X. Automodel module in Modeller 9v7 (Sali and Blundell, 1993) was applied to obtain the final refined structures. All structures were analyzed and visualized in PyMol (The PyMol Molecular Graphic System, Version 1.5.0.3, Schrodinger, LLC). The qualities of the models were evaluated by PDBsum (Laskowski, 2001).
Supplementary Material
Acknowledgments
This research was supported by Public Health Service grants (AI073354 and AI078958) to C. Li, GM072004 to C. Wolgemuth; J. Liu was supported in part by grants AI087946 from the National Institute of Allergy and Infectious Diseases (NIAID) and AU-1714 from the Welch Foundation.
Reference List
- Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 2010;18:494–503. doi: 10.1016/j.tim.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre G, Zhulin IB. Different evolutionary constraints on chemotaxis proteins CheW and CheY revealed by heterologous expression studies and protein sequence analysis. J Bacteriol. 2003;185:544–552. doi: 10.1128/JB.185.2.544-552.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker RG, Li C, Miller MR, Cunningham C, Charon NW. Identification of specific chemoattractants and genetic complementation of a Borrelia burgdorferi chemotaxis mutant: flow cytometry-based capillary tube chemotaxis assay. Appl Environ Microbiol. 2007;73:1180–1188. doi: 10.1128/AEM.01913-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- Bono JL, Elias AF, Kupko JD, III, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhvalova M, VanBruggen R, Stewart RC. CheA kinase and chemoreceptor interaction surfaces on CheW. J Biol Chem. 2002a;277:23596–23603. doi: 10.1074/jbc.M202288200. [DOI] [PubMed] [Google Scholar]
- Boukhvalova MS, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar. Importance for chemotaxis signaling in Escherichia coli. J Biol Chem. 2002b;277:22251–22259. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease, a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Butler SM, Camilli A. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat Rev Microbiol. 2005;3:611–620. doi: 10.1038/nrmicro1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SM, Nelson EJ, Chowdhury N, Faruque SM, Calderwood SB, Camilli A. Cholera stool bacteria repress chemotaxis to increase infectivity. Mol Microbiol. 2006;60:417–426. doi: 10.1111/j.1365-2958.2006.05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo MJ, Massazza DA, Parkinson JS, Studdert CA. Disruption of chemoreceptor signalling arrays by high levels of CheW, the receptor-kinase coupling protein. Mol Microbiol. 2010;75:1171–1181. doi: 10.1111/j.1365-2958.2009.07032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Cockburn Andrew, Li Chunhao, Liu Jun, Miller Kelly, Miller Michael R, et al. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Immunol. 2012 doi: 10.1146/annurev-micro-092611-150145. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spriochetes. Annu Rev Genet. 2002;36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, et al. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol. 2009;191:600–607. doi: 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nature Struct Biol. 1998;5:446–450. doi: 10.1038/nsb0698-446. [DOI] [PubMed] [Google Scholar]
- Dombrowski C, Kan W, Motaleb MA, Charon NW, Goldstein RE, Wolgemuth CW. The elastic basis for the shape of Borrelia burgdorferi. Biophys J. 2009;96:4409–4417. doi: 10.1016/j.bpj.2009.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias AF, Bono JL, Kupko JJ, III, Stewart PE, Krum JG, Rosa PA. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J Mol Microbiol Biotechnol. 2003;6:29–40. doi: 10.1159/000073406. [DOI] [PubMed] [Google Scholar]
- Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Norris SJ, Weinstock CM, White O, Sutton GG, Dodson R, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- Ge Y, Charon NW. Molecular characterization of a flagellar/chemotaxis operon in the spirochete Borrelia burgdorferi. FEMS Microbiol Lett. 1997;153:425–431. doi: 10.1111/j.1574-6968.1997.tb12606.x. [DOI] [PubMed] [Google Scholar]
- Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- Goldstein SF, Charon NW, Kreiling JA. Borrelia burgdorferi swims with a planar waveform similar to that of eukaryotic flagella. Proc Natl Acad Sci U S A. 1994;91:3433–3437. doi: 10.1073/pnas.91.8.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold IJ, Dahlquist FW. The dynamic behavior of CheW from Thermotoga maritima in solution, as determined by nuclear magnetic resonance: implications for potential protein-protein interaction sites. Biophys Chem. 2002;101–102:359–373. doi: 10.1016/s0301-4622(02)00157-6. [DOI] [PubMed] [Google Scholar]
- Hamblin PA, Bourne NA, Armitage JP. Characterization of the chemotaxis protein CheW from Rhodobacter sphaeroides and its effect on the behaviour of Escherichia coli. Mol Microbiol. 1997a;24:41–51. doi: 10.1046/j.1365-2958.1997.3241682.x. [DOI] [PubMed] [Google Scholar]
- Hamblin PA, Maguire BA, Grishanin RN, Armitage JP. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol Microbiol. 1997b;26:1083–1096. doi: 10.1046/j.1365-2958.1997.6502022.x. [DOI] [PubMed] [Google Scholar]
- Harman MW, Dunham-Ems SM, Caimano MJ, Belperron AA, Bockenstedt LK, Fu HC, et al. The heterogeneous motility of the Lyme disease spirochete in gelatin mimics dissemination through tissue. Proc Natl Acad Sci U S A. 2012;109:3059–3064. doi: 10.1073/pnas.1114362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Bakker RG, Motaleb MA, Sartakova ML, Cabello FC, Charon NW. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc Natl Acad Sci U S A. 2002;99:6169–6174. doi: 10.1073/pnas.092010499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu H, Zhang K, Liang FT. Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol Microbiol. 2010;75:1563–1576. doi: 10.1111/j.1365-2958.2010.07078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hu Y, Fu W, Xia B, Jin C. Solution structure of the bacterial chemotaxis adaptor protein CheW from Escherichia coli. Biochem Biophys Res Commun. 2007;360:863–867. doi: 10.1016/j.bbrc.2007.06.146. [DOI] [PubMed] [Google Scholar]
- Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A. 2012 May 3; doi: 10.1073/pnas.1200781109. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Martin AC, Wadhams GH, Armitage JP. The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol Microbiol. 2001;40:1261–1272. doi: 10.1046/j.1365-2958.2001.02468.x. [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci U S A. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Miller MR, Li C, Bakker RG, Goldstein SF, Silversmith RE, et al. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J Bacteriol. 2005;187:7963–7969. doi: 10.1128/JB.187.23.7963-7969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Pitzer JE, Sultan SZ, Liu J. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol. 2011a;193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Sal MS, Charon NW. The decrease in FlaA observed in a flaB mutant of Borrelia burgdorferi occurs posttranscriptionally. J Bacteriol. 2004;186:3703–3711. doi: 10.1128/JB.186.12.3703-3711.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motaleb MA, Sultan SZ, Miller MR, Li C, Charon NW. CheY3 of Borrelia burgdorferi is the key response regulator essential for chemotaxis and forms a long-lived phosphorylated intermediate. J Bacteriol. 2011b;193:3332–3341. doi: 10.1128/JB.00362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- Parkinson JS. Behavioral genetics in bacteria. Annu Rev Genet. 1977;11:397–414. doi: 10.1146/annurev.ge.11.120177.002145. [DOI] [PubMed] [Google Scholar]
- Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol. 2011;9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- Radolf JD, Caimano MJ, Stevenson B, Hu LT. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol. 2012;10:87–99. doi: 10.1038/nrmicro2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao CV, Glekas GD, Ordal GW. The three adaptation systems of Bacillus subtilis chemotaxis. Trends Microbiol. 2008;16:480–487. doi: 10.1016/j.tim.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa PA, Tilly K, Stewart PE. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat Rev Microbiol. 2005;3:129–143. doi: 10.1038/nrmicro1086. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Samuels DS. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol. 1995;47:253–259. doi: 10.1385/0-89603-310-4:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels DS. Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol. 2011;65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- Sarkar MK, Paul K, Blair D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:9370–9375. doi: 10.1073/pnas.1000935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, et al. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A. 2004;101:5646–5651. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CM, Chao LL, Yu CP. Chemotactic migration of the Lyme disease spirochete (Borrelia burgdorferi) to salivary gland extracts of vector ticks. Am J Trop Med Hyg. 2002;66:616–621. doi: 10.4269/ajtmh.2002.66.616. [DOI] [PubMed] [Google Scholar]
- Shiomi D, Zhulin IB, Homma M, Kawagishi I. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J Biol Chem. 2002;277:42325–42333. doi: 10.1074/jbc.M202001200. [DOI] [PubMed] [Google Scholar]
- Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J Clin Invest. 2004;113:1093–1101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc Natl Acad Sci U S A. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Morado DR, Liu J, Charon NW, Xu H, Li C. Carbon storage regulator A (CsrA(Bb)) is a repressor of Borrelia burgdorferi flagellin protein FlaB. Mol Microbiol. 2011;82:851–864. doi: 10.1111/j.1365-2958.2011.07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze CW, Zhang K, Kariu T, Pal U, Li C. Borrelia burgdorferi needs chemotaxis to establish infection in mammals and to accomplish its enzootic cycle. Infect Immun. 2012 Apr 16; doi: 10.1128/IAI.00145-12. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu A, Wang X, Zhou H, Dahlquist FW. The receptor-CheW binding interface in bacterial chemotaxis. J Mol Biol. 2012;415:759–767. doi: 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Xu H, Raddi G, Liu J, Charon NW, Li C. Chemoreceptors and flagellar motors are subterminally located in close proximity at the two cell poles in spirochetes. J Bacteriol. 2011;193:2652–2656. doi: 10.1128/JB.01530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Huber G, Wolgemuth CW. Forces and torques on rotating spirochete flagella. Phys Rev Lett. 2011;107:268101. doi: 10.1103/PhysRevLett.107.268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li C. Transcription and genetic analyses of a putative N-acetylmuramyl-L-alanine amidase in Borrelia burgdorferi. FEMS Microbiol Lett. 2009;290:164–173. doi: 10.1111/j.1574-6968.2008.01416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Bos E, Heymann J, Gnaegi H, Kessel M, Peters PJ, Subramaniam S. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J Microsc. 2004;216:76–83. doi: 10.1111/j.0022-2720.2004.01395.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.