Abstract
Xylella fastidiosa, which causes Pierce's disease of grapevine and other important plant diseases, is a xylem-limited bacterium that depends on insect vectors for transmission. Although many studies have addressed disease symptom development and transmission of the pathogen by vectors, little is known about the bacterial mechanisms driving these processes. Recently available X. fastidiosa genomic sequences and molecular tools have provided new routes for investigation. Here, we show that a diffusible signal molecule is required for biofilm formation in the vector and for vector transmission to plants. We constructed strains of X. fastidiosa mutated in the rpfF gene and determined that they are unable to produce the signal activity. In addition, rpfF mutants are more virulent than the wild type when mechanically inoculated into plants. This signal therefore directs interaction of X. fastidiosa with both its insect vector and plant host. Interestingly, rpfF mutants can still form in planta biofilms, which differ architecturally from biofilms in insects, suggesting that biofilm architecture, rather than a passive response to the environment, is actively determined by X. fastidiosa gene expression. This article reports a cell-cell signaling requirement for vector transmission. Identification of the genes regulated by rpfF should elucidate bacterial factors involved in transmission and biofilm formation in the insect.
The Gram-negative bacterium Xylella fastidiosa colonizes the xylem, a water transport network of interconnected vessels composed of dead cells, forming an aggregated biofilm inside the plant. Vessels can become occluded by dense colonization, and high frequencies of blocked vessels are associated with disease symptom development (1-3). X. fastidiosa is responsible for diseases that cause economic loss in many agricultural plants; however, it can also live in symptomless hosts that serve as a source of inoculum (4). In susceptible host plants, X. fastidiosa spreads throughout the xylem network from the site of inoculation. Although X. fastidiosa cells likely move along individual xylem vessels with the sap flow, it is currently thought that X. fastidiosa moves from one vessel to another through the bordered pits (intervessel channels) only after degrading the membranes that guard them (1, 5). Xylem sap-feeding insects can acquire and transmit X. fastidiosa, which forms a biofilm of polarly attached cells inside their foreguts (6, 7). Once infected with X. fastidiosa, insects remain infective with the pathogen, which multiplies in the foregut (8, 9). X. fastidiosa depends on interactions with both of these organisms for survival. Cell-cell communication among bacteria is a crucial component of many pathogenic or symbiotic interactions with plant and animal hosts (10) but has never been linked to bacterial interactions with insect vectors. Moreover, with notable exceptions (11, 12), bacterial genes required for vector colonization have not been identified in most interactions.
The plant pathogen Xanthomonas campestris pv. campestris (Xcc), which is closely related to X. fastidiosa (13) but is not insect vectored, requires rpfF for synthesis of a diffusible signaling factor (DSF) (14). rpfF encodes a protein similar to enoyl-CoA hydratases that synthesizes DSF (14), an α,β unsaturated fatty acid (15). Other Rpf proteins sense DSF and transduce the signal, leading to transcriptional regulation of genes required for pathogenic traits, such as exopolysaccharide and exoenzyme biosynthesis (16). The predicted RpfF proteins of Xcc and X. fastidiosa share 66% identical amino acid sequence, suggesting conserved functions. Indeed, X. fastidiosa produces a diffusible signal that is recognized by Xcc (17). These data, together with the presence of the other key rpf genes in the X. fastidiosa genome, established evidence for an X. fastidiosa cell-cell signaling system and suggested that it might regulate expression of traits required for interactions with the plant and insect. In this work, we sought to understand what role cell-cell signaling plays in interactions of X. fastidiosa with an insect vector and a plant host.
Materials and Methods
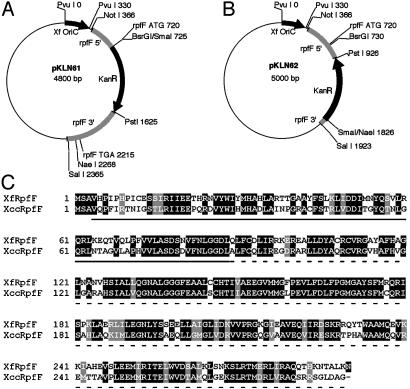
X. fastidiosa rpfF Mutants. Allelic exchange plasmids pKLN61 and pKLN62 (Fig. 1) were constructed as follows. A 1.4-kb region of the X. fastidiosa Temecula (ATCC 700964) genome including most of the rpfF coding sequence was amplified by using primers “rpfFKOFNotI” (5′-AGCGGCCGCGATGACGACGCGATACGGAAGT-3′) and “rpfFKORSalI” (5′-CGCGTCGACTGGCAGTTGCAATTGGAGTGGTG-3′), digested with NotI and SalI and ligated into the same sites of pKLN2 (1) to make pKLN60. The Tn903 kanamycin resistance gene (KanR) was digested from pKLN56 (1) with PstI and SmaI and ligated into pKLN60 cut with BsrGI, blunted, then cut with PstI, to make pKLN61, or ligated into pKLN60 cut with PstI, partially cut with NaeI and gel purified away from a 680-bp fragment containing most of the rpfF coding sequence to make pKLN62. Amplifications were performed with Pfu polymerase (Stratagene). Insert and junction sequences of all plasmids were determined. pKLN61 and pKLN62 were electroporated into X. fastidiosa, and transformants were selected as described (1). Disruption of the rpfF locus was confirmed by PCR and Southern blotting (data not shown) to yield rpfF mutant strains KLN61 and KLN62.
Fig. 1.
(A and B) pKLN61 and pKLN62 plasmids used to create rpfF mutant strains KLN61 and KLN62, respectively. The vector backbones are pGEM-5Zf(+) (Promega). The gray-shaded region represents the rpfF coding sequence and flanking DNA (start and stop codons are indicated, if present). Regions of the coding sequence were removed before insertion of the KanR gene. Double recombination with the X. fastidiosa genome occurs in the gray-shaded regions flanking the KanR gene, resulting in exchange of the wild-type allele for the deletion-harboring KanR allele. No genes are cotranscribed with rpfF in X. fastidiosa; therefore, disruption of expression of genes other than rpfF in strains KLN61 and KLN62 is highly unlikely. We note that homologous DNA in allelic exchange construct pKLN62 was only 97 bp long on one end, indicating that very short stretches of sequence are sufficient for homologous recombination in this organism. (C) An alignment of the predicted RpfF proteins from X. fastidiosa and Xcc. The sequences are 290 aa in length and are 66% identical and 80% similar overall. The coding sequence for the solid underlined region is deleted in strain KLN61 (codons 4-89). The coding sequence for the dashed underlined region is deleted in strain KLN62 (codons 89-290). The sequence encoding amino acid 99, a glutamine, is deleted in both strains. Sequences were aligned by using clustalw 1.8 (and presented in boxshade 3.21).
DSF Reporter. The DSF-inducible promoter region of engXCA (16) from Xcc strain 8004 (18) genomic DNA was amplified with primers “XccengpFEcoR I” (5′-GGA AT TCCGATCACAAACGACGCGA-3′) and “XccengpRBamHI” (5′-CGGGATCCCATGGTGATCTCCCTAGA-3′), cut with EcoRI and BamHI and ligated into those sites of pPROBE-KT′ (19), which are upstream of a promoterless gfp gene, to make pKLN51. The omega cassette conferring spectinomycin and streptomycin resistance was cut from plasmid pUC1318Ω (19) with BamHI and ligated into the pKLN51 BglII site to make pKLN55. pKLN55 was mated into rpfF mutant strain 8523 (20) of Xcc, which is unable to synthesize DSF (14) and selected on King's B agar with 100 μg/ml rifampicin, 100 μg/ml spectinomycin, and 50 μg/ml streptomycin.
DSF Assays. X. fastidiosa strains were grown to OD600 0.045 (stationary phase) in 50 ml of periwinkle wilt medium (PW) (21) at 28°, shaking at 160 rpm, then 50 ml of water-saturated ethyl acetate was added to the culture supernatant, vortexed, and centrifuged. The ethyl acetate phase was concentrated by evaporation to 20 μl and spotted onto a paper disk, which was placed on a King's B agar (KB) plate. Ten microliters of an OD600 0.25 suspension of 8523 (pKLN55) was pipetted near the disk, and plates were incubated for 48 h. Xcc strains were streaked on KB plates, incubated 1 day at 28°C. Resulting colonies were oversprayed with an OD600 0.25 suspension of 8523 (pKLN55) by using an airbrush and incubated 2 additional days. In these assays, signal diffuses from the sample to the reporter, resulting in green fluorescence. Fluorescence was viewed on a Zeiss SV11 stereoscope with Kramer epifluorescence/Optronix Color DEI450.
Xcc rpfF Rescue. rpfF was amplified from the Xcc and X. fastidiosa genomes by using primers “XccrpfFFXhoI” (5′-ACTCGAGATAAGGAGGAAAAACATATGTCTGCAGTTCAACCCTTC-3′) and “XccrpfFRSpeI” (5′-GACTAGTCAGCCCGCGTCGAGCCCTGAGCGACGCGACT-3′), or “XfrpfFFXhoI” (5′-ACTCGAGATAAGGAGGAAAAACATATGTCCGCTGTACATCCCATT-3′) and “XfrpfFRSpeI” (5′-GACTAGTCAGTTTTTTAGTGCTGTGTTTTTGTGAGTCT-3′), respectively, and ligated into pGEM-T (Promega) to make pXccrpfF-T and pXfrpfF-T, respectively. rpfF was cut from pXccrpfF-T and pXfrpfF-T with XhoI and SpeI and ligated into the same sites of pBBR1MCS-3 (22) to make pKLN69 and pKLN70, respectively. pKLN69, pKLN70, and pBBR1MCS-3 were mated into Xcc strains 8004 (18) and 8523 (20).
Transmission Tests. Blue-green sharpshooter leafhoppers, Graphocephala atropunctata (Signoret) (Hemiptera: Cicadellidae) were maintained and used in transmission tests as described (1). Adults were caged on symptomatic grapevines infected with Temecula, KLN61, or mock inoculated for a 4-day acquisition access period. Each insect was then transferred to a healthy grapevine for a 7-day inoculation access period. Grapevines were observed for symptoms in the greenhouse for 6 months. X. fastidiosa infection was verified by culturing bacteria from plant macerates (23), then genotyping bacteria by PCR with primers “rpfFKOFNotI” and “rpfFRBglII” (5′-GGAAGATCTCAGCACAGCTTTAAGTGCTCAGTT-3′).
Detection of X. fastidiosa from Insects. Forty-one insects for each treatment (Temecula, KLN61, and buffer) were exposed to infected plants for 4 days. Then, 19 were killed for bacteriological culture, and 22 were moved to uninfected grapevines for 7 days before culturing. For culturing, insect heads were severed, surface sterilized, macerated in 100 μl of buffer, and dilution plated onto periwinkle wilt medium with gellan gum (PWG) (23). We calculated our limit of detection to be 25 colony-forming units per head. For statistical analysis, we analyzed six different 2 × 2 contingency tables representing all treatment combinations to determine significant differences among treatments. We used a Bonferroni correction method, α′ = α/k (24) to determine the correct α′(0.0083), where α = 0.05 and k = 6. Excrement was collected from leafhoppers during the final day of a 4-day access period on an infected grapevine, and bacterial cells were pelleted, resuspended in buffer, heated to 95°C, and used as templates in PCR with the X. fastidiosa-specific primers RST31 and RST33 (25).
Pathogenicity Assays. Greenhouse-grown Vitis vinifera “Cabernet sauvignon” grapevines were mechanically inoculated by using standard procedures (26). The number of abscised leaf blades from the first 20 nodes distal to the inoculation site was noted. Strains infecting plants were verified by culturing and PCR-based genotyping as above.
Scanning Electron Microscopy. Grapevine petiole segments and leafhopper heads were fixed and dried according to the University of California Electron Microscope Lab protocol at biology.berkeley.edu/EML/psem.html. Samples were dissected, coated with 13 nm of gold/palladium, and viewed on an ISI DS-130 (Fig. 4) or 3 nm of platinum and 13 nm of carbon and viewed on a Hitachi S-5000 (Fig. 6) scanning electron microscope at the University of California Electron Microscope Lab.
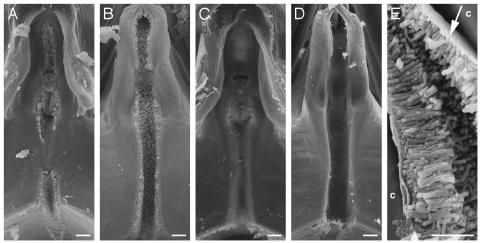
Fig. 4.
Formation of polar biofilm in insect foreguts. Scanning electron micrographs of the precibarium epipharynx (A and C) and hypopharynx (B and D) of blue-green sharpshooter leafhoppers fed on grapevines infected with the wild-type strain Temecula (A, B, and E)orthe rpfF mutant KLN61 (C and D). The stylets, which are inserted into the plant, are located above, and the cibarium (pumping chamber) is located below, the frame of the images. Xylem sap enters the precibarium from the top and runs through the canal, which is coated with a biofilm by wild-type cells (A and B) but not rpfF mutant cells (C and D). Small bits of debris are present; however, these objects were each examined at high magnification and do not represent bacterial cells, which are characteristically rod-shaped. (E) High magnification of polar biofilm that has slightly detached from cuticle (c) during fixation, revealing a mat-like structure at the attachment site (arrow). Rod-shaped structures are bacterial cells. [Bar = 10 μm (A-D) and 5 μm (E).]
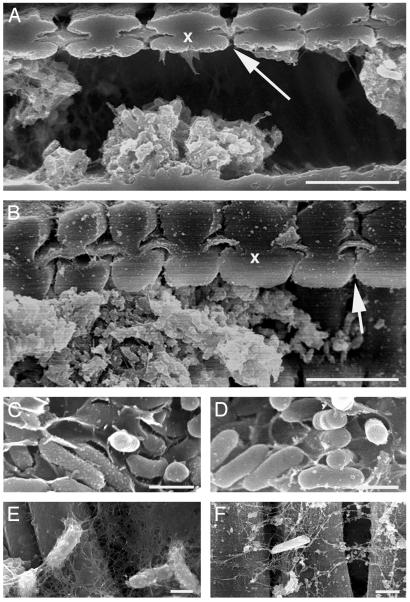
Fig. 6.
Scanning electron micrographs of cells and associated exopolysaccharide from the wild-type strain Temecula (A, C, and E), and the rpfF mutant KLN61 (B, D, and F) in the xylem of grapevine petioles from symptomatic leaves. (A and B) Wild-type and rpfF communities were similar (x, two adjacent xylem vessel walls; arrow, bordered pit with pit membrane). Cells in crowded vessels were embedded in a matrix (C and D) whereas individual cells were covered with strands of potentially the same material (E and F).
Results
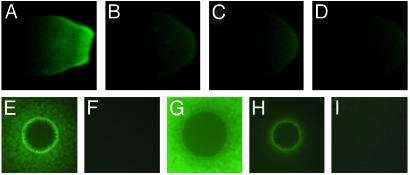
X. fastidiosa Requires rpfF to Make a Diffusible Signal Factor. We constructed X. fastidiosa strains impaired in signaling by deleting rpfF via allelic exchange mutagenesis (Fig. 1). Two independent rpfF deletion mutants were constructed, KLN61 and KLN62, linking mutant phenotypes definitively to this locus. To detect signal produced by X. fastidiosa strains, we developed a DSF reporter strain of Xcc, 8523 (pKLN55), which expresses GFP in the presence of DSF. This reporter expressed GFP in response to ethyl acetate extracts of an X. fastidiosa culture (Fig. 2A) but not sterile media (Fig. 2B), demonstrating that a DSF-like signal was extracted from the X. fastidiosa culture. No signal was detected in extracts of the rpfF mutant strains KLN61 or KLN62 (Fig. 2 C and D), indicating that rpfF is required for signal production in X. fastidiosa.
Fig. 2.
Detection of DSF from X. fastidiosa and Xcc strains. (A-D) DSF reporter strain 8523 (pKLN55) grown to the left of concentrated X. fastidiosa culture extracts of the wild-type strain Temecula (A), sterile medium (B), or rpfF mutant strains KLN61 (C) and KLN62 (D). (E-I) Strain 8523 (pKLN55) sprayed over colonies of Xcc strain 8004 (wild-type, E), strain 8523 (rpfF, F), Xcc rpfF rescue strain 8523 (pKLN69) (G), X. fastidiosa rpfF rescue strain 8523 (pKLN70) (H), empty vector control strain 8523 (pBBR1MCS-3) (I). Green fluorescence indicates detection of DSF that has diffused from the culture extract or colony.
To confirm that the X. fastidiosa RpfF protein functions as a DSF synthase, we rescued Xcc rpfF mutant strain 8523 (20) with X. fastidiosa rpfF (Fig. 2 E-I). X. fastidiosa rpfF conferred DSF production to strain 8523 (Fig. 2 F and H), although less efficiently than did Xcc rpfF (Fig. 2 F and G). The reason that rescue was less efficient with the X. fastidiosa gene than with the Xcc gene is unclear. We speculate that X. fastidiosa RpfF may have lower activity than Xcc RpfF in Xcc cells. Alternatively, the X. fastidiosa RpfF may synthesize a somewhat different signal molecule that is less active in Xcc. The Xcc rpfF gene in the rescued Xcc rpfF mutant strain produced higher levels of DSF than did the wild-type strain (Fig. 2 E and G. This finding might be expected due to an increase in copy number and transcriptional activation of the Xcc rpfF gene in the rescued strain compared with the wild type.
rpfF Mutants Are Defective in Insect Transmissibility. We investigated whether the signaling mutants were transmissible by an efficient insect vector, the blue-green sharpshooter leafhopper. Because strains KLN61 and KLN62 were equally impaired in signal production, we chose strain KLN61 for this analysis. We used a protocol that maximized insect acquisition and transmission opportunities (see Materials and Methods). After inoculation by leafhoppers, Temecula-infected grapevines had multiple symptomatic leaves by an average of 57 ± 6 SE days, whereas KLN61-infected plants were symptomless for 6 months after inoculation. Strain KLN61 was never recovered from these insect-inoculated plants whereas Temecula was always recovered (n = 8 replicates). This experiment was repeated twice with similar results (n = 11 and n = 17). The average transmission efficiency for these experiments was 0.72 ± 0.15 SE for Temecula compared with only 0.03 ± 0.03 SE for KLN61 (one case of transmission). Therefore, the rpfF mutant is highly impaired in transmissibility compared with the wild type.
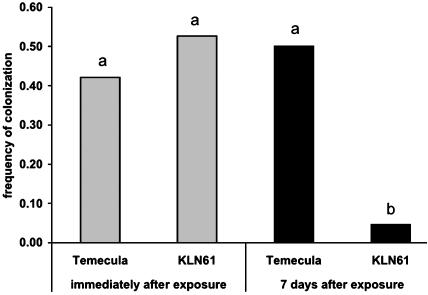
Colonization of Insects Is Impaired in rpfF mutants. Transmission of X. fastidiosa by the insect depends on uptake of bacteria by the insect during feeding. To investigate the ability of the insect to ingest KLN61 from infected source plants, we collected excrement produced during feeding on grapevines mechanically infected with either Temecula or KLN61, or mock inoculated. Both Temecula and KLN61 were detected in excrement, confirming that insects ingested both wild-type and rpfF bacteria during feeding although the frequency of detection of bacteria in excrement was extremely low [only 1 (KLN61) or 2 (Temecula) positive samples were obtained from 10 insects per treatment]. Immediately after removal from infected plants, we recovered Temecula from 8/19 (42.1%) and KLN61 from 10/19 (52.6%) leafhoppers (Fig. 3), indicating that both strains are inside leafhoppers at similar frequencies during exposure to an infected plant. When leafhoppers were then transferred to uninfected grapevines for 7 days, there was no change in the total proportion of leafhoppers retaining Temecula [11/22 (50%); χ2 = 0.26, df = 1, P = 0.6]. However, KLN61 was retained by only 1/22 leafhoppers (4.5%), revealing a significant defect in KLN61 retention by leafhoppers (Fig. 3; χ2 = 9.34, df = 1, P = 0.0005). No bacteria were recovered from insects or excrement when fed on mock-inoculated grapevines. These results suggest that the rpfF mutant is poorly transmitted because it is not retained by leafhoppers as efficiently as the wild type, as must be required for efficient vector transmission.
Fig. 3.
Colonization of leafhoppers by wild-type and rpfF mutant strains. X. fastidiosa was recovered by maceration and culturing of one set of insects directly after removal from infected plants (gray columns). For a second set of insects in the same experiment, X. fastidiosa was recovered 7 days after removal from infected plants (filled columns). Columns with the same letter are not statistically different (see Materials and Methods).
rpfF Mutants Cannot Form Biofilms in Insects. We examined leafhopper foreguts 7 days after transfer from infected to uninfected grapevines by using scanning electron microscopy. All insects fed on Temecula-infected plants (n = 8) displayed characteristic bacterial colonization throughout the precibarium, a section of the foregut, with cells attached in a polar biofilm in both the epipharynx (Fig. 4 A and E; dorsal) and hypopharynx (Fig. 4B; ventral). Although only a small number of X. fastidiosa cells are required for efficient transmission (27), we did not find any bacteria within the precibarium of insects fed on KLN61-infected plants (n = 12; Fig. 4 C and D). We conclude that polar biofilm formation in the precibarium requires rpfF, which may regulate genes involved in initial attachment or proper formation of the biofilm.
rpfF Mutants Are Hypervirulent in Plants. We compared development of Pierce's disease symptoms in grapevines mechanically inoculated with rpfF and wild-type bacteria. Pierce's disease symptoms typically progress from loss of chlorophyll in leaf margins, to leaf scorch, and finally to the highly characteristic abscission of the leaf blade from the petiole, or leaf stem (Fig. 5 A and C). Grapevines were inoculated with the X. fastidiosa strains Temecula, KLN61, or KLN62, or buffer, and symptom development was monitored. After 4 months, grapevines inoculated with KLN61 or KLN62 lost about three times as many leaves (9.83 ± 1.60 SE and 14.0 ± 1.73 SE, respectively) as grapevines inoculated with Temecula (4.14 ± 0.51 SE). Mockinoculated grapevines lost no leaves. We consistently observed that rpfF mutants caused earlier and more severe symptom onset and earlier death than did the wild type in multiple experiments (Fig. 5). This hypervirulence was surprising, given that Xcc rpfF mutants are hypovirulent and suggests that, in X. fastidiosa, cell-cell signaling tempers virulence in the plant.
Fig. 5.
Pierce's disease symptoms in grapevines. Cabernet sauvignon grapevines infected with the wild-type strain Temecula (A), the rpfF mutant KLN61 (B), or mock inoculated (C). Yellow arrows (A and B) indicate leaves with leaf scorch, an early disease symptom, and white arrows (B) indicate leaves that have detached at the blade-petiole junction, an advanced disease symptom.
To understand the mechanism of hypervirulence by the rpfF mutant, we monitored bacterial populations in mechanically inoculated grapevines before symptom expression. Plant samples 15, 30, and 60 cm distal to the inoculation site at 3, 5, and 7 weeks postinoculation were cultured from grapevines infected with Temecula, KLN61, or KLN62, or mock-inoculated (n = 8 grapevines per treatment per time point). Populations at 3 weeks postinoculation were highly variable with no apparent trend, but, at 5 and 7 weeks, populations for all strains and distances were similar, ≈108 colony-forming units/g (data not shown). These data suggested that large increases in population size or in swiftness of spread from the inoculation site are not the cause of rpfF hypervirulence although only substantial differences in populations would have been detected in our study.
We observed wild-type and rpfF mutant colonization of the grapevine xylem by scanning electron microscopy of petioles of symptomatic leaves and found that both strains formed matrixencased biofilms when found packed in crowded vessels, and both were associated with long strands of matrix when found in less crowded communities (Fig. 6). This matrix likely represents exopolysaccharide that we believe is secreted by the bacteria, suggesting that the X. fastidiosa rpfF mutant still produces exopolysaccharide, unlike the Xcc rpfF mutant, which cannot (20). Interestingly, rpfF mutations in a third related plant pathogen, Xanthomonas oryzae pv. oryzae, cause reduced virulence without affecting exopolysaccharide production (28).
Discussion
After the first identification of cell-cell signaling by way of DSF in Xcc and demonstration of its role in virulence gene expression, DSF was shown to be produced by several other bacterial species, in the Xanthomonas, Pseudomonas, Mycobacterium, and Erwinia genera (14, 15). However, with the exception of X. oryzae pv. oryzae (28, 29), the role of DSF signaling has not been investigated for any of these other species. Here, we demonstrate that a DSF activity is detected in X. fastidiosa media extracts by an Xcc-based GFP reporter (Fig. 2 A and B), suggesting that cell-cell signaling occurs in X. fastidiosa. We found that DSF reporter activation by the X. fastidiosa Temecula strain was much weaker than that by Xcc perhaps because high concentrations of the signal are produced only under specific conditions, such as in planta, or because the X. fastidiosa signal is structurally different and cannot fully activate the Xcc-based DSF reporter. These results are in agreement with those obtained for a citrus strain of X. fastidiosa (17).
We constructed two strains of X. fastidiosa Temecula in which the rpfF gene was disrupted and found that they do not have signal activity (Fig. 2 B and C), indicating that rpfF is required for DSF biosynthesis in X. fastidiosa. We are unable to rule out that RpfF may play a role in the synthesis of other molecules in addition to DSF. Expression of the X. fastidiosa rpfF gene from a constitutive promoter in an Xcc rpfF mutant restored DSF activity production (Fig. 2H). Interestingly, the X. fastidiosa gene could not confer the same level of DSF reporter activation as the Xcc gene did when expressed in either Xcc (Fig. 2G) or in Escherichia coli strain DH5α, which was used to prepare the plasmids (data not shown). That the difference in activities occurs even when both genes are placed in a heterologous host (E. coli) further suggests that the X. fastidiosa signal may have a slightly different structure than the Xcc signal. In addition, the rpfF gene alone confers production of DSF to E. coli (which does not produce DSF on its own), indicating that RpfF is sufficient for DSF biosynthesis.
We have found that the rpfF mutant was rarely transmitted by a highly efficient vector of X. fastidiosa, the blue-green sharpshooter leafhopper. This transmissibility defect was unexpected and reveals the importance of cell-cell signaling in insect transmission. Leafhoppers fed on rpfF-infected plants ingested rpfF cells but were able to rapidly clear themselves of the mutant whereas there is no evidence that the wild type can be cleared from leafhoppers except through molting (8). We show that this lack of retention (and thus transmissibility) is correlated to an inability of the rpfF mutant to form a biofilm in the insect foregut (Fig. 4). This result reveals an important and previously unappreciated connection between cell-cell signaling and transmission, as well as an apparent requirement for biofilm formation for efficient transmission. These findings may contribute to new strategies for controlling Pierce's disease by targeting transmission.
It is intriguing that biofilm formation by rpfF mutants is affected in insects (Fig. 4) but not plants (Fig. 6). In both environments, bacteria are subjected to high fluid shear force, are bathed by xylem sap, and require detachment to spread to new habitats. The main difference is in the attachment substrate (insect cuticle or xylem vessel). In plants, cells attach in any orientation to the xylem wall and aggregate in matrix-enclosed communities that frequently include space where xylem sap can flow (Fig. 6). In insects, cells attach in a polar fashion to the insect cuticle and form a mat-like structure at the point of attachment (Fig. 4E). Whether such differences in architecture result from environmental forces or bacterial activity is currently under debate (30). Impairment in only one type of biofilm formation in rpfF mutants suggests that the differences in biofilm architecture are subject to control by the organism as well as by the environment. The role of cell-cell signaling in biofilm formation and architecture has been detailed for other organisms such as Pseudomonas aeruginosa, in which it is required for maturation of the biofilm architecture (10). Motility via type IV pili and flagellae has been implicated in the achievement of mature architecture (31). Although the X. fastidiosa genome lacks genes required for flagella biosynthesis (32), it includes genes similar to those encoding type IV pili and other genes encoding putative adhesins, which may be important for attachment to the insect cuticle (33).
rpfF mutations have opposite effects on virulence to plants in X. fastidiosa and Xcc, highlighting our limited understanding of X. fastidiosa virulence. Hypervirulence in rpfF mutants suggests that X. fastidiosa employs a strategy to temper virulence to hosts. Extensive exopolysaccharide production and cell division lead to vessel plugging in the plant, presumably followed by nutrient shortage and decreased spread to new hosts as sap-feeding insects avoid blocked vessels. X. fastidiosa may use cell-cell signaling-based strategies to seek the most advantageous balance between pathogenicity and growth and spread to new hosts, as has been suggested by in planta microscopic studies (1).
DSF signaling in X. fastidiosa may regulate exoenzyme and exopolysaccharide biosynthesis, as it does in Xcc (14). Current theory proposes that X. fastidiosa expresses exoenzymes to degrade pit membranes. It is assumed, but unproven, that X. fastidiosa must break down these membranes to disperse efficiently throughout the plant and cause disease. However, if exoenzymes are positively regulated by DSF signaling, as they are in Xcc, then rpfF mutants of X. fastidiosa should be deficient in exoenzyme production in planta. In that case, one can theorize that hypervirulence of the rpfF mutants may arise from failure to escape crowded vessels into which X. fastidiosa may occasionally move by natural portals. We hypothesize that this failure would be expected to increase vessel plugging and exacerbate Pierce's disease symptoms (Fig. 7). Thus, although the bacteria may not colonize as many vessels, it may more frequently occlude those vessels to which it spreads and thus incite more disease symptoms. DSF regulation of exopolysaccharide production may also contribute to virulence and transmission in X. fastidiosa. In some systems, exopolysaccharides inhibit cells from forming initial, specific attachments required for biofilm formation, perhaps by masking attachment proteins (34, 35). In other cases, exopolysaccharide synthesis enhances biofilm formation (36, 37), perhaps at postattachment steps. In X. fastidiosa, DSF-mediated cell-cell signaling may be required for control of exopolysaccharide synthesis to allow cells to form successful biofilms on the insect cuticle.
Fig. 7.
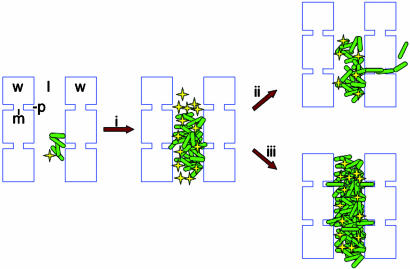
Model of X. fastidiosa movement in the plant xylem. Small colonies of cells (green rods) attach to the xylem wall (w) and grow into larger colonies (i). These larger colonies likely impede xylem sap flow, which carries fresh nutrients, through the vessel lumen (l) while accumulating DSF (yellow stars). In most crowded vessels, cells escape through the bordered pits (p) to new adjacent vessels (ii) after the bordered pit membrane (m) is breached by DSF-induced degradative enzymes. In a few cases, cells may be unable to get through the membrane, perhaps because neighboring vessels are embolized or because pit membranes are resistant to destruction, and vessel plugging and disease occurs (iii). In the rpfF mutant, the lack of DSF production may cause crowded vessels to become plugged more frequently (iii). In a few cases, rpfF mutant cells must be able to gain access to adjacent vessels through preexisting openings or broken membranes (ii).
Our finding that cell-cell signaling controls both transmission and virulence opens new avenues for exploration of host-microbe interactions. Future experiments are needed to identify the specific traits regulated by DSF signaling in X. fastidiosa and to investigate the role these traits play in interactions with insects and plants.
Acknowledgments
We thank C. Wistrom for providing insects and technical assistance; R. Hoenisch and E. Norberg for providing grapevines; K. Ho, R. Koutsoukis, S. Rashby, G. Vrdolijak, and I. Wilder for technical assistance; J. M. Dow for providing strains and helpful advice; and J. D. Newman, B. J. Staskawicz, and members of the Lindow laboratory for critical review of the manuscript. This material is based on work supported by the National Science Foundation under a grant awarded to K.L.N. in 2002 and by funding from the American Vineyard Foundation, the California Competitive Grant Program for Research in Viticulture and Enology, and the California Department of Food and Agriculture.
Abbreviations: DSF, diffusible signaling factor; Xcc, Xanthomonas campestris pv. campestris; KanR, kanamycin resistance.
References
- 1.Newman, K. L., Almeida, R. P. P., Purcell, A. H. & Lindow, S. E. (2003) Appl. Environ. Microbiol. 69, 7319-7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyson, G. E., Stojanovic, B. J., Kuklinski, R. F., Divittorio, T. J. & Sullivan, M. L. (1985) Phytopathology 75, 264-269. [Google Scholar]
- 3.Hopkins, D. L. (1981) Phytopathology 71, 415-418. [Google Scholar]
- 4.Hopkins, D. L. & Purcell, A. H. (2002) Plant Dis. 86, 1056-1066. [DOI] [PubMed] [Google Scholar]
- 5.Purcell, A. H. & Hopkins, D. L. (1996) Annu. Rev. Phytopathol. 34, 131-151. [DOI] [PubMed] [Google Scholar]
- 6.Brlansky, R. H., Timmer, L. W., McCoy, R. E. & French, W. J. (1983) Phytopathology 73, 530-535. [Google Scholar]
- 7.Purcell, A. H., Finlay, A. H. & McLean, D. L. (1979) Science 206, 839-841. [DOI] [PubMed] [Google Scholar]
- 8.Severin, H. H. P. (1949) Hilgardia 19, 190-202. [Google Scholar]
- 9.Hill, B. L. & Purcell, A. H. (1997) Phytopathology 87, 1197-1201. [DOI] [PubMed] [Google Scholar]
- 10.Parsek, M. R. & Greenberg, E. P. (2000) Proc. Natl. Acad. Sci. USA 97, 8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinnebusch, B. J., Rudolph, A. E., Cherepanov, P., Dixon, J. E., Schwan, T. G. & Forsberg, A. (2002) Science 296, 733-735. [DOI] [PubMed] [Google Scholar]
- 12.Basset, A., Tzou, P., Lemaitre, B. & Boccard, F. (2003) EMBO Rep. 4, 205-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dow, J. M. & Daniels, M. J. (2000) Yeast 17, 263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barber, C. E., Tang, J. L., Feng, J. X., Pan, M. Q., Wilson, T. J., Slater, H., Dow, J. M., Williams, P. & Daniels, M. J. (1997) Mol. Microbiol. 24, 555-566. [DOI] [PubMed] [Google Scholar]
- 15.Wang, L.-H., He, Y., Gao, Y., Wu, J. E., Dong, Y.-H., He, C., Wang, S. X., Weng, L.-X., Xu, J.-L., Tay, L., et al. (2004) Mol. Microbiol. 51, 903-912. [DOI] [PubMed] [Google Scholar]
- 16.Slater, H., Alvarez-Morales, A., Barber, C. E., Daniels, M. J. & Dow, J. M. (2000) Mol. Microbiol. 38, 986-1003. [DOI] [PubMed] [Google Scholar]
- 17.Scarpari, L. M., Lambais, M. R., Silva, D. S., Carraro, D. M. & Carrer, H. (2003) FEMS Microbiol. Lett. 222, 83-92. [DOI] [PubMed] [Google Scholar]
- 18.Turner, P., Barber, C. & Daniels, M. (1984) Mol. Gen. Genet. 195, 101-107. [Google Scholar]
- 19.Miller, W. G., Leveau, J. H. J. & Lindow, S. E. (2000) Mol. Plant-Microbe Interact. 13, 1243-1250. [DOI] [PubMed] [Google Scholar]
- 20.Tang, J. L., Liu, Y. N., Barber, C. E., Dow, J. M., Wootton, J. C. & Daniels, M. J. (1991) Mol. Gen. Genet. 226, 409-417. [DOI] [PubMed] [Google Scholar]
- 21.Davis, M. J., French, W. J. & Schaad, N. W. (1981) Phytopathology 71, 869-870. [Google Scholar]
- 22.Kovach, M. E., Elzer, P. H., Hill, D. S., Robertson, G. T., Farris, M. A., Roop, R. M. I. & Peterson, K. M. (1995) Gene 166, 175-176. [DOI] [PubMed] [Google Scholar]
- 23.Hill, B. L. & Purcell, A. H. (1995) Phytopathology 85, 209-212. [Google Scholar]
- 24.Sokal, R. R. & Rolf, F. J. (1995) Biometry: The Principles and Practice of Statistics in Biological Research (Freeman, New York).
- 25.Minsavage, G. V., Thompson, C. M., Hopkins, D. L., Leite, R. & Stall, R. E. (1994) Phytopathology 84, 456-461. [Google Scholar]
- 26.Hill, B. L. & Purcell, A. H. (1995) Phytopathology 85, 1368-1372. [Google Scholar]
- 27.Almeida, R. P. P. & Purcell, A. H. (2003) J. Econ. Entomol. 96, 265-271. [DOI] [PubMed] [Google Scholar]
- 28.Chatterjee, S. & Sonti, R. V. (2002) Mol. Plant Microbe Interact. 15, 463-471. [DOI] [PubMed] [Google Scholar]
- 29.Tang, J. L., Feng, J. X., Li, Q. Q., Wen, H. X., Zhou, D. L., Wilson, T. J., Dow, J. M., Ma, Q. S. & Daniels, M. J. (1996) Mol. Plant-Microbe Interact. 9, 664-666. [DOI] [PubMed] [Google Scholar]
- 30.Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. (2002) Annu. Rev. Microbiol. 56, 187-209. [DOI] [PubMed] [Google Scholar]
- 31.Klausen, M., Heydorn, A., Ragas, P., Lambertsen, L., Aaes-Jorgensen, A., Molin, S. & Tolker-Nielsen, T. (2003) Mol. Microbiol. 48, 1511-1524. [DOI] [PubMed] [Google Scholar]
- 32.Simpson, A. J. G., Reinach, F. C., Arruda, P., Abreu, F. A., Acencio, M., Alvarenga, R., Alves, L. M. C., Araya, J. E., Baia, G. S., Baptista, C. S., et al. (2000) Nature 406, 151-157. [DOI] [PubMed] [Google Scholar]
- 33.Van Sluys, M. A., Monteiro-Vitorello, C. B., Camargo, L. E., Menck, C. F., Da Silva, A. C., Ferro, J. A., Oliveira, M. C., Setubal, J. C., Kitajima, J. P. & Simpson, A. J. (2002) Annu. Rev. Phytopathol. 40, 169-189. [DOI] [PubMed] [Google Scholar]
- 34.Parkar, S. G., Flint, S. H., Palmer, J. S. & Brooks, J. D. (2001) J. Appl. Microbiol. 90, 901-908. [DOI] [PubMed] [Google Scholar]
- 35.Hanna, A., Berg, M., Stout, V. & Razatos, A. (2003) Appl. Environ. Microbiol. 69, 4474-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammer, B. K. & Bassler, B. L. (2003) Mol. Microbiol. 50, 101-104. [DOI] [PubMed] [Google Scholar]
- 37.Haugo, A. J. & Watnick, P. I. (2002) Mol. Microbiol. 45, 471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]