SUMMARY
Cardiovascular homeostasis is maintained in part by the rapid desensitization of activated heptahelical receptors that have been phosphorylated by G protein-coupled receptor kinase 2 (GRK2). However, during chronic heart failure GRK2 is upregulated and believed to contribute to disease progression. We have determined crystallographic structures of GRK2 bound to an RNA aptamer that potently and selectively inhibits kinase activity. Key to the mechanism of inhibition is the positioning of an adenine nucleotide into the ATP-binding pocket and interactions with the basic αF-αG loop region of the GRK2 kinase domain. Constraints imposed on the RNA by the terminal stem of the aptamer also play a role. These results highlight how a high affinity aptamer can be used to selectively trap a novel conformational state of a protein kinase.
INTRODUCTION
In all eukaryotic cells, the activation of G protein-coupled receptors (GPCRs) leads to profound physiological change, and the rapid desensitization of these receptors is crucial for an appropriate temporal response. One key desensitization mechanism is initiated by a family of GPCR kinases (GRKs) that phosphorylates serine and threonine residues in the intracellular loops and C-terminal tails of activated GPCRs (Gurevich et al., 2011). Receptor phosphorylation recruits arrestin, which decreases heterotrimeric G protein coupling, promotes receptor endocytosis, and initiates new signaling cascades (DeWire et al., 2007).
Although GRKs are critical for the desensitization of GPCRs, there are pathologies in which GRK activity is maladaptive (Metaye et al., 2005). One well-studied example is that of GRK2 in the cardiovasculature (Dorn, 2009). In the normal heart, activation of myocyte β-adrenergic receptors (β-ARs) by norepinephrine strengthens and increases the rate of contractions. GRK2 regulates signaling through cardiac β-ARs (Koch et al., 1995; Kong et al., 1994; Pippig et al., 1993) and other cardiac receptors such as the angiotensin and α1-adrenergic receptors (Cohn et al., 2008; Oppermann et al., 1996), in addition to regulating catecholamine release in adrenal chromaffin cells (Lymperopoulos et al., 2008). During the early stages of congestive heart failure, adenylyl cyclase uncouples from β2ARs, an event that coincides with increased activity and expression of GRK2. The importance of GRK2 in this process is underscored by the fact that cardiac-restricted expression of a GRK2 inhibitor in a mouse model of cardiomyopathy reduces heart failure in these animals (Rockman et al., 1998).
Therapeutic targeting of GRK2 is complicated by the fact that it is closely related to six other vertebrate GRKs that belong to the protein kinase A, G and C (AGC kinase) family. Moreover, its catalytic mechanism and active site pocket are highly conserved among over 500 protein kinases (Johnson, 2009; Manning et al., 2002). Nevertheless, two compounds from a series of molecules developed by Takeda Pharmaceuticals Company Ltd. exhibited high potency and selectivity towards GRK2 (Ikeda, 2007) by binding to the active site of GRK2 (Thal et al., 2011) in a manner similar to that of the less selective inhibitor balanol (Tesmer et al., 2010). Surprisingly, their selectivity seemed to be dictated more by the overall shape of the GRK2 active site than by their interaction with residues that are unique to each GRK subfamily (Thal et al., 2011).
A high affinity RNA aptamer that selectively inhibits GRK2 (C13) was recently reported (Mayer et al., 2008). Aptamers have also been developed for other kinases such as protein kinase C (Conrad et al., 1994) and mitogen-activated protein kinase (Seiwert et al., 2000), but the molecular mechanism their inhibition is not known. Although RNA aptamers can serve as therapeutic agents in themselves (Bonetta, 2009), they can also be useful in identifying small molecule mimetics that displace them from their targets (Hafner et al., 2006; Mayer et al., 2009). The C13 aptamer contains twenty nucleotides of selected sequence anchored by a conserved terminal stem region (Mayer et al., 2008). C13 binds GRK2 with high nanomolar affinity in a filter binding assay and with high selectivity over other protein kinases, including a 20-fold higher IC50 against closely related GRK5 (Mayer et al., 2008), which has 45% sequence identity in the kinase domain. Herein we use structural and functional studies to show that C13 stabilizes a unique inactive conformation of GRK2 through multiple interactions, both within and outside the active site pocket of the kinase domain, and that the terminal stem of the aptamer indirectly contributes to selectivity by constraining the selected portion of the RNA.
RESULTS
Determinants of GRK2 Binding and Selectivity in the C13 Aptamer
To identify the regions of C13 that are important for GRK2 binding and to optimize the RNA for crystallographic analysis, we synthesized a series of truncations and modifications of the C13.51 variant of the aptamer (Mayer et al., 2008) (Table 1). Our designs were also guided by concurrent X-ray diffraction experiments (Supplemental Experimental Procedures and Table S1 available online). Our best structural models were of GRK2 complexes with the C13.28 and C13.18 variants (Table 2).
Table 1.
Sequence, GRK2 Binding and Inhibition Properties of C13 Variants
| Variant | RNA Sequencea | Kd (nM) | Activity IC50 (nM)b | Binding IC50 (nM)c | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 35 | 40 | 45 | 50 | 55 | 60 | 65 | 70 | 74 | ||||
| C13.40 | GGGUAGCACAGACCAUACGGGAGAGAAACUUGUGCAACCC | 1.3 ± 0.1 | ||||||||||
| C13.29 | GGGCAGACCAUACGGGAGAGAAACUUGCC | 0.91 ± 0.07 | ||||||||||
| C13.28 | GGCAGACCAUACGGGAGAGAAACUUGCC | 1.2 ± 0.6d 3.8 ± 1e |
11 ± 4 | 1.5 ± 0.1 | ||||||||
| C13.26 | GGCGACCAUACGGGAGAGAAACUGCC | 0.90 ± 0.03 | ||||||||||
| C13.22 | AGACCAUACGGGAGAGAAACUU | 46 ± 9 | ||||||||||
| C13.20 | GACCAUACGGGAGAGAAACU | 48 ± 7e | 140 ± 10 | |||||||||
| C13.18 | CCAUACGGGAGAGAAACU | 35 ± 5d | 220 ± 40 | 2,000 ± 400 | ||||||||
C13.40 corresponds to nucleotides 35 to 74 of C13, with nucleotide numbers shown over the sequence. Residues corresponding to the twenty selected nucleotides from C13 are underlined and flanking nucleotides are capable of forming an imperfect (C13.40) or perfect (C13.29, C13.28, C13.26, C13.22) base paired stem region. C13.29 was designed to include a 5′ G overhang (Reyes et al., 2009). Each of these aptamers was co-crystallized with GRK2 (see Table S1). The integrity of each aptamer was assessed by end labeling with [γ-32P]ATP and radioimaging (Figure S6).
Activity-based IC50 values ± SEM were measured in phosphorylation assays in three independent experiments.
Binding IC50 values ± SEM were measured as decreased detection of bead-bound F-C13.28-bGRK2 complex in the presence of 0.5 nM F-C13.28 (see Figure S5). The data represents at least three different experiments performed in duplicate.
Kd ± SEM was measured by a flow cytometry-based direct binding assay as a function of F-RNA-bGRK2 complex formation (see Figure S2A and B)
As in (d), but detecting F-GRK2-bRNA complex formation (see Figure S2C).
Table 2.
Crystallographic Data Statistics
| Complexa X-ray Source |
C13.28-GRK2-Gβ1γ2 APS 21-ID-G |
C13.18-GRK2 APS 21-ID-G |
|---|---|---|
| Wavelength (Å) | 0.97856 | 0.97856 |
| Dmin (Å) | 4.5 (4.58-4.50) | 3.5 (3.56-3.50) |
| Space Group | P312 | P21212 |
| Unit cell constants | a=257 Å | a=113.3 Å |
| b=257 Å | b=139.7 Å | |
| c=99.4 Å | c=60.9 Å | |
| α=β=90°; γ=120° | α=β=γ=90° | |
| No. crystals | 1 | 2 |
| Unique reflections | 12,758 (96) | 8,251 (92) |
| Average multiplicity | 3.9 (2.3) | 7.8 (8.3) |
| Rsym (%) | 6.4 (38.8) | 10.3 (49.0) |
| Completeness (%)b | 58.4 (9.0) | 65.5 (14.8) |
| <I>/<σI> | 24.5 (1.6)c | 28.9 (4.7) |
|
| ||
| Refinement resolution | 20.0-4.52 (4.63-4.52) | 25.0-3.51 (3.60-3.51) |
| Total reflections used | 11,919 (154) | 7,811 (87) |
| Protein atoms | 8,112 | 4,746 |
| Non-protein atoms | 401 | 168 |
| RMSD bond lengths (Å) | 0.005 | 0.005 |
| RMSD bond angles (°) | 0.86 | 0.91 |
| Est. coordinate error (Å) | 0.95 | 0.67 |
| Ramachandran most favored, disallowed (%) | 83.1, 0.0 | 84.7, 0.0 |
| Rwork (%) | 18.4 (26.8) | 21.6 (41.4) |
| Rfree (%) | 26.8 (37.1) | 31.7 (37.1) |
| PDB Entry | 3UZS | 3UZT |
These crystal forms represent the best diffracting crystals of the various aptamers in complex with GRK2 (see Table 1 and S1).
Data sets were elliptically truncated to the maximum resolution in each direction as described in the methods.
A I/σI cutoff of 0 was applied to this crystal form during scaling.
C13.28 was designed to shorten the terminal stem of the original aptamer to only four Watson-Crick base pairs, whereas C13.18 lacks a terminal stem and two residues from the 5′ end of the 20 nucleotide variable region. To measure the direct binding of GRK2 to C13.28 and C13.18, we used a flow cytometry-based protein-RNA interaction assay (Blazer et al., 2010). Biotinylated GRK2 (bGRK2) was first immobilized on streptavidin beads and incubated with increasing amounts of 5′ Alexa Fluor-488 labeled C13.28 (F-C13.28) or C13.18 (F-C13.18). The beads were then passed through a flow cytometer and the amount of bead-bound fluorescence was quantitated to yield a dissociation constant (Kd) of 1.2 ± 0.6 and 35 ± 5 nM for F-C13.28 (Figure S1A) and F-C13.18 (Figure S1B), respectively (Table 1). A similar binding constant for C13.28 (Kd of 3.8 ± 1.2 nM) was measured when the assay was reversed such that the 5′ end of C13.28 was biotinylated and bound to beads and incubated with fluorescently labeled GRK2 (F-GRK2) (Table 1 and Figure S1C). The Kd values for these RNAs correlate with their potency in inhibiting GRK2-mediated phosphorylation of invertebrate rhodopsin, in that C13.28 inhibited GRK2 phosphorylation 20-fold more potently than C13.18 (IC50 values of 11 ± 4 and 220 ± 40 nM, respectively) (Table 1). Furthermore, we confirmed that C13.28 shows selectivity towards GRK2 by measuring the RNA binding to GRK1 and GRK6, representative members of the other two branches of the GRK subfamily (Premont and Gainetdinov, 2007; Premont et al., 1999). In this assay, biotinylated C13.28 (bC13.28) was attached to streptavidin beads and incubated with F-GRK2 for use in competition assays with unlabeled GRKs (Figure S2). The IC50 for GRK2 (3.3 ± 0.3 nM) was ~60 and ~180-fold lower than the IC50’s measured for GRK6 and GRK1 (210 ± 10 and 590 ± 160 nM), respectively, recapitulating the selectivity properties of C13.
The relative affinity of the other RNA variants for bGRK2 was determined in competition format by mixing F-C13.28 (0.5 nM) with increasing amounts of unlabeled competitor RNA and measuring the bead-associated fluorescence (Table 1). All RNAs designed to retain a terminal stem of at least three Watson-Crick base pairs (C13.40, C13.29, C13.28 and C13.26) exhibited high affinity for bGRK2, with IC50 values ranging from 0.9 to 1.5 nM. Those that did not (C13.22, C13.20 and C13.18) exhibited lower affinity by 1–3 orders of magnitude in this assay format. Collectively, our binding data indicate that the 18 nucleotides within the selected region of the aptamer (C13.18) are sufficient for binding and inhibiting GRK2, but that the terminal stem is necessary for high potency.
Structure of the C13.18-GRK2 Complex
Many C13 variants readily crystallized with GRK2 alone and/or with the GRK2-Gβγ complex (Table S1), but all exhibited diffraction from moderate to low resolution, most likely due to the flexible nature of RNA which was commonly observed in lattice contacts. The best data were obtained from crystals of the GRK2-C13.18 complex, which diffracted anisotropically to 3.5 Å spacings (Table 2 and S1). The aptamer binds in the cleft formed between the small and large lobes of the kinase domain (Figure 1A and S3A), which differ in orientation by 8° relative to those of previously reported structures of GRK2 determined in the presence of ATP (e.g. PDB entries 1OMW (Lodowski et al., 2003b) and 2BCJ (Tesmer et al., 2005)), and by 12° relative to those of GRK6 in what is considered to be the most active conformation of a GRK determined to date (Boguth et al., 2010). A smaller degree of domain reorientation was previously observed in structures of GRK2 bound to small molecule inhibitors (Tesmer et al., 2010; Thal et al., 2011). Aptamer-induced domain remodeling has also been observed for an NF-κB p50-aptamer complex (Huang et al., 2003).
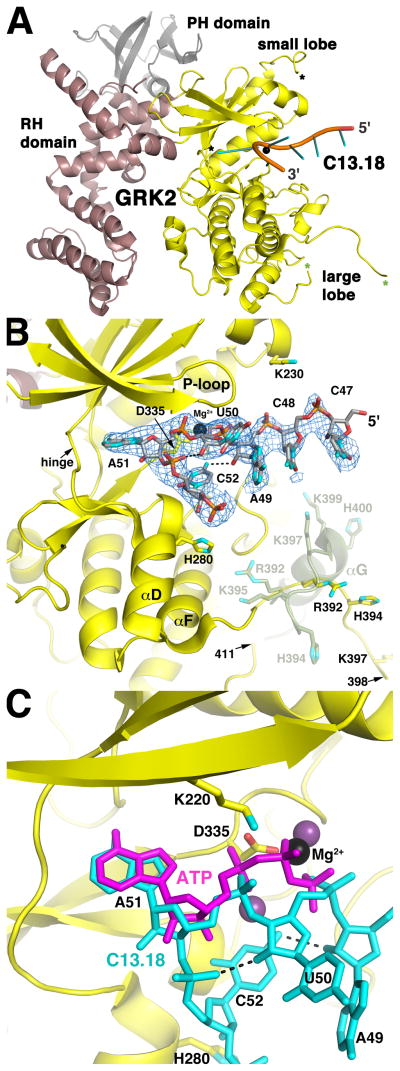
Figure 1. The C13.18 Aptamer Binds in the Active Site of GRK2.
(A) Overall structure of the C13.18-GRK2 complex. Residues 47-CCAUAC-52 of C13.18 are depicted as a cartoon ribbon with spokes indicating nucleotide bases. The kinase, RH and PH domains of GRK2 are shown in yellow, burgundy and grey, respectively. Mg2+ is a black sphere. Asterisks demark residues flanking disordered regions of the kinase domain: a segment of the active site tether (amino acids 475 and 492, black) and a segment spanning the αG helix of apo-GRK2 structures (amino acids 398 and 411, green). Aptamer binding induces an 8 change in the orientation of the large and small lobes (Figure S3A).
(B) Interactions of the C13.18 aptamer in the active site of GRK2. The blue wire cage corresponds to a 2m|Fo|−D|Fc| simulated annealing omit map contoured at 1 σ around residues 47-CCAUAC-52 of C13.18. Carbons atoms are grey for C13.18, and yellow for the kinase domain. Nitrogens are cyan, oxygens red, phosphates orange and Mg2+ black. Intramolecular hydrogen bonds in the RNA are dashed lines. The position of the αF-αG loop and αG helix in apo-GRK2 (amino acids 391–410; PDB entry 2BCJ (Tesmer et al., 2005)) is shown in green at half transparency and is reorganized in the C13.18-bound structure (see also Figure S3A). The twelve C-terminal nucleotides of the aptamer are disordered, but extend into a crystal contact lined with positively charged amino acids (Figure S3B).
(C) C13.18 mimics the binding of ATP in the GRK2 active site. GRK2-bound ATP was homology modeled based on the GRK1·2Mg2+·ATP complex (purple; PDB entry 3C4W (Singh et al., 2008)). The P-loop was omitted for clarity.
Two segments of the kinase domain are disordered. The first missing segment (amino acids 399–410) spans residues that constitute the αG helix of the large lobe in other GRK2 structures (faded green loop and helix in Figure 1B). Residues 391–398 that precede this segment adopt a novel conformation compared to previous structures (e.g. PDB entry 1OMW), such that they extend away from the large lobe and form lattice contacts with the 5′ nucleotide of a symmetry-related RNA (Figure 1A, 1B, and S3B). Conformational flexibility of the αF-αG loop/αG helix region in GRKs is consistent with its proteolytic sensitivity in GRK2 (Lodowski et al., 2005) and high temperature factors in other GRK structures (Boguth et al., 2010; Lodowski et al., 2005; Singh et al., 2008). The second missing segment (amino acids 476–491) is part of the “active site tether”, a structural element unique to AGC kinases that is expected to become more ordered as the kinase domain approaches its active state (Kannan et al., 2007). It is likewise disordered in prior GRK2 structures.
Omit maps revealed strong electron density for the first six residues of C13.18 and the 5′ phosphate of the seventh nucleotide, allowing confident modeling of 47-CpCpApUpApCp-52 (Figure 1B and S3B). The RNA makes a tight turn that projects A51 deep into the active site of the kinase such that its AMP moiety and the preceding 5′ phosphate of U50 essentially overlap with the AMP moiety and γ phosphate of the physiological substrate ATP (Figure 1C). Despite the low resolution of the map, we modeled a Mg2+ ion in persistent positive |Fo|−|Fc| electron density between the U50 5′ phosphate and GRK2-Asp335, which would help neutralize the electrostatic repulsion between these residues (Figure 1C). This Mg2+ ion is in a position analogous to one of the two Mg2+ ions that are typically observed in other AGC kinase·ATP complexes (Adams and Taylor, 1993; Singh et al., 2008).
The overall conformation of C13.18 seems to be stabilized by hydrogen bonds between N4 of C52 and the 2′-OH of A49, and between the 5′-phosphate of C52 and the 2′-OH of U50 (Figure 1C). The fold of the RNA is also probably influenced by its extensive interactions with the kinase domain, which bury 1140 Å2 of accessible surface area. The RNA recapitulates specific contacts made by the substrate ATP with the kinase domain, including hydrophobic contacts with the A51 base and hydrogen bonds to the ribose and adenine ring. Other interactions appear mostly electrostatic in nature, including interactions between the kinase and the U50 phosphate via Mg2+, as well as between His280 and the phosphodiester backbone (Figure 1B and S3B). The twelve unresolved nucleotides from the 3′ end of the C13.18 aptamer project into a cavity formed with a symmetry related kinase domain, where they likely interact electrostatically with positively charged residues that line the cavity (Figure S3B).
Interaction with the GRK2 Active Site Is Essential for C13 Binding
Both ATP and staurosporine inhibit C13 aptamer binding to GRK2 (Mayer et al., 2008), consistent with C13.18 occupying the ATP binding pocket of GRK2 in our structural model. We further demonstrated that ADP acts as a competitive inhibitor of the shorter C13.18 aptamer with an IC50 of 67 ± 22 μM (Figure 2A), a value consistent with the ~30 μM Km measured for ATP in phosphorylation reactions (Benovic et al., 1987) and the ~40 μM Ki measured for ADP in a fluorescence polarization displacement assay (J. Tesmer, unpublished data).
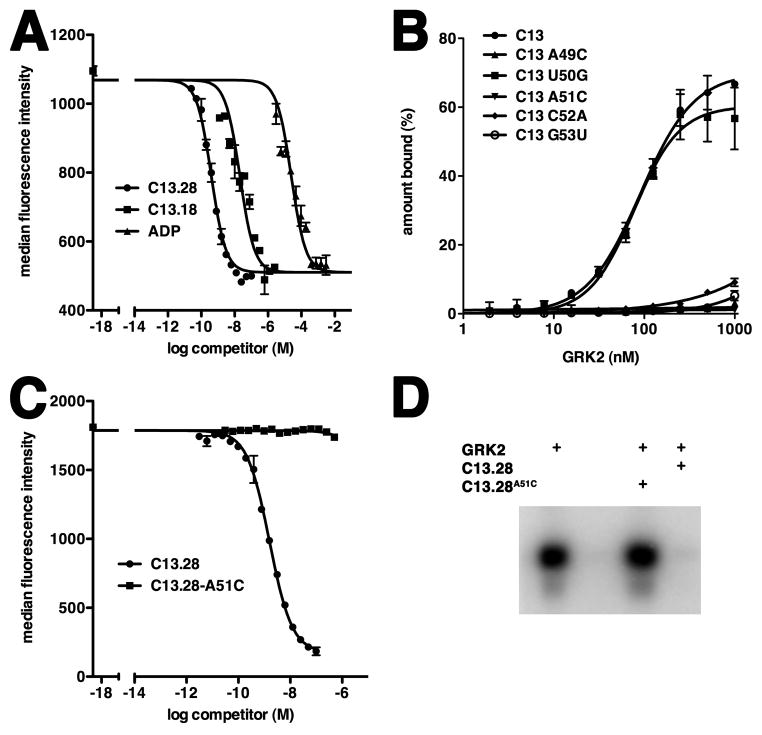
Figure 2. Nucleotides that Interact with the Active Site of GRK2 are Essential for Binding.
(A) ADP inhibits complex formation between F-C13.18 and b-GRK2. Varying concentrations of unlabeled competitor were included in binding reactions with F-C13.18 (35 nM final concentration, corresponding to the Kd) and bGRK2 bound to beads. Data from three separate experiments were fit using a single site competition model to obtain IC50 values for C13.28 (0.44 ± 0.19 nM), C13.18 (100 ± 41 nM) and ADP (67 ± 22 μM). A representative experiment is shown. ADP exhibited a 40-fold higher IC50 in competition against F-C13.28 (Figure S5).
(B) Impaired GRK2 binding by site-directed mutation of residues in C13. In this filter-binding assay, the percent of input radiolabeled wild-type C13 or its single nucleotide mutants that bound to GRK2 is plotted as a function of GRK2 concentration. The Kd± SEM for wild-type C13 and U50G were 92 ± 9 and 77 ± 9 nM, respectively. GRK2 binding to A49C, A51C, C52A and G53U was not detectable. Data represent three independent experiments.
(C) C13.28A51C does not compete for complex formation between F-C13.28 and bGRK2. A representative example from three independent flow cytometry experiments is shown.
(D) C13.28A51C does not inhibit phosphorylation of invertebrate rhodopsin by GRK2. All samples contained receptor and included GRK2 without RNA (lane 1), no GRK2 (lane 2) and GRK2 with 2 μM C13.28-A51C (lane 3) or 50 nM C13.28 (lane 4). Shown is a representative autoradiogram of the phosphorylated receptor, which was resolved on an SDS-PAGE gel.
Further validation of the C13.18-GRK2 model was gained through mutating the RNA at positions that dock in the active site of GRK2 and determining their ability to bind to GRK2 in a filter retention assay (Figure 2B). Full-length radiolabeled C13 was synthesized in vitro and bound to GRK2 with an affinity of 92 ± 9 nM, consistent with previous measurements (Mayer et al., 2008). The ~100-fold higher Kd for the aptamer in this assay compared with C13.28 in the flow-cytometry based assay (Table 1) could reflect the fact that the polyanion heparin was included as a nonspecific competitor in the filter binding experiments, or that complex formation was at 37 °C instead of 4 °C. Among the 5 mutants created, only U50G bound with an affinity similar to wild-type C13 (77 ± 9 nM). The wild-type behavior of the U50G mutant is consistent with the fact that the U50 base does not form specific interactions with either the protein or the rest of the RNA in the GRK2-C13.18 structure (Figure 1B). Four additional mutations (A49C, A51C, C52A and G53U) did not show detectable binding to GRK2 (Figure 2B). The close packing interactions of GRK2 with the cytosine base of C52, as well the specific hydrogen bond between C52 N4 and A49 2′ OH (Figure 1B), likely explain why this base cannot be substituted with a larger adenine ring. The inability to substitute A51 with cytosine is consistent with the A51 base occupying the canonical adenine binding site of the kinase domain. Because of their lack of direct base interactions with GRK2, the loss of binding of the A49C and G53U mutants is most likely explained by changes in the fold of the RNA, which could reconfigure to form a duplex stem of 46-CCC-49/55-GGG-53 (the A49C substitution) or 46-CCA-49/55-GGU-53 (the G53U substitution). These stems would place A51 in the middle of a three nucleotide hairpin loop that is unlikely to fit in the active site pocket of GRK2.
We next demonstrated that A51 plays a pivotal role in the inhibition of GRK2 activity by C13.28. C13.28 was synthesized with an A51C mutation (C13.28A51C), and shown to be incapable of binding GRK2 in our competition-based flow cytometry experiments (Figure 2C). In activity assays, C13.28A51C did not inhibit GRK2 phosphorylation of invertebrate rhodopsin at any concentration tested (up to 2 μM), in striking contrast with C13.28, which shows near complete inhibition at 50 nM (Figure 2D and Table 1).
Our structural model suggested that residues His280 and Asp335 could play a pivotal role in binding C13 in the active site, and these are among the few residues whose contacts to C13.18 are independent of A51 and not dictated by backbone atoms (e.g. the hinge of the kinase domain and the glycine rich P-loop). We individually mutated His280 and Asp335 to alanine and assayed these proteins for their ability to bind radiolabeled C13 in our filter binding assay (Figure 3A). GRK2-D335A binding to C13 was barely detectable, and the binding of GRK2-His280A (Kd of 211 ± 5 nM) was ~ 2 fold weaker than wild-type. Interestingly, these same mutant proteins did not show impaired binding to C13.28 in our flow-cytometry based assay, even when 150 mM NaCl was included in the binding buffer. We suspect that heparin, which was included in the filter binding assay to reduce non-specific RNA binding and has been shown to inhibit GRK activity (Loudon and Benovic, 1994), competes for a surface of GRK2 that is recognized by the aptamer outside of the active site. This could reduce the affinity of the aptamer for GRK2 to the extent that we can detect the contribution of residues within the active site.
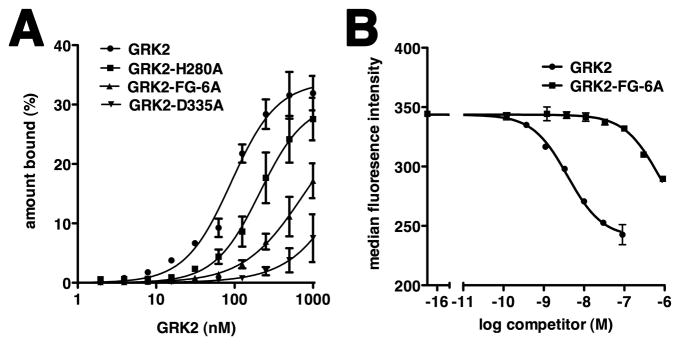
Figure 3. GRK2 Residues His280, Asp335 and the Basic αF-αG Loop Contribute to C13 Binding.
(A) Filter retention analysis of the relative amount of radioactively labeled C13 bound to wild-type GRK2 or its mutants. Results are plotted as a function of protein concentration, as described in Figure 2B. The Kd ± SEM for C13 binding to wild-type GRK2 or GRK2 H280A was 91 ± 11 nM and 211 ± 55 nM, respectively. Weaker affinity was observed for C13 binding to GRK2 FG-6A. The interaction of C13 with GRK2 D335A was barely detectable. Data represent at least two independent experiments.
(B) Mutation of basic residues in the αF-αG loop of GRK2 decreases affinity of C13.28 for GRK2 in the flow cytometry RNA-protein binding assay. IC50 ± SEM values for wild-type GRK2 and GRK2-FG-6A, measured as a function of decreased detection of F-GRK2 in complex with bead-bound C13.28, were 3.3 ± 0.3 and 510 ± 170 nM, respectively. A representative experiment is shown.
Structure of the GRK2-C13.28 Complex
Further understanding of how C13 inhibits GRK2 was gained from the crystal structure of the high affinity C13.28 variant bound to the GRK2-Gβ1γ2 complex, which was determined using anisotropic diffraction data to 4.5 Å spacings (Figure 4 and Table 2). The overall conformation of the kinase domain and lack of electron density for the αF-αG loop region in its canonical configuration is essentially the same as observed for the C13.18-GRK2 complex. The fold and shape of 49-ApUpApCp-52 is likewise consistent between the two structures (Figure 4A). Moreover, electron density for these nucleotides, and absence of interpretable density for the for the αF-αG loop region, was consistently observed in five other crystal forms of GRK2 in complex with C13 variants (Table S1), indicating that they are not an artifact of crystal packing. Outside the 49-ApUpApCp-52 sequence, the backbone of C13.28 takes a somewhat different course from that of C13.18, which is likely influenced by the presence of a terminal stem in C13.28.
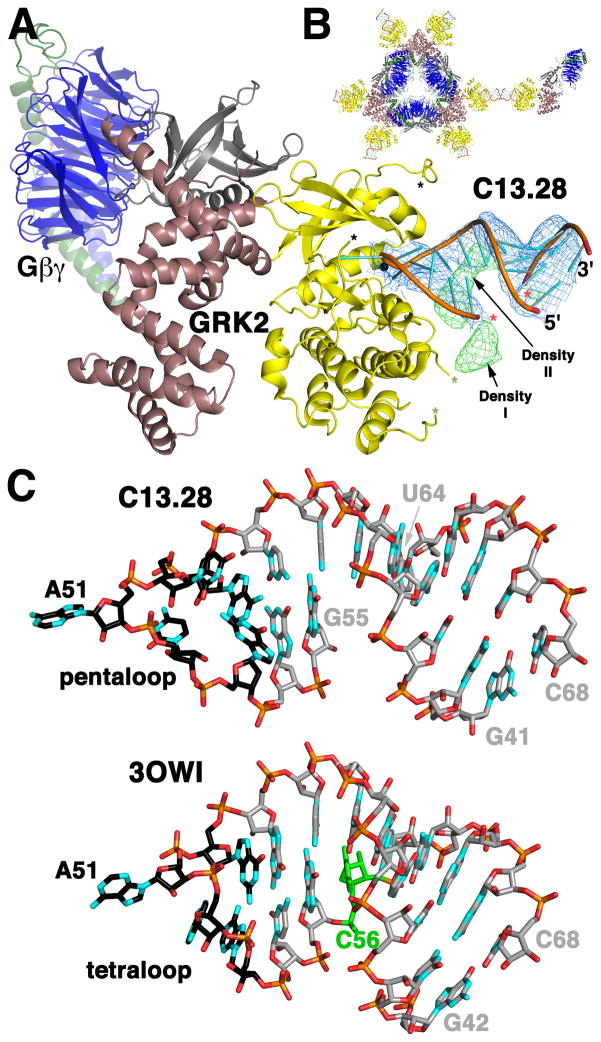
Figure 4. Structure of the C12.28-GRK2-Gβ1γ2 Complex Reveals the Topology and Contacts of C13 Outside the Active Site.
(A) Crystal structure of C13.28 bound to GRK2-Gβ1γ2. GRK2 colors and asterisks are the same as Figure 1A. Gβ1 is blue and Gγ2 green. A 5′ segment (G41–G55) and a 3′ segment (U64–C68) of C13.28 are modeled with red asterisks marking residues G55 and U64, which bracket the eight nucleotide gap. Electron density from the final 2m|Fo|−D|Fc| map contoured at 1 σ and from the final m|Fo|−D|Fc| map contoured at 3 σ are shown as blue and green wire cages, respectively. In the latter map there are two volumes of strong unaccounted for density (Density I and Density II), which are likely traversed by nucleotides 56–63 of the aptamer (Figure S4).
(B) The C13.28-GRK2-Gβ1γ2 complex crystallized in a D3 symmetric assembly at 32 centers of the crystal. The geranylgeranylated C-termini of Gγ2 project into a channel formed at the 3-fold axis of these hexamers. The kinase domain and C13.28 project outward from the hexamer, with RNA-RNA crystal contacts connecting symmetry related hexamers.
(C) Comparison of the topology of C13.28 (top) with nucleotides 42–68 of the glycine riboswitch (Huang et al., 2010) (PDB entry 3OWI; bottom). The hairpin loops (five nucleotides for C13.28 and a canonical GNRA tetraloop for the riboswitch) are drawn with black carbons. In C13.28, there is an 8 nucleotide insertion in the duplex in place of position 56 of the riboswitch (green).
We observed strong electron density consistent with an A-form double helix that was modeled as residues 41-GGCAG-44 and 64-UUGCC-68 of C13.28 (Figure 4A). This helix is positioned such that the base of the 5′ nucleotide stacks with its symmetry-related equivalent at a two-fold axis of the crystal (Figure 4B). A continuous chain could then be built from the 5′ end of the RNA through the GRK2 active site (nucleotides 41–55). The trajectory for the 5′ end of C13.28 as it exits the active site is consistent with the increasingly longer arms of electron density that project from the active site in crystal structures that contain RNA variants with progressively longer 5′ ends (i.e. the crystal structures of complexes with C13.18, C13.20, C13.22, and C13.26 respectively) (Table 1 and S1, and data not shown). In the active site cleft, the base of G53 appears to be disordered, as it is in the C13.18 structure, and G54 and G55 appear to be in position to make Watson-Crick base pairs with C48 and C47, respectively. As a result, the structure of C13.28 is similar to an RNA duplex that terminates in a standard GNRA tetraloop (Batey et al., 1999), such as that of the glycine riboswitch (Huang et al., 2010), except that the loop of C13.28 is a pentaloop (49-AUACG-53), and there is an eight nucleotide insertion within the duplex region (Figure 4C and S4).
After refining our model for C13.28-GRK2-Gβ1γ2, two regions of strong electron density (Density I and Density II) remained (Figure 4A and S4). These regions likely correspond, at least in part, to the eight unmodeled nucleotides (nucleotides 56–63), which could easily traverse both volumes (Figure S4). The first volume (Density I) is observed in all the RNA-GRK2 complexes analyzed (Table S1 and data not shown) and is centered close to the space that would have occupied by the αG helix, as it is in prior GRK2 structures. In the C13.18-GRK2 structure, GRK2 residues 391–398 extend into Density I when superimposed upon the C13.28-GRK2-Gβ1γ2 model, suggesting that Density I could correspond to both RNA and remodeled residues from the basic αF-αG loop region. Density II is adjacent to residues 350–358 of the GRK2 activation loop and was observed in all aptamer complexes except that of the shortest, C13.18. This result suggests that the conformation of the RNA that generates Density II is dependent on the nucleotide sequence immediately 5′ to C47.
Basic Residues from the GRK2 αF-αG Loop Contribute to C13 Binding
Aptamer binding correlates with structural reorganization of the αF-αG loop of GRK2, and our electron density maps from the GRK2-C12.28 structure suggest that this extremely basic loop would be positioned in a manner that could help stabilize aptamer binding. The competition profiles for F-C13.28 with b-GRK2 were also suggestive of additional contacts outside the active site, given the larger IC50 values for C13.18 and ADP (see Table 1 and Figure S5).
To test the role of basic residues in the αF-αG loop region, we simultaneously mutated GRK2-Arg392, -His394, -Lys395, -Lys397, -Lys399 and His400 to alanine. Thermal denaturation profiles of the resulting protein, GRK2-FG-6A, in the presence and absence of ATP indicated that this protein is properly folded and binds ATP (data not shown). In flow cytometry-based competition assays, the IC50 of GRK2-FG-6A for displacing F-GRK2 bound to bC13.28 was ~510 nM, which is over two orders of magnitude greater than that of wild-type GRK2 (Figure 3B). Our filter binding assay confirmed a reduced affinity of C13 for the GRK2-FG-6A variant (Figure 3A), but it was not as dramatic as in the flow cytometry assay. Perhaps heparin, which is included in the filter binding assay, competes with the aptamer for binding to this region of GRK2. Indeed, the basic residues of the αF-αG region are most likely interacting with the phosphodiester backbone of unmodeled nucleotides 56–63 of C13.28 that pass through Density I (Figure 4A).
DISCUSSION
The C13 aptamer inhibits GRK2 with high selectivity and affinity. Our crystallographic structures and biochemical data demonstrate that a small portion of this aptamer (49-AUAC-52) forms extensive interactions with the catalytic cleft between the large and small kinase domains of GRK2, stabilizing a unique conformation of the enzyme that is consistent among seven unique crystal forms. A sharp turn at A51 allows this nucleotide to insert into the canonical ATP binding site of the kinase domain, such that the RNA mimics the binding of the substrate ATP. A51 is key to binding of the aptamer, as ADP can completely inhibit aptamer binding, and mutation of A51 eliminates both C13.28 binding and kinase inhibition. Furthermore, our structure and mutagenesis studies show that a segment of the RNA backbone that mimics the γ-phosphate of ATP makes a critical contact with GRK2-Asp335 through a bridging Mg2+ ion (Figure 1C and 3A).
Contacts outside of the highly conserved active site of GRK2 also clearly stabilize formation of the C13-GRK2 complex and contribute to selectivity. In all of the GRK2-aptamer crystal structures analyzed, even those in which the aptamer lacks a terminal stem, the αF-αG loop and αG helix is reconfigured from its position in the apo-GRK2 structure to accommodate the 3′ end of the aptamer as it exits the active site. Our mutagenesis data show that basic residues in this reconfigured region are essential for high affinity RNA binding, suggesting that they make favorable electrostatic interactions with the RNA. Because all GRKs possess a similarly basic αF-αG region, but adopt distinct inactive conformations, we propose that the inactive conformation of GRK2 best allows for efficient and simultaneous binding of the RNA to the ATP binding site, the basic αF-αG loop region (Density I) and the activation loop region (Density II). In this way, the malleability of the small and large lobes and of the αF-αG loop region all contribute to aptamer selectivity. The terminal stem that tethers the selected region of the aptamer also contributes to selectivity and is critical for high affinity binding to GRK2, most likely by limiting the number of possible conformations for the selected RNA.
We conclude that although the positioning of A51 in the GRK2 active site is key for C13 binding, shape and charge complementarity between C13 and its large interaction surface on GRK2 contribute strongly to RNA affinity and selectivity. The aptamer is therefore distinct from small molecule compounds, such as balanol and those developed by Takeda Pharmaceuticals, which achieve selectivity solely by reading out differences in conformation from within the active site of the kinase domain (Tesmer et al., 2010; Thal et al., 2011). Our research opens the possibility that the C13 aptamer can be used in a high throughput displacement assay to identify novel compounds that not only bind GRK2 with high selectivity and affinity, but also that target surfaces of GRK2 outside the active site important for aptamer binding (Hafner et al., 2006; Hafner et al., 2008).
EXPERIMENTAL PROCEDURES
Reagents
C13 variants listed in Table 1, C13.28A51C and C13.28 5′-labeled with Alexa Fluor-488 (F-C13.28) and 5′ amine labeled C13.18, C13.20 and C13.28 were synthesized by Integrated DNA Technologies (Coralville, IA). All but the amine labeled RNAs were also HPLC-purified. Prior to flow cytometry and phosphorylation assays, these RNAs were treated with a folding protocol (Supplemental Experimental Procedures). Proteins expressed in baculovirus-infected insect cells were purified to homogeneity essentially as described previously. These include bovine GRK2-S670A (Lodowski et al., 2003a; Lodowski et al., 2003b), bovine Gβ1γ2 (Inglese et al., 1992; Lodowski et al., 2003a; Lodowski et al., 2003b) (composed of untagged Gβ1 and N-terminally His-tagged γ2), the palmitoylation-deficient C561S/C562S/C565S variant of human GRK6 (Lodowski et al., 2006; Loudon and Benovic, 1997) and C-terminally His-tagged bovine GRK1 truncated at residue 535 which removes the farnesylation site (Singh et al., 2008) (Figure S2).
Flow Cytometry Protein-RNA Binding Assays
Either GRK2 or 5′ amine-labeled RNA was biotinylated with biotinamidohexanoic acid N-hydroxysuccinimide (Sigma-Aldrich) and attached to SPHERO streptavidin-coated particles (Spherotech). 5′ Alexa Fluor-488-labeled RNA (F-RNA) was synthesized either directly (F-C13.28) or with a 5′ amine that was subsequent labeled with Alexa Fluor-488 carboxylic acid, 2, 3, 5, 6-tetrafluorophenyl ester, 5-isomer (Invitrogen), the same reagent used to fluorescently label GRK2 (F-GRK2). Increasing amounts of F-RNA (or F-GRK2) were incubated with bGRK2- or bRNA-beads, respectively, in a binding buffer consisting of 20 mM Tris pH 7.0, 50 mM NaCl, 5 mM MgCl2, 2 mM DTT and 0.1% Lubrol (without non-specific RNA) for at least 1 hr at 4 °C in the dark. Samples were loaded through a Hypercyt (Intellicyt Corporation) onto an Accuri C6 Flow Cytometer to measure the median fluorescence intensity at ambient temperature. The data was fit in Prism 5.0a. with a background correction corresponding to the fluorescence signal associated with unbound beads. Despite this correction, the best fit for C13.28 and C13.20 data was a total binding site model. Background-corrected C13.18 data fit best with a one site specific binding model. At least three independent experiments of duplicate samples were analyzed.
To determine the selectivity of C13.28 for GRK2, F-GRK2 (2 nM final concentration) was mixed with varying concentrations of unlabeled protein competitor (GRK1, GRK2 variants, or GRK6) prior to the addition of bC13.28-beads (Figure S2). For each RNA, the data were fit in Prism 5.0a with non-linear regression using a single binding site competition model with common top and bottom constraints to calculate IC50 values. Three independent experiments were performed in duplicate or triplicate.
To compare the binding affinity of ADP and different RNA variants to GRK2, competition analysis was performed using bGRK2 and either F-C13.28 (0.5 nM (Table 1) or 1.2 nM final concentration (Figure S5)) or F-C13.18 (35 nM final concentration). Data were processed as described above.
Phosphorylation Assay
GRK2 phosphorylation was measured by modification of a standard protocol (Pronin et al., 2002). GRK2 (50 nM final concentration) was preincubated with varying amounts of RNA on ice for 15 min at twice their final concentration in a sample containing 2.5 μl of GRK2 (in 20 mM Tris pH 8.0 and 50 mM NaCl) and 2.5 μl of RNA (in 20 mM Tris pH 8.0, 50 mM NaCl, and 5 mM MgCl2). To this sample, 5 μl of a 2× phosphorylation mix was added to give a final concentration of 20 mM Tris pH 8.0, 25 mM NaCl, 2.5 mM MgCl2, 100 μM [γ-32P] ATP and 5 μM cholate insoluble Sepia rhodopsin. Samples were incubated for 5 min at room temperature in light. The reaction was stopped with an equal volume of 2× SDS sample buffer. Quenched samples were incubated for 30 min and resolved on a 10–15% SDS-PAGE gel. The gel was dried and exposed to a phosphorimager screen, which was scanned by a Typhoon 9410 imager (GE Healthcare, Waukesha, WI). Bands were quantitated with ImageQuant (version 5.2) software and data was fit in Prism 5.0a with non-linear regression using a single binding site competition model to calculate IC50 values. Three independent experiments were performed.
C13 Mutants and Filter Retention Assay
Point mutants of C13 were obtained by PCR-mediated overlap extension (Heckman and Pease, 2007), as described in Supplemental Experimental Procedures. For the filter retention assay, radiolabeled C13 or point mutants were obtained by in vitro transcription using α-(32P)-GTP, purified with a Nucleo Spin Extract II kit (Machery-Nagel, Germany), and analyzed by gel for their integrity. To measure GRK2-RNA interactions, C13 or its point mutants were incubated with increasing concentrations (0–1000 nM) of human GRK2 in a binding buffer (phosphate buffered saline with 3 mM MgCl2) supplemented with 1 mg ml−1 heparin and 1 mg ml−1 bovine serum albumin, for 30 min at 37 °C. Subsequently, the incubation mix was passed through a 0.45 μm nitrocellulose membrane, washed three times with 200 μl binding buffer, and dried. RNA retained on the membrane was quantified using a FLA-3000 with AIDA Image software. Data were fitted to a one-site specific binding curve with a 4 parameter Hill slope using GraphPad Prism 5. Using this non-linear curve fit, a Kd was determined from at least two independent measurements.
Crystallization and Harvesting Conditions
More than seven truncations of the C13 aptamer were crystallized in a variety of space groups, either in complex with GRK2 or GRK2-Gβ1γ2, using the hanging drop method and different crystallization conditions (Table S1, Supplemental Experimental Procedures, and data not shown).
The C13.28-GRK2-Gβ1γ2 complex was isolated by gel filtration in 20 mM HEPES pH 8.0, 60 mM NaCl, 10 mM CHAPS, 5 mM MgCl2 and 2 mM DTT. The complex gave an A260/A280 ratio of 1.36 ± 0.01 (n=2), which corresponds to approximately 90% protein and 10% nucleic acid (Sambrook and Russell, 2001), as predicted for a 1:1 complex. The protein was concentrated to 6 mg ml−1 and mixed 1:1 (v/v) at 4 °C with a well solution consisting of 100 mM Tris pH 8.5, 200 mM NaCl and 3% PEG 8000. Crystals appeared within approximately one week and grew to dimensions of 180×180×60 μm.
The C13.18-GRK2 complex was isolated by gel filtration in 20 mM HEPES pH 7.0, 50 mM NaCl, 5 mM MgCl2 and 2 mM DTT. The complex had an A260/A280 ratio of 1.33 ± 0.05 (n=2), which corresponds to approximately 90% protein and 10% nucleic acid (Sambrook and Russell, 2001), slightly higher than the predicted 94% protein and 6% RNA. Diffraction-quality crystals were formed by microseeding (Ramsland et al., 2001). The C13.18-GRK2 complex (2 μl of 2 mg ml−1) was mixed with 2 μl of a well solution consisting 100 mM MES pH 5.9, 100 mM NaCl and 5% PEG 3350 supplemented with 0.1 M glycine and 0.2 μl of microseed stock at 4 °C. Thin plates appeared in about a week and grew to dimensions of 200×100×10 μm.
Both crystal forms were harvested by adding a cryoprotective solution containing 30% glycerol directly to the crystallization drop (Supplemental Experimental Procedures). Crystals were mounted on cryoloops and then plunged into liquid nitrogen for data collection.
Data Collection and Structure Determination
Diffraction from crystals of C13.28-GRK2-Gβ1γ2 and C13.18-GRK2 was collected at the Advanced Photon Source beam lines 21-ID-G on MAR CCD detectors at 110 K. The data sets were highly anisotropic, and best results in scaling and map quality were achieved by truncating the observed reflections in ellipsoidal volumes extending to 3.5 Å along a* and b* and 5.5 Å along a* for the C13.18-GRK2 data set, and to 4.5 Å along a* and b* and 7.2 Å along c* for the C13.28-GRK2-Gβ1γ2 data set. The refinement statistics in Table S1 reflect ellipsoidal truncation (Lodowski et al., 2003b), which was found to improve the quality of maps, and thus the spherical completeness statistics are atypically low at high resolution. The structures were solved by molecular replacement using either GRK2 or the GRK2-Gβγ complex from PDB entry 1OMW as a search model in the program Phaser (McCoy et al., 2007). Both structures contain one complex per asymmetric unit. The structures were refined using TLS and restrained refinement in REFMAC5 (Murshudov et al., 1997) alternating with manual model building in O (Jones et al., 1991). The final model of C13.28-GRK2-Gβ1γ2 contains amino acids 29–390, 411–475 and 490–671 of GRK2, 2–340 of Gβ1, and 2–68 of Gγ2, and nucleotides 41–55 and 64–68 of C13.28. The final model of C13.18-GRK2 contains amino acids 30–398, 411–475, 492–545, 552–568 and 576–656 of GRK2 and nucleotides 47–52 of C13.18. Atomic models and structure factors for the C13.28-GRK2-Gβ1γ2 and C13.18-GRK2 complexes have been deposited with the Protein Data Bank as entries 3UZS and 3UZT, respectively.
Supplementary Material
Highlights.
C13 RNA aptamer variants stabilize GRK2 in a unique and remodeled conformation.
The aptamer positions an adenine nucleotide into the ATP binding pocket of GRK2.
Basic residues on an exterior surface of GRK2 facilitate aptamer binding.
The terminal stem of the aptamer indirectly contributes to affinity.
Acknowledgments
The authors thank Dr. John Northup for Sepia cholate insoluble rhodopsin and Dr. David Thal for his contributions to the development of the aptamer-based flow cytometry assay and for purification of human GRK2 used in the C13 filter retention assays. We also thank Nicole Florin for technical assistance. This work was supported by the National Institute of Health grants HL071818, HL086865 and GM081655 (to J.T.). Our research used the Cell and Molecular Biology Core of the Michigan Diabetes Research and Training Center supported by DK20572. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). S.L. acknowledges financial support by the NRW Graduate School LIMES Chemical Biology. The work was further supported by grants from the German research Council (DFG, Ma 3442/1-2) to G.M. V.M.T. contributed to the experimental design, purification and crystallization of aptamer-GRK2 complexes, and flow cytometry-based binding and phosphorylation assays. S.L. performed C13 site-directed mutagenesis and filter retention assays. G.M contributed to the experimental design. J.J.G.T. contributed to the experimental design and structure determinations. V.M.T., J.J.G.T, G.M., and S.L. wrote the manuscript.
Footnotes
Supplemental Information includes one table, six figures, experimental procedures and references.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JA, Taylor SS. Divalent metal ions influence catalysis and active-site accessibility in the cAMP-dependent protein kinase. Protein Sci. 1993;2:2177–2186. doi: 10.1002/pro.5560021217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Doudna JA. Tertiary Motifs in RNA Structure and Folding. Angew Chem Int Ed Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Benovic JL, Mayor F, Jr, Staniszewski C, Lefkowitz RJ, Caron MG. Purification and characterization of the β-adrenergic receptor kinase. J Biol Chem. 1987;262:9026–9032. [PubMed] [Google Scholar]
- Blazer LL, Roman DL, Muxlow MR, Neubig RR. Use of flow cytometric methods to quantify protein-protein interactions. Curr Protoc Cytom. 2010;Chapter 13(Unit 13):11–15. doi: 10.1002/0471142956.cy1311s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguth CA, Singh P, Huang CC, Tesmer JJ. Molecular basis for activation of G protein-coupled receptor kinases. EMBO J. 2010;29:3249–3259. doi: 10.1038/emboj.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetta L. RNA-based therapeutics: ready for delivery? Cell. 2009;136:581–584. doi: 10.1016/j.cell.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Cohn HI, Harris DM, Pesant S, Pfeiffer M, Zhou RH, Koch WJ, Dorn GW, 2nd, Eckhart AD. Inhibition of vascular smooth muscle G protein-coupled receptor kinase 2 enhances α1D-adrenergic receptor constriction. Am J Physiol Heart Circ Physiol. 2008;295:H1695–1704. doi: 10.1152/ajpheart.00564.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Keranen LM, Ellington AD, Newton AC. Isozyme-specific inhibition of protein kinase C by RNA aptamers. J Biol Chem. 1994;269:32051–32054. [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. β-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- Dorn GW., 2nd GRK mythology: G-protein receptor kinases in cardiovascular disease. J Mol Med (Berl) 2009;87:455–463. doi: 10.1007/s00109-009-0450-7. [DOI] [PubMed] [Google Scholar]
- Gurevich EV, Tesmer JJ, Mushegian A, Gurevich VV. G protein-coupled receptor kinases: More than just kinases and not only for GPCRs. Pharmacol Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Schmitz A, Grune I, Srivatsan SG, Paul B, Kolanus W, Quast T, Kremmer E, Bauer I, Famulok M. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- Hafner M, Vianini E, Albertoni B, Marchetti L, Grune I, Gloeckner C, Famulok M. Displacement of protein-bound aptamers with small molecules screened by fluorescence polarization. Nat Protoc. 2008;3:579–587. doi: 10.1038/nprot.2008.15. [DOI] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, 3rd, Ghosh G. Crystal structure of NF-κB (p50)2 complexed to a high-affinity RNA aptamer. Proc Natl Acad Sci U S A. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S, Keneko M, Fujiwara S. In: Cardiotonic agent comprising GRK inhibitor. Patent W, Ikeda S, Keneko M, Fujiwara S, editors. Tadenda Pharmaceutical Company Ltd; 2007. [Google Scholar]
- Inglese J, Koch WJ, Caron MG, Lefkowitz RJ. Isoprenylation in regulation of signal transduction by G-protein-coupled receptor kinases. Nature. 1992;359:147–150. doi: 10.1038/359147a0. [DOI] [PubMed] [Google Scholar]
- Johnson LN. Protein kinase inhibitors: contributions from structure to clinical compounds. Q Rev Biophys. 2009;42:1–40. doi: 10.1017/S0033583508004745. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 (Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci U S A. 2007;104:1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch WJ, Rockman HA, Samama P, Hamilton RA, Bond RA, Milano CA, Lefkowitz RJ. Cardiac function in mice overexpressing the β-adrenergic receptor kinase or a βARK inhibitor. Science. 1995;268:1350–1353. doi: 10.1126/science.7761854. [DOI] [PubMed] [Google Scholar]
- Kong G, Penn R, Benovic JL. A β-adrenergic receptor kinase dominant negative mutant attenuates desensitization of the β2-adrenergic receptor. J Biol Chem. 1994;269:13084–13087. [PubMed] [Google Scholar]
- Lodowski DT, Barnhill JF, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Purification, crystallization and preliminary X-ray diffraction studies of a complex between G protein-coupled receptor kinase 2 and Gβ1γ2. Acta Crystallogr D Biol Crystallogr. 2003a;59:936–939. doi: 10.1107/s0907444903002622. [DOI] [PubMed] [Google Scholar]
- Lodowski DT, Barnhill JF, Pyskadlo RM, Ghirlando R, Sterne-Marr R, Tesmer JJ. The role of Gβγ and domain interfaces in the activation of G protein-coupled receptor kinase 2. Biochemistry. 2005;44:6958–6970. doi: 10.1021/bi050119q. [DOI] [PubMed] [Google Scholar]
- Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gβγ. Science. 2003b;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- Lodowski DT, Tesmer VM, Benovic JL, Tesmer JJ. The structure of G protein-coupled receptor kinase (GRK)-6 defines a second lineage of GRKs. J Biol Chem. 2006;281:16785–16793. doi: 10.1074/jbc.M601327200. [DOI] [PubMed] [Google Scholar]
- Loudon RP, Benovic JL. Expression, purification, and characterization of the G protein-coupled receptor kinase GRK6. J Biol Chem. 1994;269:22691–22697. [PubMed] [Google Scholar]
- Loudon RP, Benovic JL. Altered activity of palmitoylation-deficient and isoprenylated forms of the G protein-coupled receptor kinase GRK6. J Biol Chem. 1997;272:27422–27427. doi: 10.1074/jbc.272.43.27422. [DOI] [PubMed] [Google Scholar]
- Lymperopoulos A, Rengo G, Zincarelli C, Soltys S, Koch WJ. Modulation of adrenal catecholamine secretion by in vivo gene transfer and manipulation of G protein-coupled receptor kinase-2 activity. Mol Ther. 2008;16:302–307. doi: 10.1038/sj.mt.6300371. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mayer G, Faulhammer D, Grattinger M, Fessele S, Blind M. A RNA-based approach towards small-molecule inhibitors. Chembiochem. 2009;10:1993–1996. doi: 10.1002/cbic.200900325. [DOI] [PubMed] [Google Scholar]
- Mayer G, Wulffen B, Huber C, Brockmann J, Flicke B, Neumann L, Hafenbradl D, Klebl BM, Lohse MJ, Krasel C, et al. An RNA molecule that specifically inhibits G-protein-coupled receptor kinase 2 in vitro. RNA. 2008;14:524–534. doi: 10.1261/rna.821908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaye T, Gibelin H, Perdrisot R, Kraimps JL. Pathophysiological roles of G-protein-coupled receptor kinases. Cell Signal. 2005;17:917–928. doi: 10.1016/j.cellsig.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Oppermann M, Freedman NJ, Alexander RW, Lefkowitz RJ. Phosphorylation of the type 1A angiotensin II receptor by G protein-coupled receptor kinases and protein kinase C. J Biol Chem. 1996;271:13266–13272. doi: 10.1074/jbc.271.22.13266. [DOI] [PubMed] [Google Scholar]
- Pippig S, Andexinger S, Daniel K, Puzicha M, Caron MG, Lefkowitz RJ, Lohse MJ. Overexpression of β-arrestin and β-adrenergic receptor kinase augment desensitization of β2-adrenergic receptors. J Biol Chem. 1993;268:3201–3208. [PubMed] [Google Scholar]
- Premont RT, Gainetdinov RR. Physiological roles of G protein-coupled receptor kinases and arrestins. Annu Rev Physiol. 2007;69:511–534. doi: 10.1146/annurev.physiol.69.022405.154731. [DOI] [PubMed] [Google Scholar]
- Premont RT, Macrae AD, Aparicio SA, Kendall HE, Welch JE, Lefkowitz RJ. The GRK4 subfamily of G protein-coupled receptor kinases. Alternative splicing, gene organization, and sequence conservation. J Biol Chem. 1999;274:29381–29389. doi: 10.1074/jbc.274.41.29381. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Loudon RP, Benovic JL. Characterization of G protein-coupled receptor kinases. Methods Enzymol. 2002;343:547–559. doi: 10.1016/s0076-6879(02)43157-6. [DOI] [PubMed] [Google Scholar]
- Ramsland PA, Upshaw JL, Shultz BB, DeWitt CR, Chissoe WF, III, Raison RL, Edmundson AB. Interconversion of different crystal forms of Fabs from human IgM cryoglobulins. Journal of Crystal Growth. 2001;232:204–214. [Google Scholar]
- Reyes FE, Garst AD, Batey RT. Strategies in RNA crystallography. Methods Enzymol. 2009;469:119–139. doi: 10.1016/S0076-6879(09)69006-6. [DOI] [PubMed] [Google Scholar]
- Rockman HA, Chien KR, Choi DJ, Iaccarino G, Hunter JJ, Ross J, Jr, Lefkowitz RJ, Koch WJ. Expression of a β-adrenergic receptor kinase 1 inhibitor prevents the development of myocardial failure in gene-targeted mice. Proc Natl Acad Sci U S A. 1998;95:7000–7005. doi: 10.1073/pnas.95.12.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Seiwert SD, Stines Nahreini T, Aigner S, Ahn NG, Uhlenbeck OC. RNA aptamers as pathway-specific MAP kinase inhibitors. Chem Biol. 2000;7:833–843. doi: 10.1016/s1074-5521(00)00032-6. [DOI] [PubMed] [Google Scholar]
- Singh P, Wang B, Maeda T, Palczewski K, Tesmer JJ. Structures of rhodopsin kinase in different ligand states reveal key elements involved in G protein-coupled receptor kinase activation. J Biol Chem. 2008;283:14053–14062. doi: 10.1074/jbc.M708974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer JJ, Tesmer VM, Lodowski DT, Steinhagen H, Huber J. Structure of human G protein-coupled receptor kinase 2 in complex with the kinase inhibitor balanol. J Med Chem. 2010;53:1867–1870. doi: 10.1021/jm9017515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer VM, Kawano T, Shankaranarayanan A, Kozasa T, Tesmer JJ. Snapshot of activated G proteins at the membrane: the Gαq-GRK2-Gβγ complex. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- Thal DM, Yeow RY, Schoenau C, Huber J, Tesmer JJ. Molecular mechanism of selectivity among G protein-coupled receptor kinase 2 inhibitors. Mol Pharmacol. 2011;80:294–303. doi: 10.1124/mol.111.071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.