Abstract
Small molecules produced in Nature continue to be an inspiration for the development of new therapeutic agents. These natural products possess exquisite chemical diversity, which gives rise to their wide range of biological activities. In their host organism, natural products are assembled and modified by dedicated biosynthetic pathways that Nature has meticulously developed. Often times, the complex structures or chemical modifications instated by these pathways are difficult to replicate using traditional synthetic methods. An alternative approach for creating or enhancing the structural variation of natural products is through combinatorial biosynthesis. By rationally reprogramming and manipulating the biosynthetic machinery responsible for their production, unnatural metabolites that were otherwise inaccessible can be obtained. Additionally, new chemical structures can be synthesized or derivatized by developing the enzymes that carry out these complicated chemical reactions into biocatalysts. In this review, we will discuss a variety of combinatorial biosynthetic strategies, their technical challenges, and highlight some recent (since 2007) examples of rationally designed unnatural metabolites, as well as platforms that have been established for the production and modification of clinically important pharmaceutical compounds.
Introduction
Small molecules produced by plants, microorganisms and animals play a pivotal role in drug discovery programs. These natural products have evolved over time to interact with diverse biological targets and represent some of the most important pharmaceutical agents in human health care [1]. Nevertheless, due to the emergence of new diseases and the increasing resistance of bacteria to clinically used antibiotics, new therapeutic agents possessing novel modes of action are urgently needed. Semi-synthetic modifications of natural products aimed at enhancing their biological properties or the total synthesis of analogues serve as great starting points for generating such compounds [2]. Unfortunately, these desired molecules or modifications are sometimes commercially unfeasible due to restrictions affiliated with complex structures, pre-existing functional groups, low yields, and environmentally unfriendly byproducts generated in the reactions. Synthetic biology represents an alternative platform for developing new potential drug candidates. By genetically manipulating the biosynthetic machinery involved in the assembly of natural products and exploiting Nature’s strategies for synthesizing structurally diverse metabolites, compounds with enhanced biological features can be produced that were otherwise inaccessible using traditional synthetic methods. As this is a mini-review, we will discuss synthetic biology in the context of constructing tailor made molecules by rationally mixing and matching genetic information from biosynthetic pathways (combinatorial biosynthesis) and regret not being able to focus on small molecule production using mutasynthesis and chemobiosynthesis [3], epigenetics [4], or the overexpression of transcription factors in biosynthetic pathways [5].
Natural product building blocks: supply and demand
Based on their structural composition, most natural products can be classified as polyketides; peptides; terpenoids; oligosaccharides, including deoxysugars and aminoglycosides; alkaloids or hybrids, which contain two or more of these structural features. Recent advances in genome sequencing and bioinformatics have shown that bacteria and fungi are not only prolific producers of natural products, but that the genes responsible for the assembly of a metabolite are generally clustered together within the chromosome [6,7]. By having access to this full ensemble of genes, the chemical logic governing the assembly of a metabolite can be deduced by correlating genetic information to protein function. This logic can then be used to rationally manipulate genes for the synthesis of new metabolites, such as reprogramming the biosynthetic assembly line, altering the specificity of tailoring enzymes, or expressing a collection of genes in a surrogate host (Figure 1).
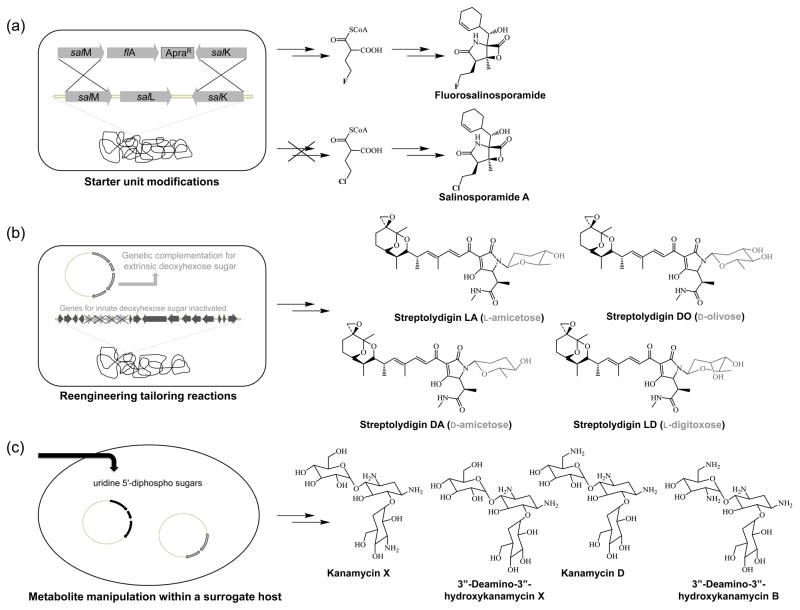
Figure 1.
Strategies for combinatorial biosynthesis. A) Engineering the production of fluorosalinosporamide in Salinospora tropica by replacing the chlorinase gene salL with the fluorinase gene flA from Streptomyces catteleya; B) Formation of glycosylated streptolvdigin derivatives by expressing genes coding for several deoxyhexoses in a mutant strain lacking the ability to produce its innate deoxyhexose moiety; C) Production of kanamycin derivatives by heterologously expressing different combinations of biosynthetic genes from Streptomyces kanamyceticus in the non-aminoglycoside-producing Streptomyces venezuelae host.
Polyketides and nonribosomal peptides represent two diverse classes of small molecules possessing a wide range of biological activities and are typically biosynthesized in an assembly-line fashion by multidomain and multimodular polyketide synthases (PKS) and nonribosomal peptide synthetases (NRPS), respectively. These megasynthases act as biosynthetic templates in which the catalytic domains embedded in the enzyme dictate the number and type of building blocks that are to be incorporated into the final product, as well as any tailoring modifications [6–10]. In polyketide biosynthesis, the general building blocks arise from simple acyl-coenzyme A (acyl-CoA) or acyl carrier protein (ACP) linked precursors [9,11], whereas in nonribosomal peptide biosynthesis, proteinogenic and non-proteinogenic amino acids can be incorporated into the growing peptide chain via aminoacyl thioesters [7,10]. While PKSs primarily choose malonyl-coenzyme A (malonyl-CoA) and methylmalonyl-coenzyme A (methylmalonyl-CoA) as their chain-extending units, specialized acyl-CoA monomers can also be incorporated into the molecule giving rise to its increased chemical diversity [9,12]. These specialized monomers, as well as the non-proteinogenic amino acids in NRPS systems, are generally synthesized from primary metabolic building blocks using a sub-collection of enzymes embedded within the corresponding gene cluster [7]. Therefore, in a re-engineered biological system, the supply of a specialized monomer must be rationalized by either providing the genes coding for its biosynthesis in trans or supplementing the system with exogenous precursors. For example, to increase the supply of unique PKS extender units for in vitro systems, a promiscuous malonyl-CoA synthetase, which is used to convert diacids into CoA-linked PKS extender units, was created and analyzed for its increased substrate tolerance [13]. Using the crystal structure of the malonyl-CoA synthetase MatB from Streptomyces coelicolor as a guide [14], two key residues predicted to control substrate specificity were selected for saturation mutagenesis experiments within the analogous MatB synthetase from Rhizobium trifolii. From the constructed mutagenesis library, two mutants were shown to possess increased substrate tolerance toward non-traditional malonate analogues such as ethylmalonate and allylmalonate. The dedicated pathway responsible for the biosynthesis of the atypical extender unit allylmalonyl-CoA, which is incorporated in the clinically important immunosuppressant FK506, was also recently elucidated [15].
Re-programming natural product assembly lines
Re-programming PKS synthases for enhanced chemical diversity has been a major focus in combinatorial biosynthesis. Rational re-programming of the avermectin PKS in Streptomyces avermitilis M1 was used to produce the veterinary antiparasitic drug doramectin [16]. While avermectin selects isobutyryl-CoA as its starting molecule, the biosynthesis of doramectin requires initiation with a unique cyclohexanecarboxylic (CHC) unit. Therefore, in an attempt to generate doramectin for industrial applications, the loading module of the avermectin PKS was replaced with the CHC loading module from Streptomyces platensis’ phoslactomycin PKS (Figure 2a). Additionally, in order to supply the starter molecule to the host, five putative genes responsible for forming CHC-CoA were also cloned from S. platensis and transformed into the mutant.
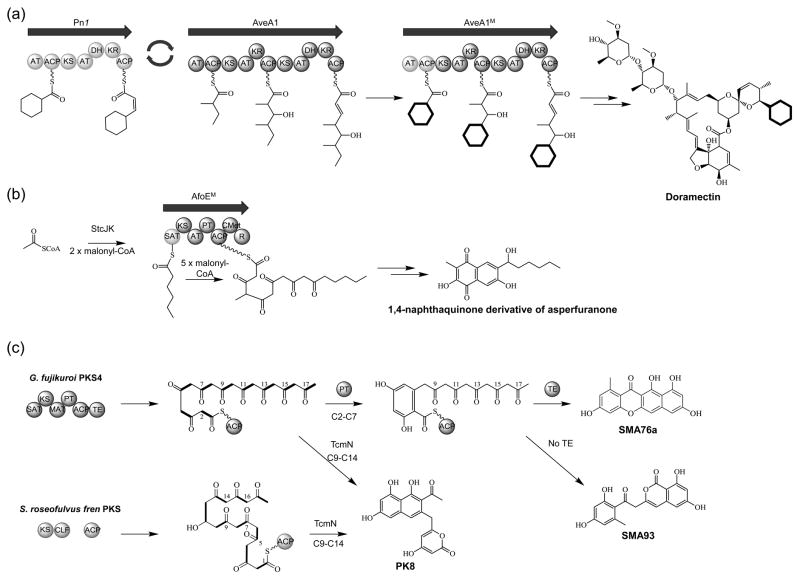
Figure 2.
Reprogramming polyketide megasynthases through rational domain swapping. A) Formation of doramectin in Streptomyces avermitilis by replacing the loading module of the avermectin PKS with the cyclohexanecarboxylic (CHC) loading module from phoslactomycin PKS; B) Production of an unnatural natural product in Aspergillus nidulans by swapping the SAT domain of the asperfuranone PKS with the sterigmatocystin SAT; C) Biosynthesis of aromatic bacterial polyketides in Escherichia coli using an engineered fungal PKS and bacterial cyclase.
Recent advances in genome sequencing have shown that bacteria and fungi contain a large number of PKS biosynthetic clusters in their genomes, and this wealth of information can be applied toward engineering the biosynthesis of novel polyketides [6]. Unfortunately from a synthetic biology perspective (i.e. standardized parts), in contrast to their well-studied bacterial counterparts, fungal megasynthases are structurally and mechanistically distinct. Fungal polyketides are assembled by iterative PKS (IPKS), which contain single copies of catalytic domains arranged on a single polypeptide and function by repeatedly using these domains in different combinations to synthesize a structurally diverse molecule [17]. As a result, the programming rules dictating the assembly of a metabolite remain relatively unresolved and limit the ability to engineer novel fungal polyketides. However, the biochemical functions governing these megasynthases are becoming more clear thanks to recent technological advances, such as algorithms for delineating domain boundaries [18]; crystallographic studies on mammalian and fungal fatty acid synthases [19,20]; and biochemical characterization of intact and dissected domains such as starter unit selection domains in non-reducing PKS (NR-PKS) [21], product template domains in NR-PKS [22,23], catalytic functions of highly-reducing PKS (HR-PKS) [24,25], and the protein-protein interactions between PKS and NRPS modules of hybrid PKS-NRPS [24,26]. Using this information as a guide, Wang and co-workers created a chimeric fungal PKS in Aspergillus nidulans by replacing the starter unit ACP transacylase (SAT) domain within the asperfuranone NR-PKS AfoE with the SAT domain from the sterigmatocystin NR-PKS StcA [27]. After swapping the loading domains, the AfoE hybrid was able to load the hexanoyl starter unit produced by StcJ/StcK and extend it to the octaketide intermediate (Figure 2b). Interestingly, the new naphthaquinone derivative that was synthesized contains the same chain length as the native AfoE product despite being initiated with a different length starter unit. Another example of fungal PKS domain swapping was used for the production of bacterial polyketides in E. coli [28]. Pharmaceutically important aromatic polyketides, such as the tetracyclines and anthracyclines, are produced by bacterial type II PKS. The ability to reconstitute their biosynthetic pathway in E. coli is inhibited because the bacterial minimal PKS, which generates the poly-β-ketone backbone, forms inclusion bodies. Therefore, to circumvent the inability to heterologously express these enzymes in E. coli, the fungal NR-PKS PKS4 from Gibberella fujikuroi was dissected and reassembled into a synthetic PKS. When expressed in E. coli, the synthetic PKS was shown to biosynthesize a variety of products not observed among fungal metabolites. Furthermore, when expressed with bacterial tailoring enzymes, complex bacterial aromatic polyketides were biosynthesized (Figure 2c). Therefore, with increasing knowledge of the different biosynthetic machineries, constructing tailored complex megasynthases from individual domains has becoming attainable.
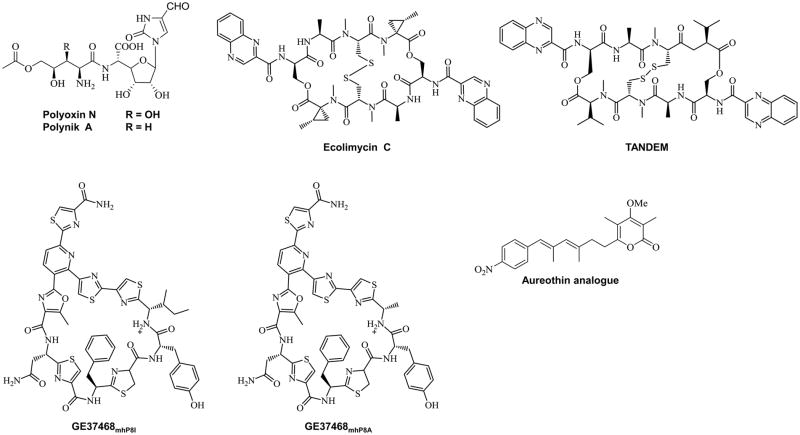
As demonstrated with the aminocoumarins [29], the development of λ RED-mediated recombination has also drastically impacted the success of combinatorial biosynthesis [30]. Having the technology to rapidly inactivate key biosynthetic genes or replace regions of DNA has greatly enhanced the ability to generate new natural product derivatives and interrogate substrate specificity of downstream tailoring enzymes. Using this technology, the biosynthesis of fluorosalinosporamide was rationally engineered in Salinispora tropica [31]. In order to produce new fermentation-based variants of the novel anticancer agent salinosporamide A for SAR studies, the chlorinase gene salL was chromosomally replaced with the fluorinase gene flA from Streptomyces cattleya using λ-RED-mediated recombination (Figure 1A). After inorganic fluoride was supplemented to the culture medium, the resulting S. tropica mutant was able to biosynthesize the 4-fluorocrotonic-Co-A building block, which was further processed into the fluorinated metabolite. λ-RED-mediated recombination was also used for the construction of the hybrid peptidyl nucleoside antibiotics polyoxin N and polynik A (Figure 3) in Streptomyces ansochromogenes [32]. In an effort to create more potent antibiotics, genes coding for the biosynthesis of the dipeptidyl moiety of polyoxin from Streptomyces cacaoi were assembled into the pSET152-derived plasmid pPOL2 by λ-Red-mediated recombination and introduced into the S. ansochromogenes sanN mutant, which is incapable of producing the peptidyl moiety of the nikkomycins. By meriting biological properties from both parents, the two hybrid metabolites exhibited improved antibiotic activities. It should be noted that the genetic manipulations discussed in this section were made possible because the biochemistry of the native natural product pathway was completely understood. Therefore, it should be taken into consideration for anyone who wants to dive into combinatorial biosynthesis that modified natural product structures are conceived by rationally manipulating a biosynthetic pathway and not merely from a “black box” approach.
Figure 3.
Examples of unnatural natural products created through combinatorial biosynthesis.
Evaluating the substrate specificity of tailoring enzymes
Ultimately, developing a successful biosynthetic platform for synthesizing unnatural natural products requires broadening the substrate specificity of tailoring enzymes so that they recognize and act on a newly formed molecule. For instance, when implementing a highly integrative approach for engineering aureothin analogues, Hertweck and collaborators examined the substrate specificity of the α-pyrone O-methyltransferase EncK from the enterocin biosynthetic pathway [33]. When the endogenous γ-regiospecific pyrone methyltransferase AurI was replaced with the with the α-regiospecific EncK homologue, iso-deoxyaureothin derivatives were produced (Figure 3) in the heterologous Streptomyces albus host. However, aurI could not complement a ΔencK mutant, which demonstrated that EncK possesses a broader substrate tolerance than AurI and could be further developed as a biocatalyst for modifying pyrone moieties.
Many biologically active natural products are also decorated with deoxysugars, which are attached by dedicated glycotransferases. For some of these molecules, their biological activities are attributed to the sugars moieties and efforts have been made at modifying these groups in hopes of enhancing their activities. While glycoside modifications are difficult using traditional synthetic methods, enzymatic glycorandomization is proving to be a powerful alternative [34]. The dienoyltetramic acid streptolydigin produced by Streptomyces lydicus NRRL2433 is a potent inhibitor of bacterial RNA polymerase and its biosynthetic cluster was recently identified and characterized [35]. A final step in streptolydigin biosynthesis involves the attachment of the deoxyhexose sugar L-rhodinose to the aglycon moiety. To interrogate the specificity of its glycosyltransferase toward deoxysugars and generate novel glycosylated derivatives, the sub-gene cluster responsible for the production of L-rhodinose was inactivated and the resulting mutant was genetically complemented with plasmids harboring genes involved in the biosynthesis of D-amicetose, L-olivose, D-olivose, and L-digitoxose (Figure 1b). New glycosylated streptolydigin derivatives were generated, which implies that the glycosyltransferase has a certain degree of substrate flexibility toward deoxysugars and sets the stage for further engineering studies aimed at generating more potent analogs. Deoxyhexopyranose analogues of the antitumor antibiotic gilvocarcin were also engineered in a similar manner [36]. Gilvocarcin’s natural D-fucofuranose pathway was inactivated and genetically complemented with plasmids coding for neutral deoxysugars. New gilvocarcin analogues possessing altered saccharide moieties were produced and its C-glycosyltransferase was determined to possess moderate flexibility because it was able to transfer both D- and L-hexopyranose sugars onto gilvocarcin’s angucyclinone scaffold.
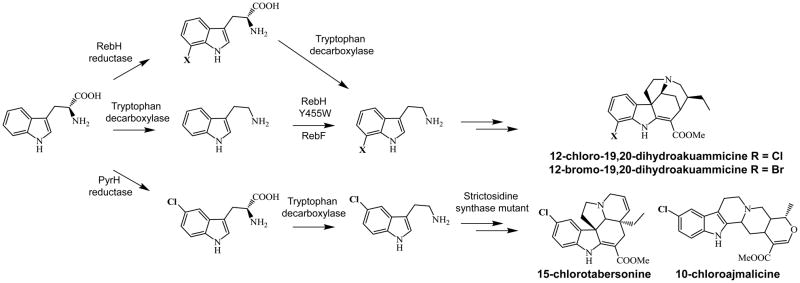
In addition to microorganisms, plants are also copious producers of natural products, many of which possess desired biological activities [37]. But, because the genes coding for a metabolite are generally scattered throughout the genome and not clustered together, the ability to identify all biosynthetic genes and ultimately modify key intermediates and building blocks or broaden the substrate specificity of downstream enzymes has hindered in planta biosynthesis. However, recent pioneering efforts by the O’Connor lab have been aimed at establishing the medicinal plant Catharanthus roseus as a biosynthetic platform [38,39]. In an endeavor to increase the biologically activity of plant metabolites by incorporating halogen atoms into their structures, the chlorinating flavoenzymes PyrH and RebH, as well as their partner reductases from soil bacteria, were introduced into C. roseus [38]. These halogenases chlorinate tryptophan in a regioselective manner and functioned in planta to generate their 5-chloro and 7-chloro tryptophan precursors, respectively (Figure 4). In alkaloid biosynthesis, one of the early biosynthetic enzymes is strictosidine synthase and because of its inability to turn over 5-chlorotryptamine, a mutant synthase (STRvm) was also introduced. After cultivation, chlorinated alkaloids were observed indicating that not only are flavin halogenases transportable among kingdoms, but that the tryptophan decarboxylase, albeit with low efficiency, can turn over halogenated tryptophan building blocks. To circumvent this bottleneck, the halogenase RebH was engineered and shown to preferentially chlorinate tryptamine, a downstream alkaloid precursor, rather than tryptophan (Figure 4) [39]. Additional protein engineering experiments, as well as directed evolution methods, have been developed for natural product engineering and the reader is referred to a recent review on this topic for further information [40].
Figure 4.
Engineered production of halogenated metabolites in Catharanthus roseus.
Heterologous expression and pathway manipulation in a surrogate host
In many cases, because the native natural product producer does not grow well or is genetically intractable, a heterologous host must be used for the reconstitution and genetic manipulation of its biosynthetic pathways [41]. Depending on the source that the genes are being transferred from and what type of a metabolite is to be biosynthesized, some hosts are more suitable for pathway reconstitution than others. Because of its quick growth rate and the ease of performing genetic manipulations, E. coli is the most commonly used heterologous host. Many pharmaceutically important natural products have been successfully reconstituted in E. coli and can now serve as platforms for the engineering of more bioactive or less toxic derivatives, such as the antibiotic erythromycin C [42] and its 6-deoxyerythronolide B macrocyclic core [43], the antimalarial artemisinin precursor artemisinic acid [44], the anticancer drugs epothilone C and D [45], the taxol precursor taxadien-5a-ol [46], as well as the ginkgolide precursor levopimaradiene [47]. Recently, a new quinomycin antibiotic was synthesized in E. coli after the plasmid-based echinomycin biosynthetic pathway was re-engineered to incorporate a NRPS gene from the SW-163 quinomycin cluster [48]. Using E. coli as a flexible platform, the unnatural hybrid nonribosomal peptide ecolimycin C (Figure 3) was produced and shown to be a more potent antibiotic than its parent compound. The same echinomycin plasmid-borne pathway was also rationally engineered to biosynthesize the antibiotic TANDEM (Figure 3), which is a synthetic analogue of triostin A [49].
While E. coli has proven to a powerful heterologous expression host; especially for the reconstitution of polyketides [50], alternative hosts are occasionally required if the metabolite being biosynthesized is toxic or if the building blocks are not available. Streptomyces can serve as more advantageous hosts because they already biosynthesize many secondary metabolites and thus have the pathways necessary for many of the essential building blocks [41,51]. For example, Streptomyces fradiae and Streptomyces roseosporus, which are commonly used for the expression and manipulation of lipopetides [51], were used to generate derivatives of the antibiotics daptomycin [52] and A54145 factors [53]. On the other hand, the non-aminoglycoside-producing host Streptomyces venezuelae was used for the synthesis of glycosylated doxorubicin analogs [54]. Doxorubicin is one of the most widely used anticancer drugs and its biological activity is attributed to its deoxysugar L-daunosamine. However, due to its severe side effects, the semisynthetic derivative epirubicin, which contains the deoxysugar with the opposite configuration at the C-4 hydroxyl group, was developed and shown to be less toxic. In an attempt to create the next generation of anticancer drugs, S. venezuelae, which lacks the ability to produce the polyketide pikromycin and its desosamine sugar, was transformed with various plasmids coding for different nucleotide deoxysugars, as well as their glycosyltransferases and tailoring enzymes, and ultimately engineered as a platform for biosynthesizing doxorubicin analogs containing unnatural sugar moieties. S. venezuelae was also used for unraveling the biosynthesis of the aminoglyoside antibiotic kanamycin [55]. By heterologously expressing different combinations of genes from the natural kanamycin producer Streptomyces kanamyceticus in S. venezuelae, Yoon and co-workers elucidated the biochemistry and genetics underlying the biosynthesis of the 4,6-disubstituted aminoglyoside antibiotics and were able to use this knowledge for engineering of kanamycin congeners with improved biological activity (Figure 1c). Finally, in addition to expressing clusters coding for polyketides, nonribosomal peptides, and terpenes, Streptomyces lividans was established as a tractable genetic host for expressing the ribosomally synthesized thiazolyl peptide GE37468 and its preprotein derivatives [56]. Altogether, having a well-established genetic system for manipulating thiazolyl peptide or thiopeptides biosynthesis will enable the genetic manipulation of these metabolites for further structure activity relationships. Additionally, given the recent advances in metabolic engineering [57], it will be interesting to see how the maturation of unnatural amino acid incorporation technology will lead to new building blocks for ribosomal peptides [58].
Perspective
Given their unprecedented chemical diversity and novel modes of action, natural products continue to function as lead compounds in many drug discovery programs. However, to keep up with the growing demand for new therapeutic agents, new strategies for producing target compounds are needed. Synthetic biology serves as a powerful platform for this endeavor by generating unnatural natural products with enhanced biological features. By rationally reprogramming and manipulating the biosynthetic machinery responsible for their production, unnatural metabolites that were otherwise inaccessible through traditional methods are brought to light. As the fields of genomics and biotechnology rapidly advance, new resources capable of biosynthesizing natural products with unparalleled structural features are constantly revealed. With the relative low-cost of genome sequencing, many organisms known to carry out intriguing chemical reactions can be sequenced by individual laboratories so that the corresponding gene cluster can be readily identified [59,60]. By uncovering the biosynthetic machinery responsible for these metabolites, we increase our basic understanding on how Nature synthesizes these chemical entities and can apply this newfound knowledge toward enhancing the structural variation of known molecules. Furthermore, the enzymes responsible for these chemical reactions can be harvested as biocatalysts and used in the synthesis and derivatization of biologically active compounds and intermediates. It is our hope that by exploiting Nature’s molecular blueprint for synthesizing structurally diverse metabolites, synthetic biology can be used as a powerful platform for creating new therapeutic agents.
Acknowledgments
Due to space constraint, we apologize for omission of other relevant works. Research on combinatorial biosynthesis in our lab is supported by funds from NIH, the Alfred Sloan Foundation and the David and Lucile Packard Foundations.
References and Recommended Reading
- 1.Li JW, Vederas JC. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 2.Carter GT. Natural products and Pharma 2011: strategic changes spur new opportunities. Nat Prod Rep. 2011;28:1783–1789. doi: 10.1039/c1np00033k. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy J. Mutasynthesis, chemobiosynthesis, and back to semi-synthesis: combining synthetic chemistry and biosynthetic engineering for diversifying natural products. Nat Prod Rep. 2008;25:25–34. doi: 10.1039/b707678a. [DOI] [PubMed] [Google Scholar]
- 4.Brakhage AA, Schroeckh V. Fungal secondary metabolites - strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Smanski MJ, Shen B. Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl Microbiol Biotechnol. 2010;86:19–25. doi: 10.1007/s00253-009-2428-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winter JM, Behnken S, Hertweck C. Genomics-inspired discovery of natural products. Curr Opin Chem Biol. 2011;15:22–31. doi: 10.1016/j.cbpa.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Walsh CT, Fischbach MA. Natural products version 2. 0: connecting genes to molecules. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulder TA, Freeman MF, Piel J. The catalytic diversity of multimodular polyketide synthases: natural product biosynthesis beyond textbook assembly rules. Top Curr Chem. 2011:1–53. doi: 10.1007/128_2010_113. [DOI] [PubMed] [Google Scholar]
- 9.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 10.Schwarzer D, Finking R, Marahiel MA. Nonribosomal peptides: from genes to products. Nat Prod Rep. 2003;20:275–287. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- 11.Chan YA, Podevels AM, Kevany BM, Thomas MG. Biosynthesis of polyketide synthase extender units. Nat Prod Rep. 2009;26:90–114. doi: 10.1039/b801658p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson MC, Moore BS. Beyond ethylmalonyl-CoA: the functional role of crotonyl-CoA carboxylase/reductase homologs in expanding polyketide diversity. Nat Prod Rep. 2011 doi: 10.1039/c1np00082a. [DOI] [PubMed] [Google Scholar]
- 13.Koryakina I, Williams GJ. Mutant malonyl-CoA synthetases with altered specificity for polyketide synthase extender unit generation. Chembiochem. 2011;12:2289–2293. doi: 10.1002/cbic.201100383. [DOI] [PubMed] [Google Scholar]
- 14.Hughes AJ, Keatinge-Clay A. Enzymatic extender unit generation for in vitro polyketide synthase reactions: structural and functional showcasing of Streptomyces coelicolor MatB. Chem Biol. 2011;18:165–176. doi: 10.1016/j.chembiol.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Mo S, Kim DH, Lee JH, Park JW, Basnet DB, Ban YH, Yoo YJ, Chen SW, Park SR, Choi EA, Kim E, et al. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J Am Chem Soc. 2011;133:976–985. doi: 10.1021/ja108399b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JB, Pan HX, Tang GL. Production of doramectin by rational engineering of the avermectin biosynthetic pathway. Bioorg Med Chem Lett. 2011;21:3320–3323. doi: 10.1016/j.bmcl.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Cox RJ. Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 18.Udwary DW, Merski M, Townsend CA. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J Mol Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenni S, Leibundgut M, Maier T, Ban N. Architecture of a fungal fatty acid synthase at 5 A resolution. Science. 2006;311:1263–1267. doi: 10.1126/science.1123251. [DOI] [PubMed] [Google Scholar]
- 20.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 21.Crawford JM, Dancy BC, Hill EA, Udwary DW, Townsend CA. Identification of a starter unit acyl-carrier protein transacylase domain in an iterative type I polyketide synthase. Proc Natl Acad Sci U S A. 2006;103:16728–16733. doi: 10.1073/pnas.0604112103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Xu W, Tang Y. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. J Biol Chem. 2010;285:22764–22773. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fisch KM, Bakeer W, Yakasai AA, Song Z, Pedrick J, Wasil Z, Bailey AM, Lazarus CM, Simpson TJ, Cox RJ. Rational domain swaps decipher programming in fungal highly reducing polyketide synthases and resurrect an extinct metabolite. J Am Chem Soc. 2011;133:16635–16641. doi: 10.1021/ja206914q. [DOI] [PubMed] [Google Scholar]
- 25.Ma SM, Li JW, Choi JW, Zhou H, Lee KK, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326:589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu W, Cai X, Jung ME, Tang Y. Analysis of intact and dissected fungal polyketide synthase-nonribosomal peptide synthetase in vitro and in Saccharomyces cerevisiae. J Am Chem Soc. 2010;132:13604–13607. doi: 10.1021/ja107084d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu T, Chiang YM, Somoza AD, Oakley BR, Wang CC. Engineering of an “unnatural” natural product by swapping polyketide synthase domains in Aspergillus nidulans. J Am Chem Soc. 2011;133:13314–13316. doi: 10.1021/ja205780g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Li Y, Tang Y. Engineered biosynthesis of bacterial aromatic polyketides in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:20683–20688. doi: 10.1073/pnas.0809084105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heide L. Genetic engineering of antibiotic biosynthesis for the generation of new aminocoumarins. Biotechnol Adv. 2009;27:1006–1014. doi: 10.1016/j.biotechadv.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 30.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A. 2003;100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eustaquio AS, O’Hagan D, Moore BS. Engineering fluorometabolite production: fluorinase expression in Salinispora tropica yields fluorosalinosporamide. J Nat Prod. 2010;73:378–382. doi: 10.1021/np900719u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Li L, Tian Y, Niu G, Tan H. Hybrid antibiotics with the nikkomycin nucleoside and polyoxin peptidyl moieties. Metab Eng. 2011;13:336–344. doi: 10.1016/j.ymben.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Werneburg M, Busch B, He J, Richter ME, Xiang L, Moore BS, Roth M, Dahse HM, Hertweck C. Exploiting enzymatic promiscuity to engineer a focused library of highly selective antifungal and antiproliferative aureothin analogues. J Am Chem Soc. 2010;132:10407–10413. doi: 10.1021/ja102751h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gantt RW, Peltier-Pain P, Thorson JS. Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules. Nat Prod Rep. 2011;28:1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]
- 35.Olano C, Gomez C, Perez M, Palomino M, Pineda-Lucena A, Carbajo RJ, Brana AF, Mendez C, Salas JA. Deciphering biosynthesis of the RNA polymerase inhibitor streptolydigin and generation of glycosylated derivatives. Chem Biol. 2009;16:1031–1044. doi: 10.1016/j.chembiol.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 36.Shepherd MD, Liu T, Mendez C, Salas JA, Rohr J. Engineered biosynthesis of gilvocarcin analogues with altered deoxyhexopyranose moieties. Appl Environ Microbiol. 2011;77:435–441. doi: 10.1128/AEM.01774-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollier J, Moses T, Goossens A. Combinatorial biosynthesis in plants: A (p)review on its potential and future exploitation. Nat Prod Rep. 2011;28:1897–1916. doi: 10.1039/c1np00049g. [DOI] [PubMed] [Google Scholar]
- 38.Runguphan W, Qu X, O’Connor SE. Integrating carbon-halogen bond formation into medicinal plant metabolism. Nature. 2010;468:461–464. doi: 10.1038/nature09524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glenn WS, Nims E, O’Connor SE. Reengineering a tryptophan halogenase to preferentially chlorinate a direct alkaloid precursor. J Am Chem Soc. 2011 doi: 10.1021/ja2089348. [DOI] [PubMed] [Google Scholar]
- 40.Zabala AO, Cacho RA, Tang Y. Protein engineering towards natural product synthesis and diversification. J Ind Microbiol Biotechnol. 2011 doi: 10.1007/s10295-011-1044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickens LB, Tang Y, Chooi YH. Metabolic Engineering for the Production of Natural Products. Annu Rev Chem Biomol. 2011;2:211–236. doi: 10.1146/annurev-chembioeng-061010-114209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peiru S, Menzella HG, Rodriguez E, Carney J, Gramajo H. Production of the potent antibacterial polyketide erythromycin C in Escherichia coli. Appl Environ Microbiol. 2005;71:2539–2547. doi: 10.1128/AEM.71.5.2539-2547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science. 2001;291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 44.Chang MC, Eachus RA, Trieu W, Ro DK, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3:274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- 45.Mutka SC, Carney JR, Liu Y, Kennedy J. Heterologous production of epothilone C and D in Escherichia coli. Biochemistry. 2006;45:1321–1330. doi: 10.1021/bi052075r. [DOI] [PubMed] [Google Scholar]
- 46.Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leonard E, Ajikumar PK, Thayer K, Xiao WH, Mo JD, Tidor B, Stephanopoulos G, Prather KL. Combining metabolic and protein engineering of a terpenoid biosynthetic pathway for overproduction and selectivity control. Proc Natl Acad Sci U S A. 2010;107:13654–13659. doi: 10.1073/pnas.1006138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe K, Hotta K, Nakaya M, Praseuth AP, Wang CC, Inada D, Takahashi K, Fukushi E, Oguri H, Oikawa H. Escherichia coli allows efficient modular incorporation of newly isolated quinomycin biosynthetic enzyme into echinomycin biosynthetic pathway for rational design and synthesis of potent antibiotic unnatural natural product. J Am Chem Soc. 2009;131:9347–9353. doi: 10.1021/ja902261a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe K, Hotta K, Praseuth AP, Searcey M, Wang CC, Oguri H, Oikawa H. Rationally engineered total biosynthesis of a synthetic analogue of a natural quinomycin depsipeptide in Escherichia coli. Chembiochem. 2009;10:1965–1968. doi: 10.1002/cbic.200900260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X, Wang P, Tang Y. Engineered polyketide biosynthesis and biocatalysis in Escherichia coli. Appl Microbiol Biotechnol. 2010;88:1233–1242. doi: 10.1007/s00253-010-2860-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baltz RH. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol. 2010;37:759–772. doi: 10.1007/s10295-010-0730-9. [DOI] [PubMed] [Google Scholar]
- 52.Alexander DC, Rock J, He X, Brian P, Miao V, Baltz RH. Development of a genetic system for combinatorial biosynthesis of lipopeptides in Streptomyces fradiae and heterologous expression of the A54145 biosynthesis gene cluster. Appl Environ Microbiol. 2010;76:6877–6887. doi: 10.1128/AEM.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen KT, He X, Alexander DC, Li C, Gu JQ, Mascio C, Van Praagh A, Mortin L, Chu M, Silverman JA, Brian P, et al. Genetically engineered lipopeptide antibiotics related to A54145 and daptomycin with improved properties. Antimicrob Agents Chemother. 2010;54:1404–1413. doi: 10.1128/AAC.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han AR, Park JW, Lee MK, Ban YH, Yoo YJ, Kim EJ, Kim E, Kim BG, Sohng JK, Yoon YJ. Development of a Streptomyces venezuelae-based combinatorial biosynthetic system for the production of glycosylated derivatives of doxorubicin and its biosynthetic intermediates. Appl Environ Microbiol. 2011;77:4912–4923. doi: 10.1128/AEM.02527-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park JW, Park SR, Nepal KK, Han AR, Ban YH, Yoo YJ, Kim EJ, Kim EM, Kim D, Sohng JK, Yoon YJ. Discovery of parallel pathways of kanamycin biosynthesis allows antibiotic manipulation. Nat Chem Biol. 2011;7:843–852. doi: 10.1038/nchembio.671. [DOI] [PubMed] [Google Scholar]
- 56.Young TS, Walsh CT. Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc Natl Acad Sci U S A. 2011;108:13053–13058. doi: 10.1073/pnas.1110435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park HS, Hohn MJ, Umehara T, Guo LT, Osborne EM, Benner J, Noren CJ, Rinehart J, Soll D. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. Ribosomal Route to small molecule diversity. J Am Chem Soc. 2011 doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chooi YH, Cacho R, Tang Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rath CM, Janto B, Earl J, Ahmed A, Hu FZ, Hiller L, Dahlgren M, Kreft R, Yu F, Wolff JJ, Kweon HK, et al. Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem Biol. 2011 doi: 10.1021/cb200244t. [DOI] [PMC free article] [PubMed] [Google Scholar]