Abstract
The spontaneous autosomal recessive mouse mutant for hydrocephaly with hop gait (hyh) exhibits dramatic cystic dilation of the ventricles at birth and invariably develops hopping gait. We show that the gene for soluble N-ethylmaleimide sensitive factor attachment protein α, also known as α-SNAP, is mutated in hyh mice. α-SNAP plays a key role in a wide variety of membrane fusion events in eukaryotic cells, including the regulated exocytosis of neurotransmitters. Homozygous mutant mice harbor a missense mutation M105I in a conserved residue in one of the α-helical domains. We demonstrate that the hyh mutant is not a null allele and is expressed; however, the mutant protein is 40% less abundant in hyh mice. The hyh mutant provides a valuable in vivo model to study vesicle/membrane trafficking and provides insight into the potential roles of α-SNAP in embryogenesis and brain development.
Hydrocephaly with hop gait (hyh) is a recessive mouse mutation that arose spontaneously in the C57BL/10J strain (1, 2). All affected mice develop hydrocephalus and hop gait and usually die by 2 months of age. Congenital hydrocephalus is a heterogeneous collection of disorders that affect brain development, resulting in increased amounts of cerebrospinal fluid within the cerebral cavities and in the space surrounding the brain. Although congenital hydrocephalus is frequent in laboratory animals and in humans, the underlying etiology is poorly understood. A significant portion of human congenital hydrocephalus is genetic in origin. In mice, a number of classical (i.e., spontaneous) mutations of congenital hydrocephalus are known, and additional mouse models can be ascertained through recent efforts of phenotype-driven large-scale N-ethyl-N-nitrosourea mutagenesis screens that focus on neurological and developmental defects (3, 4). Of the classical mutations, the molecular genetics have been recently elucidated in the autosomal recessive forms congenital hydrocephalus (Foxc1ch) (5, 6) and hydrocephalus 3 (hy3) (7). Mutations that remain to be identified are hop-sterile (hop) (8), obstructive hydrocephalus (oh) (9), and hydrocephaly with hop gait (hyh) (2).
In hyh mutant mice, hydrocephalus develops in utero and progressively becomes more severe as the mice age; a dome-shaped head becomes apparent during the second week after birth. The primary site of blockage seems to be at the caudal aqueduct (Sylvius), resulting in dilation of the aqueduct and the third and lateral ventricles (2). However, more recent detailed investigations have shown that hyh mice appear to have an open communication between all ventricles at birth and, with age, communication between the caudal aqueduct and the fourth ventricle is sealed (10, 11). The following abnormalities were also noted: complete absence of central canal in the spinal cord, ependymal denudation of the ventricle, and disorganization of neurons in the rostral vermis of the cerebellum. hyh has been mapped to the proximal end of chromosome 7 close to the Gpi-1 locus (glucose phosphate isomerase-1) at a distance of 3.8 ± 5.5 cM (2). Recently, hyh has been further localized to an interval of 1.6 Mb, distal to the cone-rod homeobox (Crx) gene and proximal to the anonymous DNA marker D7Mit56 (12).
To identify the hyh gene, we have undertaken positional cloning by first constructing a genetic map of the hyh interval by using simple sequence length polymorphisms (SSLPs) in 724 meiosis. We further localized hyh to a 250-kb interval and have investigated all known genes that map to this region by sequence and expression analyses. In this report, we show that hyh mutant mice harbor a missense mutation in one of the seven candidate genes that map to this region, the soluble N-ethylmaleimide sensitive factor (NSF) attachment protein α gene, also known as α-SNAP.
In eukaryotes, α-SNAP is an essential component of the protein machinery responsible for diverse types of membrane fusion events, including intercompartmental transport such as Golgi transport (13) and neuronal exocytosis (14-16). SNAPs act as adaptor proteins that interact with the core membrane-bound proteins, soluble NSF attachment protein receptors/SNAP receptors (SNAREs) and ATPase NSF (15, 17, 18). Membrane fusion is best understood in the context of neuronal exocytosis (19, 20-22). During neurotransmitter release, a vesicle-bound (v-SNARE) protein [synaptobrevins/vesicle-associated membrane proteins (VAMPs)] and target-membrane-bound proteins (t-SNAREs), syntaxin 1 and SNAP (synaptosome-associated protein)-25, form a highly stable 7S ternary parallel helical bundle known as SNAREs (15, 23). This complex pulls opposing membranes together and promotes fusion. α-SNAP binds to the SNARE complex and recruits NSF, forming a larger 20S complex that is subsequently disassembled by the ATPase activity of NSF (15, 23-26). It is thought that SNAPs function as lever arms, transmitting forces generated by changes in NSF driven by hydrolysis to trigger disassembly. Thus, SNAPs play a critical role in recycling of the SNARE proteins.
In mammals, there are three forms of SNAPs, α-, β-, and γ-(16, 17). Sequence comparison of the three SNAPs showed that α- and β-SNAPs are most similar to each other, with amino acid sequence identity of 83% and nucleotide sequence identity of 76% in bovine (16). γ-SNAP is less related but may have similar overall structure (16). In the mouse, α-, β-, and γ-SNAPs map to chromosomes 7, 2, and 18, respectively (27). In mice, both α- and γ-SNAP are expressed ubiquitously, with α-SNAP expression being highest in the brain (16). β-SNAP is expressed in adult brain only, whereas α-SNAP is expressed both during embryonic development and in adulthood (16, 28, 29).
Studies of temperature-sensitive mutants in yeast (30, 31) and Drosophila (32-34) and in vitro mutagenesis and binding studies of recombinant SNARE proteins (35-38) have illuminated many features of membrane fusion and trafficking. In mammals, only two knockout mouse models have been described: a v-SNARE protein synaptobrevin/VAMP2 (39) and a regulatory protein munc 18-1 (40). Synaptobrevin/VAMP2 knockout mice die immediately after birth and exhibit a shoulder hump and round body shape due to excess brown fat in the upper back (39). Analysis of brain sections did not show any apparent abnormalities or neurodegeneration. However, homozygous null mice exhibited decreased spontaneous synaptic vesicle fusion and fusion induced by hypertonic sucrose, suggesting that synaptobrevin/VAMP2 is essential for catalyzing fusion reactions but is not required for synaptic fusion (39). Munc18-1-deficient mice are completely paralyzed and die immediately after birth (40). Ablation of munc18-1 results in complete loss of neurotransmitter secretion from synaptic vesicles throughout development. Null mutants show normal brain assembly during development; however, their neurons undergo apoptosis, leading to wide-spread neurodegeneration later in development (40). Thus, hyh mice provide an excellent in vivo model for further understanding of membrane fusion and trafficking events. Additionally, hyh mutants provide insight into the potential roles of α-SNAP in embryogenesis and brain development and congenital hydrocephalus.
Materials and Methods
Animals and Matings. An inbred mouse strain carrying hyh (B6C3Fe-a/a-hyh) was purchased from The Jackson Laboratory. We conducted three types of matings: (i) brother-sister matings of hyh carriers [B6C3Fe-a/a-hyh × B6C3Fe-a/a-hyh] and (ii) intersubspecific crosses between a hyh carrier from B6C3Fe-a/a-hyh and WT Mus musculus castaneus (CAST) [B6C3Fe-a/a-hyh × CAST/Ei]; and (iii) intercrosses of F1 hyh carriers from the intersubspecific crosses to obtain homozygous hyh/hyh offspring in the CAST genetic background. WT M. m. castaneus mice were also obtained from The Jackson Laboratory, and all mice were maintained in accordance with animal usage guidelines.
Genotyping and Screening of Bacterial Artificial Chromosome (BAC) Libraries. DNA was obtained from tail clippings and extracted by using the Qiagen Dneasy 96 Tissue Kit (Qiagen, Valencia, CA). Primer sets of SSLP markers were purchased from Research Genetics (Huntsville, AL). For each SSLP, one primer was 5′ end-labeled with [γ-33P] ATP (Perkin-Elmer Applied Biosystems) by using T4 kinase according to standard protocols. PCR was performed on a PTC-225 thermal cycler (MJ Research, Watertown, MA) in 20-μl volumes containing 100 ng of genomic DNA by using 1.3 pmol of labeled primer, 6.6 pmol of unlabeled primer, 200 μM each dNTP, and 0.5 units of Taq polymerase (Perkin-Elmer Applied Biosystems) with the recommended assay buffer. Amplified products were separated on 6% denaturing polyacrylamide gels followed by drying and autoradiography. Alternatively, nonradioactively labeled PCR products were analyzed on a 4% agarose gel containing a 3:1 ratio of standard agarose and Nusieve GTG agarose (BMA Biomedicals), respectively.
Mouse BAC clones arrayed on high-density membranes, RPCI-23 and -24 BAC libraries (BACPAC Resources, Oakland, CA) were screened with appropriate sequence-tagged sites or oligonucleotides by hybridization, as recommended (http://bacpac.chori.org/protocols.htm). Additional BAC clones were identified from Mouse Genome Mapping Project data (www.ensembl.org/Mus_musculus).
Genomic Analysis of Mouse Candidate Genes. Genomic DNA isolation and PCR reactions were carried out as described. Samples were processed through an initial denaturation (96°C for 4 min), followed by 35 cycles of denaturation (94°C, 30 sec), annealing (55°C, 30 sec), and elongation (72°C, 30 sec), and terminated by a 10-min elongation at 72°C. Unincorporated dNTPs were removed by using Montage PCR Cleanup Kit (Millipore). Nucleotide sequencing was performed by using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit and analyzed on an ABI377 automated sequencer (Applied Biosystems). The M105I missense mutation was detected by using primers asnap.4F (5′-CAGTCTGCCTCCTGTCTTCA-3′) and asnap.4R (5′-CACACCCCGTTCTGTAGCTC-3′).
Northern Analysis. Total RNA was isolated from whole mouse brain tissue by using the guanidinium/cesium chloride method. RNA was denatured, size-fractionated by electrophoresis on formaldehyde agarose gels, and capillary-blotted to nylon membranes. Membranes were hybridized with probes labeled with [α-32P]dCTP at 42°C, washed to a final stringency of 0.1× SSC, 0.1% SDS at 65°C (40 min), and exposed to autoradiography. As a control for RNA integrity, membranes were hybridized with a mouse GAPDH cDNA probe. Blots were washed in sterile water containing 0.5% SDS at 90-100°C for 15 min and exposed to x-ray film to confirm removal of the probe between hybridizations. Multiple tissue blots, Mouse Brain Aging Blot (MBAB 1009-1), and Mouse Embryo Full Stage Blot (MESB 1002-1), were purchased from Seegene (Seoul, Korea). Reverse transcription reactions were performed by using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen). The following primers were used to generate a cDNA probe specific to the 3′ end of α-SNAP: aSNAP.3′F (5′-AGTGGCTTACCACCATGCTT-3′) and aSNAP.3′R (5′-AACCTGAGAGGTTGGCATTG-3′).
Protein Extraction and Immunoprecipitation (IP). Co-IP was performed essentially as described (41) with some modification. Brains were homogenized in a buffer containing 25 mM Hepes (pH 7.4), 150 mM NaCl, and either containing 1 mM ATPγS (Roche Applied Science, Indianapolis) and 2 mM EDTA or 1 mM ATP (Roche Applied Science) and 3 mM MgCl2. All buffers contained a fresh mix of protease inhibitors: 0.2 mM phenylmethylsulfonyl fluoride/1 μg/ml aprotinin/1 mM benzamidine/10 μg/ml leupeptine/10 μg/ml pepstatine. All homogenization was done on ice by using a Teflon-glass Potter-Elvehjem tissue grinder (Wheaton Scientific). Protein concentration was measured by the Bradford assay by using Bio-Rad Protein Assay (Bio-Rad). TX-100 was added to 1%, and a final protein concentration was adjusted to 1 mg/ml. Extracts were incubated on ice for 20 min and centrifuged at 10,000 × g for 15 min. One milligram of this extract was used per IP experiment. For each IP, 2 μl of antisyntaxin mAb (MAB336, Chemicon) was first incubated with 40 μl of protein A/G PLUS-Agarose (Santa Cruz Biotechnology) in a 1-ml volume of buffer (ATP or ATPγS, plus 1% TX-100) for 2 h at 4°C. Agarose beads were then collected by centrifugation at 10,000 × g for 5 min and 1 mg (1 ml) of protein extract was added and incubated for 4 h at 4°C. Finally, beads were washed four times with appropriate buffer, and proteins were detected by Western blotting by using Enhanced Chemiluminescence (Amersham Pharmacia). Proteins were detected by Western blotting by using anti-NSF (sc-15917, Santa Cruz Biotechnology), anti-SNAP-25 (MAB331, Chemicon). anti-α-SNAP (sc-13991, Santa Cruz Biotechnology; clone 77.2, Synaptic System, Göttingen, Germany). The mean density of α- and β-SNAP was obtained by using the nih image program, Ver. 1.62 (http://rsb.info.nih.gov/nih-image).
Results
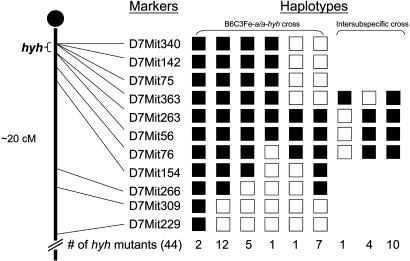
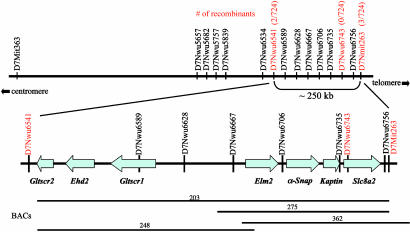
Fine-Resolution Mapping of hyh and Identification of Markers in the hyh Critical Region. The mutation hyh arose spontaneously in a C57BL/10J background and was later outcrossed onto a B6C3Fe-a/a (C57BL/6J female × C3HeB/FeJ-a/a male) F1 hybrid background (2). Therefore, we initially identified heterozygous breeder pairs that carry the mutant chromosome from the C57BL/10J strain and the WT chromosome from the C3HeB/FeJ strain by test breeding and genotypic analysis of SSLP markers from chromosome 7. Because most of the SSLP markers would not be informative between the C57BL/10J and C57BL/6J strains, this strategy allowed us to start with tested breeder pairs where their linkage phase would be known in intercrosses (i.e., yielding two informative meiosis). We first tested markers spanning the proximal 20 cM of chromosome 7, and our initial analysis of 28 hyh mutants from the B6C3Fe genetic background allowed us to define the proximal flanking marker as D7Mit363 and the distal flanking marker as D7Mit76 (Fig. 1). Thus, we localized hyh to a 3-cM interval. Next, we constructed interspecific crosses between carrier females from the B6C3Fe strain and normal CAST/Ei males. Fifteen additional hyh mutant mice from the brother-sister matings of the interspecific cross further defined the new distal flanking marker as D7Mit263, thus reducing the hyh critical region to <1 cM (Fig. 1). With the availability of the draft of the mouse genome sequence available (27, 42), we generated 13 SSLP markers between the interval D7Mit363-D7Mit263 (Table 1, which is published as supporting information on the PNAS web site), which we show to be informative between C57BL/10J, C57BL/6J, C3HeB/FeJ-a/a, and CAST/Ei. In addition, we generated a total of 362 additional mutants, which include 216 mutants from the B6C3Fe genetic background and 146 mutants from the interspecific cross with CAST/Ei. A subset of hyh mutants, which were shown to be recombinant between D7Mit363 and D7Mit263, was genotyped with these 13 SSLPs. By this procedure, we were able to further delineate the hyh critical interval to be ≈250 kb. In a total of 724 informative meioses, we were able to identify the closest proximal flanking marker to be D7Nwu6541, with two recombinants, and the most distal flanking marker to be D7Mit263, with three recombinants (Fig. 2).
Fig. 1.
Haplotype analysis of hyh mutants. Black boxes represent homozygosity for the C57BL/10J allele, and white boxes represent heterozygosity for the C57BL/10J and C3HeB/FeJ-a/a alleles or for the C57BL/10J and CAST/Ei alleles in intersubspecific intercross. The number of each progeny inheriting each haplotype is listed at the bottom. Haplotype analysis of mutants indicates that the most proximal and distal flanking markers are D7Mit363 and D7Mit263, respectively.
Fig. 2.
Physical map and gene content of the hyh critical region. The number of recombinants and total meioses is shown in red for the SSLP markers. The hyh critical region is further defined by the proximal marker D7Nwu6541, with two recombinants of 724 meiosis, and the distal flanking marker D7Mit263, with three recombinants. BAC clones are indicated by horizontal lines; their sizes were determined by sequence-tagged site content and by aligning their end sequences with the reference sequence from the public database www.ensembl.org/Mus_musculus. Gene content in the 250-kb hyh critical region is shown below. Arrows indicate the transcription orientation of each gene; the length of each arrow depicts the genomic length of each gene.
Gene Content of the Candidate Interval. We obtained the genomic sequence of the 250-kb interval from Ensembl (27) and from the Celera Discovery System (42) by using the markers D7Nwu6541 and D7Mit263 as the anchoring points. The sequence gaps not covered by the Mouse Genome Sequencing Consortium or the Celera Discovery System were later obtained by directly sequencing of BAC clones, which we identified as spanning this interval (Fig. 2). Thus, we obtained the complete sequence of the hyh critical region. We then identified genes contained in this region by sequence analysis by using the blast program (43) and the gene prediction programs genscan (44) and twinscan (45). Our analysis predicted seven genes within this 250-kb interval (Fig. 2): glioma tumor suppressor candidate region protein 2 (Gltscr2), EH-domain-containing protein 2 (Ehd2), glioma tumor suppressor candidate region protein 1 (Gltscr1), an Egl-27 and MTA1 homology 2 (ELM)-domain containing protein 2 (Elm2), NSF attachment protein α (Napa, also known as α-SNAP), Kaptin (Kptn), and a sodium/calcium exchanger, solute carrier family 8 member 2 (Slc8a2). The existence of all seven candidate genes is supported by EST or cDNA clone information (www.ensembl.org/Mus_musculus). With the exception of α-SNAP, the remaining six candidate genes are still not well characterized, and their biological functions remain to be determined. The human orthologs of these seven genes map to the conserved syntenic region, chromosome 19q13.3 (www.ensembl.org/Mus_musculus/syntenyview). Three genes, GLTSCR2, EHD2, and GLTSCR1, were originally cloned from deletion mapping studies in human diffuse gliomas in which allelic loss of the chromosome 19q arm is frequent (46). Both GLTSCR2 and GLTSCR1 have no identifiable structural motifs or homology to any known proteins, and EHD2 contains a calcium-binding EF hand motif (46). An ELM2-containing protein in the hyh critical region has homology to a human hypothetical protein DKFZp434I1930 (GenBank accession no. NM_032255) of unknown function. The ELM2 domain was first described in the Caenorhabditis elegans gene egl-27 and human protein MTA1 (47, 48). ELM2-containing proteins are thought to be regulatory factors of transcription acting by DNA-binding or protein-protein interaction. Recent investigations of egl-27 mutants suggest it has a role in embryonic patterning in C. elegans (47, 49). Kaptin (also known as 2E4), originally isolated from human blood platelets (50), is localized in the sensory epithelium of the embryonic inner ear and is thought to have a unique role in the actin rearrangements that accompany platelet activation and stereocilia formation of the inner ear (50). Slc8a2 (also known as NCX2), a member of the Na+/Ca2+ exchanger family, is implicated to play a regulatory role in intracellular Ca2+ levels (51) and is predominantly expressed in brain and skeletal muscle (51, 52). Of the seven candidate genes that lie in the hyh critical region, the function of α-SNAP is best characterized. SNAP proteins play an essential role in membrane trafficking in eukaryotic cells as reviewed earlier (14-26). In the following, we show that a missense mutation in the α-SNAP gene causes the hyh mutation.
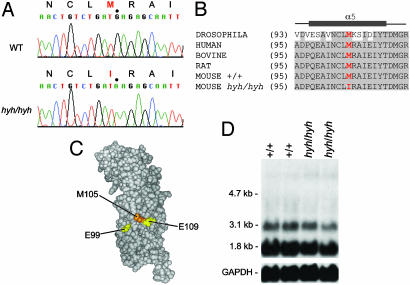
Identification of the hyh Mutation. We began to evaluate each candidate gene by Northern analysis in brain tissues. However, Northern blot analysis did not detect any gross alteration in either levels or in size in any of the seven transcripts in hyh mutants, as compared to the normal WT littermates (data not shown). Subsequently, we searched for the hyh mutation by sequencing the coding regions of all seven candidate genes by delineating their genomic structure and identifying exon-intron boundaries. We compared the sequences from hyh mutants and their normal littermates, as well as the other normal WT inbred strains, C57BL/6J, C57BL/10J, C3HeB/FeJ, and CAST/Ei. This sequence analysis detected numerous nucleotide changes within the coding regions in all candidate genes. However, all of these variants were strain specific polymorphisms; one exception was an amino acid substitution in the α-SNAP gene observed only in hyh mice (Table 2, which is published as supporting information on the PNAS web site). Homozygous hyh mutants harbored a G→A transition at amino acid position 105 in exon 4, leading to a substitution of a highly conserved methionine to isoleucine (M105I) (Fig. 3 A-C). This M105I change was not present in the founder strain C57BL/10J or in any of the other WT inbred strains C57BL/6J, C3HeB/FeJ, and CAST/Ei, thus indicating that this missense substitution is highly likely to be the basis of hyh. In addition, we sequenced exon 4 from genomic DNAs of the following WT inbred strains: AKR/J, BALB/cJ, BALB/cByJ, DBA/2J, NZB/B1NJ, SF/CamEi, SK/CamEi, and 129S1/SvImJ. Our sequence analysis showed that all of these various WT inbred strains have the predicted methionine at amino acid position 105. Significantly, species comparison of α-SNAP amino acid sequences reveals that this methionine at position 105 is evolutionarily conserved (Fig. 3B and Fig 5, which is published as supporting information on the PNAS web site). Taken together, the lack of any amino acid changes in six of the candidate genes and the unique occurrence of the M105I missense mutation in α-SNAP of hyh mice strongly argue that this mutation is responsible for the hyh phenotype in mice.
Fig. 3.
(A) Sequence data obtained from hyh/hyh and WT (WT) DNA. Note the G→ A substitution in hyh/hyh at residue position 105 of α-SNAP. This base change results in replacement of a highly conserved methionine by isoleucine in the mutant. (B) Evolutionary conservation of α-SNAP. Alignment of the fifth helical domain of α-SNAP amino acid sequences from Drosophila, human, bovine, rat, and mouse. Identical residues are shaded. The misssense mutation M105I is highlighted in red. The α-helical domain is indicated by the gray bar (54). SWISSPROT accession nos.: Drosophila, Q23983; human α-SNAP, P54920; bovine α-SNAP, P81125; mouse α-SNAP, Q9DB05; and rat α-SNAP, P54921. (C) 3D model of α-SNAP back face. The homology model was generated by using SWISSPROT and a molecular modeling program, chimera (56, 57), based on the structure of the yeast homolog SEC17p (Protein Data Bank ID 1QQE). The residues (positions 99 and 109) that are shown to have normal binding activity to the SNARE complex but enhanced disassembly by Marz et al. (38) are colored in yellow. The residue position 105, which we show to be mutated in hyh, is colored in orange. (D) Northern blot analysis of α-SNAP. Total RNA was extracted from adult brain tissues of hyh mutants and their normal WT littermates. The mutant α-SNAP is expressed in hyh mice, and the expression level is comparable to that of the WT α-SNAP in normal mice. As a control for RNA integrity, membranes were also hybridized with a GAPDH cDNA probe.
Evaluation of the α-SNAP in hyh Mutants. Because sequence analysis suggests that the hyh mutation is not a null allele, we explored whether the mutant protein is expressed. We evaluated the transcript by Northern analysis and the protein by Western analysis from the whole brain tissues. We showed that the hyh mutation does not affect expression level of the transcript (Fig. 3D). Because α- and β-SNAPs have a high sequence similarity, we had to ensure that no cross hybridization occurs; we generated a 499-bp probe from the 3′ end of the cDNA, which included 105 bp of the coding sequence and 394 bp of the 3′untranslated region. Northern analysis shows that this probe is α-SNAP-specific, hybridizing to two messages, a major species of 1.8 kb and a minor species of 3.1 kb. This observation concurs with published results for bovine α-SNAP (16). Indeed, Northern analysis showed no cross hybridization with a brain-specific β-SNAP message of 5.6 kb. In addition, we have sequenced the α-SNAP transcript by RT-PCR from brain tissues and confirmed that the M105I missense mutation is present in the transcript from hyh homozygous mutants and not present in WT littermates. We also examined α-SNAP expression during embryonic development, as well as in brains from normal aged adult mice. Temporal and spatial expression pattern studies show that α-SNAP is expressed as early as embryonic day 10 in the developing nervous system, including in the brain and spinal cord, the tissues affected in hyh mutants (28). Gross examination revealed that α-SNAP is expressed at least by embryonic day 10.5 and probably earlier (see Fig. 6A legend, which is published as supporting information on the PNAS web site) and throughout adult life (Fig. 6 A and B). We show that the level of expression is relatively consistent across the development and during adult life. We also noted a transcript size of ≈4 kb during embryogenesis (Fig. 6A). This transcript expression level decreases in the brain tissue as mice age. Whether the 4-kb transcript is another minor transcript of α-SNAP or nonspecific cross hybridization is currently unknown and needs to be investigated further. Due to the high sequence similarity, it is possible that β-SNAP may play a compensatory role for α-SNAP in hyh mice; however, this is unlikely, because β-SNAP is not expressed until after birth, from postnatal day 0 and into adulthood (28). Furthermore, defects in hyh mutants can be detected at early stages of embryonic development (11). Although both α- and β-SNAP can support intercisternal transport in Golgi membranes isolated from Chinese hamster ovary cells (18) and catecholamine release from permeabilized adrenal chromaffin cells (53), only α-SNAP can rescue the yeast ortholog (Sec17) mutant, indicating that there are functional differences between α- and β-SNAP (18).
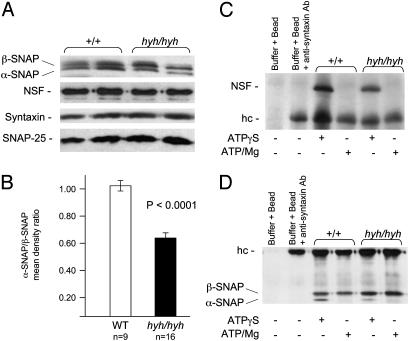
To evaluate whether the mutant protein was expressed, we performed Western blot analysis on whole-brain extracts from homozygous hyh mutants by using an antibody against α-SNAP. Our results show that steady-state levels of the mutant protein are expressed in hyh mice; however, the level of the mutant protein is not as abundant as the normal protein in WT mice (Fig. 4A). Thus the missense mutation may affect the stability of the mutant protein. Both α- and β-SNAPs are detected in mutant brain lysate (Fig. 4A); the available antibody against α-SNAP recognizes both α and β isoforms because of their considerable sequence similarity. α-SNAP has an apparent molecular mass of 35 kDa and β-SNAP, 36 kDa (16, 29). Therefore, we measured the abundance of α-SNAP in comparison to β-SNAP in each of the WT and hyh mice (Fig. 4B). The mean density ratio (±SE) of α-/β-SNAP was ≈1.040 ± 0.0315 in WT and 0.639 ± 0.0417 in hyh mice (Student's t test, P < 0.0001). Therefore, we show that the mutant α-SNAP protein is ≈40% less abundant. This result was confirmed by using two independent α-SNAP antibodies. We also evaluated the expression levels of NSF, syntaxin, and SNAP-25 proteins, which are known to interact with α-SNAP (15, 24-26, 36-38). The expression levels of these proteins in the hyh mutants were comparable to that of normal WT mice (Fig. 4A).
Fig. 4.
Western blot analysis of α-SNAP. (A) Analysis of α-SNAP, NSF, syntaxin, and SNAP-25 in brain lysate from the hyh mutants and normal WT littermates. (B) Mean density ratio of α-/β-SNAP. The mean density values of α- and β-SNAPs were obtained from each brain lysate. The mean α-/β-SNAP ratio was compared between WT and hyh mice by using Student's t test (H0: meanwt-meanhyh = 0). (C and D) Brain lysates prepared either in the ATPγS-IP buffer or ATP-IP buffer were immunoprecipitated with antisyntaxin antibody. Both NSF (C) and α-SNAP (D) were identified as a band of appropriate molecular weight that selectively coprecipitated in the ATPγS-IP buffer in the hyh and normal WT brain lysates. NSF and α-SNAP were disrupted by ATP hydrolysis in the ATP-IP buffer in the hyh and normal WT brain lysates. However, only about half of the mutant α-SNAP coprecipitated, as compared with the WT protein. hc, heavy chain IgG fragments.
Consequently, we explored the ability of the mutant α-SNAP to promote SNARE complex disassembly by NSF via a co-IP-based binding assay in hyh brain lysate. We immobilized antisyntaxin mAb to protein A/G PLUS-Agarose and allowed the SNARE complex to form and disassemble. The SNARE—NSF-SNAP complex exists in a stable association only under conditions of using hydrolysis-resistant ATP analogs (e.g., 1 mM nonhydrolyzable γ-thio ATP [ATPγS] and 2 mM EDTA) (15, 23). To test whether NSF and α-SNAP binding is modulated by ATP hydrolysis, the IP buffer contained either (i) 1 mM γ-thio ATP [ATPγS] and 2 mM EDTA (ATPγS-IP buffer) or (ii) 1 mM ATP and 3 mM MgCl2 (ATP-IP buffer). Indeed, both NSF and α-SNAP coprecipitated in the hyh mutant brain lysate in the presence of ATPγS and was also disrupted by ATP hydrolysis (Fig. 4 C and D). However, as indicated earlier, only about a half of the mutant α-SNAP coprecipitated in hyh mice. Nevertheless, our gross examination showed that SNARE complexes do assemble and disassemble in hyh mice as they do in WT mice.
Discussion
The data presented here describe high-resolution genetic and physical maps that reduced the hyh locus to a 250-kb region. Genome sequence analysis predicted seven candidate genes in this region, and comparative sequencing of hyh mutants revealed a point mutation in a highly conserved residue of α-SNAP. Although the possibility of there being a second mutation in the region genetically linked to the hyh phenotype cannot be ruled out absolutely without sequencing the entire 250 kb, we show compelling evidence that α-SNAP is the causative gene. First, all seven genes in this region were sequenced, and no differences in their expression levels were observed between hyh and control littermates; only a single coding point mutation was observed in the α-SNAP gene. Second, the hyh mutation occurred in one of the most highly conserved regions, one of the α-helix regions, suggesting that the residue mutated in hyh mice is of critical importance to the function of the protein. Third, temporal and spatial expression studies (28, 29) and our Northern analysis have shown that the normal expression pattern of α-SNAP coincides with the abnormal physiological features in hyh mice.
An important result from our study is that the mutant α-SNAP protein is expressed in hyh mice and, therefore, is not a null allele. However, Western analyses show that the mutant protein is significantly less abundant, indicating that the M105I missense mutation may affect the stability of the protein. Biochemical experiments probing the interaction of α-SNAP with syntaxin and NSF demonstrate that the SNARE complex can assemble and disassemble in hyh mutants, at least qualitatively. However, the reduction of α-SNAP in hyh mutants does appear to reduce its interaction with SNARE complex (Fig. 4D). Interestingly, this result is consistent with recent mutagenesis and in vitro studies that have shown that residues that are on the back face of the α-SNAP twisted sheet may contribute to the SNAP-NSF interface (38).
The crystal structure of SEC17, the yeast ortholog of α-SNAP, has been recently determined (54). α-SNAP consists of 14 α-helical hairpins organized into two principal domains whose overall structure is related to several other known α-helical proteins known to mediate protein-protein interactions. The N-terminal region consists of a twisted sheet of nine α-helices and the C-terminal region contains a globular bundle formed by five α-helices (ref. 54; Fig. 3 B and C; Fig. 5). In addition, the superhelical arrangement of α-helical repeats give rise to two distinct surfaces or faces, one concave and the other convex (54). It has been proposed that the concave face, whose curvature complements that of the convex surface of the SNARE complex, constitutes the binding surface for interactions. Interactions of α-SNAP/SEC17 and SNAREs have been partially mapped by using deletion mutagenesis and in vitro binding studies. Extreme N- or C-terminal regions are essential for protein-protein interaction; both domains are required for tight binding to syntaxin (24, 26, 35, 37). The extreme C-terminal region also plays a key role in stimulating the ATPase activity of NSF; deletion of C-terminal residues abolished the ability of α-SNAP to bind NSF (35, 36). More recently, site-directed mutagenesis and in vitro binding assays have further defined the SNARE complex binding surface of α-SNAP (38). Generally, point mutations in charged residues distributed over a concave surface strongly affect SNARE complex binding and subsequently affect SNARE complex disassembly by NSF (38). In contrast, mutations in the convex face (or the back face) did not affect SNARE complex binding; however, some did enhance disassembly significantly as compared to the WT protein. It is hypothesized that residues on the back face of α-SNAP play a role in coupling conformational changes in NSF to SNARE complex disassembly (38). By examining the 3D model structure of the α-SNAP-SNARE complex binding (38, 54), we suspect that the M105I missense mutation in hyh mice has similar kinetics as the group of mutations on the back face. Residue 105, at which the missense mutation is found in hyh, would lie on the back face of α-SNAP, between two residues (residues 99 and 109) that have been shown to enhance disassembly significantly when mutated (38) (Fig. 3C). Finally, it is interesting to note that deletion of the yeast ortholog SEC17 is lethal (55), suggesting that a null allele of α-SNAP in mice would also likely to be lethal because α-SNAP is highly conserved evolutionarily.
Taken together, our analysis shows that hyh is a valuable in vivo model to study the conformational changes in NSF to SNARE complex disassembly and thus further elucidate our understanding of recycling of the SNARE complex. In the past, much of our attention to α-SNAP has been focused on its interaction with the SNARE complex by using recombinant proteins. However, with identification of mutation in α-SNAP, hyh mutant mice serve as a valuable entry point to evaluate the role of α-SNAP in embryogenesis and development of the nervous system.
Supplementary Material
Acknowledgments
We thank Dr. Kathleen Millen for helpful discussions; Dr. Jui-Ming Lin for technical advice; Dr. Theodore Jardetzky and Austin Kirschner for creating the 3D images of α-SNAP; and Wei-Min Song, Elaine Tsao, Amanda Falk, and Debbie Fenner for technical assistance. This work was supported by National Institutes of Health Grant HD34857 (to A.C.). H.-K.H. is an Associate and J.S.T. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations: NSF, N-ethylmaleimide sensitive factor; α-SNAP, soluble NSF attachment protein α; SNAREs, SNAP receptors; SSLPs, simple sequence length polymorphisms; BAC, bacterial artificial chromosome; IP, immunoprecipitation.
See Commentary on page 1431.
References
- 1.Lane, P. W. (1985) Mouse News Lett. 73, 18. [Google Scholar]
- 2.Bronson, R. T. & Lane, P. W. (1990) Dev. Brain Res. 54, 131-136. [DOI] [PubMed] [Google Scholar]
- 3.Justice, M., Noveroske, J. K., Weber, J. S., Zheng, B. & Bradley, A. (1999) Hum. Mol. Genet. 8, 1955-1963. [DOI] [PubMed] [Google Scholar]
- 4.Brown, S. D. M. & Balling, R. (2001) Curr. Opin. Genet. Dev. 11, 268-273. [DOI] [PubMed] [Google Scholar]
- 5.Kume, T., Deng, K.-Y., Winfrey, W., Gould, D. B., Walter, M. A. & Hogan, B. L. M. (1998) Cell 93, 985-996. [DOI] [PubMed] [Google Scholar]
- 6.Hong, H.-K., Lass, J. H. & Chakravarti, A. (1999) Hum. Mol. Genet. 8, 625-637. [DOI] [PubMed] [Google Scholar]
- 7.Davy, B. E. & Robinson, M. L. (2003) Hum. Mol. Genet. 12, 1163-1170. [DOI] [PubMed] [Google Scholar]
- 8.Handel, M. A., Park, C. & Sotomayor, R. (1985) Mouse News Lett. 72, 124. [Google Scholar]
- 9.Borit, A. & Sidman, R. L. (1972) Acta Neuropathol. 21, 316-331. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Figares, J. M., Jimenez, A. J., Perez-Martin, M., Fernandez-Llebrez, P. & Cifuentes, M. (1998) J. Neuropathol. Exp. Neurol. 57, 188-202. [DOI] [PubMed] [Google Scholar]
- 11.Jimenez, A. J., Tome, M., Paez, P., Wagner, C. & Rodriguez, S. (2001) J. Neuropathol. Exp. Neurol. 60, 1105-1119. [DOI] [PubMed] [Google Scholar]
- 12.Chae, T. H., Allen, K. M., Davisson, M. T., Sweet, H. O. & Walsh, C. A. (2002) Mamm. Genome 13, 293-244. [DOI] [PubMed] [Google Scholar]
- 13.Rothman, J. E. & Wieland, F. T. (1996) Science 272, 227-234. [DOI] [PubMed] [Google Scholar]
- 14.Rothman, J. E. (1994) Nature 372, 55-62. [DOI] [PubMed] [Google Scholar]
- 15.Söllner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P. & Rothman, J. E. (1993) Nature 362, 318-324. [DOI] [PubMed] [Google Scholar]
- 16.Whiteheart, S. W., Griff, I. C., Brunner, M., Clary, D. O., Mayer T., Buhrow, S. A. & Rothman, J. E. (1993) Nature 362, 353-355. [DOI] [PubMed] [Google Scholar]
- 17.Clary, D. O. & Rothman, J. E. (1990) J. Biol. Chem. 265, 10109-10117. [PubMed] [Google Scholar]
- 18.Clary, D. O., Giff, I. C. & Rothman, J. E. (1990) Cell 61, 709-721. [DOI] [PubMed] [Google Scholar]
- 19.Lin, R. C. & Scheller R. H. (2000) Annu. Rev. Cell Dev. Biol. 16, 19-49. [DOI] [PubMed] [Google Scholar]
- 20.Rizo, J. & Südhof, T. C. (2002) Nat. Rev. Neurosci. 3, 641-653. [DOI] [PubMed] [Google Scholar]
- 21.Jahn, R., Lang, T. & Südhof, T. C. (2003) Cell 112, 519-533. [DOI] [PubMed] [Google Scholar]
- 22.Söllner, T. H. (2003) Mol. Membrane Biol. 20, 209-220. [DOI] [PubMed] [Google Scholar]
- 23.Söllner, T., Bennett, M. K., Whiteheart, S. W., Scheller, R. H. & Rothman, J. E. (1993) Cell 75, 409-418. [DOI] [PubMed] [Google Scholar]
- 24.Hanson, P. I., Otto, H., Barton. N. & Jahn, R. (1995) J. Biol. Chem. 270, 16955-16961. [DOI] [PubMed] [Google Scholar]
- 25.Hohl, T. M., Parlati, F., Wimmer, C. & Rothman, J. E. (1998) Mol. Cell 2, 539-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi, T., Yamasaki, S., Nauenburg, S., Binz, T. & Niemann, H. (1995) EMBO J. 14, 2317-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouse Genome Sequencing Consortium (2002) Nature 420, 520-561.12466850 [Google Scholar]
- 28.Püschel, A. W., O'Connor, V. & Betz, H. (1994) FEBS Lett. 347, 55-58. [DOI] [PubMed] [Google Scholar]
- 29.Nishiki, T., Nihonmatsu, I., Tsuhara, Y., Kawasaki, M., Sekiguchi, M., Sato, K., Mizoguchi, A. & Takahashi, M. (2001) Neuroscience 107, 363-371. [DOI] [PubMed] [Google Scholar]
- 30.Kaiser, C. A. & Schekman, R. (1990) Cell 61, 723-733. [DOI] [PubMed] [Google Scholar]
- 31.Griff, I. C., Schekman, R., Rothman, J. E. & Kaiser, C. A. (1992) J. Biol. Chem. 267, 12106-12115. [PubMed] [Google Scholar]
- 32.Pallanck, L., Ordway, R. W. & Ganetzky, B. (1995) Nature 376, 25. [DOI] [PubMed] [Google Scholar]
- 33.Siddiqi, O. & Benzer, S. (1976) Proc. Natl. Acad. Sci. USA 73, 3253-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horsnell, W. G., Steel, G. J. & Morgan, A. (2002) Biochemistry 41, 5230-5235. [DOI] [PubMed] [Google Scholar]
- 35.Barnard, R. J. O., Morgan A. & Burgoyne, R. D. (1996) Mol. Biol. Cell 7, 693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnard, R. J. O., Morgan A. & Burgoyne, R. D. (1997) J. Cell Biol. 139, 875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kee, Y., Lin, R. C., Hsu, S.-C. & Scheller, R. H. (1995) Neuron 14, 991-998. [DOI] [PubMed] [Google Scholar]
- 38.Marz, K. E., Lauer, J. M. & Hanson, P. I. (2003) J. Biol. Chem. 278, 27000-27008. [DOI] [PubMed] [Google Scholar]
- 39.Schoch, S., Deák, F., Königstorfer, A., Mozhayeva, M., Sara, Y. & Südhof, T. C. (2001) Science 294, 1117-1122. [DOI] [PubMed] [Google Scholar]
- 40.Verhage, M., Maia, A. S., Plomp, J. A., Brussard, A. B., Heeroma, J. H., Vermeer, H., Toonen, R. F., Hammer, R., E., van den Berg, T. K., Missler, M., et al. (2000) Science 287, 864-869. [DOI] [PubMed] [Google Scholar]
- 41.Osten, P., Srivastava, S., Inman, G. J., Vilim, F. S., Khatri, L., Lee, L. M., States, B. A., Einheber, S., Milner, T. A., Hanson, P. I. & Ziff, E. B. (1998) Neuron 21, 99-110. [DOI] [PubMed] [Google Scholar]
- 42.Kerlavage, A., Bonazzi, V., di Tommmaso, M., Lawrence, C., Li, P., Mayberry, F., Mural, R., Nodell, M., Yandell, M., Zhang, J. & Thomas, P. D. (2002) Nucleic Acids Res. 30, 129-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul, S. F., Gish, W., Miller W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 44.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78-94. [DOI] [PubMed] [Google Scholar]
- 45.Korf, I., Flicek, P., Duan, D. & Brent, M. R. (2001) Bioinformatics 17, S140-S148. [DOI] [PubMed] [Google Scholar]
- 46.Smith, J. S., Tachibana, I., Pohl, U., Lee, H. K., Thanarajasingam, U., Portier, B. P., Ueki, K., Ramaswamy, S., Billings, S. J., Mohrenweiser, H. W., et al. (2000) Genomics 64, 44-50. [DOI] [PubMed] [Google Scholar]
- 47.Solari, F., Bateman, A. & Ahringer, J. (1999) Development (Cambridge, U.K.) 126, 2483-2494. [DOI] [PubMed] [Google Scholar]
- 48.Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Cote, J. & Wang, W. (1998) Mol. Cell 2, 851-861. [DOI] [PubMed] [Google Scholar]
- 49.Ch'ng, Q. & Kenyon, C. (1999) Development (Cambridge, U.K.) 126, 3303-3312. [DOI] [PubMed] [Google Scholar]
- 50.Bearer, E. L. & Abraham, M. T. (1999) Eur. J. Cell Biol. 78, 117-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, A., Matsuoka S., Hryshko, L. V., Nicoll, D. A., Bersohn, M. M., Burke, E. P., Lifton, R. P. & Philipson, K. D. (1994) J. Biol. Chem. 269, 17434-17439. [PubMed] [Google Scholar]
- 52.Canitano, A., Papa, M., Boscia, F., Castaldo, P., Sellitti, S., Taglialatela, M. & Annunziato, L. (2002) Ann. NY Acad. Sci. 976, 394-404. [DOI] [PubMed] [Google Scholar]
- 53.Sudlow, A. W., McFerran, B. W., Bodill, H., Barnard, R. J., Morgan, A. & Burgoyne, R. D. (1996) FEBS Lett. 393, 185-188. [DOI] [PubMed] [Google Scholar]
- 54.Rice, L. M. & Brunger, A. T. (1999) Mol. Cell 4, 85-95. [DOI] [PubMed] [Google Scholar]
- 55.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 56.Huang, C. C., Couch, G. S., Pettersen, E. F. & Ferrin, T. E. (1996) Pac. Symp. Biocomput. 1, 724. [PubMed] [Google Scholar]
- 57.Sanner, M. F., Olson, A. J. & Spehner, J. C. (1996) Biopolymers 38, 305-320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.