Abstract
Background
We sought to investigate whether variants in genes involved in bacterial sensing and autophagy (NOD2, TLR5, IRGM, ATG16L1) and the interleukin-23 signalling pathway (IL12B, IL23R, STAT3) were associated with development of antimicrobial antibodies in patients with Crohn’s disease (CD).
Methods
A cohort of 616 CD patients from a tertiary referral hospital (Mount Sinai Hospital, Toronto) was evaluated. DNA was tested for three CD-associated NOD2 variants (3020insC, G908R, R702W), variants in IRGM, ATG16L1, IL12B, IL23R, STAT3, and a TLR5-stop mutation. Serum was analyzed by ELISA for anti-Saccharomyces cervesiase (ASCA) IgG and IgA, anti-outer membrane protein C (anti-ompC), anti-Cbir1 flagellin, and anti-Pseudomonas fluorescens (anti-I2).
Results
NOD2 3020insC was associated with cumulative seroreactivity by quartile sum (p=0.003) and number of positive antibodies (p=0.02). NOD2 G908R was also associated with quartile sum (p=0.05). Increased ASCA seropositivity was associated with NOD2 3020insC (odds ratio (OR)= 1.9, p=0.02) and G908R (OR=1.8, p=0.05), and ATG16L1 T300A (OR=1.4, p=0.01) variants; ASCA positive patients had an increased cumulative number of NOD2 3020insC and ATG16L1 T300A variants (p=0.007). TLR5-stop mutation abrogated development of anti-flagellin in a dominant-negative fashion (OR=0.5, p=0.009). The IRGM CD risk variant was associated with increased anti-flagellin seropositivity (OR=1.5, p=0.03). IL12B, IL23R, and STAT3 variants did not contribute to development of anti-microbial antibodies.
Conclusions
Variants in innate immune genes involved in pattern recognition and autophagy but not the IL-23 signaling pathway influence antimicrobial seroreactivity in CD. In particular, the additive effect of NOD2 3020insC and ATG16L1 T300A suggests a role for autophagy in development of ASCA.
Keywords: Inflammatory bowel disease, biomarkers, ASCA, genetics, NOD2
Introduction
Genome wide association studies have led to significant advances in our understanding of inflammatory bowel disease (IBD). In particular, the discovery of susceptibility genes involved in innate and adaptive immune pathways has contributed to the model of IBD pathogenesis. Positional cloning of the IBD1 locus on chromosome 16 led to discovery of the first Crohn’s disease (CD) susceptibility gene, NOD2 (1, 2), which encodes a cytoplasmic pattern recognition receptor that recognizes the bacterial moiety dimuramyl peptide. Two groups have recently observed that NOD2’s product instructs autophagy (3, 4), an innate immune process controlled by two other CD susceptibility genes, ATG16LI and IRGM (5, 6); this suggests that autophagy is a central innate immune pathway involved in CD.
In 2000, Oppman et al.’s discovery of interleukin-23 (IL-23), which shares its p40 subunit with IL-12 (7), led to a reassessment of disorders once thought to be driven by IL-12, the canonical Th1 cytokine. Most previous work implicating IL-12 in inflammatory disorders, including IBD, had targeted the p40 subunit (8). Murine models subsequently suggested a key role for IL-23 driven immune responses, in particular via the Th17 lineage, in IBD pathogenesis (9, 10). The discovery of IL-23 pathway components as IBD susceptibility genes (8, 11–16) has further reinforced the importance of IL-23 driven inflammation in disease ontogeny. The IL-23 signaling pathway is orchestrated by a heterodimeric receptor encoded in part by IL23R, and downstream by the transcription factor STAT3 (17). Variants in IL23R, STAT3, and IL12B (encoding the p40 subunit) have all been implicated in CD (11, 12, 15).
It has been argued that IBD results in part from an innate immune deficit leading to adaptive immune hyperactivity against luminal antigens (17, 18), a model substantiated by the enrichment of anti-microbial antibodies found in CD (19). The greatest body of evidence surrounds anti-Saccharomyces cervesiase antibodies (ASCA). Despite its name, there is uncertainty surrounding the source of antigen driving ASCA response, with Candida albicans as a potential immunogen (20). A number of other antibodies enriched in CD have been more recently described including anti-cBir1 flagellin (anti-flagellin), anti-outer membrane protein C (anti-ompC), and anti-Pseudomonas fluorescens (anti-I2) (21).
The influence of IBD gene variants on development of anti-microbial antibodies is incompletely understood. A number of studies have suggested an association between NOD2 variants and development of antimicrobial antibodies, especially ASCA (19, 22–25). Devlin et al. demonstrated that NOD2, but not TLR2, TLR4, or TLR9 variants, was positively associated with cumulative seroreactivity against a panel of anti-microbial antibodies including anti-I2, anti-flagellin, anti-ompC and ASCA (19). Another study suggested that a CD protective TLR5-stop mutation was associated with decreased anti-flagellin response in healthy controls (26). Flagellin is the natural ligand of the TLR5 product Toll-like receptor 5 (TLR5). However, to our knowledge no studies have examined the interaction between autophagy or IL-23 pathway genes and development of anti-microbial antibodies. In the present study, we investigated the association between CD gene variants involved in bacterial sensing and autophagy (NOD2, TLR5, IRGM, ATG16L1) and the IL-23 signaling pathway (IL12B, IL23R, STAT3), with the presence of anti-microbial antibodies in CD patients.
Methods
Patients
Serum and DNA were collected from a cohort of CD patients (n=616) recruited at Mount Sinai Hospital in Toronto, Canada. Diagnosis and classification of CD was made using standard clinical, endoscopic, and histologic criteria (27). Ethics approval was obtained from the Mount Sinai Hospital Research Ethics Board, and informed consent was obtained from all subjects. Samples and data were anonymized. Clinical characteristics are summarized in Table 1.
Table 1.
Clinical characteristics of the study population
| Clinical characteristics | Crohn’s disease subjects (n=616) |
|---|---|
| Male (%) | 312 (50.6) |
| Median age at diagnosis in yrs (range) | 19 (2 – 56) |
| Caucasian (%) | 480 (77.9) |
| Disease location (%) | |
| L1 ileal | 184 (30.5) |
| L2 colonic | 119 (19.7) |
| L3 ileocolonic | 300 (49.8) |
| Disease behaviour (%) | |
| B1 non-stricturing, non- penetrating | 300 (49.4) |
| B2 stricturing | 165 (19.7) |
| B3 penetrating | 142 (49.6) |
| Prevalence of antibodies (%) | |
| ASCA IgA | 264 (42.9) |
| ASCA IgG | 283 (45.9) |
| anti-OmpC | 190 (30.8) |
| anti-flagellin (CBir1) | 302 (49.0) |
| anti-I2 | 310 (50.3) |
Genotyping
DNA was extracted from venous blood of patients. Single nucleotide polymorphisms (SNPs) were genotyped using the Illumina Golden Gate platform (28). SNPs (NCBI SNP IDs in square brackets) included those in NOD2 (3020insC [rs2066847]; R702W [rs2066844]; and G908R [rs2066845]), ATG16L1 (T300A [rs2241880]), IRGM ([rs11747270]), TLR5 (TLR5-stop [rs5744168]), IL23R ([rs11465804]), STAT3 ([rs744266]), and IL12B ([rs10045431])(1, 2, 11, 14, 15, 26). Utilized SNPs including genotype distributions are summarized in the Supplementary Table. There was a failure rate of less than 5% in genotyping each locus, thus accounting for variation in the number of reported results by locus. SNPs/genes were chosen for analysis based on their previous associations with Crohn’s disease and immunological function; NOD2 and TLR5 encode pattern-recognition receptors, NOD2, ATG16L1, and IRGM encode products involved in autophagy, and IL23R, IL12B, and STAT3 encode components of the IL-23 signalling pathway (17, 26).
Serological analysis
Sera were analyzed for ASCA IgA and IgG, anti-flagellin (anti-CBir1), anti-ompC, and anti-I2 by enzyme-linked immunosorbent assay (ELISA) at Cedars-Sinai Medical Center in Los Angeles, as previous described (19, 29). Antibody levels are expressed in ELISA units (EU/mL) in relationship to established standards, derived from a pool of patient sera with well-characterized disease found to have reactivity to these antigens. Prevalence of the antimicrobial antibodies in the study population is included in Table 1.
Statistical analysis
Quartile sum scores were tabulated as a semi-quantitative measure of cumulative seroreactivity. Antibody levels were given a score of 1 to 4 based on their quartile within the distribution, 4 denoting the highest. ASCA IgA and IgG values were log-transformed and standardized, and the higher standardized unit was utilized for determination of ASCA quartile, as previously shown (19). Quartile sums of 4 to 16 were determined by adding scores from each of the four antibodies. The number of patients in each quartile appeared normally distributed (data not shown). Associations between SNPs and quartile sums were evaluated using linear regression, assuming an additive genetic model. In addition, the number of positive antibodies was used as a second measure of cumulative seroreactivity and analyzed in the same fashion.
For analysis comparing SNPs and specific antibodies, antibody levels were dichotomized using pre-specified cutoffs. ASCA IgG was considered positive if >40 EU/mL, ASCA IgA if >20 EU/mL, anti-ompC if >23 EU/mL, anti-flagellin if >30 EU/mL, and anti I2 if >20 EU/mL. Patients were considered ASCA-positive if either IgA or IgG was positive. Screening for associations between CD risk variants and antibody positivity was undertaken using an additive genetic model and univariate logistic regression. Associations discovered by this method were further analyzed using multivariate logistic regression (with age at diagnosis, and disease behaviour and location as co-variates), and comparing mean antibody titers by linear regression. These subsequent analyses were undertaken using the best-fitting genetic model (i.e. dominant or recessive), as shown in results section. Quantitative data is expressed as mean +/− standard error of mean. All analyses were undertaken using SAS 9.2 (SAS Institute Inc., Cary). Statistical significance was considered to be P<0.05.
Results
NOD2 3020insC is associated with cumulative seroreactivity to microbial antigens
Analysis confirmed a positive association between NOD2 3020insC and quartile sum (p=0.003) (Supplementary Table). The association between NOD2 G908R and quartile sum also approached statistical significance (p=0.05). There was no association between quartile sum and NOD2 R702W (p=0.85) or the other tested SNPs. A second surrogate measure of cumulative seroreactivity utilized was the number of positive antibodies. NOD2 3020insC (p=0.02), but no other tested SNP was associated with an increased number of positive antibodies.
NOD2 3020insC and ATG16L1 T300A variants are associated with increased ASCA seropositivity
There was a significant positive association between NOD2 3020insC and ASCA seropositivity (Odds ratio (OR)= 1.9 (95% confidence interval 1.1–3.2); p=0.02) (Figure 1A). NOD2 G908R was also associated with ASCA seropositivity (OR=1.8 (1.0–3.4); p=0.05). In contrast, NOD2 R702W (p=0.48) was not associated with ASCA. There was no association between NOD2 variants and anti I2, anti-ompC and anti-flagellin in this cohort.
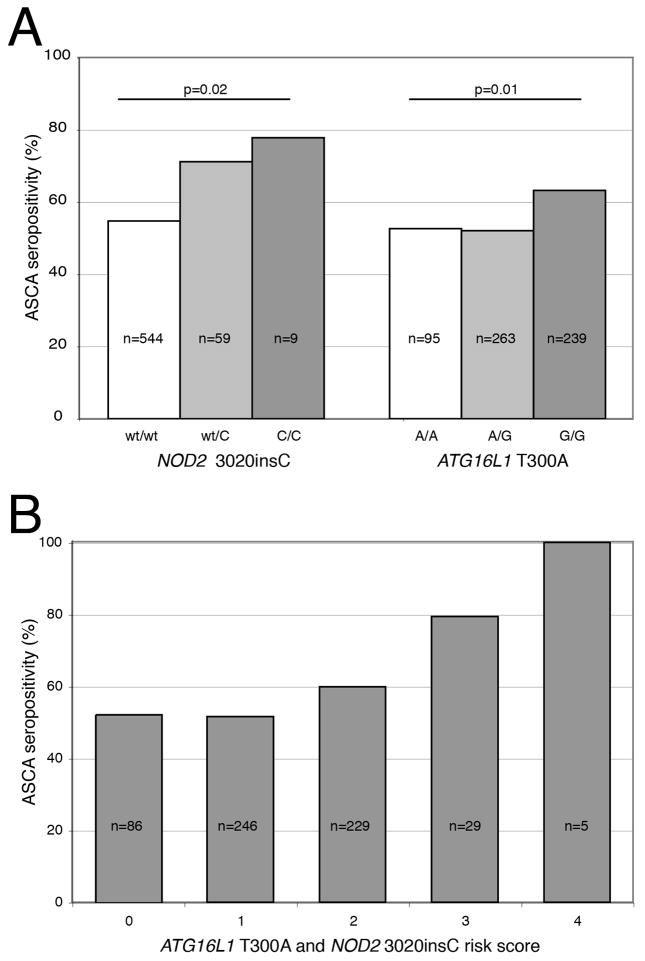
Figure 1.
(A) NOD2 3020insC (allele C) and ATG16L1 T300A (allele G) are associated with increased ASCA seropositivity. P-value by logistic regression assuming additive genetic model. (B) ASCA seropositivity, stratified by 3020insC and ATG16L1 T300A risk score
Like NOD2 3020insC, the ATG16L1 T300A variant was associated with increased ASCA seropositivity (OR=1.4 (1.1–1.8); p=0.01) (Figure 1A), but not other antibodies.
Multivariate logistic regression, adjusting for disease location, behavior, and age at diagnosis, confirmed the independence of associations between NOD2 3020insC and ASCA (p=0.03, dominant model), and ATG16L1 T300A and ASCA (p=0.02, recessive model).
To explore the cumulative effect of NOD2 3020insC and ATG16L1 T300A on development of ASCA, a risk score was created; CD patients were stratified from 0 to 4 by number of NOD2 3020insC and ATG16L1 T300A risk alleles. The risk score was significantly higher in ASCA positive patients (1.45 versus 1.24; p=0.007). There was an apparent additive effect of risk score on ASCA seropositivity (Figure 1B). Of note, all five patients homozygous for NOD2 3020insC and ATG16L1 T300A were ASCA positive.
Analysis of NOD2 3020insC and ATG16L1 T300A variants and ASCA titers
Analysis comparing antibody titers was also undertaken to confirm the above associations. CD patients carrying at least one 3020insC variant (i.e. dominant model) had higher antibody titers of ASCA IgA (43.3 +/− 4.7 versus 26.3 +/− 1.3 EU/mL; p<0.0001) and IgG (65.0 +/− 6.2 versus 47.2 +/− 1.9 EU/mL; p=0.002); carriage of at least one G908R allele was only correlated with increased ASCA IgA levels (36.6 +/− 4.6 versus 27.4 +/− 1.4 EU/mL; p=0.03). However, ATG16L1 T300A was not associated with elevated ASCA IgA or IgG titers (recessive model, not shown).
TLR5 and IRGM mutations are associated with anti-flagellin response
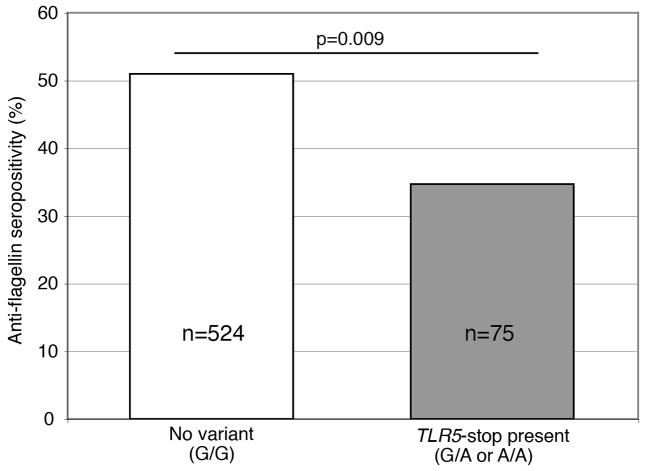
Using an additive genetic model, there was no significant association between TLR5-stop and anti-flagellin in the present study (p=0.10). However, this variant is known to act in a dominant fashion (30), and CD patients with at least one copy of TLR5-stop were less likely to be anti-flagellin positive (OR=0.51 (0.31–0.85); p=0.009 using dominant model) (Figure 2). The IRGM risk variant at rs11747270 (allele G) was associated with increased anti-flagellin seropositivity (OR 1.45 (CI 1.03–2.03); p=0.03). There were no significant associations between IRGM or TLR5 variants and anti-flagellin titers or other antibody seropositivity.
Figure 2.
TLR5-stop mutation (allele A) is associated with reduced anti-flagellin seropositivity in a dominant pattern in Crohn’s disease patients.
IL-23 axis gene variants do not influence development of specific anti-microbial antibodies
Variants in IL23R (encoding a subunit of the IL-23 receptor), IL12B (encoding the p40 subunit shared by IL-23 and IL-12), and STAT3 (encoding the downstream transcription factor STAT3) were not associated with antibody seropositivity (data not shown).
Discussion
Although previous work has investigated the molecular basis of the heritability of anti-microbial antibodies, it has mostly focused on the influence of pattern recognition receptors such as NOD2 and TLRs (19, 25). The present study explored the role of pattern recognition receptors, autophagy, and IL-23 signaling pathway genes on the humoral response against luminal antigens in Crohn’s disease. We demonstrate that innate immune genes involved in pattern recognition and autophagy are important in development of ASCA and anti-flagellin in a cohort of CD patients.
Quartile sum analysis is commonly used as a measure of general antimicrobial seroreactivity in IBD; however, the fact that CD patients react to different subsets of antigens (21) suggests that this approach may obscure more specific relationships, in our case between gene variants and antibody development. In our cohort, only NOD2 variants were associated with elevated quartile sums. No other tested gene variants appeared to influence this measure. Devlin et al. previously found that quartile sum was equally increased in patients with each of the three common NOD2 variants (19). This difference is possibly a reflection of sample size (n=732 versus n=616) or population heterogeneity.
The two largest studies specifically investigating the heritability of antimicrobial antibodies (19, 25) both described the association between pooled NOD2 variants, i.e. the presence of any NOD2 variant and increased ASCA seropositivity. Neither reported on whether the ASCA association was driven by a specific NOD2 variant. A smaller study (n=316) showed an association between G908R and 3020insC, but not R702W, and ASCA seropositivity (31). Our data suggests that the NOD2 3020insC mutation influences ASCA seropositivity and IgA/IgG titers. The G908R variant was also associated with ASCA seropositivity and IgA levels, though not at the same level of significance. We would suggest that the 3020insC variant, and to a lesser degree G908R, are largely driving the associations previously reported between pooled NOD2 variants and ASCA.
It is becoming increasingly apparent that an inability of the innate immune system to recognize and clear intracellular bacteria is central to the pathogenesis of CD, supported by the association of CD with NOD2, IRGM, and ATG16L1 polymorphisms (17). One model for understanding the development of anti-microbial antibodies in CD is that persistence of microbial products in the lamina propria because of defective clearance mechanisms results in loss of tolerance and hyperactive humoral response to microbial products. We therefore expected to find a broad association between NOD2 polymorphisms and development of a number of antibodies, but instead found a predominant association with ASCA. It is not immediately apparent why a pattern recognition receptor recognizing the breakdown product of bacterial peptidoglycan (NOD2) would be principally associated with development of antibodies against a yeast polysaccharide (ASCA) versus bacterial antigens (anti-ompC, anti-flagellin, anti-I2). NOD2 has no known role in recognition of C. albicans (32), an immunogen for ASCA (20). One possibility is that this effect is mediated by intestinal barrier dysfunction; the 3020insC mutation, but not R702W or G908R, has been associated with increased intestinal permeability (33). However, it has been suggested that ASCA does not result from defects in intestinal permeability (34, 35), and this possibility would furthermore not explain the prominence of the association between the 3020insC mutation and ASCA. We instead postulate that autophagy is an important mechanism controlling the development of ASCA, supported by the association between ATG16L1 T300A and ASCA, and the additive effect of NOD2 3020insC and ATG16L1 T300A loci on ASCA seropositivity. Dendritic cells from CD patients with NOD2 3020insC or ATG16L1 T300A variants are defective in both autophagy and presentation of antigen on MHC II (3). Autophagy defects may also influence development of anti-flagellin, as evidenced by the association described here between the IRGM CD risk variant and anti-flagellin seropositivity.
Following the observation that a dominant-negative TLR5-stop mutation abolishes TLR5 signalling (30), Gewirtz et al. demonstrated that this mutation reduces humoral response to TLR5’s natural ligand, flagellin, in healthy controls, but not CD patients (26). They suggested that flagellin was likely dispensable as an adjuvant in the proinflammatory environment of CD. We demonstrated here, in a larger cohort of CD patients, that TLR5-stop is associated with reduced response to flagellin. It is appealing to suggest that flagellin acts as both antigen and adjuvant, inducing dendritic cell maturation via TLR5 (36) to orchestrate the anti-flagellin humoral response, and that loss of functional TLR5 thereby minimizes anti-flagellin immunoglobulin development in healthy and inflammatory (shown here) states. An additional hypothesis is that loss of functional TLR5 results in a change in composition of the intestinal microbiota, as has been shown in TLR5-deficient mice (37), and thus modulates the composition of luminal antigens.
We found no associations between any IL-23 signaling pathway gene polymorphisms and development of anti-microbial antibodies. A key action of IL-23 is in promoting immunopathology mediated by Th17 cells (38), a CD4+ T-cell subset important in mucosal proinflammatory responses. IL-23 driven pathways are classically invoked as an arm of the adaptive immune system(17). Within this paradigm, perhaps the association between innate but not IL-23 related genes and development of antimicrobial antibodies in CD is a reflection of Janeway’s hierarchical model of innate instructing adaptive immunity (39). Nonetheless, it is becoming increasingly clear that IL-23 is a mediator of the intestinal innate immune system (40, 41).
In summary, we demonstrate in the present study that variants in innate immune genes involved in pattern recognition (NOD2 and TLR5) and autophagy (NOD2, ATG16L1, and IRGM) influence antimicrobial seroreactivity in CD, in contrast to variants in the IL-23 signalling pathway (IL12B, IL23R, and STAT3). We confirm the previous association between NOD2 variants and the presence of ASCA. Variants in TLR5 and IRGM were associated with changes in anti-flagellin reactivity. NOD2 and ATG16L1 variants appeared to synergistically influence ASCA development. Future research should elucidate the role of autophagy in the development of antimicrobial antibodies in CD. Understanding the mechanism of generation of antimicrobial antibodies may shed light on their role in CD pathogenesis.
Supplementary Material
Acknowledgments
Dr. Silverberg was supported by The National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) Grant DK62423, the Crohn’s and Colitis Foundation of Canada, and the Gale and Graham Wright Research Chair in Digestive Diseases. Dr. Murdoch was supported by a Canadian Association for Gastroenterology Resident Research Award. Work at Cedars-Sinai was supported by NIDDK Grant DK046763. Dr Targan is supported in part by the Feintech Family Chair in Inflammatory Bowel Disease and Dr. Rotter by the Cedars-Sinai Board of Governors' Chair in Medical Genetics.
Footnotes
Conflicts of interest: The authors do not declare any conflicts of interest.
References
- 1.Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 2.Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 3.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 4.Travassos L, Carneiro L, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2009;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 5.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oppmann B, Lesley R, Blom B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 8.McGovern D, Powrie F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut. 2007;56:1333–6. doi: 10.1136/gut.2006.115402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahern PP, Izcue A, Maloy KJ, et al. The interleukin-23 axis in intestinal inflammation. Immunol Rev. 2008;226:147–59. doi: 10.1111/j.1600-065X.2008.00705.x. [DOI] [PubMed] [Google Scholar]
- 10.Elson CO, Cong Y, Weaver CT, et al. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology. 2007;132:2359–70. doi: 10.1053/j.gastro.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 11.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGovern DP, Rotter JI, Mei L, et al. Genetic epistasis of IL23/IL17 pathway genes in Crohn's disease. Inflamm Bowel Dis. 2009;15:883–9. doi: 10.1002/ibd.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor KD, Targan SR, Mei L, et al. IL23R haplotypes provide a large population attributable risk for Crohn's disease. Inflamm Bowel Dis. 2008;14:1185–91. doi: 10.1002/ibd.20478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 15.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubinsky MC, Wang D, Picornell Y, et al. IL-23 receptor (IL-23R) gene protects against pediatric Crohn's disease. Inflamm Bowel Dis. 2007;13:511–5. doi: 10.1002/ibd.20126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 18.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–29. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 19.Devlin SM, Yang H, Ippoliti A, et al. NOD2 variants and antibody response to microbial antigens in Crohn's disease patients and their unaffected relatives. Gastroenterology. 2007;132:576–86. doi: 10.1053/j.gastro.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Standaert-Vitse A, Jouault T, Vandewalle P, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn's disease. Gastroenterology. 2006;130:1764–75. doi: 10.1053/j.gastro.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Landers CJ, Cohavy O, Misra R, et al. Selected loss of tolerance evidenced by Crohn's disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–99. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 22.Papp M, Altorjay I, Norman GL, et al. Seroreactivity to microbial components in Crohn's disease is associated with ileal involvement, noninflammatory disease behavior and NOD2/CARD15 genotype, but not with risk for surgery in a Hungarian cohort of IBD patients. Inflamm Bowel Dis. 2007;13:984–92. doi: 10.1002/ibd.20146. [DOI] [PubMed] [Google Scholar]
- 23.Marrakchi R, Bougatef K, Moussa A, et al. 3020insC insertion in NOD2/CARD15 gene, a prevalent variant associated with anti-Saccharomyces cerevisiae antibodies and ileal location of Crohn's disease in Tunisian population. Inflamm Res. 2009;58:218–23. doi: 10.1007/s00011-008-8139-x. [DOI] [PubMed] [Google Scholar]
- 24.Dassopoulos T, Frangakis C, Cruz-Correa M, et al. Antibodies to Saccharomyces cerevisiae in Crohn's disease: higher titers are associated with a greater frequency of mutant NOD2/CARD15 alleles and with a higher probability of complicated disease. Inflamm Bowel Dis. 2007;13:143–51. doi: 10.1002/ibd.20031. [DOI] [PubMed] [Google Scholar]
- 25.Henckaerts L, Pierik M, Joossens M, et al. Mutations in pattern recognition receptor genes modulate seroreactivity to microbial antigens in patients with inflammatory bowel disease. Gut. 2007;56:1536. doi: 10.1136/gut.2007.125468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gewirtz AT, Vijay-Kumar M, Brant SR, et al. Dominant-negative TLR5 polymorphism reduces adaptive immune response to flagellin and negatively associates with Crohn's disease. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1157–63. doi: 10.1152/ajpgi.00544.2005. [DOI] [PubMed] [Google Scholar]
- 27.Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19 (Suppl A):5–36. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 28.Shen R, Fan J-B, Campbell D, et al. High-throughput SNP genotyping on universal bead arrays. Mutat Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 29.Landers CJ, Cohavy O, Misra R, et al. Selected loss of tolerance evidenced by Crohn's disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–99. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 30.Hawn TR, Verbon A, Lettinga KD, et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J Exp Med. 2003;198:1563–72. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Annese V, Lombardi G, Perri F, et al. Variants of CARD15 are associated with an aggressive clinical course of Crohn's disease--an IG-IBD study. Am J Gastroenterol. 2005;100:84–92. doi: 10.1111/j.1572-0241.2005.40705.x. [DOI] [PubMed] [Google Scholar]
- 32.van der Graaf CAA, Netea MG, Franke B, et al. Nucleotide oligomerization domain 2 (Nod2) is not involved in the pattern recognition of Candida albicans. Clin Vaccine Immunol. 2006;13:423–5. doi: 10.1128/CVI.13.3.423-425.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buhner S, Buning C, Genschel J, et al. Genetic basis for increased intestinal permeability in families with Crohn's disease: role of CARD15 3020insC mutation? Gut. 2006;55:342–7. doi: 10.1136/gut.2005.065557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrer M, Reinisch W, Dejaco C, et al. Do high serum levels of anti- Saccharomyces cerevisiae antibodies result from a leakiness of the gut barrier in Crohn's disease? Eur J Gastroenterol Hepatol. 2003;15:1281–5. doi: 10.1097/00042737-200312000-00005. [DOI] [PubMed] [Google Scholar]
- 35.Vermeire S, Peeters M, Vlietinck R, et al. Anti-Saccharomyces cerevisiae antibodies (ASCA), phenotypes of IBD, and intestinal permeability: a study in IBD families. Inflamm Bowel Dis. 2001;7:8–15. doi: 10.1097/00054725-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 36.Means TK, Hayashi F, Smith KD, et al. The Toll-like receptor 5 stimulus bacterial flagellin induces maturation and chemokine production in human dendritic cells. J Immunol. 2003;170:5165–75. doi: 10.4049/jimmunol.170.10.5165. [DOI] [PubMed] [Google Scholar]
- 37.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGeachy MJ, Cua DJ. Th17 cell differentiation: the long and winding road. Immunity. 2008;28:445–53. doi: 10.1016/j.immuni.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Janeway CA, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 40.Uhlig HH, McKenzie BS, Hue S, et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–18. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 41.Buonocore S, Ahern PP, Uhlig HH, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–5. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.