Abstract
In Aspergillus nidulans the global regulatory gene veA is necessary for the biosynthesis of several secondary metabolites, including the mycotoxin sterigmatocystin (ST). In order to identify additional veA-dependent genetic elements involved in regulating ST production, we performed a mutagenesis on a deletion veA (ΔveA) strain to obtain revertant mutants (RM) that regained the capability to produce toxin. Genetic analysis and molecular characterization of one of the revertant mutants, RM3, revealed that a point mutation occurred at the coding region of the rtfA gene, encoding a RNA-pol II transcription elongation factor like protein, similar to Saccharomyces cerevisiae Rtf1. The A. nidulans rtfA gene product accumulates in nuclei. Deletion of rtfA gene in a ΔveA background restored mycotoxin production in a medium-dependent manner. rtfA also affects the production of other metabolites including penicillin. Biosynthesis of this antibiotic decreased in the absence of rtfA. Furthermore, rtfA is necessary for normal morphological development. Deletion of the rtfA gene in wild-type strains (veA+) resulted in a slight decrease in growth rate, drastic reduction in conidiation, and complete loss of sexual development. This is the first study of an Rtf1 like gene in filamentous fungi. We found rtfA putative orthologs extensively conserved in numerous fungal species.
Keywords: gene regulation, mutagenesis, veA, velvet, Rtf1, mycotoxin, penicillin, morphogenesis
Introduction
Species of the genus Aspergillus produce a variety of secondary metabolites (Adrio and Demain, 2003; Reverberi et al., 2010; Brakhage and Schroeckh, 2011). Some of these compounds are beneficial to humankind, for example antibiotics and other medical drugs, whereas others are deleterious, such as mycotoxins (Bennett and Klich, 2003). The model filamentous fungus Aspergillus nidulans produces sterigmatocystin (ST). This mycotoxin is similar to carcinogenic compounds called aflatoxins (AF) (Payne and Yu, 2010; Sweeney and Dobson, 1999; Payne and Brown, 1998). Furthermore, ST is the penultimate precursor in the conserved AF biosynthetic pathway found in related species such as A. flavus, A. parasiticus, and A. nomius (Cole and Cox, 1981). The genes involved in the ST/AF biosynthetic pathway are clustered and contain a regulatory gene called aflR, that encodes a transcription factor necessary for cluster activation (Keller and Hohn, 1997; Yu et al., 1996; Fernandes et al., 1998).
Regulation of secondary metabolism presents common genetic links with regulatory mechanisms influencing fungal morphogenesis (Calvo et al., 2002). We have previously shown that a global regulatory gene called veA, known to control sexual and asexual development in A. nidulans (Kim et al., 2002), is also required for the synthesis of numerous secondary metabolites, including ST (Kato et al., 2003). Deletion of the veA gene in A. nidulans results in loss of aflR expression and absence of ST biosynthesis. In this fungus, the production of other metabolites, including penicillin (PN), is also dependent on veA (Kato et al., 2003). In other fungal species, veA homologs regulate the synthesis of PN production in Penicillium chrysogenum (Hoff et al., 2010) and cephalosporin C in Acremonium chrysogenum (Dreyer et al., 2007). It also controls the biosynthesis of other mycotoxins, such as AF, cyclopiazonic acid and aflatrem in Aspergillus flavus (Duran et al., 2007; Calvo et al., 2004; Duran et al., 2009), fumonisins and fusarins in Fusarium spp, including F. verticillioides and F. fujikuroi (Myung et al., 2012; Niermann et al., 2011), and trichothecenes in F. graminerum (Merhej et al., 2011).
veA is found broadly conserved in Ascomycetes (Myung et al., 2012). Substantial insight on the veA regulatory mechanism of action has been gained using A. nidulans as a model system. The VeA protein interacts with the KapA α-importin and is transported to the nucleus, particularly in the absence of light, a condition that favors ST production (Stinnett et al., 2007, Araujo-Bazan et al., 2009). In the nucleus, VeA interacts with other protein such as light-responsive proteins, FphA, LreA and LreB that also have an effect on development and mycotoxin production (Purschwitz et al., 2008). FphA represses sexual development and synthesis of ST, while LreA and LreB have an antagonistic effect. VeA also interacts with VelB and LaeA (Bayram et al., 2008; Bayram and Braus, 2012). LaeA, a putative methyl transferase involved in chromatin conformation (Reyes-Dominguez et al., 2007), is also required for normal mycotoxin biosynthesis (Bok and Keller, 2004). Deletion of velB results in reduced and delayed ST production, indicating a role in the modulation of ST biosynthesis (Bayram et al., 2008; Bayram and Braus, 2012).
In addition to the regulatory effect of VeA on morphogenesis and secondary metabolism, recent studies have shown VeA homologs to be involved in plant pathogenicity by different mycotoxigenic fungal species, for example in Aspergillus flavus (Duran et al., 2009), Fusarium verticillioides (Myung et al., 2012), F. fujikuroi (Wiemann et al., 2010), and F. graminearum (Merhej et al., 2011). Deletion of veA homologs in these fungi resulted in a reduction in virulence as well as in a reduction in mycotoxin biosynthesis.
With the initial goal of uncovering new veA-dependent genetic elements mediating ST biosynthesis in the model organism A. nidulans, we performed mutagenesis in search of revertant mutants that regain the capacity to produce this mycotoxin in a veA deletion genetic background. To facilitate a visual screening, the subject of this mutagenesis was a strain that also lacked stcE, a gene in the ST gene cluster. Deletion of stcE in strains with a functional veA allele results in accumulation of norsolorinic acid (NOR), an orange colored intermediate. Several revertant mutants (RM) able to accumulate NOR were obtained. Following genetic approaches, we further explored one of the selected revertants, RM3. This revertant presented a point mutation in a gene that we denominated rtfA, encoding a putative RNA polymerase transcription elongation factor like protein with homology to Rtf1 in S. cerevisiae. In this study we show that rtfA differentially affects ST biosynthesis in a veA-dependent and in a medium-dependent manner. rtfA also regulates the production of other secondary metabolites, such as PN. Furthermore, rtfA is also necessary for normal sexual and asexual development in A. nidulans.
Results
Revertant mutants recovered mycotoxin production
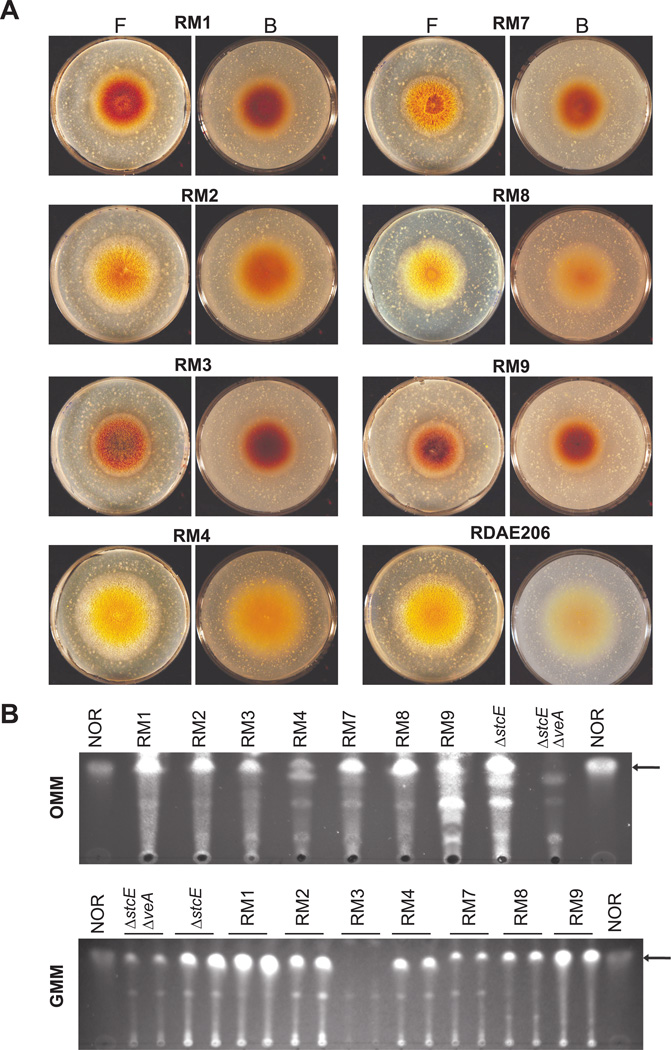
Deletion of the stcE gene blocks the conversion of NOR into averantin, resulting in accumulation of the orange colored NOR. Production of this compound is especially visible when the fungus is growing on oatmeal medium (OMM). In addition, it is known that veA is necessary for normal production of ST mycotoxin (Kato et al., 2003). The double mutant RDAE206 (ΔveA, ΔstcE) does not produce NOR or produces trace amounts on OMM or GMM. After chemical mutagenesis of RDAE206, we searched for revertant mutants in which NOR was biosynthesis was restored to normal levels on OMM. Although the RDAE206 strain still produces an additional pale orange pigment on OMM (different from NOR as verified by TLC analysis), recovery of NOR production levels after mutagenesis resulted in an identifiably darker orange color (Fig. 1A). In order to isolate revertant mutants that could be involved in the regulation of the ST biosynthetic pathway, we screened approximately 100,000 colonies after the chemical mutagenesis treatment and analyzed for production of NOR. We obtained seven revertant mutants (RM strains, Fig. 1A). NOR production in all the selected strains was further analyzed by TLC (Fig. 1B). These RMs accumulated NOR at different levels both on OMM and GMM. When cultured on OMM, RM1, RM2, RM3, RM7, RM8 and RM9 presented abundant levels of NOR, whereas RM4 accumulated this compound at lower levels (Fig. 1B). RM1 and RM9 produced the highest levels of NOR on GMM, whereas other mutants, RM2, RM4, RM7 and RM8, produced moderate amounts. Interestingly, the mutation in RM3 does not remediate NOR production on GMM (Fig. 1B).
Fig. 1. Revertant mutant (RM) strains produce NOR on OMM and GMM.
A. RM point-inoculated OMM cultures incubated at 37°C in the dark for five days. F, front. B, back.
B. TLC analysis of NOR production by RM strains. Extracts were obtained from top-agar inoculated cultures incubated at 37°C in the dark for five days. NOR was analyzed as described in experimental procedures. Extracts from ΔstcE (RAV1) and ΔstcE, ΔveA (RDAE206) strains were included as controls. NOR standard was included on both sides (location of NOR on the TLC plates is indicated with arrows).
The revertant mutants isolated also presented a slight decrease in colony growth and alterations in conidiation compared to its isogenic wild-type RDAE206 strain on OMM (Fig. 1A) and on GMM (data not shown). Mutants RM1, RM2, RM3 and RM9 appeared almost aconidial whereas mutants RM4, RM7 and RM8 showed a slight reduction in conidiation compared to that of RDAE206.
RM3 presents a mutation in the rtfA gene
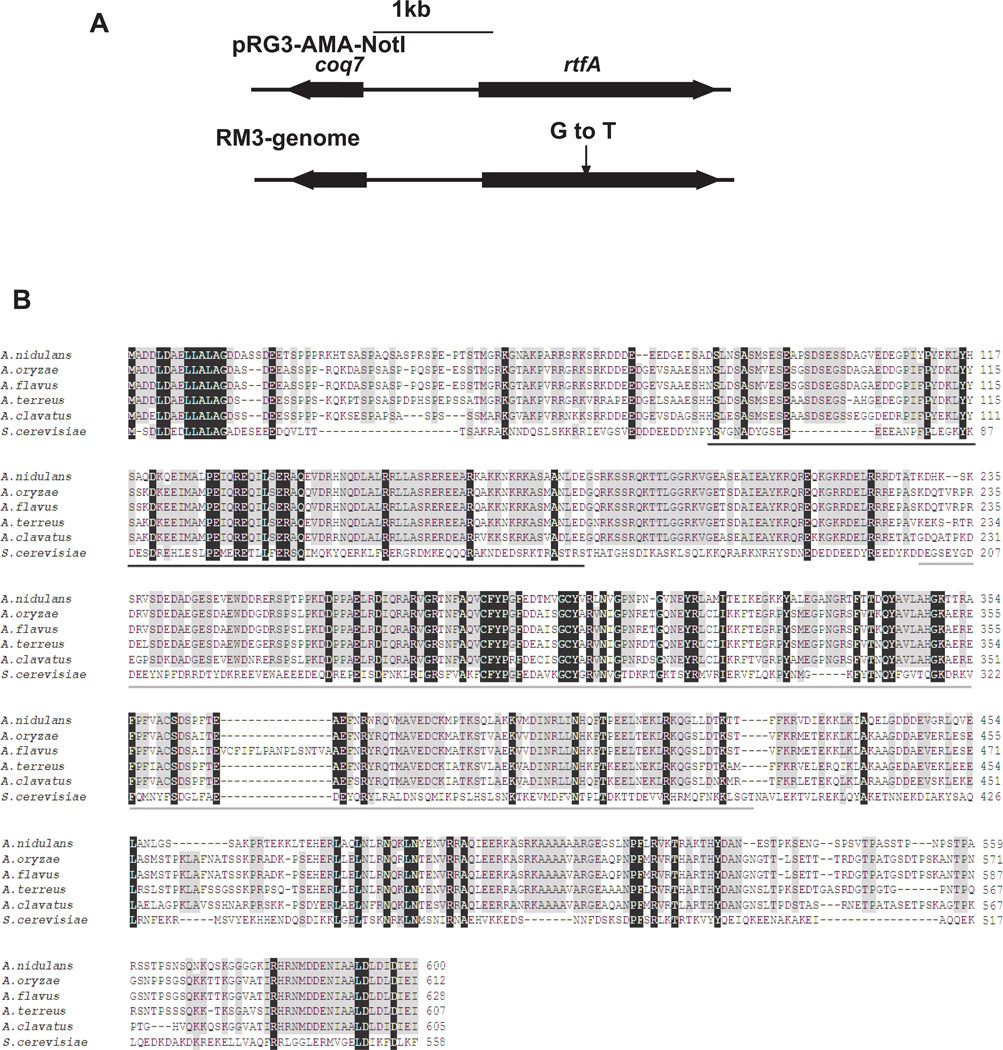
In our study, we further analyzed one the RM revertant mutants, RM3. In order to determine whether RM3 was mutated in one or more genes as a result of the mutagenesis, RM3 was crossed with RAV2 (Table 1), in which stcE is deleted. Information on the progeny segregation pattern is included in the experimental procedure section. Progeny analysis of the meiotic recombination between RM3 and RAV2 mutants indicated that the mutation in RM3 occurs at a single locus (data not shown). We also tested for dominance or recessiveness of this mutation by crossing RM3-R2 progeny (obtained from the cross between RM3 and RAV2, Table 1) with RAV1. The resultant heterokaryons and the diploid strain presented RAV1 phenotype indicating that the mutation in this strain was recessive. RM3-R2 progeny with pyrG89 auxotrophy were selected and used to identify the mutated gene by complementation with the pRG3-AMA1-NOT1 A. nidulans genomic library (Osherov and May, 2000). Upon transformation of RM3-R2 progeny with this genomic library, we obtained 5 positive transformants showing wild-type phenotype. Plasmid DNA were extracted from the transformants, and sequenced. Sequencing of the plasmids from independent transformants indicated that the plasmids contained the same insert, and that and the insert included two open reading frames encoding a putative homolog of Saccharomyces cerevisiae ubiquinone biosynthesis protein, Coq7, and a protein with similarity to an RNA polymerase II transcription elongation factor like protein, that we denominated RtfA. In order to determine where the inflicted mutation was located in the RM3 mutant, the corresponding DNA fragment was PCR amplified from genomic DNA samples of RM3-R2 and RM3 using primers RM3-f1 and RM3-r2 (Table 2), and the PCR products were sequenced. This analysis indicated that the gene encoding RNA polymerase II transcription elongation factor like protein presented a G-T transversion at nucleotide 913 of the rtfA ORF, leading to a change from codon GGA (glycine) to TGA (stop codon) corresponding to position 305 of the deduced amino acid sequence of RtfA (Fig. 2A).
Table 1.
Fungal strain used in the study.
| Strain name | Pertinent genotype | Source |
|---|---|---|
| FGSC4 | Wild type (veA+) | FGSC1 |
| RDAE206 | yA2, pabaA1, pyrG89; argB2, ΔstcE::argB, ΔveA::argB | This study |
| RDAEp206 | yA2; ΔstcE::argB, ΔveA::argB | This study |
| RAV1 | yA2, pabaA1, pyrG89; wA3; argB2, ΔstcE::argB; veA1 | This study |
| RAV1p | yA2; wA3; ΔstcE::argB; veA1 | This study |
| RAV2 | yA2; wA3; argB2, ΔstcE::argB; pyroA4; veA1 | This study |
| RAV3 | wA3; argB2, ΔstcE::argB, ΔveA::argB; pyroA4 | This study |
| RM mutants | yA2, pabaA1, pyrG89; argB2, ΔstcE::argB, ΔveA ::argB, sup− | This study |
| RM3 | yA2, pabaA1, pyrG89; argB2, ΔstcE::argB, ΔveA ::argB, rtfA− | This study |
| RM3p | yA2, ΔstcE::argB, ΔveA ::argB, rtfA− | This study |
| RM3-R2 | yA2, pyrG89; wA3; argB2, ΔstcE::argB, rtfA−, veA1 | This study |
| RM3p-R2 | yA2; wA3; ΔstcE::argB, rtfA−, veA1 | This study |
| RM3-R2-com | yA2, pyrG89; wA3; argB2, ΔstcE::argB, rtfA−, pRG3− AMA-NOT1-rtfA::pyr4; veA1 | This study |
| RJMP1.49 | pyrG89; argB2, ΔnkuA::argB; pyroA4; veA+ | Shaaban et al., 2010 |
| TRV50 | pyrG89, pyrG+; argB2, ΔnkuA::argB; pyroA4; veA+ | This study |
| TRVΔrtfA | pyrG89; argB2, ΔnkuA::argB; ΔrtfA:: pyrGA.fum; pyroA4; veA+ | This study |
| TRVΔrtfA-com | pyrG89; argB2, ΔnkuA::argB, ΔrtfA::pyrGA.fum; pyroA::rtfA; pyroA4; veA+ | This study |
| TRV51 | pyrG89; argB2, ΔnkuA::argB, alcA(p)::rtfA::pyr4; pyroA4; veA+ | This study |
| RM3-com | yA2, pabaA1, pyrG89; argB2, ΔveA::argB, ΔstcE::argB, Δsup3, pRG3-Ama1-Not-rtfA | This study |
| RM3p-com | yA2; ΔveA::argB, ΔstcE::argB, Δsup3, pRG3-Ama1-Not-rtfA | This study |
| FGSC89 | biA1, argB2; veA1 | FGSC |
| TDAEΔrtfA | yA2,pabaA1, pyrG89; pyrGA.fum::Δrft1,ΔstcE::argB, ΔveA::argB | This study |
| TDAEpΔrtfA | yA2; pyrGA.fum::Δrft1,ΔstcE::argB, ΔveA::argB | This study |
| RSS3.A.2ΔrtfA | ΔrtfA; veA1 | This study |
| RDIT2.3 | veA1 | Bok et al., 2004 |
| RJW46.4 | methG1, ΔlaeA::methG; veA1 | Bok et al., 2004 |
| RJW41.A | methG1, ΔlaeA::methG; veA+ | Bayram et al., 2008 |
| RJW34-1 | pyrG89; wA3; ΔstcE::argB; ΔlaeA::methG; trpC801; veA1 | Bok et al., 2004 |
| RSD18.1 | pyrG89; wA3; argB2, ΔnkuA::argB; ΔrtfA::pyrGA.fum; ΔlaeA::methG; veA+ | This study |
| RSD18.2 | pyrG89; wA3; argB2, ΔnkuA::argB; ΔrtfA::pyrGA.fum; ΔlaeA::methG; veA+ | This study |
| RSD19.1 | pyrG89; wA3; argB2, ΔnkuA::argB; ΔrtfA::pyrGA.fum; ΔlaeA::methG; veA1 | This study |
| RSD19.2 | pyrG89; wA3; argB2, ΔnkuA::argB; ΔrtfA::pyrGA.fum; ΔlaeA::methG; veA1 | This study |
| TSD21.1 | pyrG89; ΔnkuA::argB; rtfA::gfp::pyrGA.fum; pyroA4; hhoA::mCherry::pyroAA.fum; veA+ | This study |
| SNA3 | pyrG89; ΔnkuA::argB; pyroA4; veA+::S-Tag::pyrGA.fum | This study |
| TSD22.4 | pyrG89; ΔnkuA::argB; rtfA::gfp::pyroAA.fum; pyroA4; veA+::S-Tag::pyrGA.fum | This study |
FGSC, Fungal Genetics Stock Center. sup, suppressor gene
Table 2.
PCR primers used in the study
| Name | Sequence (5’ → 3’) |
|---|---|
| TF1 | TCAACTCGTCGTCCAGCTCTTCTCC |
| TF2 | AGTGAATGAATTTGCTGACAGCCGACG |
| TF3 | GTGACTGAGACGGTGCAGCATC |
| TF4 | TTCCGTGGTAATGTTGAGTAGACGC |
| Afp1 | CGTCGGCTGTCAGCAAATTCATTCACTGCCTCAAACAATGCTCTTCACCC |
| Afp2 | GATGCTGCACCGTCTCAGTCACCTGTCTGAGAGGAGGCACTGATGC |
| RM3-f1 | CACTACATGAACGTGTTGGTGGCG |
| RM3-r2 | TCACGTACTTTGTCGCCTTAGGGC |
| TF5 | AAGGTTGGCGAGGCTTCAGAGG |
| TF6 | TCTCCCAACTTCATCGTCATCACCG |
| pyrGAngsp1 | CAGCCATCCCACTTCCAGCTTC |
| pyrGAngsp2 | CTGGTAATACTATGCTGGCTGC |
| RtfAcom1 | AAAAAACCGCGGTCGCAAGCATATCCTTCAACTCG |
| RtfAcom2 | AAAAAAGGTACCTTAGGCAGTGGGTATGATGTTGG |
| Rtf-OE1 | AAAAAAGGTACCATGGCCGATGATCTCGACG |
| RtfA-OE2 | AAAAAATTAATTAA CAAGTTCTAAATTTCAATATCGATGTCC |
| FusionMarkerLinkerR | GTCTGAGAGGAGGCACTGATGCG |
| RM33UTRFusionF | CTTGAATATATTCGGTAAAAGATGCTTTTATTCTGC |
| RM33UtrFusionR | GCCTTCTTCTCCGCTTCAGCC |
| RM35UTRFusionR | AGCATACAAGAGACAGCGAGAGC |
| RM3GFPFusionF | CCTGGATCTGGACATCGATATTGAAATTGGAGCTGGTGCAGGCGCTGGAGCC |
| RM3GFPFusionR | GCAGAATAAAAGCATCTTTTACCGAATATATTCAAGGTCTGAGAGGAGGCACTGATGCG |
| actin-F | ATGGAAGAGGAAGTTGCTGCTCTCGTTATCGACAATGGTTC |
| actin-R | CAATGGAGGGGAAGACGGCACGGG |
| nsdD-F | CATCTCACCAGCCACAATTACAGGCGGAACCATCAC |
| nsdD-R | TTGCGAGCCAGACACAGAGGTCATAACAGTGCTTGC |
| steA-F | TCCAGCAAATGGAACCGTGGAATCAGGTGCTC |
| steA-R | GAAGGGATGGGGCAAGAATGAGACTTCTGCGGGTAA |
| brlA-F | AGCTGCCTGGTGACGGTAGTTGTTGTTGGTGTTGC |
| brlA-R | CAGGAACGAATGCCTATGCCCGACTTTCTCTCTGGA |
| S1-822 | TGACCACGAGCACCCCCATC |
| S2-823 | ACGCATGGTGGCAGGCTTTG |
| S3-824 | TCTGCCGGCGTTTATTTGTCA |
| S4-825 | TCGAGAGGCTGAAGCAGAAGACA |
| S5-826 | CAAAGCCTGCCACCATGCGTGGAGCTGGTGCAGGCGCTGGAG |
| S6-827 | TGACAAATAAACGCCGGCAGACTGTCTGAGAGGAGGCACTGAT |
| hhoAF | GCCTCCCAAGAAAGCTCCCACTACGGCC |
| hhoAR | GGCCTTCTTGTTCTTAGCAGTCTTCTTGGTCGCC |
| hhoA_mRFP_F | GGCGACCAAGAAGACTGCTAAGAACAAGAAGGCCGGAGCTGGTGCAGGCGCTGGAGCC |
| hhoA_mRFP_R | CGCTCAAACAATCTCCAAGCCAGGCTGCCTGTTTAGGCGCCGGTGGAGTGGCGGCCC |
| ANhhoA_3UTR_F | ACAGGCAGCCTGGCTTGGAGATTGTTTGAGCG |
| ANhhoA_3UTR_R | AAAAAAGGTACCGGGGGAACAACGGAGCCGGTCCAGTGG |
| rtfADHfor | AAAAAAAGATCTGTATGGCCGATGATCTCGAC |
| rtfADHrev | AAAAAAAGATCTCTAAATTTCAATATCGATGTCCA |
Fig. 2. RM3 presents a mutation in the rtfA gene.
A. Complementation insert from the genomic library plasmid in the RM3 mutant and corresponding genomic fragment in the RM3 mutant. Arrow indicates the point mutation at rtfA in RM3, a G-T transversion at nucleotide 915 of the rtfA ORF (from GGA to TGA resulting in a non-sense mutation. coq7 is an adjacent putative ubiquinone biosynthetic gene.
B. Alignment of deduced amino acid sequences of A. nidulans RtfA and putative orthologs in A. oryzae, A. flavus, A. terreus, A. clavatus and S. cerevisiae. The alignment was performed using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.htm) multiple sequence alignment software program. Conserved residues are highlighted in black (identical) and grey (similar). Black and gray underlining indicates two conserved domains present in S. cerevisiae Rtf1 (amino acid residues, 62 – 152 and 201 – 395), demonstrated to be involved in histone methylation, telomeric silencing (black and grey) and ORF association (gray).
rtfA is conserved in numerous fungal species
The amino acid sequence of A. nidulans RtfA revealed significant identity with orthologs from other Aspergillus spp such as A. oryzae (70% identity, 82% similarity), A. terreus (70%, 92%), A. flavus (69% 80%), A. clavatus (69%, 80%), A. niger (72%, 84%) and A. fumigatus (68%, 79) and lower identity with the putative homologous protein in Saccharomyces cerevisiae, Rtf1 (26%, 43%). Rtf1 homologs are also found in species of other fungal genera (Suppl. Fig.1), as well as in complex eukaryotes (Warner et al., 2007; reviewed by Jaehning, 2010). The RtfA protein has highly conserved domains among the putative orthologs from different Aspergillus spp. (Fig. 2B). These domains are also conserved in RtfA/Rtf1 homologs in other eukaryotes (Warner et al., 2007). Putative orthologs from other fungal genera are listed in Suppl. Table 1. An extensive alignment and phylogenetic tree is shown in Suppl. Fig. 1 and Suppl. Fig. 2. Our analysis showed that RtfA is conserved across fungal genera. The RtfA tree topology was consistent with established fungal taxonomy.
rtfA differentially influences mycotoxin production in a medium-dependent manner
In order to confirm that recovery of NOR production in the absence of veA was due to rtfA loss-of-function, we deleted rtfA in the RDAE206 strain (ΔstcE, ΔveA), obtaining TDAEΔrtfA (ΔstcE, ΔveA, ΔrtfA) (Table 1). In addition, we also deleted rftA in a strain with a veA+ wild-type allele resulting in TRVΔrtfA (Table 1). These deletion mutants, TDAEΔrtfA and TRVΔrtfA, were confirmed by PCR (data not shown) followed by Southern blot analysis (Suppl. Fig. 3). Hybridization analysis using upstream flanking 5’UTR as probe 1, revealed 0.8 and 3.0 kb PstI fragments in the wild-type FGSC4 strain and 0.8 and 1.7 kb PstI fragments in TDAEΔrtfA and TRVΔrtfA mutants as expected. This confirmed that the rtfA coding region was deleted in these mutants. Also, hybridization with AfpyrG specific probe 2 (Suppl. Fig. 3) revealed 1.2 and 1.7 kb PstI fragments in the ΔrtfA mutants that were absent in the wild-type FGSC4, further confirming that rtfA was replaced by AfpyrG in the deletion mutants. Prototrophs were generated in order to compare NOR production using isogenic strains (Fig. 3).
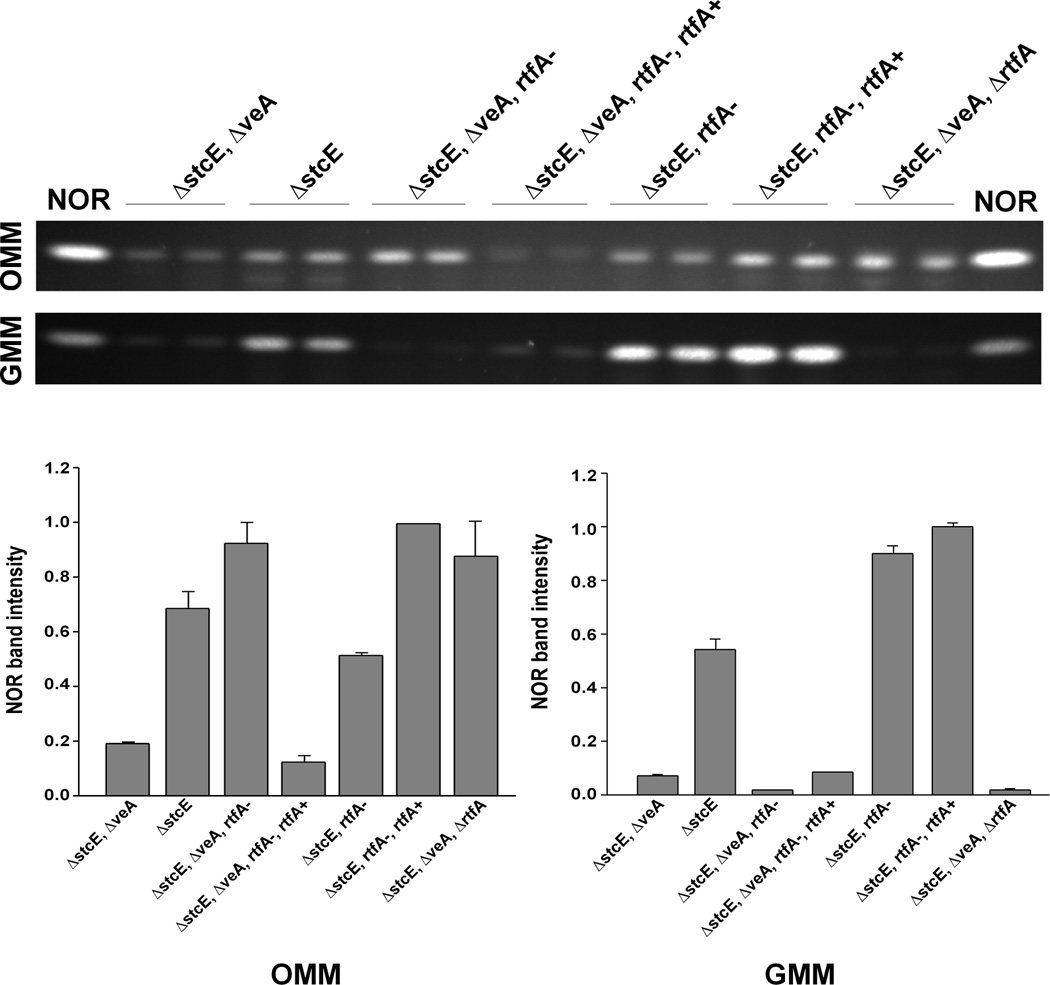
Fig. 3. TLC analysis of NOR production.
Extracts were obtained from top-agar inoculated OMM and GMM cultures of the strains ΔstcE ΔveA (RDAEp206), ΔstcE (RAV1p), ΔstcE ΔveA rtfA− (RM3p), ΔstcE ΔveA rtfA− rtfA+ (RM3p-com), ΔstcE rtfA− (RM3p-R2), ΔstcE ΔveA rtfA+ (RM3-R2-com), ΔstcE ΔveA ΔrtfA (TDAEp206)(Table 1), incubated at 37°C in the dark for five days. NOR was analyzed as described in experimental procedures. NOR standard was included on both sides. Asterisk indicates not detected. Densitometry was carried out with the Scion Image Beta 4.03 software. The NOR band intensity values were normalized with respect to the highest intensity considered as 1.
The TDAEpΔrtfA (ΔstcE, ΔveA, ΔrtfA) mutant produced NOR, as did the RM3p mutant (ΔstcE, ΔveA, rtfA−) on OMM medium (Fig. 3). However, on GMM, both the TDAEpΔrtfA mutant and the RM3p strain produced only trace amounts of NOR. This further supports that the rtfA gene differentially modulates mycotoxin biosynthesis depending on medium composition. When the RM3 strain is complemented with a wild-type allele of the rtfA gene, the resultant RM3p-com strain, as in the case of RDAEp206 (ΔstcE, ΔveA), only produced trace amounts of NOR on OMM or GMM media.
The rtfA effect on mycotoxin biosynthesis depends on the veA allele
Differently from RM3p (ΔstcE, ΔveA, rtfA−) and TDAEpΔrtfA (ΔstcE, ΔveA, ΔrtfA), RM3p-R2 (ΔstcE, rtfA−, veA1) produced lower levels of NOR on OMM, but higher amounts on GMM, compared to RM3p mutant or TDAEpΔrtfA (Fig. 3). Furthermore, deletion of rftA in a veA1 strain (RSS3A.4ΔrtfA obtained by meioitic recombination between TRVΔrtfA and FGCS89, Table 1) also accumulated higher levels of ST compared to the control strain (Suppl. Fig. 4). These results suggest that the function of rtfA in ST biosynthesis is dependent on the presence or absence of veA, also altering the effect of the nutritional environment on toxin production. Restoration of the rtfA gene in RM3-R2 (complemented strain RM3-R2-com) restored normal levels of NOR production, similar as those in the control strain RAV1p (Fig. 3).
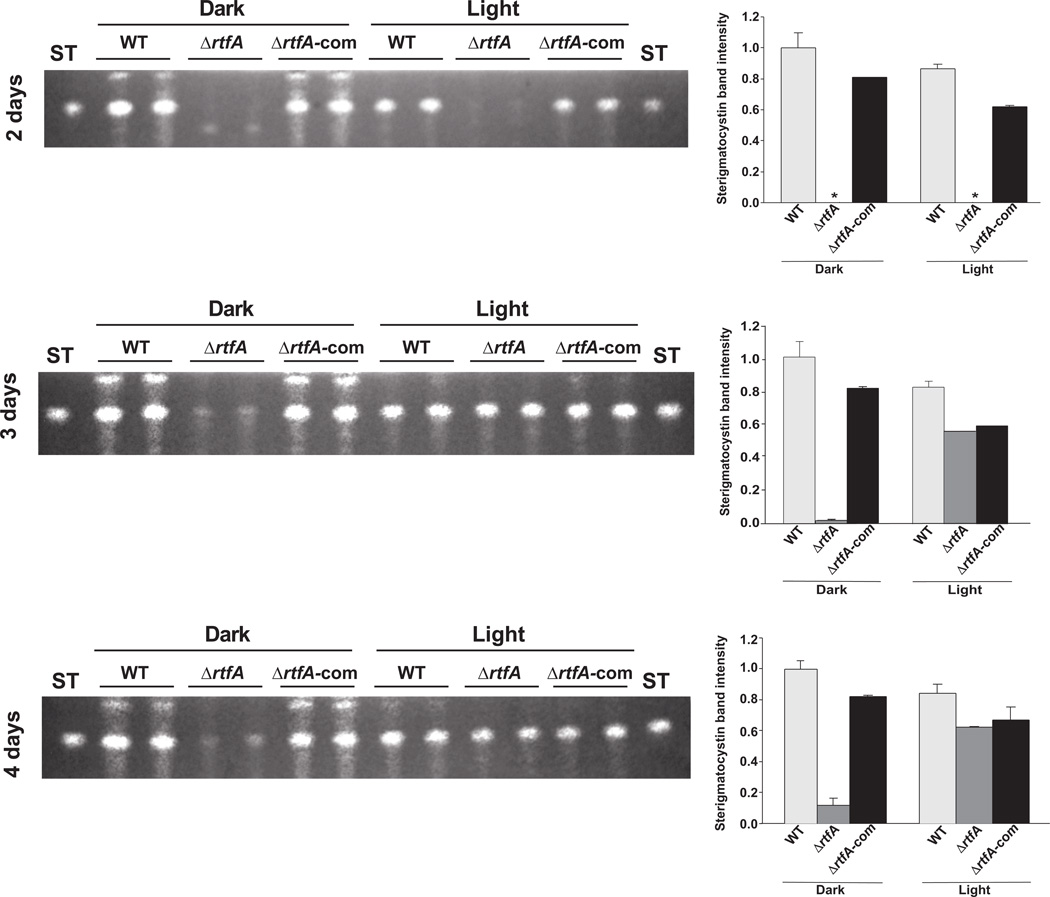
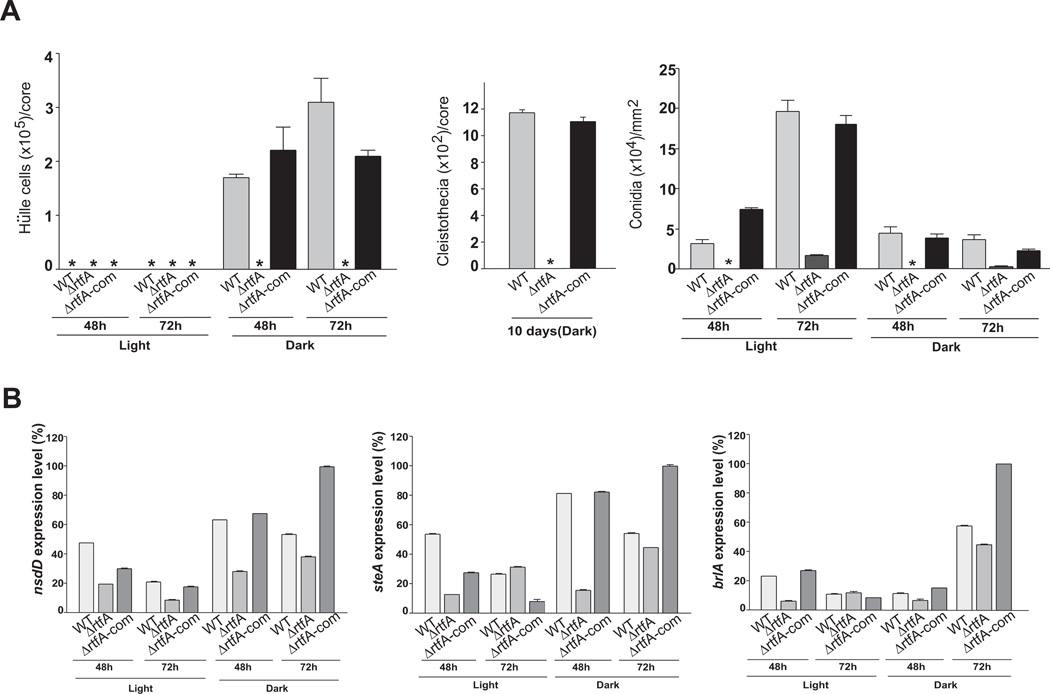
We also examined the role of rtfA on ST biosynthesis in a veA+ genetic background, and whether the presence or absence of light modulates its effect on toxin production over time (Fig. 4). Our TLC analyses indicated that ΔrtfA, in the presence of a veA+ wild-type allele, produces only trace amounts of ST after two days of incubation on GMM in both light and dark conditions compared to the isogenic wild-type strain and complementation strain. On the third day and fourth day of incubation, the ΔrtfA mutant produced detectable amounts of ST but at lower levels compared with the controls. Notably, the production of toxin in the rtfA deletion mutant was favored by light. Similar results were also obtained from cultures grown on OMM (Suppl. Fig. 5). These results contrast with those observed in strains in which rtfA is mutated or deleted in a ΔveA or veA1 background.
Fig. 4. Effects of rtfA deletion and light on ST production in A. nidulans veA+ strains.
TLC analysis of ST production from cultures grown on GMM. Wild type (WT) veA+ control (TRV50), ΔrtfA (TRVΔrftA) and complementation strain (TRVΔrftA-com) were top-agar inoculated and incubated at 37°C either in the dark or the light for 2, 3 and 4 days. ST was extracted and analyzed as described in the experimental procedures section. Asterisk indicates not detected. The densitometry was carried out with the Scion Image Beta 4.03 software. ST band intensity values were normalized with respect to the highest intensity considered as 1.
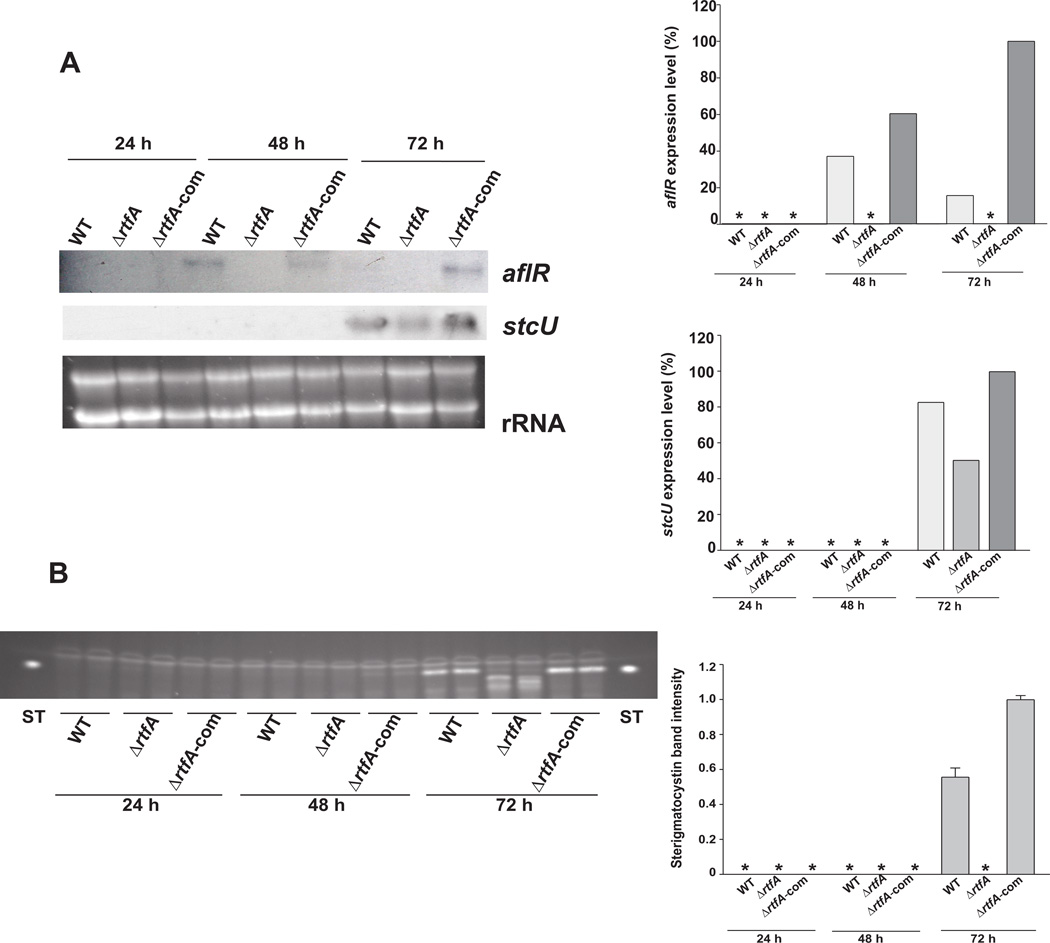
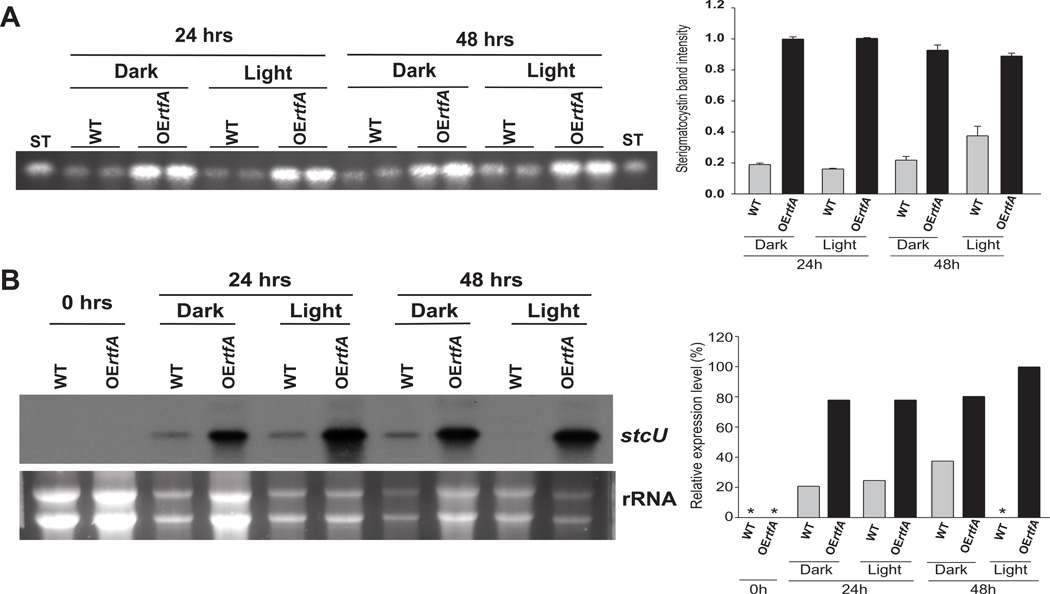
Parallel to the decrease in ST production as a result of the absence of rtfA in the presence of veA+, the expression of the regulatory aflR gene and structural stcU, a gene in the ST cluster frequently used as indicator for cluster activation (i.e. Kato et al., 2003; Hicks et al., 1997), was also decreased in the ΔrtfA mutant compared to the expression levels detected in the wild-type and complementation controls (Fig. 5). Forced over-expression of rtfA (alcA(p)::rftA, veA+) increased ST production over time compared to the wild-type strain, both under light and dark conditions (Fig. 6). This was accompanied by an increase in gene cluster transcription, as indicated by the greater accumulation of stcU transcript in the over-expression rtfA compared to the control strain.
Fig. 5. Effect of the rftA deletion on the expression of aflR and stcU over time.
A. Wild type (WT) veA+ control (TRV50), ΔrtfA (TRVΔrftA) and complementation strain (TRVΔrftA-com) were inoculated in liquid GMM. Mycelia were collected after 24, 48 and 72 hrs of culture in a shaker incubator at 250 rpm at 37°C. Expression of aflR and stcU was analyzed by Northern blot. rRNA serves as loading control. The densitometry was carried out with the Scion Image Beta 4.03 software. Asterisk indicates not detected. ST band intensity values were normalized with respect to the highest intensity considered as 1.
B. TLC showing accumulation of ST in the cultures described in (A).
Fig. 6. Overexpression of rtfA increases ST production.
A. TLC analysis of ST production. Wild-type (WT)(veA+ TRV50) and overexpression rtfA (TRV51) strains were inoculated in liquid GMM medium (106 conidia ml−1) and incubated for 16 hrs. Then, mycelia were harvested and transferred onto TMM agar, medium. The cultures were further incubated for 24 and 48 hrs under either light or dark conditions. Toxin analysis was carried out as described in the experimental procedure. Densitometry was performed using Scion Image Beta 4.03 software. Asterisk indicates not detected.
B. Expression analysis of stcU by Northern blot. Total RNA was extracted from samples obtained from cultures grown as described in (A). rRNA stained with ethidium bromide is shown to indicate RNA loading.
Deletion of rtfA does not rescue ST production in a deletion laeA strain
rtfA loss-of-function recovers ST production in a deletion veA strain. Due to the fact that the VeA and LaeA proteins interact and are functionally dependent, we tested whether deletion of rtfA could also rescue ST production in a deletion laeA strain. Double ΔrtfAΔlaeA mutants in veA1 and veA+ backgrounds were generated by meiotic recombination by crossing RJW34-1 and TRVΔrtfA (Table 1). Our TLC results indicated that deletion of rtfA was not sufficient to recover ST biosynthesis in the laeA deletion mutants (Suppl. Fig. 6).
rtfA affects penicillin production
Results from our chemical analysis indicated that rtfA is also necessary for the synthesis of other metabolites (i.e. Fig. 4, Suppl. Fig. 5, and data not shown). For this reason, we also examined whether rtfA has a role in biosynthesis of PN. We analyzed production of this antibiotic in TRVΔrtfA and compared it with PN production levels in the isogenic control TRV50 (strained derived from the complementation of RJMP1.49 with A. nidulans pyrG wild-type allele) and complementation strain TRVΔrtfA-com (Fig. 7). With this in mind we performed a well established bioassay using a strain of B. calidolactis as the testing organism. Deletion of rtfA decreased PN production with respect to the control strains (Fig. 7), indicating that rftA is necessary for normal levels of PN biosynthesis. However, overexpression of rtfA did not increase production of this antibiotic (data not shown).
Fig. 7. rtfA regulates penicillin production.
A. Effect of fungal extracts on the growth of B. calidolactis C953. The wells in the solid medium contained extracts from wild-type strain (WT), (veA+ TRV50), ΔrtfA (TRVΔrtfA) and complementation strain (TRVΔrtfA-com). Arrows indicate the edge of the inhibition halo. Additional controls with the same extracts mixed with 5 U of penicillase were also analyzed (data not shown),
B. Penicillin concentrations are represented as micrograms per milliliter of culture supernatant. Commercial Penicillin G (Sigma) was used as the standard. Values are means of four replicates. Standard error is shown.
rtfA controls growth and hyphal branching pattern
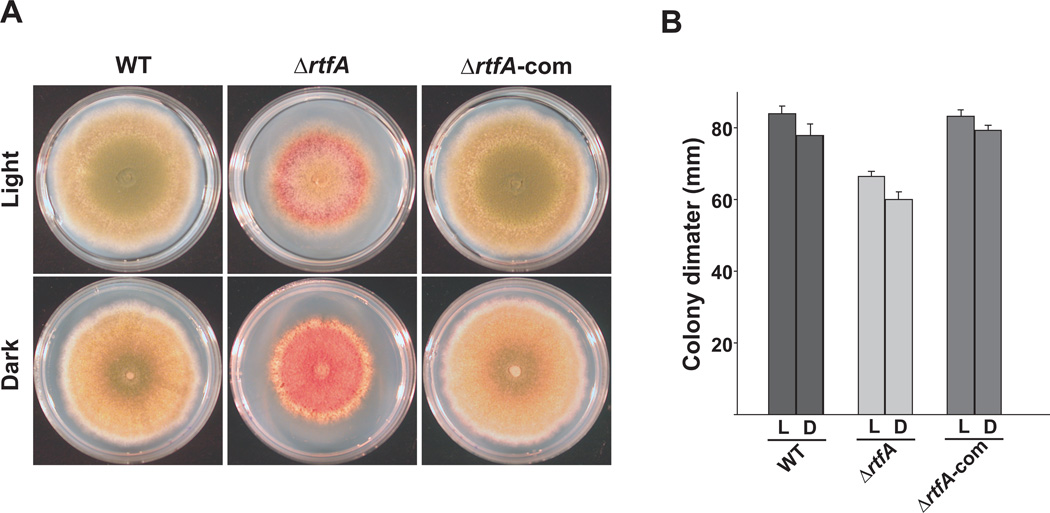
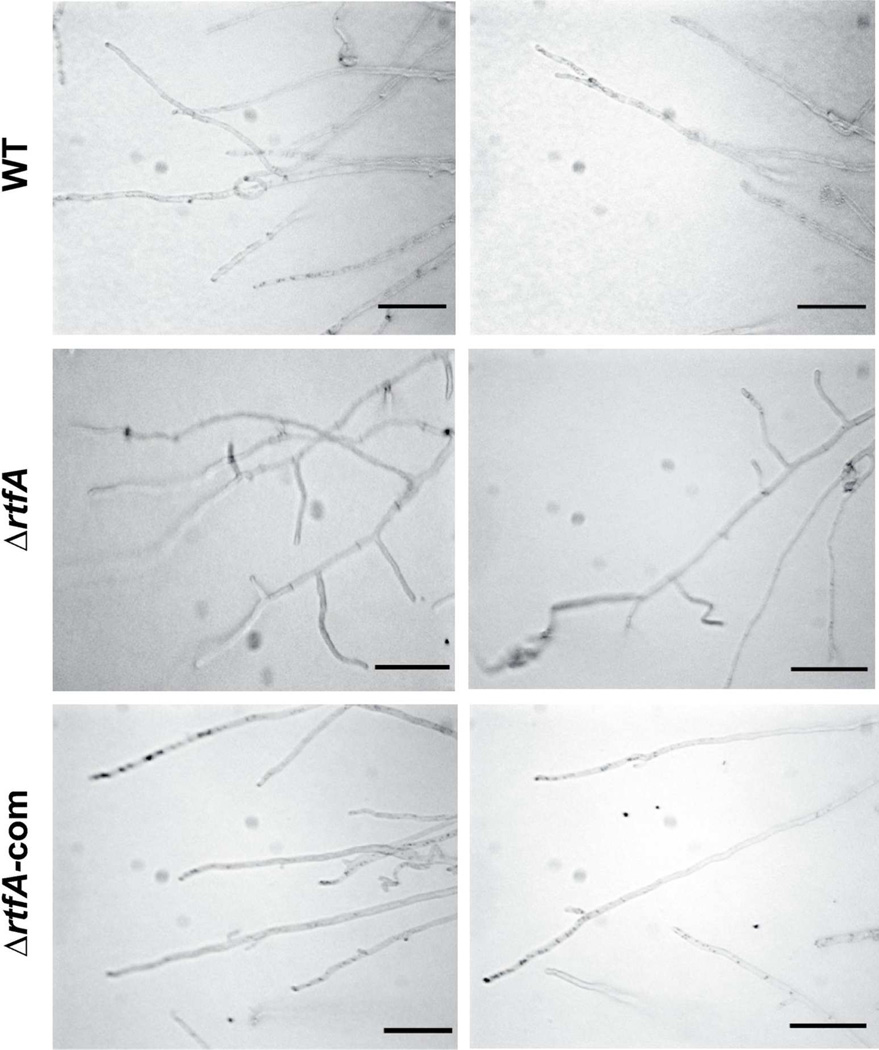
In addition to the effects of rtfA deletion on secondary metabolism, deletion of this gene also altered colony growth. Radial growth of TRVΔrtfA was slightly reduced compared to its isogenic wild-type strain under both light and dark conditions (Fig. 8). In addition, the mutant presented a dark pink pigment on top of the colony (Fig. 8) and a dark brown color at the bottom (data not shown). Colonies of the TRVΔrtfA mutant appeared compact. The ΔrtfA deletion mutant presented a hyperbranching hyphal pattern with respect to the control strains (Fig. 9). Complementation with the rtfA wild-type allele restored wild-type phenotype.
Fig. 8. Deletion of rtfA affects fungal growth and colony pigmentation.
A. Aspergillus nidulans strains, wild type (WT) (veA+ TRV50), ΔrtfA (TRVΔrtfA) and complementation (TRVΔrtfA-com) were point inoculated on GMM plates and incubated at 37°C in either dark or light for 6 days.
B. Fungal growth measured as colony diameter. Values are means of four replicates. Standard error is shown.
Fig. 9. rtfA is necessary for normal A. nidulans hyphal branching pattern.
Wild type (WT) (veA+ TRV50), ΔrtfA (TRVΔrtfA) and complementation (TRVΔrtfA-com) strains were point-inoculated on GMM and incubated at 37°C in the light for 6 days. Two micrographs from the growth front edge of each culture are shown. Deletion rtfA mutant presents hyperbranching pattern in comparison to the control strains. Images were captured with a Nikon E600 Eclipse light microscope. Scale bar represents 500 micrometers.
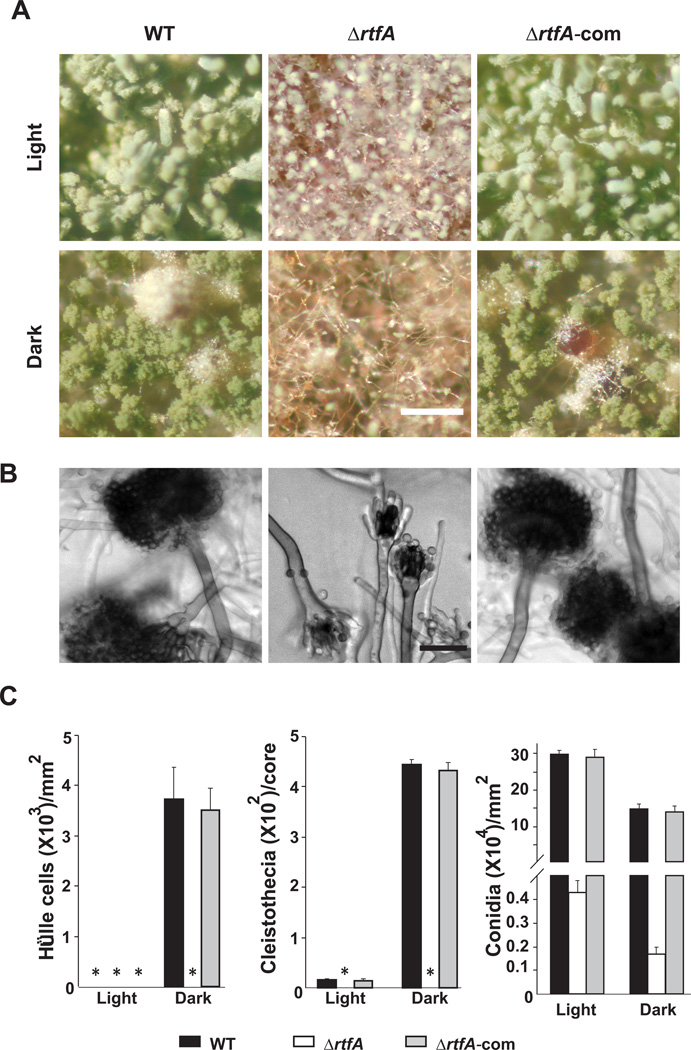
rtfA is necessary for normal sexual and asexual development
The TRVΔrtfA mutant did not produce Hülle cells, nursing cells involved in the formation of cleistothecia (Yager, 1992). Fruiting bodies were also absent in the rtfA deletion mutant cultures in both light and dark conditions (Fig. 10 and Fig 11A). Complementation of the TRVΔrtfA mutant with the wild-type allele of rtfA restored both asexual and sexual development (Fig. 10). Expression of nsdD and steA, both encoding transcription factors involved in the activation of sexual development in A. nidulans (Han et al., 2001; Vallim et al., 2000), was reduced in the rtfA deletion mutant in comparison with expression levels in the control strains (Fig. 11B), indicating that rtfA influences the expression of genes involved in the formation of sexual structures in A. nidulans.
Fig. 10. Deletion of rtfA mutant results in defective conidiogenesis and loss of sexual development.
A. Micrographs of point-inoculated cultures of wild type (WT) (veA+ TRV50), ΔrtfA (TRVΔrtfA) and complementation (TRVΔrtfA-com) strains grown in the dark for 6 days. Microscopy samples were collected 2 cm from the point of inoculation. Images were captured using upright Leica MZ75 stereomicroscope. Scale bar represents 200 micrometers.
B. Micrographs of conidiophores from wild type, ΔrtfA and complementation strain. Microscopy samples were collected 2 cm from the point of inoculation. Samples were observed with an upright Nikon Eclipse 80i microscope. Scale bar represents 20 micrometers.
C. Quantitative analysis of sexual and asexual reproduction. Strains grown as described above were used for quantification of Hülle cells, cleistothecial production, and conidial production. Samples were collected 2 cm from the point of inoculation and homogenized in water for cell counts. Cleistothecia were also collected 2 cm from the point of inoculation and were counted after spraying the mycelial cores (15 mm diameter) with 70% ethanol to improve visualization under dissecting microscope. Asterisk indicates not detected. Values are means of three replications. Error bar indicates standard errors.
Fig. 11. Effects of the rtfA deletion on A. nidulans development over time.
A. Quantitative analysis of asexual and sexual reproduction in a time-course experiment. Wild type (WT) (veA+ TRV50), ΔrtfA (TRVΔrtfA) and complementation (TRVΔrtfA-com) strains were top agar-inoculated on GMM and incubated at 37°C either in the dark or in the light. Production of Hülle cells, cleistothecia, and conidia were examined. Asterisk indicates not detected. Values are means of three replications. Error bar indicates standard errors.
B. Expression analysis of genes implicated in morphological development in A. nidulans. The transcriptional pattern of nsdD, steA, and brlA was evaluated by qRT-PCR. Total RNA was isolated at 48 and 72 h from top-agar inoculated GMM cultures incubated either in the dark or in the light. The relative expression levels were calculated using the 2−ΔΔCt method described by Livak and Schmittgen (2001), and all values were normalized to the expression of the A. nidulans actin gene. The error bars indicate the ranges for three replicates.
Deletion of rtfA also resulted in defective asexual development, especially at early developmental stages of conidiogenesis under both light and dark conditions, resulting in a delay and a reduction of conidiophore formation, which also presented abnormal morphology. This was accompanied by reduced conidial production (Fig. 10A, B, and C and Fig. 11A). Expression of brlA, which encodes a transcription factor necessary for conidiophore formation (Adams et al., 1988), was altered in the rtfA deletion mutant. This was observed 48h after inoculation on solid GMM medium (Fig. 11B), showing a delay and a reduction of brlA transcript accumulation in the absence of rtfA. Complementation of the rtfA deletion mutant with the wild-type allele of rtfA also remediated the asexual development defects. Deletion of rtfA in a strain with a veA1 genetic background, showed the same sexual and asexual development defects observed in the ΔrtfA, veA+ strain (Suppl. Fig. 7).
Subcellular localization of RtfA
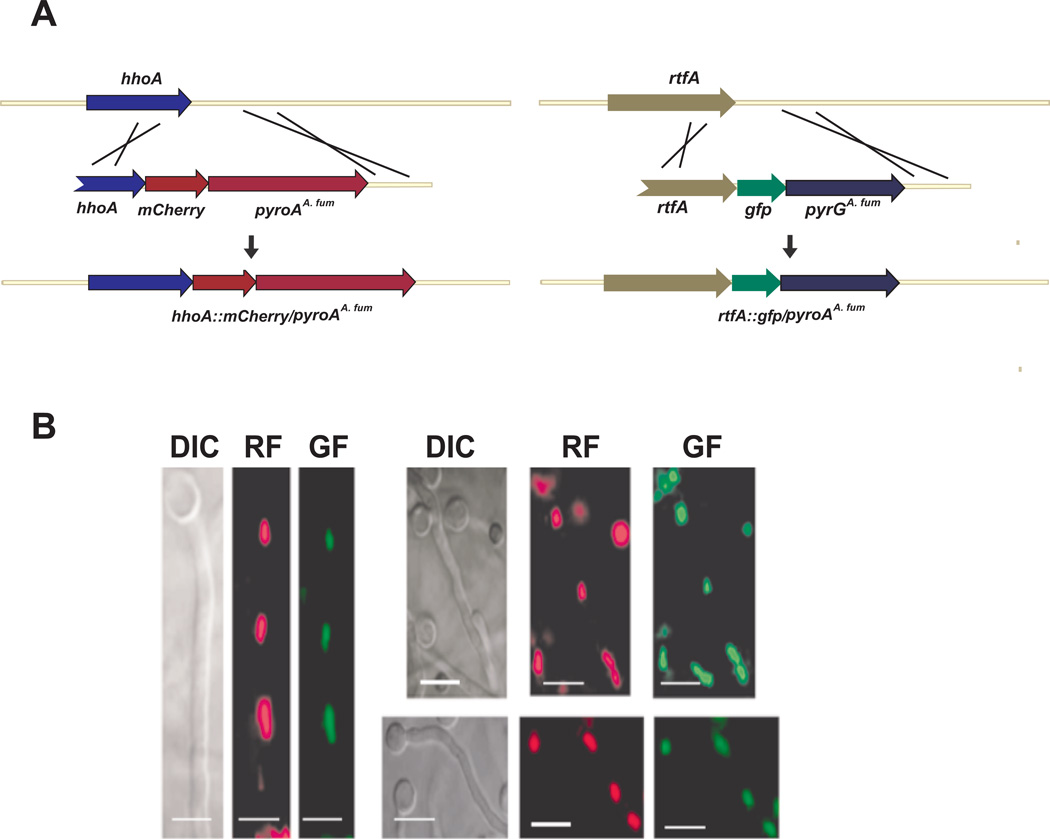
To further characterize the function of A. nidulans rtfA, we examined its subcellular localization. For this purpose we generated a strain containing RtfA fused to GFP. Observations of this strain using fluorescence microscopy indicated that RftA localizes in nuclei, as revealed when compared with the localization of an HhoA::mCherry fusion, constructed in the same strain to serve as a reference to indicate nuclear location (Fig. 12).
Fig. 12. RftA localizes in nuclei.
A. Representation of the strategy followed to fuse mCherry to A. nidulans HhoA, and GFP to RtfA respectively. The tagged constructs were introduced at the hhoA and rtfA loci, respectively, by double-recombination events.
B. Micrographs showing the subcellular localization of the RtfA::GFP fusion described in (A) in A. nidulans. From left to right, Normaski images, red fluorescence images, and green fluorescence images are included. Red fluorescence images correspond to HhoA::RFP fusion used as nuclear localization internal control.
Analysis of possible RtfA-VeA protein interactions
rtfA was identified through a deletion veA-revertant screening. In addition, our findings indicate that the role of rftA in ST production is dependent on the veA allele as well as on light. For this reason, we explored the possibility that RftA and VeA could interact at protein level. To test this hypothesis we generated a strain with two fusions, VeA::Stag and RftA::GFP. Affinity purification (as described in Liu et al., 2010) followed by Western blot analysis using a GFP antibody did not show a positive result for RtfA::GFP in the bound fraction corresponding to VeA-interacting proteins, while RtfA::GFP was detected in the unbound fraction (data not shown). In addition, interaction between RtfA and VeA was evaluated by the yeast two-hybrid system. The results showed no interaction between these two proteins (data not shown). This suggests that RftA function might not involve a direct physical interaction with the VeA protein under the experimental conditions assayed.
Discussion
Regulation of fungal secondary metabolism is often linked to morphological differentiation (Calvo et al., 2002; Calvo, 2008; Bayram and Braus, 2011). One of these links is mediated by the global regulator VeA. This protein controls development in response to light. In A. nidulans strains with a wild-type veA allele, while under illumination the fungus mainly develops asexually forming air-borne conidia, in the dark conidiation is reduced and sexual development takes place forming abundant ascospore-producing cleistothecia (Yager, 1992). Deletion of veA blocks formation of cleistotecia in A. nidulans (Kim et al., 2002) and sclerotia in A. flavus and A. parasiticus (Calvo et al., 2004; Duran et al., 2007). Our laboratory showed that VeA is also required for the production of diverse secondary metabolites (Calvo, 2008), including mycotoxins such as ST in A. nidulans (Kato et al., 2003) and AF in A. flavus and A. parasiticus (Calvo et al., 2004; Duran et al., 2007). Several studies from our group and others have contributed to gain insight into the VeA mechanism of action (i.e. Kato et al., 2003; Calvo et al., 2004; Duran et al., 2007; Purschwitz et al., 2008; Bayram et al., 2008; Cary et al., 2007; Bayram et al., 2010). VeA is necessary for the transcription of aflR in A. nidulans and A. flavus and A. parasiticus (Kato et al., 2003; Calvo et al., 2004; Duran et al., 2007). The transcription factor AflR activates the gene clusters responsible for the synthesis of the mycotoxins ST and AF (Yu et al., 1996; Chang et al., 1995). Another gene in the A. nidulans ST cluster, stcE, encodes a dehydrogenase responsible for the conversion of norsolorinic acid, NOR, to averantin. Accumulation of the orange NOR in the deletion stcE mutants facilitates visual detection of an active ST biosynthetic pathway (Butchko et al., 1999). In our study we generated an A. nidulans ΔveAΔstcE double mutant that was exposed to chemical mutagenesis to obtain reventant mutants capable of producing abundant ST intermediates in the absence of the VeA regulator. This powerful genetic technique yielded several NOR-producer mutants. The revertant mutants also displayed morphological defects to different extents, indicating that, as in the case of veA, the genes affected in the revertant mutants are involved in both development and mycotoxin biosynthesis.
We further investigated the selected mutant RM3. The mutation in RM3 was located in a gene that we denominated rtfA, encoding a protein with homology to S. cerevisiae Rtf1, a subunit of the evolutionary conserved Paf1 complex that promotes optimal gene expression by controlling co-transcriptional histone modification (Warner et al., 2007; Rosornina and Manley, 2005; Jahening, 2011). Rtf1 is required for ubiquitination of histone H2B (Ng et al., 2003) and di-methylation (me2) and tri-methylation (m3) of histone H3 (Warner et al., 2007; Briggs et al., 2002; Sun and Allis, 2002). Addition or removal of modifications such as methylation, ubiquitination, acetylation, phosphorylation and sumoylation of histones (i.e. Jenuwein and Allis, 2001; Shilatifard 2006) governs chromatin structure, conditioning activation or repression of gene transcription (Lieb and Clarke, 2005). In S. cerevisiae, ubiquitination of histone H2B at lysine 123 is a prerequisite for H3 methylation (Briggs et al., 2002; Sun and Allis, 2002). Histone methylation has been associated with actively transcribed genes (Ng et al., 2003). In addition to promoting histone modifications, as part of the Paf1 complex Rtf1 has been described as being involved in TATA site selection by TATA box-binding proteins (TBP) (Stolinski et al., 1999), in interaction with active open reading frames (ORFs), in proper attachment of components from RNA polymerase II, and in binding of chromatin remodeling proteins such as the ATP-dependent chromatin-remodeling protein Chd1 (Warner et al., 2007; Jahening, 2011), a component of the SAGA protein complex that includes a deubiquitination enzyme as well as a acetyltransferases affecting H2B and H3 (Grant et al., 1997; Grant et al., 1997; Suka et al., 2001). The studies mentioned above indicate that Rtf1 is a multifunctional protein that utilizes several independent mechanisms to coordinate modifications of chromatin to achieve proper gene expression.
In S. cerevisiae Rft1 is also necessary for gene silencing in subtelomeric regions (Ng et al., 2003; Mueller et al., 2006) through H3 methylation at lysines K4 and K79 by the Set1-containing COMPASS complex and Dot1, respectively (Krogan et at., 2003; Mueller et al., 2006). In A. nidulans, the ST gene cluster is located in a subtelomeric region in Chromosome IV. Regulation of ST and AF cluster activation has been associated with histone modifications, such as methylation and acetylation (Reyes-Dominguez et al., 2010; Shwab et al., 2007; Roze et al., 2007). For example, histone modification such as methylation of K9 in H3 negatively affects toxin production by silencing the ST gene cluster. H3K9 methylation is dependent on LaeA (Reyes-Dominguez et al., 2010), a VeA-interacting protein (Bayram and Braus, 2012). Furthermore, A. nidulans H3K4 methylation also affects ST gene cluster silencing. Loss-of-function of the A. nidulans CclA, a Bre2 homolog part of the COMPASS complex, reduced levels of H3K4me2/3, which was also found to be associated with lower levels of H3K9me2/3 (Bok et al., 2009), both chromatin marks conductive to silencing. Based on the previous findings from studies carried out in S. cerevisiae and other eukaryotic organisms it is likely that in A. nidulans the role of rtfA in ST production involves changes in chromatin landscape. Considering the multifunctional properties of Rtf1 homologs in eukaryotes, future research is necessary to elucidate with precision the RtfA mechanism of action in A. nidulans and efforts in our laboratory will be focused on furthering this goal. One likely scenario is that recovery of ST in the veA deletion strains could be due to a loss of RtfA-mediated silencing. However, our results revealed that the function of rtfA affecting ST biosynthesis is dependent on the veA allele. Deletion of rtfA in the presence of the veA+ allele, had the opposite effect on toxin production compared to its effect in the absence of veA, leading to delayed and decreased accumulation of ST in comparison to the wild type. This effect was influenced by light, since the reduction in ST production caused by the deletion of rtfA was more notable in dark cultures compared to cultures grown in the light (Fig. 4). Furthermore, over-expression of rtfA resulted in an increase in gene cluster activity and ST production, suggesting that RtfA is a positive regulator of ST production in the presence of wild-type VeA.
Interestingly, rtfA deletion allowed toxin production in veA1 mutants genetic background, commonly used in A. nidulans research laboratories. Although the influence of culture media on ST production varied in the case of ΔrtfA ΔveA and ΔrtfA veA1, both double mutants resulted in biosynthesis of toxin, and at higher levels in the case of ΔrtfA veA1 compared to the veA1 control. This coincides with the increase of ST production found in the absence of proper heterochomatin marks such as H3K9 methylation in the A. nidulans clrD mutant (Reyes-Dominguez et al., 2010) or absence of H3K4 metylation leading to an increase in the synthesis of other secondary metabolites observed in A. nidulans cclA mutant (Bok et al., 2009). Both, clrD and cclA studies, were carried out in a veA1 genetic background, and the effect of these mutations in a veA wild-type genetic background remains to be determined. The clearly different effects caused by the presence or absence as well as the type of veA allele on ST biosynthesis in the rtfA mutant further supports that rtfA is functionally veA-dependent. These results also suggest that the first 36 amino acids, absent in the VeA1 mutant protein, are important for normal RtfA function. In addition, our study showed that deletion of rtfA did not rescue ST production in the absence of laeA, indicating that laeA is still required in this process.
RtfA also affects the synthesis of other secondary metabolites. Our study revealed differences in the metabolic profiles between the rtfA deletion mutant and the control strains. For this reason, we also analyzed the role of rtfA in the production of the beta-lactam antibiotic PN. Deletion of rtfA resulted in a reduction in PN production compared to that of the wild-type strain (Fig. 7), indicating that rtfA is necessary for production of wild-type levels of this antibiotic in A. nidulans. Unlike ST biosynthesis, PN production was not increased with over-expression of rtfA, suggesting that perhaps a balanced stoichiometry with other factors could also play a role in the production of PN at wild-type levels.
In addition to its effect on secondary metabolism, A. nidulans RtfA is also necessary for normal morphological differentiation. The absence of rtfA in an A. nidulans strain resulted in a decrease in colony growth, hyphal hyperbranching, and formation of aberrant conidiophores with a remarkable reduction in conidiation rate. This coincided with reduced and delayed brlA expression in the rtfA mutant. Furthermore, we found that rtfA is also required for the formation of sexual structures. Gene expression analysis indicated that rtfA is necessary for normal transcription levels of nsdD and steA, essential for the formation of fruiting bodies in A. nidulans (Han et al., 2001; Vallim et al., 2000).
It is possible that the mechanisms through which VeA and VeA-interacting proteins regulate the expression of numerous genes under their control (Cary et al., 2007) could be mediated by RtfA, since its homolog RTF1 contributes to the coordinated regulation of gene transcription by the RNA polymerase II. Our results indicated that the A. nidulans RtfA ortholog is localized in the nucleus (Fig. 12), where RtfA could have a similar role in selective gene transcriptional activation in this fungus. We explored the possibility of a direct interaction between RtfA and VeA, however our study did not indicate a direct interaction at protein level. Nevertheless, it is possible that RftA could be associated with additional regulatory transcriptional machinery elements functionally connected to the VeA protein complex.
Overall, our study describes a deletion veA-revertant mutant with a mutation in the rtfA gene, encoding a nuclear regulator with similarity to S. cerevisiae Rtf1. Our study also revealed that rtfA controls asexual and sexual reproduction as well as secondary metabolism; affecting the production of ST and PN in A. nidulans. The fact that rtfA is necessary for normal growth, morphological development and the synthesis of several secondary metabolites, together with the fact that rtfA putative orthologs are present in several species from different fungal genera, suggest that rtfA could have a conserved role in these organisms, which in a VeA-functionally connected manner also responds to environmental cues. This is the first study of an Rtf1 putative homolog in filamentous fungi.
Experimental procedures
Fungal strains and growth conditions
Aspergillus nidulans strains used in this study are listed in Table 1. Strains were cultured on YGT medium (0.5% yeast extract, 2% dextrose, and 1 mL of trace elements prepared as described (Käfer, 1977), glucose minimal media (GMM) (Käfer, 1977), and oat meal medium (OMM) (Butchko et al., 1999), plus the appropriate supplements for the corresponding auxotrophic markers (Käfer, 1977). Glucose was replaced with threonine (TMM) for the induction of alcA promoter in gene overexpression experiments. Solid medium was prepared by adding 10 g/liter agar. Strains were stored as 30% glycerol stocks at −80°C.
Mutagenesis
Conidia of A. nidulans RDAE206 strain (yA1, pabA1, pyrG89, argB2, ΔstcE::argB, ΔveA ::argB) were mutagenized with 4-nitro-1-quinoline oxide (NQO) as previously described by (Wieser et al., 1994). Approximately 100,000 surviving colonies were screened on oatmeal agar and colonies showing orange pigmentation were selected and further confirmed by thin layer chromatography (TLC) analysis of NOR.
Genetic techniques
Meiotic recombination between A. nidulans strains was obtained following protocols previously described (Pontecorvo et al., 1953). RAV2 (wA1, yA2, pyroA4, argB2, ΔstcE::argB) was crossed with the RM3 strain, isolated after chemical mutagenesis treatment. Progeny with ΔstcE, pyroA4, ΔveA genotype (strain RAV3) was used for diploid analysis. Progeny from RM3 x RAV2 were analyzed for the presence or absence of veA by PCR. Colony morphology and NOR production were also examined. The progeny from this cross included the following four genotypes: 1. ΔveA, ΔstcE, X- (RM3 parental type); 2. pyroA4, ΔstcE (RAV2 parental type); 3. recombinant ΔveA, ΔstcE (RM3-R1); and 4. recombinant ΔstcE, X- (RM3-R2).
The dominance test was carried out by constructing diploids using the RM3-R2 strain and RAV1 strain. The resulting strains were analyzed for growth, conidial production and NOR accumulation.
Identification of the revertant mutation in RM3
A. nidulans genomic library pRG3-AMA1-NOT1 was used to transform RM3-R2 (ΔstcE, X−) strain. Positive transformants that restored wild-type phenotype were selected. Plasmid DNA was rescued by transforming E. coli cells with total DNA from the selected transformants. Ampicillin-resistant colonies were isolated and plasmid DNA was extracted and analyzed by restriction digestion and by PCR as described previously (Osherov and May, 2000). Finally, both end regions of the DNA insert in the isolated plasmids were sequenced and the complete insert sequences were found in the A. nidulans genome database (http://www.aspgd.org) by BLAST analysis. The exact location of the mutation in RM3 was identified by PCR amplification of the same genomic DNA region from RM3, and by sequencing of the identified locus.
Sequence search and alignment
The protein sequence of RtfA (ANID_04570) was compared against databases from different fungal genera, using the BLAST (blastp) tool provided by National Center for Biotechnology Information (NCBI), (http://www.ncbi.nlm.nih.gov/), with default settings. The gene entry with the highest percentage of identity and the lowest e-value for each of the species was selected (Suppl. Table S1). Pairwise sequence alignment of the proteins was performed using the EMBOSS Needle tool (http://www.ebi.ac.uk/Tools/psa/emboss_needle/) from EMBL-EBI (European Molecular Biology Laboratory's European Bioinformatics Institute). Percentage of similarities and percentage of identities were tabulated for each of the alignments (Suppl. Table 1). Multiple sequence alignment was performed with RtfA (A. nidulans) and orthologs found across various fungal species using the MultAlgin tool (Corpet, 1998), followed by shading using BoxShade v3.2.1 (http://www.ch.embnet.org/software/BOX_form.html) for presentation purposes.
Phylogenetic analysis
The phylogenetic analysis included putative RtfA orthologs from 20 different fungal species (Suppl. Fig. 2). Orthologs of RtfA (A.nidulans) were identified in the described databases (Suppl. Table 1). Multiple sequence alignment performed with MUSCLE v3.8.31 (Edgar, R.C, 2004) was used to build a Hidden Markov model (HMM), followed by realignment of sequences against the generated HMM, using the hmmbuild and hmmalign tools in HMMER v3.0b2 (http://hmmer.org/). A maximum likelihood phylogeny reconstruction method implemented in the software PhyML v3.0 (Guindon et al., 2010; Guindon and Gascuel, 2003), whose workflow is available at iPlant collaborative™ (http://www.iplantcollaborative.org/) was used for tree construction with default settings. The resulting tree was viewed using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/). Midpoint rooting (Boykin et al., 2010) of the tree was done in order to minimize the large distances from the root to any leaf. The numbers on the branches indicate the approximate likelihood branch support values in percentages (Anisimova et al., 2006).
Generation of the rtfA deletion strain
The DNA cassette used to delete rftA by gene replacement was obtained by fusion PCR following a method previously described (Szewczyk et al., 2006). The 1.1 kb 5’ UTR flanking region of rtfA was amplified with TF1 and TF2 primers (Table 2) from wild-type strain FGSC4 genomic DNA. Similarly, the 0.8 kb 3’ UTR flanking region of rtfA was also amplified with TF3 and TF4 primers. The A. fumigatus pyrG selectable marker was amplified with Afp1 and Afp2 primers from plasmid p1439 (Stinnett et al., 2007). 5’ and 3’ UTR fragments were fused to AfpyrG to generate the rtfA replacement construct using TF1 and TF4 primers. Polyethylene glycol-mediated transformation of RDAE206 and RJMP1.49 host strains (Table 1) was carried out as described previously (Szewczyk et al., 2006). The entire rtfA coding region was replaced. Transformants were selected on appropriate selection medium, screened for gene replacement by PCR using TF5 and TF6 primers and confirmed by Southern blot analysis. The deletion strains were designated as TDAEΔrtfA and TRVΔrtfA, respectively. In addition, RJMP1.49 was transformed separately with the wild-type pyrG allele amplified by PCR from A. nidulans FGSC 4 genomic DNA with pyrGAngsp1 and pyrGangsp2 primers (Table 2) to generate a pyrG prototroph strain to be utilized as isogenic control. The resultant transformant was designated as TRV50.
Generation of complementation strain
To obtain the complementation strain, TRVΔrtfA was transformed to re-introduce the rtfA wild-type allele. The entire rtfA gene, including 1.1 kb of upstream and 0.5kb downstream regions was amplified with the primers RtfAcom1 and RtfAcom2, digested with SacII and KpnI and cloned into pSM3 vector, containing pyroA, resulting in pSM3-rtfAcom. The complementation vector, pSM3-rtfAcom was then transformed into TRVΔrtfA and RM3-R2 strains, and selected on appropriate medium. Complementation was confirmed by PCR and Southern blot analysis (data not shown). The complemented strains were designated as TRVΔrtfA-com and RM3-R2-com respectively.
rtfA over-expression
To over-express rtfA, the coding region of rtfA was amplified from A. nidulans FGSC4 genomic DNA with primers Rtf-OE1 and RtfA-OE2 (Table 2), digested with KpnI and PacI and cloned into pmacro plasmid, containing the A. nidulans alcA promoter and trpC terminator, resulting in the plasmid pMacroRtfOE. This plasmid was transformed into RJMP1.49 resulting in TRV51. Transformants were screened by PCR and confirmed by Southern blot analysis (data not shown).
For over-expression analysis, 400 mL of liquid GMM was inoculated with approximately 106 conidia/mL from TRV50 or TRV51, and incubated for 16 hrs at 37°C and 250 rpm. Then, mycelia were collected, and equal amounts were spread onto induction medium, threonine minimal medium (TMM), where the cultures were further incubated in the light or in the dark. Cultures were collected for analysis at 24 and 48 hrs after shift onto the induction medium.
Toxin analysis
Plates containing 25mL of solid GMM or OMM with appropriate supplements were inoculated with 5 mL of top agar containing approximately 106 spores/mL. The cultures were incubated in either dark or light conditions. Three cores (16 mm diameter) from each replicate plate were extracted with CHCl3. The extracts were dried overnight and then resuspended in 200 µl of CHCl3. Samples were fractionated by thin-layer chromatography (TLC) using benzene and glacial acetic acid [95:5 (v/v)] as solvent system for ST analysis, and chloroform:acetone:n-hexane (85:15:20) as solvent system for NOR analysis. The plates were then sprayed with aluminum chloride (15% in ethanol) and baked for 10 min at 80°C. ST/NOR bands on TLC silica plates were viewed by exposing the plates under UV light (375-nm).
Penicillin analysis
The PN bioassay was performed as previously described (Brakhage et al., 1992) with some modifications, using Bacillus calidolactis C953 as the test organism. Briefly, strains were inoculated with approximately 106 spores/mL in 25 mL of seed culture medium, and incubated at 26°C for 24 hours at 250 rpm. Mycelia were then transferred to PN-inducing medium (Brakhage et al., 1992). The experiment was carried out with three replicates. After 96 hours, the cultures were filtered using Miracloth (Calbiochem, USA) and the supernatants were collected for analysis. Three hundred milliliters of Tryptone-Soy Agar was supplemented with 20 ml of Bacillus calidolactis C953 culture and plated on three 150 mm-diameter Petri dishes. Twenty microliters of each culture supernatant was added to 7 mm-diameter wells. Bacteria were allowed to grow at 55°C for 16 hours and inhibition halos were visualized and measured. To confirm that the observed antibacterial activity was due to the presence of PN and not to the presence of different fungal compounds in the supernatant, additional controls containing commercial penicillinase from Bacillus cereus (Sigma, MO, USA) were used. A standard curve using various concentrations of PN G (Sigma, MO, USA) was used to determine PN concentration in the samples.
Fluorescence microscopy
Aspergillus nidulans RJMP1.49 strain (Table 1) was co-transformed with rtfA::gfp::pyrGA.fum and hhoA::mCherry::pyroAA.fum fusion PCR products as described by Szewczyk et al., (2006). Primers used for fusion PCR are listed in Table 2. Plasmids p1439 (Stinnett et al., 2007) and pHL85 (FGSC) were used as template for the PCR amplification of the intermediate fragment, respectively. Directed integration was confirmed by PCR and Southern blot analysis (data not shown). Conidia from the selected transformants (i.e. TSD21.1, Table 1) were inoculated as described previously (Stinnett et al., 2007). Briefly, conidia were allowed to germinate on the surface of the coverslip immersed in Watch minimal medium (Peñalva, 2005) with 1% glucose as carbon source for 16 hours at 30°C in strict darkness. Samples were observed with a Nikon Eclipse E-600 microscope with a 60× oil-immersion objective, Nomarski optics and fluorochromes for GFP (ex 470, em 525) and mCherry (ex 545, em 620) detection. Micrographs were taken using Hamamatsu ORCA-ER high-sensitivity monochrome digital CCD camera with MicroSuite5 image capture and optimization software. DIC images were taken with an exposure of 50 ms, mCherry with 200 ms, and GFP with 700 ms.
Morphological studies
A. nidulans strains TRV50, TRVΔrtfA, TRVΔrtfA-com (Table 1) were point inoculated on GMM and incubated at 37°C under either light or dark conditions for 6 days, when the colony diameters were measured. Experiments were performed triplicate.
In addition, plates containing 25 ml of solid GMM with the appropriate supplements were inoculated with 5 ml of top agar containing 106 spores/mL. The cultures were incubated at 37°C in either dark or light conditions. Cores from both top-agar and point inoculated cultures were harvested from each plate and homogenized in water. Conidia and Hülle cells were counted using a hemacytometer. Identical cores were taken to examine cleistothecial production under a dissecting microscope. To increase visualization of cleistothecia the cores were spread with 70% ethanol to remove conidiophores.
Gene expression analysis
Total RNA was extracted from lyophilized mycelial samples using RNeasy Mini Kit (Qiagen) or Triazol (Invitogen), following the manufacturer’s instructions. Northern blots or quantitative reverse transcription-PCR (qRT-PCR) were used to evaluate gene expression levels. For qRT-PCR, 2 micrograms of total RNA was treated with RQ1 RNAse-Free DNAse (Promega) and cDNA was synthesized with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega). Applied Biosystems 7000 Real-Time PCR System was used for qRT-PCR and SYBR green dye was used for fluorescence detection. The primer pairs used for qRT-PCR are listed in Table 1.
Affinity purification and Western blot
The A. nidulans strain RJMP1.49 was transformed with veA::Stag::pyrGA. fum to generate SNA3 (Table 1). The cassette was generated by fusion PCR (Szewczyk et al., 2006) using the primers S1–S6 listed in Table 2. pAO81 (FGSC) was used as template for the PCR amplification of the intermediate fragment. In a second transformation, SNA3 was transformed with rtfA::gfp::pyroA4A fum, generated from p1831 (provided by E. Espeso) and primers listed in Table 2 by fusion PCR, resulting in the strain TSD22.4 (Table 1). Plates containing liquid GMM inoculated with TSD22.4 were incubated at 37°C without shaking for 40 hours. Mycelia was filtered through Miracloth, washed twice with distilled water and lyophilized. Power lyophilized mycelia was extracted with 4ml of modified HK buffer containing 25mM Tris-Cl (pH7.5), 0.5% NP-40, 300mM NaCl and 5mM EDTA. One µg/ml of Pepstatin A and 10µg/ml of Leupeptin was added to inhibit protease activity. Additionally, PMSF and DTT were added just before use to a final concentration of 250µg/ml and 1mM respectively. One-hundred µl of S-beads (Novagen, USA) were added to a solution containing 50 µg of fungal protein, incubated for 2 hours, and centrifuged to pellet the beads. The beads were washed six times with a wash buffer containing 25mM Tris-Cl (pH7.5), 300mM NaCl and 5mM EDTA, 1mM DTT and 50µg/ml PMSF. Proteins were resuspended in 100µl of 2X Sample buffer containing 6M Urea, 0.125M Tris-Cl (pH 6.8) 10% β-Mercaptoethanol, 20% Glycerol and 4% SDS and bromophenol blue to color and separated by SDS_PAGE. Bound and unbound fractions were analyzed. Then, proteins were transferred to a nitrocellulose membrane and incubated with mouse polyclonal primary antibody against GFP (1:5000 dilution) (ICL Laboratories, USA). Hybridization with anti-mouse IgG secondary antibody (1:4000 dilution) (ICL Laboratories, USA) followed. Detection was carried out using enhanced chemiluminescence substrate (Amersham, USA).
Yeast-two hybrid system
The plasmid pAMC46, containing veA in the bait vector pGBKT7 (Clonetech), and pEE1 plasmid, containing kapA in the prey vector pACT2 (Clonetech), were used as positive control in this experiment, and empty vectors were used as negative controls as previously described (Stinnett et al., 2007). To test possible RtfA-VeA interaction, rtfA cDNA was amplified using primers rtfADHfor and rtfADHrev with engineered BglII restriction sites, and the PCR product was digested with BglII and ligated into pACT2, previously digested with BamHI, resulting in plasmid pNA8. Bait vectors were introduced in pJ69-4a (MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14A ga18OA LYSZ::GALl-HIS3 GAL2-ADE2 metZ::GAL7-lacZ) and prey vectors were introduced in YM706 (MATα ga14-542 ura3-52 his3-200 ade2-101 lys2-801 tql-901 tyrl-501). Yeast transformation (using high efficiency LiAc method), mating and selection were carried out as described previously (James et al., 1996).
Supplementary Material
Acknowledgements
This work was supported by NIH grant 1R15AI081232-01. We thank Nansalmaa Amarsaikhan, Lauren Jurkowski and Vikas Belamkar for their technical support.
References
- Adams TH, Boylan MT, Timberlake WE. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988;54:353–362. doi: 10.1016/0092-8674(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Adrio JL, Demain AL. Fungal biotechnology. Int Microbiol. 2003;6:191–199. doi: 10.1007/s10123-003-0133-0. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Systematic Biology. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Araujo-Bazan L, Dhingra S, Chu J, Fernandez-Martinez J, Calvo AM, Espeso EA. Importin alpha is an essential nuclear import carrier adaptor required for proper sexual and asexual development and secondary metabolism in Aspergillus nidulans. Fungal Genet Biol. 2009;46:506–515. doi: 10.1016/j.fgb.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Bayram O, Braus GH, Fischer R, Rodriguez-Romero J. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet Biol. 2010;47:900–908. doi: 10.1016/j.fgb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Bayram O, Braus GH. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev. 2012;36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- Bayram O, Krappmann S, Ni M, Bok JW, Helmstaedt K, Valerius O, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- Bennett JW, Klich M. Mycotoxins. Clin Microbiol Rev. 2003;16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Keller NP. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CC, Keller NP. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boykin LM, Kubatko LS, Lowrey TK. Comparison of methods for rooting phylogenetic trees: A case study using Orcuttieae (Poaceae: Chloridoideae) Molecular Phylogenetics and Evolution. 2010;54:687–700. doi: 10.1016/j.ympev.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Brakhage AA, Browne P, Turner G. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J Bacteriol. 1992;174:3789–3799. doi: 10.1128/jb.174.11.3789-3799.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakhage AA, Schroeckh V. Fungal secondary metabolites - strategies to activate silent gene clusters. Fungal Genet Biol. 2011;48:15–22. doi: 10.1016/j.fgb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Butchko RAE, Adams TH, Keller NP. Aspergillus nidulans mutants defective in stc gene cluster regulation. Genetics. 1999;153:715–720. doi: 10.1093/genetics/153.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM. The VeA regulatory system and its role in morphological and chemical development in fungi. Fungal Genet Biol. 2008;45:1053–1061. doi: 10.1016/j.fgb.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Calvo AM, Bok J, Brooks W, Keller NP. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl Environ Microbiol. 2004;70:4733–4739. doi: 10.1128/AEM.70.8.4733-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo AM, Wilson RA, Bok JW, Keller NP. Relationship between secondary metabolism and fungal development. Microbiol Mol Biol Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary JW, Obrian GR, Nielsen DM, Nierman W, Harris-Coward P, Bhatnagar JYD, et al. Elucidation of veA-dependent genes associated with aflatoxin and sclerotial production in Aspergillus flavus by functional genomics. Appl Microbiol Biotechnol. 2007;76:1107–1118. doi: 10.1007/s00253-007-1081-y. [DOI] [PubMed] [Google Scholar]
- Chang PK, Ehrlich KC, Yu J, Bhatnagar D, Cleveland TE. Increased expression of Aspergillus parasiticus aflR, encoding a sequence-specific DNA-binding protein, relieves nitrate inhibition of aflatoxin biosynthesis. Appl Environ Microbiol. 1995;61:2372–2377. doi: 10.1128/aem.61.6.2372-2377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sokorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cole RJ, Cox RH. Handbook of Toxic Fungal Metabolites. New York: Academic Press; 1981. [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Research. 1998;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer J, Eichhorn H, Friedlin E, Kurnsteiner H, Kuck U. A homologue of the Aspergillus velvet gene regulates both cephalosporin C biosynthesis and hyphal fragmentation in Acremonium chrysogenum. Appl Environ Microbiol. 2007;73:3412–3422. doi: 10.1128/AEM.00129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran RM, Cary JW, Calvo AM. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA a gene necessary for sclerotial formation. Appl Microbiol Biotechnol. 2007;73:1158–1168. doi: 10.1007/s00253-006-0581-5. [DOI] [PubMed] [Google Scholar]
- Duran RM, Cary JW, Calvo AM. The role of veA on Aspergillus flavus infection of peanuts, corn and cotton. Open Mycology journal. 2009;3:27–36. [Google Scholar]
- Ebersberger I, Strauss S, von Haeseler A. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evol Biol. 2009;9:157. doi: 10.1186/1471-2148-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M, Keller NP, Adams TH. Sequence-specific binding by Aspergillus nidulans AflR, a C-6 zinc cluster protein regulating mycotoxin biosynthesis. Mol Microbiol. 1998;28:1355–1365. doi: 10.1046/j.1365-2958.1998.00907.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Han KH, Han KY, Yu JH, Chae KS, Jahng KY, Han DM. The nsdD gene encodes a putative GATA-type transcription factor necessary for sexual development of Aspergillus nidulans. Mol Microbiol. 2001;41:299–309. doi: 10.1046/j.1365-2958.2001.02472.x. [DOI] [PubMed] [Google Scholar]
- Hicks JK, Yu JH, Keller NP, Adams TH. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G alpha protein-dependent signaling pathway. EMBO J. 1997;16:4916–4923. doi: 10.1093/emboj/16.16.4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff B, Kamerewerd J, Sigl C, Mitterbauer R, Zadra I, Kurnsteiner H, et al. Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum. Eukaryot Cell. 2010;9:1236–1250. doi: 10.1128/EC.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1977;19:33–131. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- Kato N, Brooks W, Calvo AM. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA a gene required for sexual development. Eukaryot Cell. 2003;2:1178–1186. doi: 10.1128/EC.2.6.1178-1186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller NP, Hohn TM. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- Kim HS, Han KY, Kim KJ, Han DM, Jahng KY, Chae KS. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet Biol. 2002;37:72–80. doi: 10.1016/s1087-1845(02)00029-4. [DOI] [PubMed] [Google Scholar]
- Liu HL, Osmani AH, Ukil L, Son S, Markossian S, Shen KF, Govindaraghavan M, Varadaraj A, Hashmi SB, De Souza CP, Osmani SA. Single-step affinity purification for fungal proteomics. Eukaryot Cell. 2010;9:831–833. doi: 10.1128/EC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Merhej J, Urban M, Dufresne M, Hammond-Kosack KE, Richard-Forget F, Barreau C. The velvet gene, FgVe1, affects fungal development and positively regulates trichothecene biosynthesis and pathogenicity in Fusarium graminearum. Mol Plant Pathol. 2011 doi: 10.1111/j.1364-3703.2011.00755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–567. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Zitomer NC, Duvall M, Glenn AE, Riley RT, Calvo AM. The conserved global regulator VeA is necessary for symptom production and mycotoxin synthesis in maize seedlings by Fusarium verticillioides. Plant Pathology. 2012;61:152–160. doi: 10.1111/j.1365-3059.2011.02504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem. 2003;278:33625–3368. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Osherov N, May G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics. 2000;155:647–656. doi: 10.1093/genetics/155.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne GA, Brown MP. Genetics and physiology of aflatoxin biosynthesis. Annu Rev Phytopathol. 1998;36:329–362. doi: 10.1146/annurev.phyto.36.1.329. [DOI] [PubMed] [Google Scholar]
- Payne GA, Yu J. Ecology, development and gene regulation in Aspergillus flavus. In: Masayuki M, gomi K, editors. Aspergillus: Molecular Biology and Genomics. Norfolk, UK: Caister Academic Press; 2010. pp. 157–171. [Google Scholar]
- Penalva MA. Tracing the endocytic pathway of Aspergillus nidulans with FM4–64. Fungal Genet Biol. 2005;42:963–975. doi: 10.1016/j.fgb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM, Mackdonald KD, Bufton AWJ. The genetics of Aspergillus nidulans. Adv. Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Purschwitz J, Mueller S, Kastner C, Schoser M, Haas H, Espeso EA, et al. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Current Biology. 2008;18:255–259. doi: 10.1016/j.cub.2008.01.061. [DOI] [PubMed] [Google Scholar]
- Reverberi M, Ricelli A, Zjalic S, Fabbri AA, Fanelli C. Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl Microbiol Biotechnol. 2010;87:899–911. doi: 10.1007/s00253-010-2657-5. [DOI] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol Microbiol. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roze LV, Arthur AE, Hong SY, Chanda A, Linz JE. The initiation and pattern of spread of histone H4 acetylation parallel the order of transcriptional activation of genes in the aflatoxin cluster. Mol Microbiol. 2007;66:713–726. doi: 10.1111/j.1365-2958.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- Shaaban M, Palmer J, EL-Naggar WA, EL-Sokkary MA, Habib EE, Keller NP. Involvement of transposon-like elements in penicillin gene cluster regulation. Fungal Genet Biol. 2010;47:423–432. doi: 10.1016/j.fgb.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban M, Bok JW, Lauer C, Keller NP. Suppressor mutagenesis identifies a velvet complex remediator of Aspergillus nidulans secondary metabolism. Eukaryot Cell. 2010;9:1816–1824. doi: 10.1128/EC.00189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, Hesse C, O'Rourke B, Serdani M, et al. A five-gene phylogeny of Pezizomycotina. Mycologia. 2006;98:1018–1028. doi: 10.3852/mycologia.98.6.1018. [DOI] [PubMed] [Google Scholar]
- Stinnett SM, Espeso EA, Cobeno L, Araujo-Bazan L, Calvo AM. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol Microbiol. 2007;63:242–255. doi: 10.1111/j.1365-2958.2006.05506.x. [DOI] [PubMed] [Google Scholar]
- Stolinski LA, Eisenmann DM, Arndt KM. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4490–4500. doi: 10.1128/mcb.17.8.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MJ, Dobson AD. Molecular biology of mycotoxin biosynthesis. FEMS Microbiol Lett. 1999;175:149–163. doi: 10.1111/j.1574-6968.1999.tb13614.x. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, et al. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Vallim MA, Miller KY, Miller BL. Aspergillus SteA (sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol Microbiol. 2000;36:290–301. doi: 10.1046/j.1365-2958.2000.01874.x. [DOI] [PubMed] [Google Scholar]
- Warner MH, Roinick KL, Arndt KM. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol. 2007;27:6103–6115. doi: 10.1128/MCB.00772-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P, Brown DW, Kleigrewe K, Bok JW, Keller NP, Humpf HU, Tudzynski B. FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07263.x. MMI7263 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser J, Lee BN, Fondon JW, Adams TH. Genetic requirements for initiating asexual development in Aspergillus nidulans. Curr Genet. 1994;27:62–69. doi: 10.1007/BF00326580. [DOI] [PubMed] [Google Scholar]
- Yager LN. Early developmental events during asexual and sexual sporulation in Aspergillus nidulans. Biotechnology. 1992;23:19–41. [PubMed] [Google Scholar]
- Yu JH, Butchko RAE, Fernandes M, Keller NP, Leonard TJ, Adams TH. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A flavus. Curr Genet. 1996;29:549–555. doi: 10.1007/BF02426959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.