Abstract
We previously reported the presence of a mtDNA mutation hotspot in UV-induced premalignant and malignant skin tumors in hairless mice. We have modeled this change (9821insA) in murine cybrid cells and demonstrated that this alteration in mtDNA associated with mtBALB haplotype can alter the biochemical characteristics of cybrids and subsequently can contribute to significant changes in their behavioral capabilities. This study shows that changes in mtDNA can produce differences in expression levels of specific nuclear-encoded genes, which are capable of triggering the phenotypes such as seen in malignant cells. From a potential list of differentially expressed genes discovered by microarray analysis, we selected MMP-9 and Col1a1 for further studies. Real-time PCR confirmed up-regulation of MMP-9 and down-regulation of Col1a1 in cybrids harboring the mtDNA associated with the skin tumors. These cybrids also showed significantly increased migration and invasion abilities compared to wild type. The non-specific MMP inhibitor, GM6001, was able to inhibit migratory and invasive abilities of the 9821insA cybrids confirming a critical role of MMPs in cellular motility. Nuclear factor-κB (NF-κB) is a key transcription factor for production of MMPs. An inhibitor of NF-κB activation, Bay11-7082, was able to inhibit the expression of MMP-9 and ultimately decrease migration and invasion of mutant cybrids containing 9821insA. These studies confirm a role of NF-κB in the regulation of MMP-9 expression and through this regulation modulates the migratory and invasive capabilities of cybrids with mutant mtDNA. Enhanced migration and invasion abilities caused by up-regulated MMP-9 may contribute to the tumorigenic phenotypic characteristics of mutant cybrids.
Keywords: mtDNA mutation, reactive oxygen species, microarrays, gene expression, MMP-9, migration and invasion
Introduction
There is increasing awareness of a role of mtDNA alterations in the development of cancer since mtDNA point mutations are found at high frequency in a variety of human tumors [1-2]. Most of these mutations were often found to be homoplasmic in the nature, which has important implications as to the role of these DNA changes in the development of various cancers [3]. mtDNA mutations have been identified in epithelial tumors, cutaneous tumors, tumors of musculoskeletal, central nervous, and endocrine system [4-12; 3; 13-17].
We have previously observed somatic alterations in the mitochondrial tRNAArg gene in UV induced mouse skin tumors and modeled the effects of somatic variation at mt-Tr by harvesting mitochondria from brain synaptosomes of B6 and BALB mice and transferring them to a mouse fibroblast ρ0 cell line (LMEB3ρ0) that lacked its own mtDNA [18]. The resulting cybrid cell lines LMEB3(mtBALB) and LMEB3(mtB6) contain the same nuclear genotype and differ in their mitochondria at three nucleotides. The locations of the mtDNA differences between B6 (the mouse reference sequence) and BALB are a T to C polymorphism at 9461 and a 9348G to A change resulting in the amino acid change V248I which is thought to be a neutral polymorphism. The final difference between the two strains is an additional A insertion in the mt-Tr locus resulting in the expansion of a homopolymeric A tract in the pseudouridine loop of the tRNAArg molecule (from 8 consecutive A residues (the B6 reference sequence) to 9 consecutive As (9821insA)). An acquired somatic alteration at the locus would produce heteroplasmy of both B6 and BALB mitochondrial tRNAArg alleles [19]. As documented in Table 1, mtBALB cybrid cells with genetic alteration in mt-Tr gene (9821insA) showed significant biochemical changes (diminished levels of complex I protein, less cellular oxygen consumption, and lower ATP levels) [18] and exhibited increased levels of ROS, resistance to UV-induced apoptosis and changes in cell growth, migratory and invasive capabilities supporting the tumorigenic phenotypes [19].
Table 1. Biochemical and phenotypic differences between wild type (mtB6) and mutant (mtBALB) cybris cells.
mtBALB cybrids with genetic alteration in mt-Tr gene (9821insA) showed significant biochemical changes such as diminished levels of complex I protein, less cellular oxygen consumption, and lower ATP levels. These cybrids exhibited increased levels of ROS, resistance to UV-induced apoptosis and changes in cell growth, migration and invasion capabilities [18, 19].
| mtBALB relative to mtB6 | ||
|---|---|---|
| Biochemical changes | ATP production | ↓ |
| Complex I levels | ↓ | |
| Oxygen consumption (baseline) | ↓ | |
| Oxygen consumption (CII substrates) | ↑ | |
| Levels of mitochondrial ROS | ↑ | |
| Phenotypic changes | Proliferation rate | ↑ |
| Resistance to UV-induced apoptosis | ↑ | |
| Migration | ↑ | |
| Invasion | ↑ | |
Although there is an intensive cross-talk between mitochondria and nucleus, the detailed mechanisms of these interactions still remain unclear. A proposed mechanism of mutual influence between these organelles relies on ROS [20]. ROS are natural byproducts of oxidative phosphorylation that can damage lipids, proteins and DNA and contribute to the development of malignant tumors [21]. It has been suggested that the most of the mtDNA damage is due to imbalance between oxidative stress and free radical scavenging enzymes. Mitochondrial ROS generation is increased in cells with abnormal function as well as under physiological and pathological conditions. The presence of higher ROS levels in the cells is the ultimate result of diminished mitochondrial function and is often observed in mitochondria with mtDNA mutations [19]. Furthermore, mtDNA integrity may even influence the rate of apoptosis by regulating ROS production [22]. Simultaneously, oxidative damage affects nucleic acids, and in particular mtDNA, by the induction of single- and double-strand breaks, base damage, and modification resulting in the generation of point mutation and deletions [23-25]. Interestingly, complex I was proposed to be a major component in the formation of superoxide radicals and a decline in complex I activity was found to dramatically increase ROS production [26]. Besides the influence of ROS on mitochondria by directly inducing mtDNA changes, it has also been suggested that ROS changes affect the expression of nuclear encoded genes. ROS generated by mitochondria can affect nuclear transcriptional events by serving as signal transduction intermediates to induce transcription factor activation (e.g., NF-kB), gene expression, cell growth, and apoptosis [20]. In addition to direct ROS influences on the expression of nuclear encoded genes, ROS can alter nuclear gene through epigenetic mechanisms. Metabolic oxidants produced in mitochondria affect the production of glutathione (GHS) which influence DNA and histone methylation by limiting availability of S-adenosylmethionine, the cofactor utilized during epigenetic control of gene expression by DNA and histone methyltransferases [27]. Moreover, ATP and acetyl-CoA produced by mitochondria phosphorylate and acetylate chromatin which opens nuclear DNA for transcription and replication while the lack of these components reduces chromatin phosphorylation and acetylation resulting in the suppression of gene expression [28].
Despite the fact that the accumulation of mtDNA point mutations has been observed in numerous types of human tumors, the molecular mechanism as well as functional significance of mtDNA mutations in this complex process is not fully understood. We seek to elucidate the role of mtDNA changes in the multistep process of carcinogenesis. In our study, we report that nucleotide changes in mtDNA can trigger alterations in expression levels of specific nuclear-encoded genes associated with malignancy, contributing to tumorigenic phenotypes observed in our cybrid cells. The identification of nuclear genes which can interact with mtDNA variants and the elucidation of their mode of action will ultimately increase the understanding of a role of mtDNA alterations in complex genetic disorders.
Materials and methods
Cell lines and media
LMEB3(mtBALB) and LMEB3(mtB6) cybrid cells were generated by harvesting the mitochondria from brain synaptosomes of B6 and BALB mice and electrofusing them to a mouse fibroblast LMEB3ρ0 cell line that lacked its own mtDNA [29]. All cybrid cell lines were grown in high glucose (4.5g/L) GIBCO® Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% GIBCO® fetal bovine serum (FBS; Invitrogen, Carlsbad, CA).
RNA extraction
Total RNA from three clones of mtB6 and mtBALB cybrid cells was isolated individually using Qiagen RNeasy Mini Kit (Qiagen Sciences, Gaithersburg, MD) according to the manufacturer's protocol. The RNA integrity was checked by the RNA 6000 Nano chip kit using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Microarray analysis
Three individual clones of mtB6 and mtBALB cybrid cells were used for microarray analysis. In vitro transcription was carried out using Agilent Qick Amp labeling kit (Agilent Technologies, Santa Clara, CA) in the presence of Cy3- and Cy5-CTP. Then, 825 ng of labeled cRNA from individual clones of mtBALB and mtB6 cybrid cells were purified separately, combined, and mixed with hybridization buffer before being applied on the microarray. The hybridization solution was prepared using Agilent Gene expression hybridization kit (Agilent Technologies, Santa Clara, CA) product. Agilent 4×44k Whole Mouse Genome Microarray (Agilent Technologies, GPL4134) containing 43,581 coding and non-coding sequences of mouse genome and 1,639 Agilent control sequences was used for hybridization performed in the hybridization oven G2545A (Agilent Technologies, Palo Alto, CA). Conditions of hybridization and washing were performed according to the protocol of Agilent Oligonucleotide Microarray Hybridization manual (Agilent Technologies, Santa Clara, CA). After washing, microarrays were scanned using the GenePix 4000B Scanner (Molecular Devices, Sunnyvale, CA) with laser excitation at 532 and 635 nm at the highest possible resolution (five pixels per micron), and saved as 16-bit grayscale TIFF images. Intensity values were extracted using GenePix Pro 6.0 (Molecular Devices, Sunnyvale, CA), and the data for each array were lowess normalized followed by ANOVA analysis using Limma package in R statistical program version R2.5.1 (http://www.r-project.org/). Original MIAMEcompliant data (detailed protocols of labeling, hybridization, data extraction and data processing) is stored at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the following locator: GSE21855.
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (RT-PCR)
Mouse MMP-9 (Mm00442991_m1), Col1a1 (Mm00801666_g1) and GAPDH (Mm99999915_m1) primer/probes were obtained from ABI (Applied Biosystems, Branchburg, NJ). cDNAs from three individual mtBALB and mtB6 cybrid cells were synthesized from 500 ng of total RNA in a 50ul reaction with master mix containing 10xRT buffer, 5.5mM MgCl2, 2mM dNTPs, 2.5μM random hexamers, 2 Units of RNase Inhibitor and 62.5 Units of Multi Scribe Reverse Transcriptase. All MasterMix reagents were purchased from ABI (Applied Biosystems, Branchburg, NJ). Reactions were performed in MJ Thermocycler PTC-200 (MJ Research, Inc., Watertown, MA) followed by these conditions: 25°C for 10 minutes, 48°C for 30 minutes and 95°C for 5 minutes. 10ng of cDNA was then used to amplify the mouse MMP-9 and Col1a1 sequences. The conditions for PCR reactions were: 10 minutes at 95°C followed by 15 seconds at 95°C, 1 minute at 60°C for 40 cycles by using ABI7000 Real-Time PCR System (Applied Biosystems, Foster City, CA). PCR amplification of the mouse GAPDH was used to control quality of the cDNA. Non-template controls were included on each PCR plate. MMP-9 and Col1a1 levels were normalized to the GAPDH control (ΔCt = Ct(gene of interest) – Ct(housekeeping gene)). Amplification plots were generated and the Ct values (cycle number at which fluorescence reaches threshold) recorded.
In experiments with Bay11-7082 (EMD Chemicals, Gibbstown, NJ), irreversible inhibitor of cytokine-induced IκB-α phosphorylation resulting in decreased expression of NF-κB, the cybrid cells were pretreated with this compound (the final concentration 5 uM) for 60 min. Then cells were allowed to grow for extra 24 hours in culture, RNA was extracted and RT-PCR was performed as described above.
Transwell Migration Assay
Eight micrometer pore size translucent transwell migration chambers (BD Biosciences, San Jose, CA) in 24-well plate were used for migration analysis. Briefly, 600ul of migration buffer (DMEM containing 0.5% FBS and 0.1% BSA) was added to the bottom of each well, and a total of 2.5 × 104 mtB6 or mtBALB cybrid cells resuspended in 150μl of migration buffer were seeded on the top of the membrane. After 18 h incubation at 37°C, 5% CO2, non-invading cells were removed by wiping the upper side of the membrane, and invading cells were fixed with methanol and stained with crystal violet (Sigma-Aldrich, St. Louis, MO) for 1 min. The number of migrating cells was quantified by counting 10 random fields per filter at 400x magnification. Three membrane filters and three individual cybrid clones were used for each condition within one experiment.
In experiments with non-specific MMP inhibitor, 10 mM stock solution of GM6001 (EMD Chemicals, Gibbstown, NJ) in dimethyl sulfoxide (DMSO), the cybrid cells were treated with this compound during the 18 h overnight incubation. The final concentration of GM6001 was 20uM. DMSO was used as a negative control for all experiments at the concentration corresponding to the one used in experiments with GM6001.
In experiments with Bay 11-7082 (EMD Chemicals, Gibbstown, NJ), irreversible inhibitor of cytokine-induced IκB-α phosphorylation resulting in decreased activation of NF-κB, the cybrid cells were pretreated with this compound (the final concentrations 1, 2.5, 5 and 10 uM) for 30-60 min prior to adding the cells on the top of the membrane. 10mM stock solution of Bay 11-7082 in 100% ethanol was used. As a negative control, 100% ethanol was used for all experiments at the concentrations corresponding to the specific concentrations used in experiments with Bay 11-7082.
Transwell Invasion Assay
Cell invasion was performed using modified Boyden chambers consisting of transwells with pre-coated Matrigel™ membrane filters inserted in 24-well tissue culture plates (BD Biosciences, San Jose, CA). A total of 2.5 × 104 cybrid cells (75% confluence) were resuspended in 300μl of serum free media containing only 0.1% BSA and placed on the top of each chamber. After 24 h incubation at 37°C, 5% CO2, non-invading cells were removed by wiping the upper side of the membrane, and invading cells were fixed with methanol and stained with crystal violet (Sigma-Aldrich, St. Louis, MO) for 1 min. The number of invading cells was quantified by counting 10 random fields per filter at 400x magnification. Three membrane filters of three independent clones were used for each condition within one experiment.
In experiments with non-specific MMP inhibitor, the cybrid cells were treated with GM6001 during the 24 h overnight incubation at final concentration 20uM. DMSO was used as a negative control for all experiments at the concentration corresponding to the one used in experiments with GM6001.
In experiments with Bay 11-7082, the cybrid cells were pretreated with this compound for 30-60 min prior to the actual transwell invasion experiment at the final concentration 5 uM. 100% ethanol was used as a negative control for all experiments at the concentration corresponding to the one used in experiments Bay 11-7082.
Statistical analysis
Data for RT-PCR, transwell migration and invasion assays are represented as mean ± SEM of three individual clones. All experiments were performed at least three times. Statistical significance between any two groups was determined by the two-tailed Student's t-test, P values less than 0.05 were considered to be significant.
Results
Changes in mtDNA produce differences in expression levels of specific nuclear encoded genes associated with tumorigenesis
We have previously demonstrated that alteration in the mt-Tr locus associated with the mtBALB haplotype can alter the biochemical characteristics of murine cybrids [18] and subsequently can contribute to significant changes in their behavioral capabilities such as proliferation, resistance to UV-induced apoptosis, migration and invasion [19] (Table 1). Based on these observations, we investigated whether these phenotypic differences could be caused by a unique spectrum of nuclear gene expression alterations induced by the mtDNA changes. Microarray analysis was conducted in order to elucidate the expression profile of three independent clones of wild type mtB6 and mutant mtBALB cybrid lines. Microarray experiments using Agilent 44K mouse whole genome chip revealed 285 transcripts up-regulated and 139 transcripts down-regulated in mtBALB cybrids compared to mtB6 cybrids with the fold change (FC) higher than 1.5. We found 50 transcripts to be up-regulated in mtBALB cybrids with the fold change higher than 2 and 11 transcripts with the fold change higher than 3, including MMP-9. On the other hand, there was found 16 transcripts to be down-regulated in mtBALB cybrids with the fold change higher than 2 and 4 transcripts with the fold change higher than 3, including Col1a1. In Table 2 is shown the potential list of genes (FC≥1.5) with lower expression in mtBALB cybrids relative to mtB6 based on function of interest, including the genes that are involved in aging, skeletal system development, cell cycle, differentiation, apoptosis, cell adhesion, ion transport and various enzymatic activities. On the other hand, Table 3 shows the list of potential targets (FC≥1.5) with higher expression in mtBALB cells relative to mtB6 containing the genes with metallopeptidase, cytokine, isomerase, transferase and oxidoreductase activity, genes playing a role in angiogenesis and cell adhesion. From the list of potential targets we focused on MMP-9 which has been reported to promote cancer cell migration and invasion and Col1a1 which plays an important role in wound healing process.
Table 2. Selected list of genes with lower expression in mtBALB cybrids relative to mtB6 cybrids based on category of interest.
Potential targets with absolute value of the fold change (FC) higher than 1.5 were selected. Col1a1 (in bold) was selected for further studies. Log (FC) means the logarithm of fold change at the base 2. Negative log (FC) values correspond with down-regulation of selected genes. Three individual clones of mtB6 and mtBALB cybrid cells were assayed for each Agilent subarray.
| Category of interest | Gene name | Public ID | Gene symbol | Log (FC) | Absolute value of FC |
|---|---|---|---|---|---|
| Aging | interferon, alpha-inducible protein 27 | NM_029803 | Ifi27 | -1.56 | 2.95 |
| Cell adhesion | collagen, type VI, alpha 1 | NM_009933 | Col6a1 | -0.87 | 1.83 |
| Skeletal system development | collagen, type I, alpha 1 | NM_007742 | Col1a1 | -1.84 | 3.58 |
| Cytoskeleton | spectrin beta 2, transcript variant 1 | NM_175836 | Spnb2 | -0.62 | 1.54 |
| Cell cycle | cyclin D1 | S78355 | Cyl1 | -0.60 | 1.52 |
| Apoptosis | tumor necrosis factor, alpha-induced protein 3 chloride channel calcium activated 2 growth arrest and DNA-damage-inducible 45 beta tumor necrosis factor receptor superfamily, member 12a |
NM_009397 NM_030601 NM_008655 NM_013749 |
Tnfaip3 Clca2 Gadd45b Tnfrsf12a |
-1.41 -1.48 -1.44 -0.65 |
2.66 2.79 2.71 1.57 |
| Transcription factor activity | interferon regulatory factor 1 forkhead box M1 interferon regulatory factor 7 signal transducer and activator of transcription 2 |
NM_008390 NM_008021 NM_016850 NM_019963 |
Irf1 Foxm1 Irf7 Stat2 |
-0.90 -0.73 -0.64 -0.58 |
1.87 1.66 1.56 1.49 |
| Cell differentiation | growth arrest and DNA-damage-inducible 45 beta four and a half LIM domains 1 |
NM_008655 NM_001077361 |
Gadd45b Fhl1 |
-1.44 -1.02 |
2.71 2.03 |
| Isomerase activity | peptidylprolyl isomerase (cyclophilin) like 5 peptidylprolyl isomerase (cyclophilin)-like 1 galactose-4-epimerase, UDP isopentenyl-diphosphate delta isomerase |
NM_001081406 NM_026845 NM_178389 NM_177960 |
Ppil5 Ppil1 Gale Idi1 |
-0.85 -0.62 -0.88 -1.03 |
1.80 1.54 1.84 2.04 |
| Oxidoreductase activity | lysyl oxidase-like 3 Cytochrome c oxidase subunit 2 dihydrofolate reductase |
NM_013586 P00405 NM_010049 |
Loxl3 Dhfr |
-0.96 -0.90 -0.58 |
1.95 1.87 1.49 |
| Transferase activity | cytochrome P450, family 51 prenyl (solanesyl) diphosphate synthase, subunit 1 aldehyde dehydrogenase 1 family, member L1 |
NM_020010 NM_019501 NM_027406 |
Cyp51 Pdss1 Aldh1l1 |
-1.01 -0.60 -0.98 |
2.01 1.52 1.97 |
| Ion transport | chloride channel calcium activated 1 chloride channel calcium activated 2 solute carrier family 25, member 1 ATPase, H+ transporting, lysosomal V0 subunit A1 ATPase, aminophospholipid transporter, class I, type 8A |
NM_009899 NM_030601 NM_153150 NM_016920 NM_001038999 |
Clca1 Clca2 Slc25a1 Atp6v0a1 Atp8a1 |
-2.10 -1.48 -0.90 -0.82 -0.71 |
4.29 2.79 1.87 1.77 1.64 |
Table 3. Selected list of genes with higher expression in mtBALB cybrids relative to mtB6 cybrids based on category of interest.
Potential targets with absolute value of the fold change (FC) higher than 1.5 were selected. MMP-9 (in bold) was selected for further studies. Log (FC) means the logarithm of fold change at the base 2. Positive log (FC) values correspond with up-regulation of selected genes. Three individual clones of mtB6 and mtBALB cybrid cells were assayed for each Agilent subarray.
| Category of interest | Gene name | Public ID | Gene symbol | Log (FC) | Absolute value of FC |
|---|---|---|---|---|---|
| Inflammatory response/Cytokine activity | chemokine (C-C motif) ligand 5 rantes, complete cds chemokine (C-C motif) ligand 20 glucose phosphate isomerase 1 CD28 antigen lymphocyte antigen 96 |
NM_013653 M77747 NM_016960 NM_008155 NM_007642 NM_016923 |
Ccl5 Rantes Ccl20 Gpi1 Cd28 Ly96 |
1.09 1.27 1.80 0.88 2.29 0.62 |
2.13 2.41 3.48 1.84 4.89 1.54 |
| Cell adhesion | camello-like 4 camello-like 5 dystonin, transcript variant b Von Willebrand factor homolog collagen, type XVI, alpha 1 vascular cell adhesion molecule 1 collagen, type VIII, alpha 1 |
NM_023455 NM_023493 NM_134448 NM_011708 NM_028266 NM_011693 NM_007739 |
Cml4 Cml5 Dst Vwf Col16a1 Vcam1 Col8a1 |
1.45 1.05 0.65 0.69 0.61 0.70 0.65 |
2.73 2.07 1.57 1.61 1.53 1.62 1.57 |
| Skeletal system development/Metalloendopeptidase activity |
matrix metallopeptidase 9 a disintegrin-like and marallopgptidase (reprolysin type) with thrombospondin type 1 motif, 7 O-sialoglycoprotein endopeptidase-like 1 |
NM_013599 NM_001003911 NM_028091 |
Mmp9 Adamts7 Osgepl1 |
1.55 0.86 0.59 |
2.93 1.82 1.51 |
| Isomerase activity | thioredoxin domain containing 10 glucuronyl C5-epimerase |
NM_198295 NM_033320 |
Txndc10 Glce |
0.60 1.11 |
1.52 2.16 |
| Angiogenesis/Cell differentiation | angiopoietin-like 6 | NM_145154 | Angptl6 | 0.69 | 1.61 |
| Transferase activity | nicotinamide nucleotide adenylyltransferase 3 nicotinamide N-methyltransferase monoacylglycerol O-acyltransferase 1 integrin beta 1 binding protein 3 glucosamine-phosphate N-acetyltransferase 1 aldehyde dehydrogenase 1 family, member L2 glutathione S-transferase, alpha 2 glutathione S-transferase, alpha 1 (Ya) |
NM_144533 NM_010924 NM_026713 NM_027120 NM_019425 NM_153543 NM_008182 NM_008181 |
Nmnat3 Nnmt Mogat1 Itgb1bp3 Gnpnat1 Aldh1l2 Gsta2 Gsta1 |
1.31 1.20 1.01 1.01 0.91 0.89 0.84 0.90 |
2.48 2.30 2.01 2.01 1.88 1.85 1.79 1.87 |
| Oxidoreductase activity | aldo-keto reductase family 1, member B7 cytochrome c oxidase, subunit VI a, polypeptide 2 cytochrome b5 reductase 1 phytanoyl-CoA dioxygenase domain containing 1 FAD-dependent oxidoreductase domain containing 1 dehydrogenase/reductase (SDR family) member 7 |
NM_009731 NM_009943 NM_028057 NM_172267 NM_172291 NM_025522 |
Akr1b7 Cox6a2 Cyb5r1 Phyhd1 Foxred1 Dhrs7 |
1.49 1.35 1.05 1.03 0.67 0.59 |
2.81 2.55 2.07 2.04 1.59 1.51 |
Nonspecific MMP inhibitor significantly decreased the enhanced migration and invasion capabilities of mtBALB cybrids caused by up-regulated MMP-9
Quantitative real-time PCR was used to confirm the up-regulation of MMP-9 and down-regulation of Col1a1 in mtBALB cybrids (Figure 1). We previously reported that cybrids harboring mtBALB mtDNA have significantly higher level of cell migration through uncoated transwell inserts and invasiveness through matrigel coated inserts compared to mtB6 cybrids [19]. Based on the reported role of MMP-9 in tumor cell invasion and migration, we examined whether the increased migratory capabilities of mtBALB haplotype are caused by the up-regulation of MMP-9 in mtBALB cybrid cells. To do so, we conducted transwell migration and matrigel invasion assays using the cybrid cells treated with GM6001, a nonspecific MMP inhibitor. As shown in Figure 2, GM6001 was able to inhibit both the migratory (Figure 2A, B) and invasive (Figure 2C, D) phenotypes of cybrids confirming a role for MMPs in invasiveness of cultured fibroblasts and also suggesting a critical role for these proteins in cellular motility. This enhanced migration and invasion capabilities caused by up-regulated MMP-9 may contribute to the malignant phenotypic characteristics of mtBALB cybrid cells.
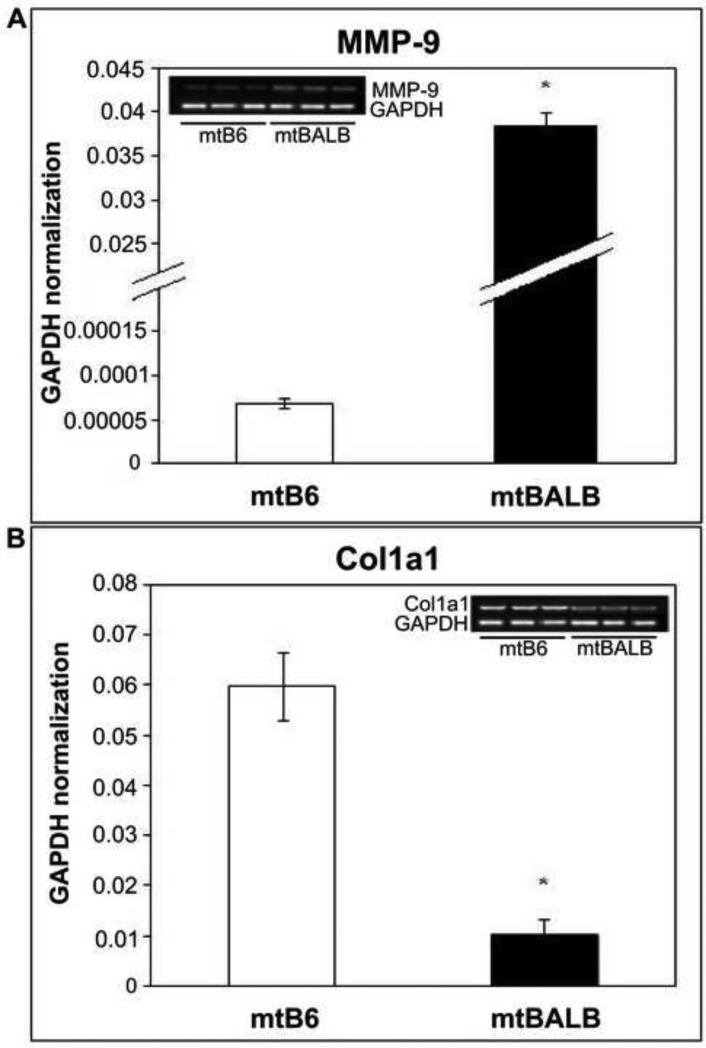
Figure 1. mRNA expression levels of two selected genes in mtB6 and mtBALB cybrid cells.
Histogram showing the expression levels of (a) MMP-9 and (b) Col1a1 genes as analyzed by quantitative real-time RT-PCR. Samples were normalized to GAPDH. Moreover, RT-PCR samples were also analyzed using agarose gel. Three individual cybrid clones were assayed for each experiment and the averages were graphed with the standard error of the mean (SEM). The asterisks indicate that mtBALB cybrid cells showed a significantly higher expression of MMP-9 but a statistically significant lower expression of Col1a1 compared to mtB6 cybrid cells. *P value < 0.001.
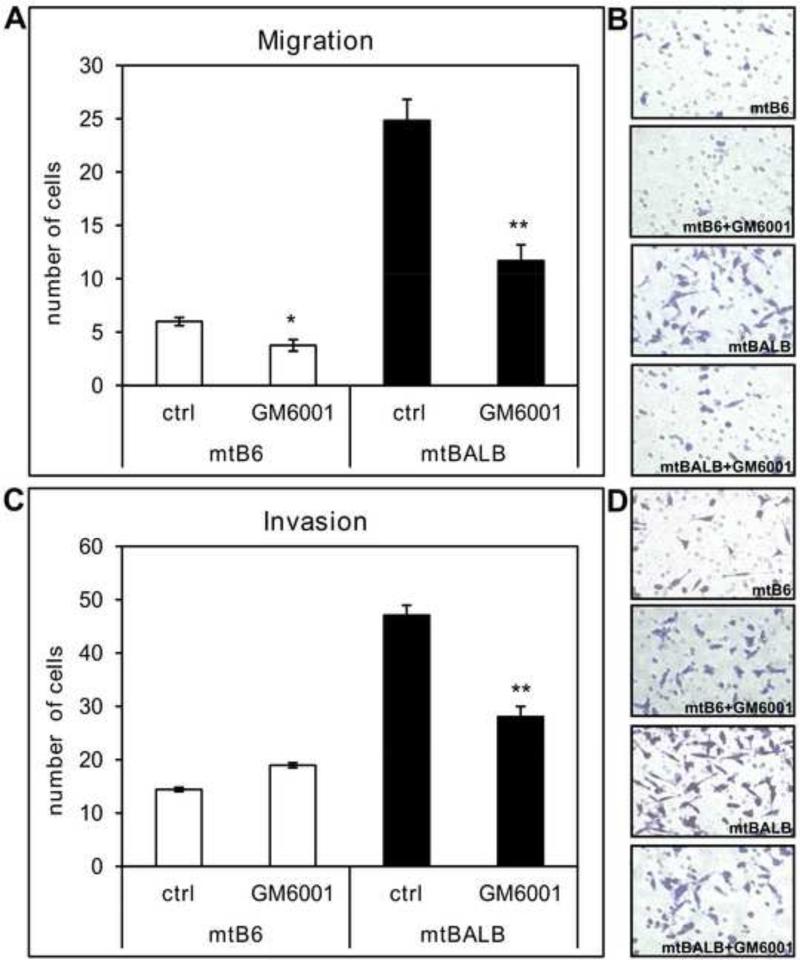
Figure 2. Migration and invasion capabilities of cybrid cells in the presence of non-specific MMP inhibitor GM6001.
Migration transwell assays were performed using uncoated inserts and invasion assay was conducted on inserts coated with matrigel. (a) The bar graph represents the average numbers of cybrid cells migrated through uncoated inserts in the presence of 20μM GM6001. (b) Representative microscopic pictures of stained cells that migrated through the pores of the uncoated inserts in the presence or absence of GM6001. (c) The bar graph shows the average of cells invading through the matrigel-coated inserts in the presence of 20μM GM6001. (d) Representative microscopic pictures of stained cells that invaded through the pores of matrigel coated inserts in the presence or absence of GM6001.The cybrid cells were assayed in triplicate transwells for each experiment, and the averages were graphed with the standard error of the mean (SEM). The asterisks indicate that mtBALB cybrid cells showed a statistically significant decrease in migration and invasion from the mtB6 in the presence of non-specific MMP inhibitor GM6001. *P value < 0.05, **P value < 0.005.
The inhibition of NF-κB activation significantly reduced the expression of MMP-9 and ultimately decreased migration and invasion capabilities of mtBALB cybrids containing 9821insA
NF-κB is a key transcription factor for production of MMPs. Although significant alterations in NF-κB expression levels were not detected in the cybrids using microarrays, we explored the role of NF-κB inhibition on MMP expression and its effect on motility in the cybrids. An inhibitor of NF-κB activation, Bay11-7082, was used to inhibit the expression of MMP-9 in cybrids. Transwell assays were performed in order to see whether decreased expression of MMP-9 caused by Bay11-7082 would diminish migratory abilities of cybrids. Our results showed a significant decrease in migration (Figure 3A) and invasion (Figure 3B) of mtBALB cybrids in the presence of NF-κB inhibitor. Furthermore, the quantitative real time PCR indicates a significant decrease in the mRNA expression levels of MMP-9 in cybrid cells after they were treated with NF-κB inhibitor, Bay11-7082 from the untreated cells (Figure 4). These studies suggest a role of NF-κB in the regulation of MMP-9 expression and through this regulation the inhibition of NF-κB could modulate the migratory and invasive capabilities of mtBALB cybrids.
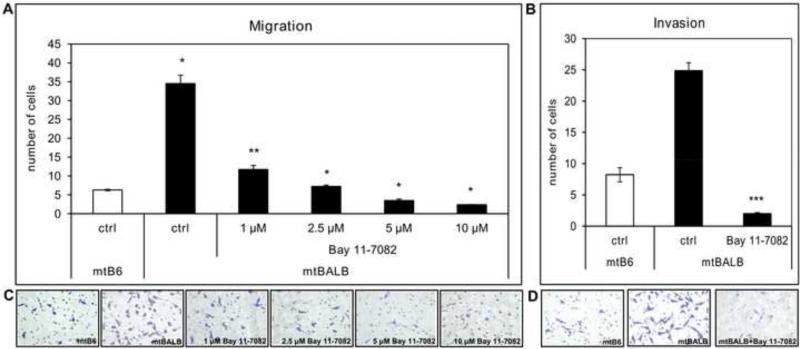
Figure 3. Migration and invasion capabilities of mtBALB cybrid cells in the presence of Bay11-7082.
Migration transwell assays were performed using uncoated inserts and invasion assay was conducted on inserts coated with matrigel. (a) The bar graph represents the average of cybrid cells migrated through uncoated inserts in the presence of various concentrations of Bay11-7082. (b) The bar graph shows the average of invading mtBALB cells through the matrigel-coated inserts in the presence of 5μM Bay11-7082. Representative microscopic pictures of stained cells that migrated through the pores of uncoated inserts (c) or invaded through the pores of matrigel coated inserts (d) in the presence or absence of Bay11-7082. The cybrid cells were assayed in triplicate transwells for each experiment, and the averages were graphed with the standard error of the mean (SE. The asterisks indicate that mtBALB cybrid cells showed a statistically significant decrease in migration and invasion from the mtB6 in the presence of NFκB inhibitor Bay11-7082. *P value < 0.0005, **P value < 0.001, ***P value < 0.0001.
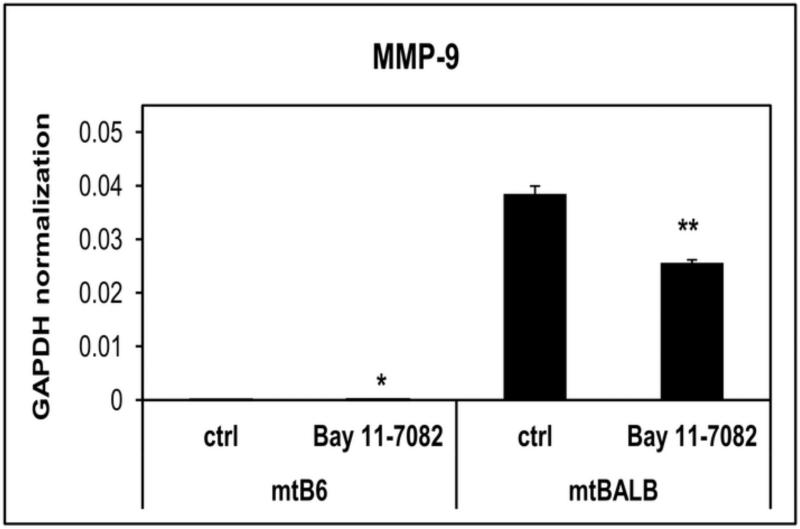
Figure 4. mRNA expression levels of MMP-9 in mtB6 and mtBALB cybrid cells treated with Bay11-7082.
Quantitative real-time RT-PCR histogram showing the levels of expression of MMP-9 in the cybrid cells after they were treated with 5μM Bay11-7082. Three individual cybrid clones were assayed for each experiment and the averages were graphed with the standard error of the mean (SEM). The results indicate that cybrid cells showed a statistically significant decrease in mRNA expression of MMP-9 after they were treated with NFκB inhibitor Bay 11-7082 from the untreated cells. *P value < 0.01, **P value < 0.001.
Discussion
For the first time, we demonstrate that nucleotide changes in mtDNA can trigger the differences in expression levels of specific nuclear encoded genes ultimately causing the previously observed biochemical and behavioral changes seen in cybrid cells (Table 1). The data presented in this study strongly support a potential contribution of mtDNA changes in cancer progression.
The mouse mitochondrial mt-Tr (tRNAArg) gene appears to be a hotspot for UV-induced mutations. Such mutations may be important in tumor progression since mtDNA changes lead to increased oxidative stress which can be mitogenic to cells [18]. We employed a mouse model for analysis of mtDNA changes in non melanoma skin cancer (NMSC) since it is a well established way to generate epidermal tumors that are similar to those seen in human squamous cell carcinoma and the mtDNA is similar in size and structure to that of humans. We previously discovered a mutation hot spot (9821insA) in mouse mt-Tr locus (tRNAArg) in approximately one third of premalignant and malignant skin tumors. To determine the functional relevance of this particular mutation in vitro, cybrid cell lines with the same nuclear background but containing different mt-Tr (tRNAArg) alleles were generated [18]. We demonstrated that mtBALB cybrid cells with genetic alteration in mt-Tr gene (9821insA) showed significant biochemical changes such as diminished levels of complex I protein, less cellular oxygen consumption, and lower ATP levels [18]. Furthermore, these cybrids exhibited resistance to UV-induced apoptosis and changes in cell growth, migratory and invasive capabilities supporting the tumorigenic phenotypes seen in malignant cells [19]. Moreover, we showed that there are mtDNA- driven differences in ROS production between wild type (mtB6) and mutant (mtBALB) cybrids [18-19]. Our findings of diminished complex I as well as increased ROS levels in mutant cybrid cells correlate with previous observations of Lenaz et al. [26]. Moreover, Ishikawa et al. previously reported that cybrids containing mtDNA mutations (G13997A and 13885insC) in the gene encoding ND6 produced a deficiency in respiratory complex I activity and were associated with overproduction of ROS leading to the up-regulation of some nuclear encoded genes associated with regulation of metastasis formation in these cells [30]. Of the three nucleotide changes between the two mtDNA types, the 9821insA in mt-Tr locus together with 9348G to A base change are likely to be the most important contributors to these diverse phenotypes since the 9461T to C change is a neutral polymorphism coding the same amino acid (methionine) in both haplotypes [19]. Given the highly pleomorphic consequences of inherited tRNA mutations in human mitochondrial disease, it is not surprising that these minimal changes in the mtDNA may be linked to such diverse cellular and organismal changes such as learning, hearing, metabolism, and neoplasia.
We sought to evaluate the interactions between the nucleus and mtDNA changes at mt-Tr locus in order to increase the understanding of a role of mtDNA alterations in complex genetic disorders similarly as it was executed previously using LHON cybrids by Danielson et al. [31]. They utilized microarray analysis to examine the altered expression of nuclear genes caused by cybridization process and by interaction with mtDNA mutations that cause LHON [31]. Microarray analysis is often used for gene expression analysis since it is a powerful genomic tool which enables to examine the expression of thousands of transcripts in parallel ultimately leading to the list of potential up- or down- regulated genes. The high-density microarrays provide the most important advantage of allowing parallel quantification of expression levels of thousands of genes available from the genome including those which greatly contribute to the tumorigenic behavior [32-33]. Microarrays serve as the first step to generate a profile of gene expression differences between two samples. Although they are a reproducible method for differential gene expression analysis, they should be considered more as a preliminary method for generation of fold change values for a larger number of genes and should be always accompanied by follow up RT-PCR confirmation of a fold-change in expression of particular gene of interest [34]. However, there is inherent variability in fold change between the microarray data and RT-PCR data due to the nature of the specific assays. Moreover, it is quite usual that many differentially expressed genes identified by microarrays (sometimes up to 10% of all genes) are not confirmed by RT-PCR. This artifact is usually explained as a consequence of using different type of probes for microarray and RT-PCR. Hence, in our study microarrays were used to obtain the list of potential candidate genes of the major significance according to our primary scientific focus. Data for individual genes of interest were then confirmed/rejected by more accurate RT-PCR analysis. Based on previously seen phenotypic characteristics of mtBALB cybrids such as increased proliferation, migration and invasiveness [19], we have focused on nuclear genes affecting the cell motility. Quantitative RT-PCR confirmed 300x higher mRNA levels of MMP-9 and 6x lower mRNA levels of Col1a1 in mtBALB cells, the genes playing a role in tumor cell invasion [35-36] and wound healing [37], respectively.
Remodeling of the extracellular matrix (ECM) catalyzed by MMPs is central to morphogenetic phenomena during wound healing as well as numerous pathologic conditions such as cancer [38]. Cancer cell motility is a complex multistep process during which the cells form new attachments and break old attachments to migrate and/or invade to the surrounding tissues. One of the characteristics of invasive cells is the ability to degrade the ECM by MMPs. MMPs have been shown to have significantly higher expression in almost all human cancers since they have ability to degrade the basal lamina and ECM [39]. MMP-9 (known also as 92-kD type IV collagenase or gelatinase B) is a unique type of proteinase that hydrolyze backbone of extracellular matrix (type IV collagen) [40] and was shown to be up-regulated in many types of cancers indicating that in addition to tumor cell invasion and angiogenesis, MMP-9 can also contribute to carcinogenesis and tumor growth [36]. Several clinical reports showed that cancer patients with higher level of MMP-9 have lower survival rate than those with low MMP-9 expression [41]. Thus, our primary focus was to determine whether the altered MMP-9 expression produced by specific mutations in mouse mtDNA corresponds with the changes in cell motility.
We found that the changes in invasiveness imparted by mtBALB haplotype were indeed correlated with MMP-9 expression levels. It was also somewhat surprising that the MMP-9 expression levels were correlated with motility through transwells without matrigel coating. Expression of MMPs and other genes involved in the processes of transformation is regulated by nuclear transcription factors, including NF-κB [42]. NF-κB is a cytoplasmic transcription factor commonly inhibited by the I B molecule. However, IκB phosphorylation and its subsequent degradation release NF-κB triggering transcription of many nuclear genes involved in carcinogenesis including metalloproteinases, integrins and cytokines [43]. Moreover, it was suggested that the activation of NF-κB could be also triggered by several stress impulses generating ROS [44]. We previously showed that there are mtDNA-driven differences in ROS production between wild type (mtB6) and mutant (mtBALB) cybrids which could also have profound influence on migratory abilities since antioxidants were able to diminish migratory capacity of the mtBALB cells and caused them to migrate at a level that was similar to the mtB6 cells [19]. Our results demonstrate that NF-κB regulates the expression of MMP-9 and through this regulation predictably modulates the invasion of mtBALB cybrids and somewhat unexpectedly modulates the migration of mtBALB cybrids. Additionally, we have found that the non-specific MMP inhibitor, GM6001 significantly inhibited migratory and invasive abilities of mtBALB cybrids which confirms a role for MMPs in invasiveness of fibroblasts and also suggests a critical role for MMPs in cellular motility, consistent with the results of Aung et al. [35]. Mechanism of proteolysis catalyzed by MMPs in controlled cell spreading, motility and invasiveness involve cell surface tethered MMPs, binding of soluble MMPs to the cell surface and in situ activation; and a diffusion based mode of interaction of the enzymes with the underling ECM substrata. MMPs utilize a remarkable surface diffusion mechanism for substrate interaction. All the components of the collagenolytic complex are capable of processive movement on a surface of the collagen fibril. Recent results establish that MMP-1, -2, -9 and trans-membrane protease (MT1-MMP) can diffuse laterally on the collagen substrate surface without noticeable dissociation [38].
Microarray gene expression analysis revealed many more potentially interesting candidate genes besides of MMP-9 and Col1a1 which are currently under the further investigation. This is the first report of how nucleotide changes in mtDNA cause alterations in expression levels of specific nuclear-encoded genes associated with malignancy. The identification of nuclear genes which can interact with mtDNA variants and the elucidation of their mode of action will ultimately increase the understanding of a role of mtDNA alterations in the development of various cancers.
Research Highlights.
Cybrids are useful models to study the role of mtDNA changes in cancer development.
mtDNA changes affect the expression of nuclear genes associated with tumorigenesis.
MMP-9 is up-regulated and Col1a1 is down-regulated in mutant cybrids.
GM6001 reduced the enhanced motility of mutant cybrids caused by up-regulated MMP-9.
The MMP-9 expression and invasiveness of mutant cybrids were reduced by Bay11-7802.
Acknowledgements
This work was supported by an NCI Cancer Center Support Grant P30 CA023074 (CCSG) and R01 AR 0501552 to JS, by the VA Merit Award to JS.
Abbreviations
- mtDNA
Mitochondrial DNA
- UV
Ultraviolet
- NF-κB
Nuclear Factor-κB
- ROS
Reactive Oxidative Species
- GHS
Glutathione
- ATP
Adenosine Triphosphate
- NMSC
Non Melanoma Skin Cancer
- FC
Fold Change
- ECM
Extracellular Matrix
- MMP
Metalloproteinases
- LHON
Leber's Hereditary Optic Neuropathy
- DMEM
Dulbecco's Modified Eagle's Medium
- FBS
Fetal Bovine Serum
- BSA
Bovine Serum Albumin
- DPBS
Dulbecco's Phosphate Buffered Saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
aUniversity of Arizona, Southern Arizona VA Healthcare System, Department of Medicine, Dermatology Division and Arizona Cancer Center, 1515 N Campbell Avenue, Tucson, AZ 857 24, United States
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Copeland WC, Wachsman JT, Johnson FM, Penta JS. Mitochondrial DNA alterations in cancer. Cancer Invest. 2002;20:557–569. doi: 10.1081/cnv-120002155. [DOI] [PubMed] [Google Scholar]
- 2.Verma M, Kumar D. Application of mitochondrial genome information in cancer epidemiology. Clin Chim Acta. 2007;383:41–50. doi: 10.1016/j.cca.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Polyak K, Li YB, Zhu H, Lengauer C, Willson JKV, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–293. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 4.Alonso A, Martin P, Albarran C, Aguilera B, Garcia O, Guzman A, Oliva H, Sancho M. Detection of somatic mutations in the mitochondrial DNA control region of colorectal and gastric tumors by heteroduplex and single-strand conformation analysis. Electrophoresis. 1997;18:682–685. doi: 10.1002/elps.1150180504. [DOI] [PubMed] [Google Scholar]
- 5.Birch-Machin MA. The role of mitochondria in ageing and carcinogenesis. Clin Exp Dermatol. 2006;31:548–552. doi: 10.1111/j.1365-2230.2006.02161.x. [DOI] [PubMed] [Google Scholar]
- 6.Eshaghian A, Vleugels RA, Canter JA, McDonald MA, Stasko T, Sligh JE. Mitochondrial DNA deletions serve as biomarkers of aging in the skin, but are typically absent in nonmelanoma skin cancers. J Invest Dermatol. 2006;126:336–344. doi: 10.1038/sj.jid.5700088. [DOI] [PubMed] [Google Scholar]
- 7.Fliss MS, Usadel H, Cabellero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–2019. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 8.Gasparre G, Lommarini L, Porcelli AM, Lang M, Ferri GG, Kurelac I, Zuntini R, Mariani E, Pennisi LF, Pasquini E, Pasquinelli G, Ghelli A, Bonora E, Ceccarelli C, Rugolo M, Salfi N, Romeo G, Carelli V. An Inherited Mitochondrial DNA Disruptive Mutation Shifts to Homoplasmy in Oncocytic Tumor Cells. Hum Mutat. 2009;30:391–396. doi: 10.1002/humu.20870. [DOI] [PubMed] [Google Scholar]
- 9.Gasparre G, Porcelli AM, Bonora E, Pennisi LF, Toller M, Iommarini L, Ghelli A, Moretti M, Betts CM, Martinelli GN, Ceroni AR, Curcio F, Carelli V, Rugolo M, Tallini G, Romeo G. Disruptive mitochondrial DNA mutations in complex I subunits are markers of oncocytic phenotype in thyroid tumors. Proc Natl Acad Sci U S A. 2007;104:9001–9006. doi: 10.1073/pnas.0703056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horton TM, Petros JA, Heddi A, Shoffner J, Kaufman AE, Graham SD, Gramlich T, Wallace DC. Novel mitochondrial DNA deletion found in a renal cell carcinoma. Gene Chromosome Canc. 1996;15:95–101. doi: 10.1002/(SICI)1098-2264(199602)15:2<95::AID-GCC3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 11.Mayr JA, Meierhofer D, Zimmermann F, Feichtinger R, Kogler C, Ratschek M, Schmeller N, Sperl W, Kofler B. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin Cancer Res. 2008;14:2270–2275. doi: 10.1158/1078-0432.CCR-07-4131. [DOI] [PubMed] [Google Scholar]
- 12.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC. mtDNA mutations increase tumorigenicity in prostate cancer. P Natl Acad Sci USA. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porcelli AM, Ghelli A, Ceccarelli C, Lang M, Cenacchi G, Capristo M, Pennisi LF, Morra I, Ciccarelli E, Melcarne A, Bartoletti-Stella A, Salfi N, Tallini G, Martinuzzi A, Carelli V, Attimonelli M, Rugolo M, Romeo G, Gasparre G. The genetic and metabolic signature of oncocytic transformation implicates HIF1alpha destabilization. Hum Mol Genet. 2010;19:1019–1032. doi: 10.1093/hmg/ddp566. [DOI] [PubMed] [Google Scholar]
- 14.Tan DJ, Bai RK, Wong LJC. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Research. 2002;62:972–976. [PubMed] [Google Scholar]
- 15.Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 16.Toro JR, Nickerson ML, Wei MH, Warren MB, Glenn GM, Turner ML, Stewart L, Duray P, Tourre O, Sharma N, Choyke P, Stratton P, Merino M, Walther MM, Linehan WM, Schmidt LS, Zbar B. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet. 2003;73:95–106. doi: 10.1086/376435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmermann FA, Mayr JA, Neureiter D, Feichtinger R, Alinger B, Jones ND, Eder W, Sperl W, Kofler B. Lack of complex I is associated with oncocytic thyroid tumours. Brit J Cancer. 2009;100:1434–1437. doi: 10.1038/sj.bjc.6605028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jandova J, Eshaghian A, Shi M, Li M, King LE, Janda J, Sligh JE. Identification of an mtDNA Mutation Hot Spot in UV-Induced Mouse Skin Tumors Producing Altered Cellular Biochemistry. J Invest Dermatol. 2012;132:421–428. doi: 10.1038/jid.2011.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jandova J, Shi M, Norman KG, Stricklin GP, Sligh JE. Somatic alterations in mitochondrial DNA produce changes in cell growth and metabolism supporting a tumorigenic phenotype. Biochim Biophys Acta. 2012;1822:293–300. doi: 10.1016/j.bbadis.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marin-Garcia J, Pi Y, Goldenthal MJ. Mitochondrial-nuclear cross-talk in the aging and failing heart. Cardiovasc Drug Ther. 2006;20:477–491. doi: 10.1007/s10557-006-0584-6. [DOI] [PubMed] [Google Scholar]
- 21.Nishikawa M. Reactive oxygen species in tumor metastasis. Cancer Lett. 2008;266:53–59. doi: 10.1016/j.canlet.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 22.Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki K, Suzuki T, Ueda M, Ichihashi M, Reguer G, Yamasaki H. CC to TT mutation in the mitochondrial DNA of normal skin: relationship to ultraviolet light exposure. Mutat Res-Gen Tox En. 2000;468:35–43. doi: 10.1016/s1383-5718(00)00038-3. [DOI] [PubMed] [Google Scholar]
- 24.Shen ZZ, Wu WJ, Hazen SL. Activated leukocytes oxidatively damage DNA, RNA, and the nucleotide pool through halide-dependent formation of hydroxyl radical. Biochemistry-Us. 2000;39:5474–5482. doi: 10.1021/bi992809y. [DOI] [PubMed] [Google Scholar]
- 25.Yakes FM, VanHouten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. P Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenaz G, D'Aurelio M, Pich MM, Geneva ML, Ventura B, Bovina C, Formiggini G, Castelli GP. Mitochondrial bioenergetics in aging. Bba-Bioenergetics. 2000;1459:397–404. doi: 10.1016/s0005-2728(00)00177-8. [DOI] [PubMed] [Google Scholar]
- 27.Hitchler MJ, Domann FE. An epigenetic perspective on the free radical theory of development. Free Radical Bio Med. 2007;43:1023–1036. doi: 10.1016/j.freeradbiomed.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace DC, Fan WW. Energetics, epigenetics, mitochondrial genetics. Mitochondrion. 2010;10:12–31. doi: 10.1016/j.mito.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trounce I, Wallace DC. Production of transmitochondrial mouse cell lines by cybrid rescue of rhodamine-6G pre-treated L-cells. Somatic Cell and Molecular Genetics. 1996;22:81–85. doi: 10.1007/BF02374379. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 31.Danielson SR, Carelli V, Tan G, Martinuzzi A, Schapira AH, Savontaus ML, Cortopassi GA. Isolation of transcriptomal changes attributable to LHON mutations and the cybridization process. Brain. 2005;128:1026–1037. doi: 10.1093/brain/awh447. [DOI] [PubMed] [Google Scholar]
- 32.Deyholos MK, Galbraith DW. High-density microarrays for gene expression analysis. Cytometry. 2001;43:229–238. [PubMed] [Google Scholar]
- 33.Schena M, Shalon D, Davis RW, Brown PO. Quantitative Monitoring of Gene-Expression Patterns with a Complementary-DNA Microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 34.Imaoka T, Yamashita S, Nishimura M, Kakinuma S, Ushijima T, Shimada Y. Gene expression profiling distinguishes between spontaneous and radiation-induced rat mammary carcinomas. J Radiat Res. 2008;49:349–360. doi: 10.1269/jrr.07126. [DOI] [PubMed] [Google Scholar]
- 35.Aung CS, Hill MM, Bastiani M, Parton RG, Parat MO. PTRF-cavin-1 expression decreases the migration of PC3 prostate cancer cells: Role of matrix metalloprotease 9. Eur J Cell Biol. 2011;90:136–142. doi: 10.1016/j.ejcb.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Beeghly-Fadiel A, Lu W, Shu XO, Long JR, Cai QY, Xiang YB, Gao YT, Zheng W. MMP9 polymorphisms and breast cancer risk: a report from the Shanghai Breast Cancer Genetics Study. Breast Cancer Res Tr. 2011;126:507–513. doi: 10.1007/s10549-010-1119-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu KQ, Carrougher GJ, Couture OP, Tuggle CK, Gibran NS, Engrav LH. Expression of collagen genes in the cones of skin in the Duroc/Yorkshire porcine model of fibroproliferative scarring. J Burn Care Res. 2008;29:815–827. doi: 10.1097/BCR.0b013e3181848141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collier IE, Legant W, Marmer B, Lubman O, Saffarian S, Wakatsuki T, Elson E, Goldberg GI. Diffusion of MMPs on the surface of collagen fibrils: the mobile cell surface-collagen substratum interface. PLoS One. 2011;6:e24029. doi: 10.1371/journal.pone.0024029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stellas D, El Hamidieh A, Patsavoudi E. Monoclonal antibody 4C5 prevents activation of MMP2 and MMP9 by disrupting their interaction with extracellular HSP90 and inhibits formation of metastatic breast cancer cell deposits. Bmc Cell Biol. 2010;11 doi: 10.1186/1471-2121-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y, Dong LJ. [Role of matrix metalloproteinases in the pathogenesis and therapy of leukemia] Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2003;11:316–320. [PubMed] [Google Scholar]
- 41.Peng CW, Liu XL, Liu X, Li Y. Co-evolution of cancer microenvironment reveals distinctive patterns of gastric cancer invasion: laboratory evidence and clinical significance. J Transl Med. 2010;8 doi: 10.1186/1479-5876-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Annunziata CM, Stavnes HT, Kleinberg L, Berner A, Hernandez LF, Birrer MJ, Steinberg SM, Davidson B, Kohn EC. Nuclear Factor kappa B Transcription Factors Are Coexpressed and Convey a Poor Outcome in Ovarian Cancer. Cancer-Am Cancer Soc. 2010;116:3276–3284. doi: 10.1002/cncr.25190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richmond A. NF-kappa B, chemokine gene transcription and tumour growth. Nat Rev Immunol. 2002;2:664–674. doi: 10.1038/nri887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobar N, Villar V, Santibanez JF. ROS-NF kappa I’ mediates TGF-beta 1-induced expression of urokinase-type plasminogen activator, matrix metalloproteinase-9 and cell invasion. Mol Cell Biochem. 2010;340:195–202. doi: 10.1007/s11010-010-0418-5. [DOI] [PubMed] [Google Scholar]