SUMMARY
The efficiency of Gd(III) contrast agents in magnetic resonance image enhancement is governed by a set of tunable structural parameters. Understanding and measuring these parameters requires specific analytical techniques. This Feature describes strategies to optimize each of the critical Gd(III) relaxation parameters for molecular imaging applications and the methods employed for their evaluation.
Molecular imaging refers to the development, testing, and implementation of diagnostic tools for the visualization of molecular processes in vivo.1–3 Deviations in gene expression, receptor abnormalities, enzymatic catalysis, or alteration of biochemical signaling environments can be observed with molecular imaging.1 The development of new molecular imaging probes focuses on circumventing the challenges of the field.3 A common limitation of contrast agents in molecular imaging applications is the discrepancy between target abundance and complex efficiency. Millimolar quantities of the agent may be required for distinguishable contrast of a target whose concentration is in the micromolar to picomolar range (a difference of nearly seven orders of magnitude).4 To overcome the signal amplification challenges of current imaging probes, researchers have pursued the development of highly efficient contrast agents to investigate biochemical processes. Amplification of the imaging probe signal at a specific target can be used to monitor physiology in addition to cellular events and genetic abnormalities preceding, during, and after treatment.
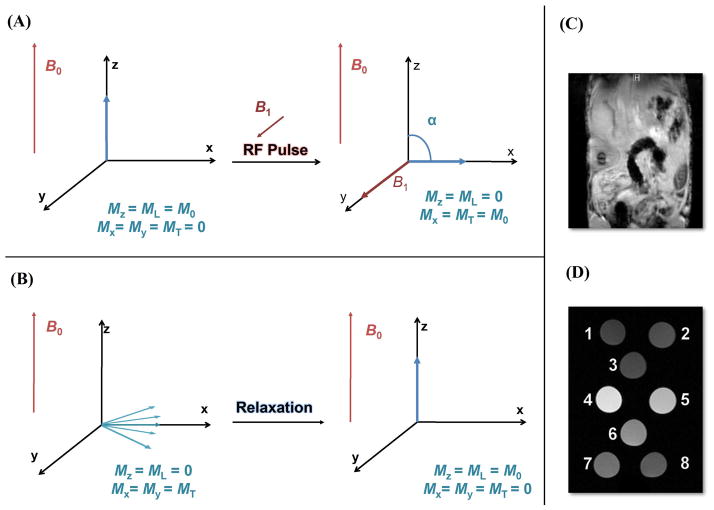
Magnetic resonance imaging (MRI) is a diagnostic and research tool that is poised to overcome a number of the challenges facing molecular imaging. MRI can be used to obtain high resolution images of the internal structure of opaque organisms. In comparison to other medical imaging techniques [positron emission tomography (PET) and single photon emission computed tomography (SPECT)], MRI utilizes no ionizing radiation allowing serial measurements to be obtained without radiation risk to the organism.5 Unlike optical imaging techniques, MRI permits deep tissue penetration resulting in tomographic images with excellent spatial resolution.6 Image acquisition is based on the perturbation of hydrogen nuclei (water protons) in a static magnetic field via a pulse sequence (Figure 1). A pulse sequence is a timed series of radio frequency and magnetic gradient exposures that manipulate the net magnetism of a sample.7 The gradient coils of the MRI scanner allow the position of the water protons to be encoded by their phase and precession frequency. This encoding can be mathematically transformed into an intensity for each voxel to generate the final image.8 The contrast of this final image corresponds to the NMR signal intensity of individual water protons. The variation in the relaxation rate of the water protons to equilibrium in different tissues after an RF perturbation is used to create image contrast. While this review focuses on the perturbation of relaxation rates to alter image contrast, it should be noted that other mechanisms such as magnetization transfer, diffusion-weighted imaging, and chemical exchange saturation transfer (CEST) imaging can all be used to generate contrast in MR imaging.9–11 Although diverse pulse sequences can modulate specific weighting in an image, MRI is ultimately limited by low sensitivity due to an intrinsically low nuclear 1H polarization.8
Figure 1. An overview of MR Physics.
Hydrogen nuclei, responsible for the inherent contrast of MR, have one proton and an odd atomic mass resulting in a spin of ½ and an overall magnetic moment. In the presence of an external magnetic field (B0) the spins align parallel to the axis of the field with a slight population preference in the direction of B0 (+ z). This induced polarization for the lower energy state (alignment with the field) is dictated by the Boltzmann distribution. The accumulation of individual proton magnetic moments results in a net nonzero magnetization in the z-direction (Mz = M0); in the xy plane, the random alignment of each of the individual spins cancel to give an overall transverse magnetization (Mxy) of zero. (A) Excitation: The polarized nuclei are perturbed by a rotating magnetic field (B1) perpendicular to B0 to deliver radio frequency electromagnetic energy into the system (RF pulse). The energy of the RF pulse is equal to the Larmor frequency to facilitate resonance with the water proton spins. The duration of the RF pulse is correlated to the flip angle (a) relative to the external field. For example, a 90° RF pulse translocates the net magnetization vector into the transverse plane (Mz = 0, Mxy = M0). (B) Relaxation: Upon removal of the RF pulse, the energized nuclear spins return to equilibrium. The nuclear spins will release this excess energy to their surroundings through molecular vibrations/rotations that occur at the Larmor frequency. The characteristic time in which the net magnetization returns to equilibrium by this mechanism is known as the spin-lattice (longitudinal) relaxation time (T1).8 Alternatively, coherence of the transverse component can be lost as energy is absorbed/transferred to nearby protons with the same Larmor frequency. These intermolecular and intramolecular vibrations and rotations cause the net magnetization to fluctuate; this results in a time dependent decay [spin-spin (transverse) relaxation time, T2]. However, inhomogeneities in the magnetic field and magnetic susceptibility differences of adjacent tissues can distort the true T2 value; these contributions are expressed as T2*. Pulse sequences enhance MR image contrast by exploiting differences in the T1 and T2 of tissues. (C) Variability in T1 and T2 resulting from the dissimilar water concentrations of different tissues contributes to the inherent grayscale contrast of a typical MR image. (D) An example of how Gd(III) contrast agents can enhance the contrast in an MR image of nanodiamonds through shorting T1. T1-weighted MR images of (1) water, (2) 1 mg/mL undecorated nanodiamonds, (3) undecorated nanodiamonds + coupling reagents, and (4)–(8) Gd(III) –nanodiamond conjugates. Reprinted with permission from Ref. 50. Copyright 2010 American Chemical Society.
Contrast agents are employed to further enhance the image contrast of tissues that are magnetically similar but histologically distinct. Coordination complexes of the electron rich Gd(III) are often used to supplement MR scans. Through a dissociative ligand exchange Gd(III) contrast agents sequentially coordinate the water molecules of a given sample increasing their inherent longitudinal and transverse relaxation rates. This alteration in relaxation corresponds to greater levels of contrast in an acquired MR image. However, despite widespread medical use, clinically available Gd (III) contrast agents have limited applicability in studying biochemical processes in a research setting.12 The applications of these small molecule chelates are restricted by rapid clearance, large dose requirements to observe sufficient contrast, poor cell uptake, and nontargeted ligand structures.
Ushering MR imaging applications into the molecular arena has involved synthetic modification of the Gd(III) contrast agent ligand to reach the required contrast intensity.13 Amplification of signal, increased targeting to a biological moiety, and activatable detection of molecular processes can all be realized through tuning the structural parameters that govern the efficiency of a Gd(III) contrast agent. Analytical methods to measure and quantify these parameters are essential to understanding the mechanism of efficiency enhancement in Gd(III) contrast agents. Furthermore, synthetic modifications to the Gd(III) contrast agent ligand structure can be developed and evaluated, maximizing targeting and delivery mechanisms. This Feature focuses on the physical parameters that govern Gd(III)-contrast agent efficiency, the design strategies for optimizing each parameter, and the methods employed for their evaluation.
RELAXATION THEORY OF Gd(III) CONTRAST AGENTS
The well-known Gd(III) paramagnetic ion is considered the optimal candidate for increasing the longitudinal relaxation rate of water protons due to its unique magnetic properties.14 The large magnetic moment (7.9 BM) conferred by the 7-unpaired f-electrons of Gd(III) allow it to interact strongly with nearby spins.14 In comparison to other lanthanides with high magnetic moments, the ground state of Gd(III) is symmetric (S7/2) resulting in a uniquely long electronic relaxation time (10−8 – 10−9 s) that is advantageous for facilitating the relaxation of neighboring spins.8, 15
The efficiency at which a Gd(III) contrast agent can shorten relaxation time is referred to as the relaxivity (r, in mM−1s−1). Relaxivity values are directly proportional to the level of signal intensity observed in an MR image. The relaxivity of a contrast agent can be determined by the slope of the plot of the longitudinal (1/T1) or transverse relaxation rates (1/T2) as function of Gd(III) concentration (Equation 1),
| (1) |
where 1/T1,2obs is the combination of the paramagnetic (1/T1,2p) and diamagnetic (1/T1,2d) relaxation rates. To measure relaxivity, a series of aqueous solutions of a Gd(III) contrast agent at different concentrations are subjected to a pulse of radio frequency energy; the return to equilibrium conditions is then monitored within a static magnetic field.14 The longitudinal relaxation time (T1) is determined by fitting the mono-exponential curve of the recovery to a net nonzero magnetization in the z-direction. T1 measurements can be performed on a NMR spectrometer or benchtop relaxometer unit with special notation of their dependencies on field strength and temperature.16–17 Concentrations of the Gd(III) contrast agent solutions should be determined to the highest level of accuracy possible, namely by inductively coupled plasma mass spectrometry (ICP-MS) or inductively coupled plasma atomic emission spectroscopy (ICP-AES).
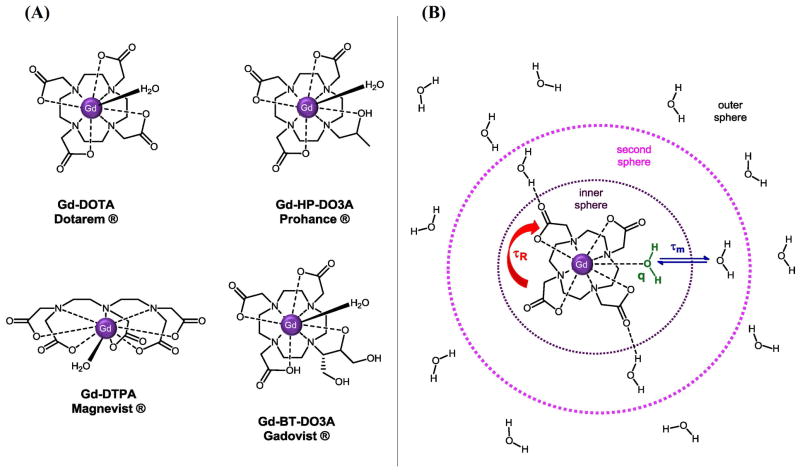
The relaxivity of current clinical Gd(III) contrast agents is low averaging 4.7 mM−1s−1 at 60 MHz, 10-fold less than the theoretical maximum.18 This inefficiency stems from a lack of optimization in one or more of the structural parameters that govern the interaction between the Gd(III) center and the bound water (Figure 2). These parameters are defined through the equations of Solomon, Bloembergen, and Morgan (SBM) and are described briefly below for longitudinal relaxation processes.8, 19–20
Figure 2. Gd(III) contrast agents: design and structural parameters.
(A) Common clinically approved Gd(III) contrast agents. Gd(III), a nine-coordinate lanthanide metal, is chelated by cyclic or linear ligands leaving at least one coordination site open for water exchange. Chelation by the organic framework mitigates toxicity concerns while simultaneously allowing additional synthetic modification near the paramagnetic center. (B) A schematic depiction of the parameters that govern Gd(III) relaxation enhancement. The relaxivity of a Gd(III) contrast agent is controlled by the rotational correlation time (τR), the number of water molecules coordinated to the Gd(III) ion (q), and the time the water remains coordinated to the metal (τM).
Gd(III) complexes are known to increase the longitudinal relaxation rate (1/T1) of water protons in their vicinity. As stated above, the observed relaxation rate for a solution supplemented by a paramagnetic solute is the sum of the relaxation rate of any diamagnetic contribution from the bulk solvent (1/T1,d) and the paramagnetic influence (1/T1,p) (Equation 2).
| (2) |
The paramagnetic contribution to the relaxation rate can be further broken down into the sum of the relaxation rate enhancements in the inner- and outer-sphere (Equation 3).
| (3) |
The outer-sphere relaxation rate contribution arises from both the hydrogen bonding of bulk solvent molecules to the acetate arms of the Gd(III) chelate and the diffusion of the bulk solvent molecules in the proximity of the Gd(III) chelate.8 While each of these mechanisms is on par with the contributions of the inner-sphere relaxation, these outer-sphere modes of relaxation are rarely the target of chemical modification. In contrast, modulation of the inner-sphere relaxation contribution is easily facilitated by synthetic modification of the core chelate. The inner-sphere relaxation rate refers to the exchange of solvent molecules directly coordinated to the paramagnetic center to the bulk environment (Equation 4).
| (4) |
In this equation, c is the concentration of the paramagnetic species in solution, q is the number of solvent molecules coordinated to the paramagnetic center, ρm is the mole fraction of the paramagnetic contrast agent, τM is the lifetime of the bound water molecule [inverse of the exchange rate (kex−1)], and T1M is the longitudinal relaxation time of the bound water.
If the conditions are such that τM ≪ T1M, the inner-sphere relaxation rate is governed by T1M. This longitudinal relaxation time is modulated by both dipole-dipole and scalar relaxation mechanisms. The dipole-dipole relaxation mechanism describes the through-space interaction between a proton nuclear magnetic point dipole and the spatially-extending magnetic field of an electronic point dipole; the scalar relaxation mechanism describes the contact interaction between a proton nuclear magnetic point dipole and the uniform magnetic field at the infinitesimal center of an electronic point dipole. In a dipole-dipole mechanism, the relaxation is controlled by changes in the orientation of the exchangeable solvent proton relative to the direction of the Gd(III) electron spin. This relationship is given by Equation 5 and Equation 6,
| (5) |
| (6) |
where γI is the nuclear gyromagnetic ratio, g is the electron g factor, μB is the Bohr magneton, rGd-H is the distance between the electron spins of the paramagnetic center and the solvent proton, S is the Gd(III) electronic spin quantum number, ωI is the nuclear Larmor frequency, ωs is the electron Larmor frequency, and τc1,2 are the correlation times. A scalar relaxation mechanism is not dependent upon orientation of the nuclear spin but only on the electron spin relaxation of the Gd(III) and water exchange of the paramagnetic ion. This dependence is described in Equation 7,
| (7) |
where (A/ħ) is the scalar coupling constant between the Larmor frequency of the Gd(III) and the water proton. The ionic nature of the Gd(III)-water coordination and the distance between the water protons and Gd(III) will cause dipole-dipole mechanisms to dominate relaxation.8
The correlation time (τc), alluded to in the dipole-dipole mechanism, is dependent upon a set of parameters inherent to Gd(III) complexes. This relationship is summarized in Equation 8,
| (8) |
where T1e is the electronic spin relaxation time, τR is the rotational correlation time, and τM is the lifetime of the bound solvent molecule to the paramagnetic center. When the inverse of the correlation time is equal to the Larmor frequency, the relaxivity is maximized. However, all parameters must be optimized simultaneously for maximum relaxivity.8
It should be noted that the maximum attainable relaxivity values are dependent upon the strength of the external magnetic field.16–17 Higher external magnetic field strength will increase spatial resolution by extending the energy gap between the aligned and anti-aligned nuclear spins. However, this increase in 1H nuclear polarization comes at the expense of τM- and τR-modulated relaxivity enhancement. At higher external magnetic fields, the benefits of slow molecular tumbling and increased water exchange rates are attenuated.14, 17
Strategies to increase the relaxivity of Gd(III) contrast agents have centered on modulation of q, τR, and τM. (Figure 2). Synthetic modification of the ligand can tune the values of these parameters to near optimal states.16 The theoretical limit of relaxivity for a single Gd(III) chelate at 20 MHz (100 mM−1s−1) has not been achieved in practice. This shortcoming is most often due to the rapid rotational motion of small molecule agents in solution (τR). Other limiting parameters may emerge once the contrast agent has been synthetically modified to optimize τR; in many cases an unoptimized τM or q are responsible for the only modest relaxivity gains of τR optimized complexes. Therefore, appropriate analytical techniques have been developed to quantify the values of these parameters in an effort to classify the cause of relaxivity enhancement for a given Gd(III) chelate. A survey of these techniques is outlined in the following sections in addition to parameter-specific strategies for relaxivity enhancement of Gd(III) contrast agents.
HYDRATION NUMBER (Q)
For in vivo applications, the cytotoxicity of Gd(III) ions is mitigated by chelation with poly(amino-carboxylate) derivatives of cyclen and diethylenetriamine. The high kinetic and thermodynamic stability of the resulting coordination complexes (kd ~1022 – 1025) eliminate the possibility of transmetallation with endogenous Ca(II) and Zn(II) coordination systems. Current FDA approved contrast agents bind the Gd(III) ion with a coordination number of eight; as Gd(III) has a coordination number of nine, the remaining site is available for water exchange.
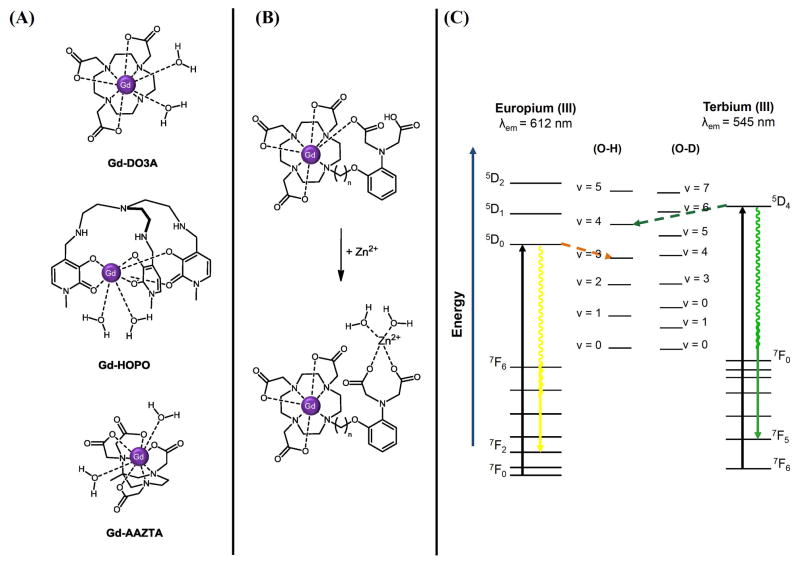
Increasing, decreasing, or controlling the value of q in response to physiological stimuli has a significant effect on the relaxivity at the expense of the dissociation constant. Removal of a single acetate arm from the Gd(III) macrocyclic chelate DOTA nearly doubles the relaxivity while simultaneously decreasing the Gd(III) dissociation constant by three orders of magnitude.18 However, this open position permits conjugation of targeting groups, nanoparticles, and biomacromolecules to the Gd(III)-DO3A ligand. Alternative ligands (e.g. DTTA, AAZTA) have been developed to increase the hydration number without sacrificing thermodynamic stability (Figure 3).21–24 Hydroxypyridinone (HOPO) ligands exhibit high hydration numbers (q = 2–3) with relaxivities three-fold higher than clinical Gd(III) contrast agents.25–27 The aqueous solubility of Gd(III)-HOPO derivatives can be enhanced through conjugation to dendrimers or viral capsids resulting in relaxivities in excess of 30 mM−1s−1 per Gd(III).28–30
Figure 3.
Strategies for q-modulated relaxation enhancement include: (A) Alternative ligand design,18, 21, 25 and (B) Ion-responsive contrast enhancement. Adapted with permission from Ref. 34. Copyright 2008 American Chemical Society. (C) Energy level diagrams for Eu(III) and Tb(III) pertaining to q measurement by luminescence lifetime decay. The denoted emission transitions are the most intense radiative transitions for each tripositive lanthanide. Radiationless decay mechanisms mediated by O-H oscillators are labeled with dotted arrows. Adapted from ref. 40. Copyright 1965, American Institute of Physics.
q-modulated Gd(III) contrast agents control relaxivity through changing the number of water molecules that can be coordinated to the paramagnetic center in response to physiological conditions. Meade and colleagues pioneered q-modulation strategies to observe changes in physiological ion (Ca2+, Zn2+) concentration via MR contrast enhancement (Figure 3).31–34 Moreover, MR detection of gene expression or oncogenic activity has been demonstrated through enzyme-mediated changes in the hydration number of the Gd(III) chelate.35–38 For example, a Gd(III)-DO3A chelate conjugated to a β-galactosidase sugar functionality was developed with q = 0 per coordination of endogenous carbonate; cleavage of the sugar by β-galactosidase permits access by one water molecule to the chelate resulting in conversion from a dark to bright signal.36, 38 This correlation of enzyme activity with MR signal intensity is an important step forward in the development of MR-based molecular imaging as the β-galactosidase enzyme is a common reporter gene in many biological fields of study.39
The luminescent properties of the 7F0 → 5D0 transition in the Eu(III) and the 7F6 →5D4 in the Tb(III) analogues of Gd(III) chelates permit q determination via time-dependent luminescence lifetime decay.40 Upon excitation, the major pathway of energy release for the tripositive lanthanide is through vibrations with nearby oscillators (Figure 3). O-H oscillators of a water molecule bond to the paramagnetic center can afford an effective route for radiationless decay; however, the O-D oscillators of D2O are too low in energy, mitigating any significant effect.40–41 Using known X-ray crystal structures of Eu(III) and Tb(III) chelates Horrocks and Sudnick derived a linear relationship between the luminescence decay lifetimes of Ln(III) in H2O and D2O and the hydration number of the Gd(III)chelate.42 The luminescence decay lifetimes of each solution [0.5 – 1 mM Ln(III)] can be determined from the mono-exponential fit of the fluorescence intensity decay as a function of time. Correcting for the inherent O-H and N-H oscillators of the chelate and any second-sphere effects, the hydration numbers for the Eu(III) chelates can be calculated using Equation 9,
| (9) |
where τH2O and τD2O are the excited state lifetimes of the Eu(III) complexes in H2O and D2O, respectively; nOH, nNH, and namide are the number of O-H, N-H, and amide oscillators of the chelating ligand, respectively.43 A similar relationship for the Tb(III) analogues has been derived (Equation 10).44
| (10) |
The solution to these equations will give a non-integer values representing a weighted average of the hydration numbers of each unique Ln(III) water exchange environment with an error of ± 0.1 water molecules.
Alternatively, q can be determined from the Ln(III)-induced shifts of the water peak by 17O NMR spectroscopy as described by Djanshvili and Peters.45 In comparison to luminescence lifetime decay measurements, the hydration number of the Gd(III) chelate can be directly measured. Therefore, only one analogue needs to be synthesized for both relaxivity and Gd(III) parameter characterization. Although a high concentration of the Gd(III) chelate is recommended (20 mM), a comparatively low volume of sample is required. Accurate measurement of samples with Gd(III) concentrations as low as 2 mM is obtainable with care. A method to determine Gd(III) concentration (such as ICP-MS) is necessary for this q measurement technique.
17O NMR spectra of the Gd(III) contrast agent (in D2O) and a control of D2O alone (both enriched with < 3% v/v water-17O) are obtained. The difference in chemical shift of the water peak corresponds to the Ln(III)-induced shift (δobs). This shift results from the sum of the diamagnetic (δdia), contact (δcon), and pseudocontact (δpc) contributions specific to the interaction of the Ln(III) complex and the bound water molecule.45 For Gd(III) systems, δpc (through-space magnetic moment interactions) and δdia contributions are negligible and not included.46 Gd(III) δcon contribution is determined by a set of lanthanide(III) and nuclei dependent values.46 These resultant factors simplify to Equation 11 and Equation 12 allowing determination of q for Gd(III) complexes,
| (11) |
| (12) |
where Pm is the mole fraction of Gd(III), F is a pre-factor characteristic of 17O used in the calculation of contact shift δcon, and T is temperature in Kelvin.45 For Gd(III), spectra must be obtained at 80 ºC to ensure fast enough water exchange on the 17O NMR timescale. Similar to luminescence lifetime decay measurements, the hydration numbers obtained by Ln(III)-induced shifts are noninteger values due to an average of multiple Ln(III) water exchange environments with an error of ± 0.2 water molecules.
ROTATIONAL CORRELATION TIME (τR)
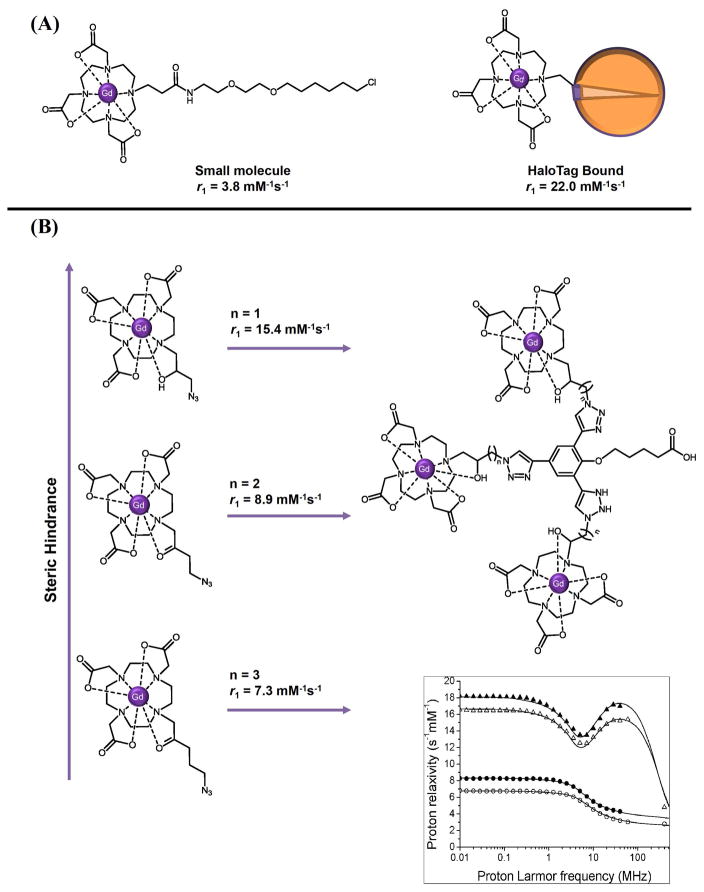
Rotational motion is the dominant factor in modulating Gd(III) relaxivity at current clinical field strengths (10–70 MHz). The rotational correlation time (τR) is dependent upon the effective radius of the contrast agent, a measure of the molecular mass of the system. Conjugation of small molecule Gd(III) chelates to nanoparticles, proteins, peptide scaffolds, and virus capsids has been shown to effectively slow global rotational motion elongating τR to optimal values (1–10 ns depending on field strength).47–52 Targeting Gd(III) contrast agents to specific biomarkers or creating multivalent nanoparticle scaffolds decorated with Gd(III) chelates can greatly increase the concentration of an agent to a region of interest in vivo.53–55 Specific biochemical processes can be monitored while simultaneously improving the signal-to-noise ratio in the acquired MR image. For example, a Gd(III)-DO3A chelate modified with a haloalkane group known to covalently bind the Halotag reporter protein showed a six-fold increase in per Gd(III) relaxivity upon binding to the intended target (Figure 4).56 Furthermore, increases in τR can be obtained through reducing the local degrees of freedom between the Gd(III) center and target substrate.57–58 Recently, we have shown a ten-fold increase in τR upon conjugating a small molecule Gd(III)-DO3A chelate to a trisubstituted benzyl scaffold; this enhancement resulted from a strong dependence on the carbon linker length between the individual chelates and the aromatic platform (Figure 4).57
Figure 4.
Strategies for τR modulation. (A) Schematic of a HaloTag-targeted Gd(III) contrast agent. The haloalkane tail of the Gd(III)-DO3A chelate is irreversibly bound to the mutated enzyme activity site buried deep in the interior of the protein. Upon binding, the effective radius of the small molecule is greatly increased slowing rotational motion. This τR-modulated relaxation enhancement strategy boosts the efficiency of the chelate almost six-fold in comparison to the unconjugated small molecule. Adapted with permission from Ref. 56. Copyright 2011 American Chemical Society. (B) Decreasing the number of carbons between a small molecule agent and its intended benzyl scaffold increases relaxivity via a local τR effect. An NMRD curve shows the characteristic bell-curve feature at field strengths corresponding to the Larmor frequency signaling a τR effect in the corresponding agents. Small molecule agents with short τR values give the relatively featureless curves at these same frequencies. The NMRD curve is reprinted with permission from Ref. 57. Copyright 2011 American Chemical Society.
Nuclear magnetic resonance dispersion (NMRD) is a common technique used to quantify τR in addition to other SBM parameters.16 NMRD is a measurement of the relaxation rate of a Gd(III) complex as a function of magnetic field (0.01 – 100 MHz) and temperature. In comparison to other techniques, large amounts of material are not required, and data collection is relatively rapid. However, the field-cycling NMR units used to perform NMRD experiments are rare with only a few operating worldwide limiting easy access. The features of the NMRD curve correspond to the exchange rate between the bound and the bulk water, the hydration number, and the reorientation time of the Gd(III) chelate.16 For example, small molecule agents with rapid rotational correlation times (100 ps) give relatively featureless curves corresponding to limited optimization of rotational influence. Conjugation of the same Gd(III) chelate to a scaffold or barycenter boosts τR to near optimal values (10 ns). This enhancement is reflected in the NMRD as characteristic bell shape feature near the proton Larmor frequencies (10–70 MHz) where rotational motion dominates the correlation time.14 Parameters inherent to the Gd(III) complexes, including τR, can be derived by fitting these NMRD curves to the SBM equations.
However, the complexity of characterizing these features highlights the disadvantages of NMRD analysis. From each NMRD curve, six possible parameters of the Gd(III) contrast agent must be quantified simultaneously. For small molecule agents, this NMRD curve can be relatively flat complicating analysis.18 Moreover, present fitting programs cannot always provide perfect agreement with experimental data due to the presence of both static and transient zero field splitting that govern Gd(III) relaxation rates at low fields. Therefore, although NMRD data cannot be the only means to characterize contrast agents, it can provide a way to assess a given Gd(III) environment. These findings can be used to classify suspected alterations to global rotational motion or water exchange events.
Other methods to measure τR have been developed that utilize NMR spectroscopy. Determining τR through proton-linewidth measurements of the Yb(III) analogues (assuming Curie relaxation mechanisms) is a new method developed by Zech and coworkers.59 The Yb(III) analogues induce significant paramagnetic broadening in the 1H NMR spectrum that linearly correlate with increasing concentrations of the τR-modulating target conjugate (protein, nanoparticle, scaffold) as long as the exchange is rapid on the NMR timescale. The transverse relaxation rate (R2) can be determined through the linear correlation of the linewidth of the proton peak as a function of the bound agent. Substitution of R2 into the equation for Curie relaxation rate can be used to estimation τR (Equation 13)
| (13) |
where μ0 is the permeability of the vacuum (4π × 10−7 T·m·A−1), ωH is the proton angular Larmor frequency (in rad·s−1), gJ is the Landé g factor for Yb(III) (8/7), μB is the electron Bohr magneton (9.274 × 10−24 J·T−1), J is the vector sum of electronic orbital and spin angular momenta (J = 7/2 for Yb(III)), kB is Boltzmann’s constant, T is the temperature in Kelvin, rYb-H is the proton-metal distance, and τc is the total correlation time. In this method, where the relaxation rates of non-exchangeable protons are being determined, τc is reduced to τR giving the simplified version in Equation 14.
| (14) |
However, to use this method bond distances between the protons of the chelate and the Yb(III) center must be known; these may be approximated if an X-ray diffraction structure is unavailable.
Zang and Xie have reported an off-resonance spin-lock technique for tR determination of Gd(III) chelates that can be performed on a NMR spectrometer or benchtop relaxometer unit.60 The off-resonance rotating frame relaxation rates (R1ρ) of several samples are measured at multiple spin-lock strengths (ωe) and angles (θ) by changing the RF amplitude and resonance offsets. The data is fitted to an mono-exponential decay extracting the initial [M(0+)] and equilibrium [M(∞)] magnetization, which can be used to calculate R1ρ (Equation 15)
| (15) |
where R1 is the longitudinal relaxation rate measured by an inversion recovery pulse sequence. The value obtained for R1ρ (ωe, θ) is fitted to SBM theory in the rotating frame to solve for τR.
LIFETIME OF THE COORDINATED SOLVENT MOLECULE (τM)
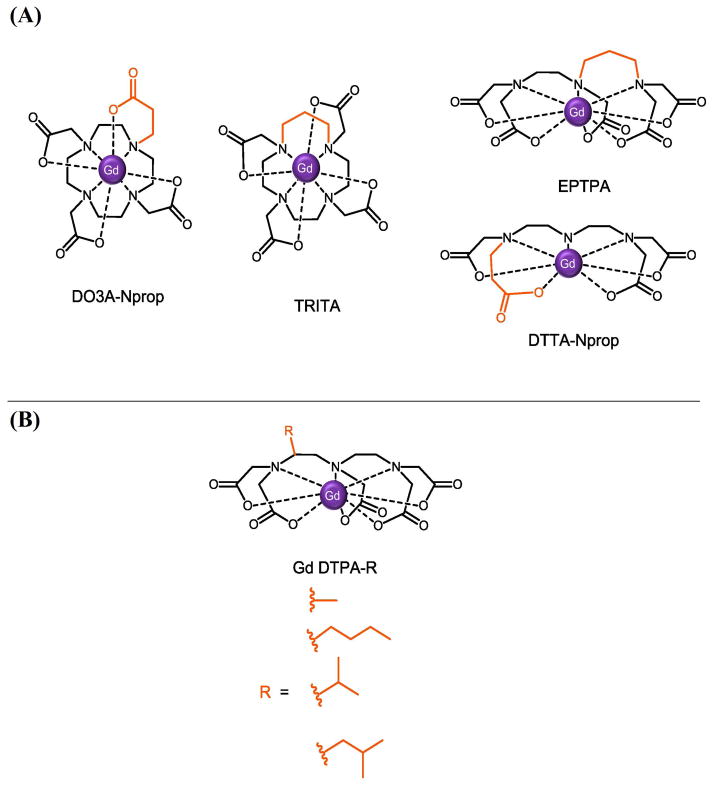
Upon optimization of τR, the lifetime of the coordinated solvent molecule (τM) becomes the dominating factor in further relaxivity modulation. Optimization of τM (10–50 ns at low field) can be achieved by increasing steric hindrance and destabilizing the interaction between the bound water and the paramagnetic center to assist in faster dissociative exchange (Figure 5).14 Gd(III) chelates with a coordinating propionate functional group substituted in place of one of the usual acetate arms (DO3A-Nprop, DTTA-Nprop) have a τM one order of magnitude shorter than their parent complexes.61 Moreover, when the ligand backbone is extended by a methylene carbon (TRITA, EPTPA), the τM decreases two orders of magnitude with respect to the parent complexes; however, this benefit is coupled with a decrease in kinetic and thermodynamic stability.61 Laurent and coworkers showed modification of the DTPA backbone with bulky methyl, butyl, isobutyl, and isopropyl functional groups improves τM two- to four-fold over the unmodified DTPA chelate.62 Finally, given the water exchange is mediated through a dissociative exchange mechanism, increasing the negative charge of the chelate increases the exchange rate.8, 63
Figure 5.
Strategies to optimize of the lifetime of the bound solvent molecule (τM) include: (A) Steric compression via the addition of a methylene group to the coordinating acetate arms or chelate backbone,61 and (B) addition of sterically bulky groups to the chelate backbone.62
The rate of water exchange between the inner coordination sphere of the Gd(III) contrast agent and the bulk water can be determined by variable temperature 17O-NMR spectroscopy.16 In this technique, the 17O transverse relaxation rate (R2) of water is measured in the presence of a Gd(III) contrast agent over a series of temperatures. The transverse relaxation rate is proportional to the linewidth of the 17O-NMR peak with correction for diamagnetic influence. A fit of R2 as a function of temperature to Equations 16–18 can simultaneously derive τM, the electronic spin relaxation time (T1e), and the change in enthalpy (ΔH).
| (16) |
| (17) |
| (18) |
In Equations 16–18, S is the spin number of Gd(III), A/ħ is the hyperfine coupling constant, and R is the ideal gas constant. With this method, scalar forces are the dominant mechanism of relaxation. However, a large concentration of the Gd(III) contrast agent (20–30 mM) is required to observe significant linewidth broadening.
The 17O NMR method for determining τM only applies to fast tumbling agents (where τR is not optimized). For agents with a long τR, measuring the transverse relaxivity (r2) of the Dy(III) analogues at high field over a range of temperatures (VT r2) can be used to derive τM.59, 64–66 Curie mechanisms dominate the relaxation of Dy(III) analogues. Compared to SBM relaxation, which describes the relaxation of a proton nuclear spin (water) upon interaction with the instantaneous fluctuations of a Ln(III) electron spin, Curie spin relaxation is an additional relaxation mechanism that occurs because of the non-zero time thermal average of a Ln(III) electron spin in the presence of a magnetic field. This creates a large static magnetic moment called Curie spin that can interact with protons to induce relaxation.67 At high magnetic fields, Curie relaxation is the most important relaxation mechanism for Dy(III) complexes because of its high magnetic moment and short electronic relaxation time. τM is found by fitting the r2 of the Dy(III) contrast agent analogues at a series of temperatures to Curie relaxation equations. The analysis is much more complex than the simplified Gd(III) method because ten parameters are fit simultaneously.
OUTLOOK
The field of molecular imaging presents new challenges to which imaging modalities are swiftly adapting. MRI is an attractive technique due to its noninvasive approach with no accompanying radiation risk. The insensitivity of MR probes in molecular imaging can be addressed through the modification of Gd(III) coordination complex ligand structure. The efficiency (relaxivity) of Gd(III) contrast agents can be increased through modulation of the parameters inherent to these complexes including: τR, the rotational correlation time, τM, the lifetime of the bound water molecule to the paramagnetic center, and q, the number of solvent molecules bond to the Gd(III) at any one time. Quantification of each of these parameters can provide confirmation of relaxation enhancement mechanisms or provide strategies for further synthetic modification. Diverse methods exist to measure these parameters, each with advantages and disadvantages. As the sensitivity and variety of instrumentation continue to advance the applicability and precision of these measurements will continue to improve.
Acknowledgments
The authors gratefully acknowledge Lauren Matosziuk for the abstract graphic and support by the National Institute of Health/National Institute of Biomedical Imaging and Bioengineering Grant R01 EB005866.
Biographies
BIOGRAPHIES
Lisa M. Manus completed her Bachelor of Science degree in Chemistry and Biochemistry and Molecular Biology at Illinois State University in 2007. She recently completed her Ph.D. studies under the direction of Prof. Thomas J. Meade at Northwestern University on Gd(III)-Nanodiamond conjugates for MRI contrast enhancement.
Renee C. Strauch completed her Bachelor of Science degree in Biochemistry at Miami University (Oxford, OH) in 2004. Renee earned her Ph.D. at Northwestern University under the direction of Prof. Thomas J. Meade studying the enhancement and optimization of MR probes.
Andy H. Hung completed his Bachelor of Applied Science degree in Engineering Science and Master of Applied Science degree in Biomedical Engineering at the University of Toronto. He is currently completing his Ph.D. studies under the direction of Prof. Thomas J. Meade at Northwestern University.
Amanda L. Eckermann received a Bachelor of Science in Chemistry at the California Institute of Technology and a Ph.D. in Chemistry from the University of Illinois at Urbana-Champaign. She is currently a Research Associate Professor at Northwestern University.
Thomas J. Meade is currently the Eileen Foell Chair in Cancer Research and Professor of Chemistry, Molecular Biosciences, Neurobiology, Biomedical Engineering and Radiology, as well as the Director of the Center for Advanced Molecular Imaging (CAMI). Professor Meade’s research focuses on bioinorganic coordination chemistry and biological molecular imaging, electron transfer processes and the development of electronic biosensors for the detection of DNA and proteins. He has founded three biotech companies developing hand-held devices for protein and DNA detection and bioactivated MR contrast agents for in vivo imaging of cancer.
References
- 1.Gore JC, Manning HC, Quarles CC, Waddell KW, Yankeelov TE. Magn Reson Imaging. 2011;29:587–600. doi: 10.1016/j.mri.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brindle K. Nat Rev Cancer. 2008;8:94–107. doi: 10.1038/nrc2289. [DOI] [PubMed] [Google Scholar]
- 3.Weissleder R, Mahmood U. Radiology. 2001;219:316–333. doi: 10.1148/radiology.219.2.r01ma19316. [DOI] [PubMed] [Google Scholar]
- 4.Driehuys B. Science. 2006;314:432–433. doi: 10.1126/science.1134532. [DOI] [PubMed] [Google Scholar]
- 5.Frullano L, Meade T. J Biol Inorg Chem. 2007;12:939–949. doi: 10.1007/s00775-007-0265-3. [DOI] [PubMed] [Google Scholar]
- 6.Brindle KM. Br J Radiol. 2003;76:S111–S117. doi: 10.1259/bjr/50577981. [DOI] [PubMed] [Google Scholar]
- 7.Jackson EF, Ginsberg LE, Schomer DF, Leeds NE. Surg Neurol. 1997;47:185–199. doi: 10.1016/s0090-3019(96)00375-8. [DOI] [PubMed] [Google Scholar]
- 8.Merbach A, Toth E. The Chemistry of Contrast Agents in Medical Magnetic Resonance Imaging. John Wiley & Sons, Ltd; New York: 2001. [Google Scholar]
- 9.Patterson DM, Padhani AR, Collins DJ. Nat Clin Prac Oncol. 2008;5:220–233. doi: 10.1038/ncponc1073. [DOI] [PubMed] [Google Scholar]
- 10.Wolff SD, Balaban RS. Magn Reson Med. 1989;10:135–144. doi: 10.1002/mrm.1910100113. [DOI] [PubMed] [Google Scholar]
- 11.Woods M, Woessner DE, Sherry AD. Chem Soc Rev. 2006;35:500–511. doi: 10.1039/b509907m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terreno E, Dastru W, Delli Castelli D, Gianolio E, Geninatti Crich S, Longo D, Aime S. Curr Med Chem. 2010;17:3684–3700. doi: 10.2174/092986710793213823. [DOI] [PubMed] [Google Scholar]
- 13.Major JL, Meade TJ. Acc Chem Res. 2009:893–903. doi: 10.1021/ar800245h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermann P, Kotek J, Kubicek V, Lukes I. Dalton Trans. 2008:3027–3047. doi: 10.1039/b719704g. [DOI] [PubMed] [Google Scholar]
- 15.Vlaardingerbroek MT, den Boer JA. Magnetic Resonance Imaging Theory and Practice. Springer Verlag; Germany: 1996. [Google Scholar]
- 16.Caravan P. Chem Soc Rev. 2006;35:512–523. doi: 10.1039/b510982p. [DOI] [PubMed] [Google Scholar]
- 17.Caravan P, Farrar CT, Frullano L, Uppal R. Contrast Media Mol Imaging. 2009;4:89–100. doi: 10.1002/cmmi.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Chem Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 19.Solomon I. Phys Rev. 1955;99:559–565. [Google Scholar]
- 20.Bloembergen N, Morgan LO. J Chem Phys. 1961;34:842–850. [Google Scholar]
- 21.Aime S, Calabi L, Cavallotti C, Gianolio E, Giovenzana GB, Losi P, Maiocchi A, Palmisano G, Sisti M. Inorg Chem. 2004;43:7588–7590. doi: 10.1021/ic0489692. [DOI] [PubMed] [Google Scholar]
- 22.Costa J, Tóth É, Helm L, Merbach AE. Inorg Chem. 2005;44:4747–4755. doi: 10.1021/ic0500309. [DOI] [PubMed] [Google Scholar]
- 23.Costa J, Ruloff R, Burai L, Helm L, Merbach AE. J Am Chem Soc. 2005;127:5147–5157. doi: 10.1021/ja0424169. [DOI] [PubMed] [Google Scholar]
- 24.Livramento JB, Tóth É, Sour A, Borel A, Merbach AE, Ruloff R. Angew Chem Int Ed. 2005;44:1480–1484. doi: 10.1002/anie.200461875. [DOI] [PubMed] [Google Scholar]
- 25.Raymond KN, Pierre VC. Bioconjugate Chem. 2004;16:3–8. doi: 10.1021/bc049817y. [DOI] [PubMed] [Google Scholar]
- 26.Cohen SM, Xu J, Radkov E, Raymond KN, Botta M, Barge A, Aime S. Inorg Chem. 2000;39:5747–5756. doi: 10.1021/ic000563b. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Churchill DG, Botta M, Raymond KN. Inorg Chem. 2004;43:5492–5494. doi: 10.1021/ic049028s. [DOI] [PubMed] [Google Scholar]
- 28.Floyd WC, Klemm PJ, Smiles DE, Kohlgruber AC, Pierre VrC, Mynar JL, Fréchet JMJ, Raymond KN. J Am Chem Soc. 2011;133:2390–2393. doi: 10.1021/ja110582e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klemm PJ, Floyd WC, Smiles DE, Fréchet JMJ, Raymond KN. Contrast Media Mol Imaging. 2012;7:95–99. doi: 10.1002/cmmi.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garimella PD, Datta A, Romanini DW, Raymond KN, Francis MB. J Am Chem Soc. 2011;133:14704–14709. doi: 10.1021/ja204516p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W-h, Fraser SE, Meade TJ. J Am Chem Soc. 1999;121:1413–1414. [Google Scholar]
- 32.Li W-h, Parigi G, Fragai M, Luchinat C, Meade TJ. Inorg Chem. 2002;41:4018–4024. doi: 10.1021/ic0200390. [DOI] [PubMed] [Google Scholar]
- 33.Major JL, Parigi G, Luchinat C, Meade TJ. Proc Natl Acad Sci U S A. 2007;104:13881–13886. doi: 10.1073/pnas.0706247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Major JL, Boiteau RM, Meade TJ. Inorg Chem. 2008;47:10788–10795. doi: 10.1021/ic801458u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duimstra JA, Femia FJ, Meade TJ. J Am Chem Soc. 2005;127:12847–12855. doi: 10.1021/ja042162r. [DOI] [PubMed] [Google Scholar]
- 36.Urbanczyk-Pearson LM, Femia FJ, Smith J, Parigi G, Duimstra JA, Eckermann AL, Luchinat C, Meade TJ. Inorg Chem. 2007;47:56–68. doi: 10.1021/ic700888w. [DOI] [PubMed] [Google Scholar]
- 37.Moats RA, Fraser SE, Meade TJ. Angew Chem Int Ed. 1997;36:726–728. [Google Scholar]
- 38.Louie AY, Huber MM, Ahrens ET, Rothbacher U, Moats R, Jacobs RE, Fraser SE, Meade TJ. Nat Biotech. 2000;18:321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 39.Jiang T, Xing B, Rao J. Biotechnol Genet Eng Rev. 2008;25:41–75. doi: 10.5661/bger-25-41. [DOI] [PubMed] [Google Scholar]
- 40.Kropp JL, Windsor MW. J Chem Phys. 1965;42:1599–1608. [Google Scholar]
- 41.Kropp JL, Windsor MW. J Phys Chem. 1967;71:477–482. [Google Scholar]
- 42.Horrocks WD, Sudnick DR. Acc Chem Res. 1981;14:384–392. [Google Scholar]
- 43.Supkowski RM, Horrocks WD., Jr Inorg Chim Acta. 2002;340:44–48. [Google Scholar]
- 44.Quici S, Cavazzini M, Marzanni G, Accorsi G, Armaroli N, Ventura B, Barigelletti F. Inorg Chem. 2005;44:529–537. doi: 10.1021/ic0486466. [DOI] [PubMed] [Google Scholar]
- 45.Djanashvili K, Peters JA. Contrast Media Mol Imaging. 2007;2:67–71. doi: 10.1002/cmmi.132. [DOI] [PubMed] [Google Scholar]
- 46.Bleaney B. J Magn Reson. 1972;8:91–100. [Google Scholar]
- 47.Datta A, Hooker JM, Botta M, Francis MB, Aime S, Raymond KN. J Am Chem Soc. 2008;130:2546–2552. doi: 10.1021/ja0765363. [DOI] [PubMed] [Google Scholar]
- 48.Anderson EA, Isaacman S, Peabody DS, Wang EY, Canary JW, Kirshenbaum K. Nano Lett. 2006;6:1160–1164. doi: 10.1021/nl060378g. [DOI] [PubMed] [Google Scholar]
- 49.Bull SR, Guler MO, Bras RE, Meade TJ, Stupp SI. Nano Lett. 2004;5:1–4. doi: 10.1021/nl0484898. [DOI] [PubMed] [Google Scholar]
- 50.Manus LM, Mastarone DJ, Waters EA, Zhang XQ, Schultz-Sikma EA, MacRenaris KW, Ho D, Meade TJ. Nano Lett. 2010;10:484–489. doi: 10.1021/nl903264h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karfeld LS, Bull SR, Davis NE, Meade TJ, Barron AE. Bioconjugate Chem. 2007;18:1697–1700. doi: 10.1021/bc700149u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song Y, Xu X, MacRenaris KW, Zhang XQ, Mirkin CA, Meade TJ. Angew Chem Int Ed. 2009;48:9143–9147. doi: 10.1002/anie.200904666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sukerkar PA, MacRenaris KW, Townsend TR, Ahmed RA, Burdette JE, Meade TJ. Bioconjugate Chem. 2011;22:2304–2316. doi: 10.1021/bc2003555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sukerkar PA, MacRenaris KW, Meade TJ, Burdette JE. Mol Pharm. 2011;8:1390–1400. doi: 10.1021/mp200219e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SM, Song Y, Hong BJ, MacRenaris KW, Mastarone DJ, O’Halloran TV, Meade TJ, Nguyen ST. Angew Chem Int Ed. 2010;49:9960–9964. doi: 10.1002/anie.201004867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strauch RC, Mastarone DJ, Sukerkar PA, Song Y, Ipsaro JJ, Meade TJ. J Am Chem Soc. 2011;133:16346–16349. doi: 10.1021/ja206134b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mastarone DJ, Harrison VSR, Eckermann AL, Parigi G, Luchinat C, Meade TJ. J Am Chem Soc. 2011;133:5329–5337. doi: 10.1021/ja1099616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Y, Kohlmeir EK, Meade TJ. J Am Chem Soc. 2008;130:6662–6663. doi: 10.1021/ja0777990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zech SG, Eldredge HB, Lowe MP, Caravan P. Inorg Chem. 2007;46:3576–3584. doi: 10.1021/ic070011u. [DOI] [PubMed] [Google Scholar]
- 60.Zhang H, Xie Y. J Magn Reson. 2006;183:213–227. doi: 10.1016/j.jmr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 61.Jaszberenyi Z, Sour A, Toth E, Benmelouka M, Merbach AE. Dalton Trans. 2005:2713–2719. doi: 10.1039/b506702b. [DOI] [PubMed] [Google Scholar]
- 62.Laurent S, Botteman F, Vander Elst L, Muller RN. Magn Reson Mater Phys, Biol Med. 2004;16:235–245. doi: 10.1007/s10334-004-0031-2. [DOI] [PubMed] [Google Scholar]
- 63.André JP, Maecke HR, Tóth É, Merbach AA. J Biol Inorg Chem. 1999;4:341–347. doi: 10.1007/s007750050320. [DOI] [PubMed] [Google Scholar]
- 64.Caravan P, Greenfield MT, Bulte JWM. Magn Reson Med. 2001;46:917–922. doi: 10.1002/mrm.1277. [DOI] [PubMed] [Google Scholar]
- 65.Vander Elst L, Roch A, Gillis P, Laurent S, Botteman F, Bulte JWM, Muller RN. Magn Reson Med. 2002;47:1121–1130. doi: 10.1002/mrm.10163. [DOI] [PubMed] [Google Scholar]
- 66.Pubanz D, Gonzalez G, Powell DH, Merbach AE. Inorg Chem. 1995;34:4447–4453. [Google Scholar]
- 67.Gueron M. J Magn Reson. 1975;19:58–66. [Google Scholar]