SUMMARY
The RNA-mediated disease model for myotonic dystrophy (DM) proposes that microsatellite C(C)TG expansions express toxic RNAs which disrupt splicing regulation by altering MBNL1 and CELF1 activities. While this model explains DM manifestations in muscle, less is known about the effects of C(C)UG expression on the brain. Here, we report that Mbnl2 knockout mice develop several DM-associated CNS features including abnormal REM sleep propensity and deficits in spatial memory. Mbnl2 is prominently expressed in the hippocampus and Mbnl2 knockouts show a decrease in NMDAR synaptic transmission and impaired hippocampal synaptic plasticity. While Mbnl2 loss did not significantly alter target transcript levels in the hippocampus, mis-regulated splicing of hundreds of exons was detected using splicing microarrays, RNA-seq and HITS-CLIP. Importantly, the majority of the Mbnl2-regulated exons examined were similarly mis-regulated in DM. We propose that major pathological features of the DM brain result from disruption of the MBNL2-mediated developmental splicing program.
INTRODUCTION
During mammalian development, alternative splicing of pre-mRNAs plays a critical role in the extensive remodeling of tissues throughout both embryonic and postnatal phases (Chen and Manley, 2009; Kalsotra and Cooper, 2011). The spatial and temporal expression patterns of specific protein isoforms are exquisitely controlled during each developmental window such that the unique physiological requirements of each cell type are adequately met. While >90% of human multi-exon genes produce alternatively spliced transcripts, the complex network of dynamic interactions between multiple cell types that characterizes the CNS suggests that alternative splicing regulation is particularly critical for the developing brain (Li et al., 2007; Licatalosi and Darnell, 2010; Wang et al., 2008).
The importance of alternative splicing during developmental transitions has been highlighted by studies on the autosomal dominant disease myotonic dystrophy (DM) (Cooper et al., 2009; Poulos et al., 2011). CNS function is compromised in DM with hypersomnia, cognitive/behavioral abnormalities, progressive memory problems, cerebral atrophy and, in the congenital form of the disease, mental retardation (Meola and Sansone, 2007; Weber et al., 2010). DM is caused by microsatellite CTG expansions in the DMPK gene (DM type 1, DM1) or CNBP CCTG expansions (DM type 2, DM2). Transcription of these repeats generates C(C)UG expansion [C(C)UGexp] RNAs that disrupt alternative splicing, resulting in the persistence of fetal splicing patterns in adult tissues. A current disease model suggests that splicing disruption occurs because the muscleblind-like protein 1 (MBNL1), which normally promotes adult splicing patterns, is sequestered by C(C)UGexp RNAs (Cooper et al., 2009; Du et al., 2010; Poulos et al., 2011). Additionally, expression of C(C)UGexp RNAs is reported to increase levels of CELF1, a splicing factor that promotes fetal splicing (Kuyumcu-Martinez et al., 2007). Thus, the developmental regulation of some DM-targeted exons may be achieved by modulating the levels of two antagonistic splicing factors, MBNL1 and CELF1. Although this MBNL loss-of-function model for DM1 and DM2 is supported by the splicing patterns observed in the skeletal and heart muscles of mouse Mbnl1 knockouts and Celf1 overexpression transgenics (Du et al., 2010; Kanadia et al., 2003; Koshelev et al., 2010; Ward et al., 2010), it is not clear if alternative splicing in the brain is similarly dysregulated. Moreover, the view that DM is solely a spliceopathy has been recently challenged (Sicot et al., 2011). The expression of mutant DMPK and CNBP microsatellites also results in alterations in mRNA localization, microRNA and mRNA turnover pathways and induces repeat-associated non-ATG-initiated (RAN) translation (Zu et al., 2011). These additional pathogenic mechanisms highlight the importance of discriminating direct from indirect actions of DM mutations to link specific disease manifestations to distinct pathways.
Since Mbnl1 knockout (Mbnl1ΔE3/ΔE3) mice show modest effects on alternative splicing regulation in the brain (Suenaga et al., 2012), we have now addressed the possibility that the other major MBNL protein expressed in adult tissues, MBNL2, is the principal factor dysregulated in the DM CNS. Here, we report the generation of Mbnl2 knockout mice which exhibit several phenotypes consistent with features of DM neurologic disease. Loss of Mbnl2 leads to widespread changes in postnatal splicing patterns in the brain, many of which are similarly dysregulated in the human DM1 brain, but not in skeletal muscle. Direct Mbnl2 RNA targets are identified by high throughput sequencing-crosslinking immunoprecipitation (HITS-CLIP) and the generation of an Mbnl2 splicing map. Mbnl2 knockouts should provide novel insights into the developmental regulation of splicing in the CNS and identify the molecular events that impact the brain in myotonic dystrophy.
RESULTS
Mbnl2 Knockout Mice Fail to Model DM-Associated Muscle Deficits
Previous gene trap studies have reported contradictory results on the effects of Mbnl2 allele disruption on DM-relevant muscle pathology and alternative splicing (Figure S1A). Insertion of an EN2-βgeo gene trap into Mbnl2 intron 4 (Mbnl2GT4) resulted in a decrease in Mbnl2 mRNA in Mbnl2GT4/GT4 homozygotes but no changes in muscle structure/function or in the splicing of Mbnl1 RNA targets (Lin et al., 2006). In contrast, Mbnl2GT2/GT2 mice, in which the same gene trap had inserted into Mbnl2 intron 2, were reported to develop myotonia, Clcn1 mis-splicing and skeletal muscle defects reminiscent of DM (Hao et al., 2008). To address this inconsistency, we generated Mbnl2 knockout mice (Mbnl2ΔE2/ΔE2) using a homologous recombination strategy (Figure S1B). Similar to Mbnl1 exon 3, Mbnl2 exon 2 encodes the initiation codon for the full-length Mbnl2 protein (Figure S1C) (Kanadia et al., 2003). Mbnl2 constitutive knockout lines were generated by mating chimeric males to a CMV-cre transgenic line (Figure 1A). Genomic DNA blot analysis confirmed the presence of the disrupted allele in heterozygous and homozygous knockout mice (Figure S1D). Ablation of full-length Mbnl2 mRNA expression in the homozygous knockouts was confirmed by RT-PCR (Figure 1B). While a potential alternative initiation codon exists in exon 3 (Figure S1C), immunoblot analysis using a monoclonal antibody (mAb) that recognizes all annotated Mbnl2 isoforms demonstrated a complete absence of Mbnl2 protein in all Mbnl2ΔE2/ΔE2 tissues examined (Figure 1C). Therefore, Mbnl2ΔE2/ΔE2 lines are functional nulls and will be subsequently referred to as Mbnl2 knockouts.
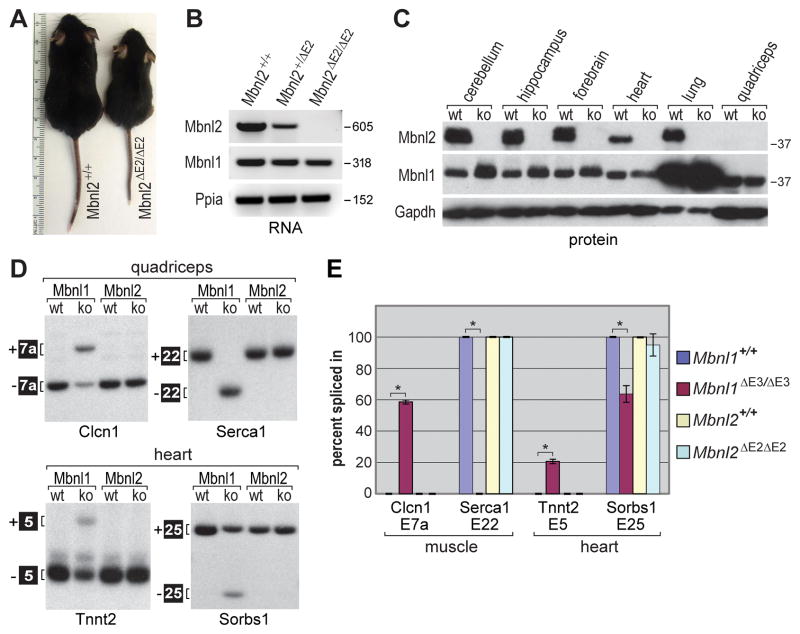
Figure 1. Mbnl2 Knockout Mice Fail to Model DM-Associated Muscle Deficits.
(A) Mbnl2ΔE2/ΔE2 knockout mice are small at weaning compared to wild-type sibs.
(B) Mbnl2 RNAs containing exon 2 are absent in Mbnl2 knockouts. RT-PCR analysis of hippocampus RNA from Mbnl2+/+, Mbnl2+/ΔE2 and Mbnl2ΔE2/ΔE2 mice using primers in Mbnl2 exons 1 and 3. Peptidylprolyl isomerase A (Ppia) was included as a loading control.
(C) Immunoblot showing that Mbnl2 protein, present in Mbnl2+/+ mice (wt), is absent in Mbnl2 knockouts (ko). Mbnl2 protein was detectable in wt quadriceps using longer film exposures. Mbnl1, which is abundantly expressed in lung and upregulated in Mbnl2 knockout brain, and Gapdh were used as family member and loading controls, respectively.
(D) Mbnl1 RNA targets are not mis-spliced in either Mbnl2 knockout skeletal (quadriceps) or heart muscles. Splicing patterns were determined using RT-PCR and Mbnl1+/+ (Mbnl1 wt), Mbnl1ΔE3/ΔE3 (Mbnl1 ko), Mbnl2+/+ (Mbnl2 wt) and Mbnl2ΔE2/ΔE2 (Mbnl2 ko) RNAs.
(E) Quantification of exon inclusion for alternative splicing of Clcn1, Serca1, Tnnt2 and Sorbs1 (Figure 1D) RNAs. Data are SEM (n=3) and only the differences between Mbnl1+/+ and Mbnl1ΔE3/ΔE3 were significant (*P<0.05).
Although Mbnl1 protein is expressed at similar levels in adult skeletal and heart muscle (Figure 1C), and throughout postnatal development (Ladd et al., 2005), Mbnl2 protein expression in adult muscle is low compared to other tissues (Figure 1C). This result is in agreement with prior studies showing that Mbnl2 protein expression is downregulated during myogenic differentiation of C2C12 myoblasts (Bland et al., 2010; Holt et al., 2009). Mbnl2 knockout adults were small at weaning (Figure 1A, postnatal day P21) but were normal in weight by P29 (Fig. S2A). Interestingly, DMSXL homozygous mice, which express a human DMPK transgene with a CTG1200-1700 expansion, are also small (Gomes-Pereira et al., 2007). Mbnl2 knockouts did not develop overt skeletal muscle pathology or motor deficits, as assayed by rotarod analysis, prior to 6 months of age (Fig. S2B). Although a few centralized nuclei were detectable in muscle cells of 3–5 month old Mbnl2 knockout mice (Figure S2C), myotonia was absent and the expression levels and spatial distribution of the muscle chloride channel Clcn1 was normal (Figure S2D). Moreover, exons controlled by Mbnl1 in skeletal muscle and heart showed normal splicing patterns in Mbnl2 knockout adult mice (Figures 1D and 1E). We conclude that if Mbnl2 functions as a splicing factor in skeletal muscle during the neonate to adult transition, its targets include few, if any, of the events known to be perturbed in DM muscle (Du et al., 2010), and that loss of Mbnl2 alone does not significantly contribute to DM-like skeletal muscle pathology. Since our studies were performed on Mbnl2 knockouts less than 7 months of age, we cannot exclude the possibility that loss of Mbnl2 expression contributes to pathology in aging skeletal muscle.
DM Disease Relevant Phenotypes in Mbnl2 Knockout Mice
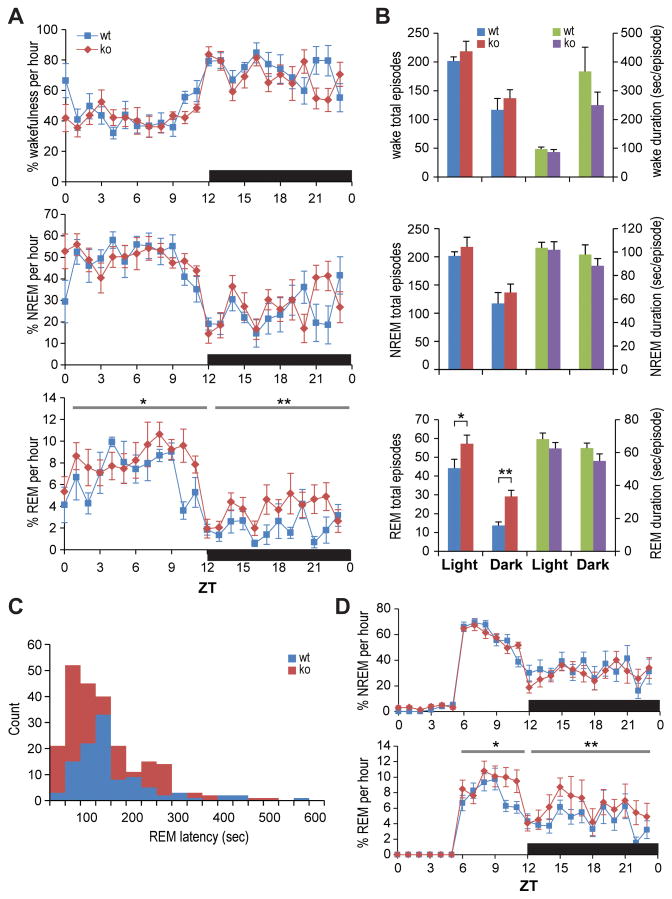
Hypersomnia, or excessive daytime sleepiness (EDS), and associated perturbations in rapid eye movement (REM) sleep patterns are among the most characteristic non-muscle features of DM (Ciafaloni et al., 2008; Yu et al., 2011). Because Mbnl2 protein was readily detected in multiple regions of the brain (Figure 1C), we performed sleep EEG/EMG evaluations on Mbnl2 knockout mice; only males were analyzed since estrous cycles affect sleep patterns. Mbnl2 knockouts had normal amounts, and natural diurnal distributions, of wakefulness and non-REM (NREM) sleep (Fig. 2A). Modest wake fragmentation during dark periods (frequent/shorter wake episodes) was also observed in the knockout mice (Fig. 2B). However, the most profound sleep phenotypes were an increase in REM sleep amounts, associated with increased numbers of REM sleep episodes (Figs. 2A and 2B) and increased EEG theta power (data not shown). This change was most notable during the dark period when mice are normally awake. Interestingly, a larger portion of these dark period REM sleep episodes in Mbnl2 knockouts exhibited a short latency (<100 sec) from the preceding wake episodes (Fig. 2C). The mean REM sleep latency of all observed episodes for the knockouts (115.8 ± 5.4 sec) was significantly shorter than that of wild-type mice (132.3 ± 8.7 sec). No direct transition from Wake to REM sleep, an EEG/EMG phenotype equivalent to behavioral cataplexy (Fujiki et al., 2009), was seen in either wild-type or knockout mice. A change in REM sleep in Mbnl2 knockouts was also observed during rebound sleep after a 6-hr sleep deprivation period initiated at ZT 0, where a more profound REM sleep rebound was observed in knockout, compared to wild-type, mice (Fig. 2D). These sleep changes were REM sleep specific as no changes in wake and NREM sleep was seen in these knockout mice at the baseline and during sleep rebound. Overall, these results indicate that Mbnl2 knockout mice exhibit increased REM sleep propensity and provide a valuable model to study the molecular basis of REM-associated sleep abnormalities in myotonic dystrophy.
Figure 2. Altered REM sleep regulation in Mbnl2 knockouts.
(A) Baseline wake (top), NREM (middle) and REM (bottom) sleep amount changes in Mbnl2 wild type (wt, blue) and knockout (ko, red) mice across 24 hr (n=8 each for Mbnl2 wt and ko mice; error bars represent SEM). The dark period is indicated by a black bar.
(B) Numbers of episodes (left) and mean episode duration (right four bars) of wake (top), NREM (middle) and REM (bottom) sleep.
(C) REM sleep latency for all REM sleep episodes in Mbnl2 wt and ko during the dark period. The mean REM sleep latency in ko mice (115.8 ± 5.4 [SEM] sec, n = 229) was significantly shorter than that of wt (132.3 ± 8.7 sec, n = 106), with p= 0.0287 by Mann-Whitney U test.
(D) Time course of NREM (top) and REM (bottom) sleep as percentage of time spent every hour during and after sleep deprivation for one day (P<0.05, by one-tailed (*) and two-tailed (**) Student’s t test).
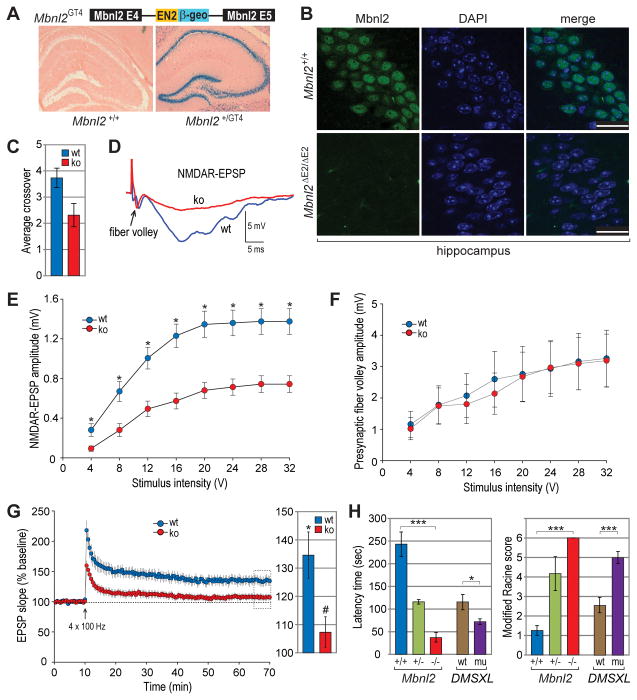
To address additional DM-linked phenotypes in Mbnl2 knockouts, we first mapped the spatial expression pattern of Mbnl2 in the brain using Mbnl2GT4 heterozygous mice and tagged allele-specific β-galactosidase expression. This analysis revealed Mbnl2 expression throughout the brain in both neurons and glia with prominent expression in neurons of the hippocampus, dentate gyrus and cerebellar Purkinje cells (Figure 3A and Figure S2E). Mbnl2 localized predominantly to the nucleus in these neuronal populations although in other regions of the brain, including the cerebral cortex, it was detectable in both the nucleus and cytoplasm (Figures 3B and S2F). Mbnl1 is primarily cytoplasmic in some neuronal populations, such as Purkinje cells (Daughters et al., 2009), but was detectable in hippocampal nuclei at a much lower level than Mbnl2 (Figure S2G). Since Mbnl2 was prominently expressed in the hippocampus, a key region of the brain involved in learning and memory, we next examined if Mbnl2 loss resulted in memory impairment as observed in DM patients. To test for spatial memory deficits, adult Mbnl2 knockout and wild-type mice were evaluated by the Morris water maze test, in which the mice are trained to locate a submerged platform using visual cues. Over the training period, both knockout and wild-type mice learned to locate the hidden platform but upon platform removal Mbnl2 knockouts crossed over the location of the platform significantly fewer times than wild-type mice, indicating a deficit in spatial reference memory (Figure 3C).
Figure 3. Learning/Memory Deficits, Abnormal Hippocampal Function and Seizure Susceptibility.
(A) Mbnl2 expression in the hippocampus and dentate gyrus. Sections were obtained from wild type (left panel) and Mbnl2GT4 (right) brains, which carry a gene trap in Mbnl2 intron 4 that expresses β-galactosidase (β-geo) and stained for β-gal activity (blue) and counterstained with eosin (pink).
(B) Immunohistochemistry of hippocampal sections from Mbnl2+/+ and Mbnl2ΔE2/ΔE2 mice using anti-Mbnl2 antibody. Nuclear DNA is indicated by DAPI (blue); scale bars = 25 μm.
(C) Spatial memory deficit by the Morris water maze test. Mbnl2+/+ (wt) and Mbnl2ΔE2/ΔE2 (ko) mice were evaluated for the number of times the missing platform area was crossed (average crossover). Data are SEM (n=10 for both experimental groups) and significant (P<0.05).
(D) NMDA receptor-mediated synaptic potentials are reduced in CA1 of the hippocampus in Mbnl2 knockouts. Representative traces of the NMDAR synaptic responses and fiber potential (arrowhead) obtained from Mbnl2+/+ (wt, blue) and Mbnl2ΔE2/ΔE2 (ko, red) male mice.
(E) Input-output curves for the mean NMDAR EPSP amplitude versus stimulus intensity. Mbnl2 male knockouts (red circles, n = 5/15 mice/slices) exhibited reduced NMDAR-mediated responses when compared to wild type (blue circles, n = 3/10 mice/slices). Stars indicate significant difference (*P<0.05).
(F) Mean presynaptic fiber volley amplitude versus stimulus intensity for Mbnl2 wild type (blue circles, n = 3/15 mice/slices) and knockout (red circles, n = 5/15 mice/slices) male mice.
(G) Time course changes in the field EPSP (left) obtained from hippocampal slices 10 min before, and 60 min after, stimulation to induce LTP for Mbnl2 male wild type (blue circles, n = 3/7 mice/slices) and knockouts (red circles, n = 5/10 mice/slices). The bar diagram (right) shows the average magnitude of LTP during the last 5 min of recording (dotted area in time course) from Mbnl2 wild type (blue) and knockouts (red). The * indicates a significant difference from baseline (100%) and # indicates difference between Mbnl2 wt and ko.
(H) Mbnl2 heterozygous (+/−, blue) and homozygous (−/−, green) knockout, as well as DMSXL polyCTG transgenic (mu, purple), mice are susceptible to seizures compared to wild type sibs (Mbnl2+/+, blue; DMSXL wt, brown) using a modified Racine scale: 0, no motor seizures; 1, freezing, staring, mouth or facial movements; 2, head nodding or isolated twitches and rigid posture; 3, tail extension, unilateral-bilateral forelimb clonus; 4, rearing, immobile state on rear haunches with one or both forelimbs extended; 5, clonic seizures with loss of posture, jumping, falling; 6, tonic seizures with hindlimb extension and death. Indicated data are for males (Mbnl2 n=4 for wt and ko, n=6 het; DMSXL n=10 each for wt and mut) and are significant (*P=0.05, ***P<0.001).
Since Mbnl2 knockouts exhibited impaired spatial memory on a hippocampal-dependent task, we performed electrophysiological recordings on hippocampal slices to evaluate the effects of Mbnl2 loss on NMDAR-mediated synaptic transmission and synaptic plasticity (long-term potentiation, LTP). Input-output curves of the NMDAR-mediated component of the field excitatory post-synaptic potentials (EPSP) indicated significant effects of stimulation intensity [F (7, 161) = 91.77, p <0.0001] exhibiting a decreased response in Mbnl2 knockouts when compared to wild type controls [F (1, 161) = 21.94, p <0.0001] (Figures 3D and 3E). The decrease in the NMDAR component of synaptic transmission was not due to a loss of synaptic input since the presynaptic fiber volley amplitude was similar across the two groups (Figure 3F). Furthermore, the input-output curves of the slope of the synaptic responses were not significantly different between Mbnl2 wild type and knockout mice (data not shown), indicating that the difference was specific for NMDAR function. For analysis of synaptic plasticity, LTP-inducing stimulation was delivered to the test pathway. Although pattern stimulation induced LTP in wild type mice compared to the non-tetanized path [F (1, 11) = 16.63, p <0.001], this LTP induction was not observed in 5/6 Mbnl2 knockout mice (wild type, 134.6 ± 8.09; Mbnl2 knockout, 104.9 ± 5.96) (Figure 3G). Thus, loss of Mbnl2 expression results in decreased synaptic NMDAR activity, impaired LTP and learning and memory deficits.
Another neurologic phenotype, which emerged with low penetrance (<10%) in Mbnl2 homozygous knockout males prior to weaning, was extreme hyperactivity followed by tonic-clonic seizures and death within 24 hr. To determine if Mbnl2 deficiency promoted seizure activity, we compared Mbnl2 wild-type with heterozygous and homozygous male knockouts for seizure susceptibility using the GABA antagonist pentylenetetrazole (PTZ). Following intraperitoneal injection of PTZ, mice were evaluated for seizure activity using a modified Racine scale and by measuring the time to the initial appearance of abnormal behavior (latency time). Remarkably, a low PTZ dose (40 mg/kg) was sufficient to generate enhanced seizure incidence, including tonic-clonic and clonic seizures, even in Mbnl2 heterozygous knockout mice (Figure 3H). Mbnl2 homozygous knockouts were more seizure-prone and generally died <5 min post-injection following a generalized tonic-clonic (GTC) seizure or were in a postictal state for >30 minutes while wild-type mice showed reduced activity following PTZ injection and then behaved normally and rarely developed seizures. The same Racine score of 6 was obtained for Mbnl2 knockout females although the mean latency time doubled (to ~100 sec). The reason for increased latency in females is unclear but sex-specific differences in brain function have been reported previously (Cosgrove et al., 2007).
Because epilepsy is not a common feature of DM, we examined the seizure-inducing effects of PTZ on DMSXL male mice, which express a human DMPK transgene carrying a CTG1200-1700 expansion (Gomes-Pereira et al., 2007). Importantly, enhanced seizure incidence was also observed in this DM1 mouse model (Figure 3H). Thus, either loss of Mbnl2 or expression of CUGexp RNAs in mice resulted in enhanced seizure susceptibility. Since Mbnl2 appeared to play a minor role in developmental splicing regulation in muscle while Mbnl2 knockout mice were affected by several neurologic abnormalities, we next tested whether Mbnl2 functions as an alternative splicing factor in the brain.
Widespread Alternative Splicing Changes in the Mbnl2 Knockout Brain
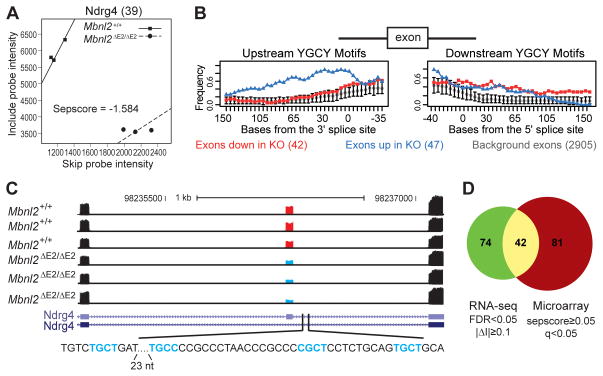
Because of high Mbnl2 expression and electrophysiological deficits in the hippocampus (Figures 1C and 3D–G), RNAs were extracted from hippocampi of Mbnl2 wild-type and knockout adults and analyzed for splicing changes using both splicing-sensitive microarrays (Du et al., 2010) and RNA-seq (Wang et al., 2008). For microarrays, alternative cassette splicing was assessed by separation score (sepscore) analysis, which measures the difference in the log2 ratio of exon skipping to inclusion in a mutant versus wild-type transcript (Table S1). RT-PCR validation rates are typically ~85% for splicing changes with |sepscore| ≥0.3 and q-value ≤0.05 (Du et al., 2010; Ni et al., 2007; Sugnet et al., 2006). Using those parameters, we identified 388 cassette exons whose splicing was significantly altered, and an additional 423 splicing changes in other splicing modes (e.g. retained introns, alternative 3’ss). One of the mis-regulated cassettes was in Ndrg4, a gene in the N-myc downregulated gene family whose expression is restricted to heart and neurons in the brain and, similar to Mbnl2 knockouts, Ndrg4−/− mice exhibit spatial memory deficits (Yamamoto et al., 2011). Microarray analysis identified a 39 nt Ndrg4 exon with enhanced skipping in Mbnl2 knockout mice (Figure 4A). Splicing-sensitive microarray analysis can predict binding motifs for splicing factors and earlier studies discovered that the preferred binding motif for Mbnl1 is YGCY (where Y is a pyrimidine) (Du et al., 2010; Goers et al., 2010). Using the top 42 exons which show enhanced skipping (sepscore ≤−0.6) and 47 with elevated inclusion (sepscore ≥0.6), we found prominent enrichment of the same motif upstream of exons whose inclusion increases in Mbnl2 knockout brain, compared with exons not significantly changed in the same dataset (Figure 4B).
Figure 4. Mbnl2 Regulates Alternative Splicing in the Brain.
(A) Sepscore analysis of splicing microarray results for Ndrg4 (39 nt alternative exon 14) in Mbnl2+/+ versus Mbnl2ΔE2/ΔE2 mice. The negative sepscore (−1.584) for Ndrg4 exon 14 (39 NT) predicts enhanced skipping in the Mbnl2 knockout.
(B) Bioinformatics analysis of splicing microarrays predicts that Mbnl2 recognizes the same binding motif as Mbnl1. Mapping of YGCY motifs upstream (left panel) and downstream (right panel) of exons that are preferentially skipped (n=42, red squares) or included (n=47, blue triangles) in Mbnl2 knockouts. The frequency of YGCY in 50 nucleotide windows shifted by 5 nucleotides are compared to a background set of 2905 exons whose inclusion does not change in the Mbnl2 knockout. Error bars mark the 95% confidence interval around the mean frequency.
(C) Wiggle tracks of tag coverage obtained from RNA-seq of the Ndrg4 locus flanking exon 14 shows enhanced skipping (three biological replicates each) in Mbnl2ΔE2/ΔE2 (bottom, blue cassette) versus Mbnl2+/+ (top, red cassette) mice. Putative Mbnl2 YGCY binding sites located downstream of the Ndrg4 exon 14 5’ss are also indicated (turquoise).
(D) Comparison of Mbnl2-dependent exons monitored by microarrays and RNA-seq. Among the 3,222 cassette exons analyzed by both platforms, 116 and 123 exons have high-confidence Mbnl2 dependent splicing, as determined by RNA-seq and splicing microarrays, respectively. Among these, 42 exons were included in the high-confidence set by both platforms (P<1.4×10−32, Fisher’s exact test).
As an alternative approach, we also examined alternative splicing using paired end (PE)-RNA-seq of Mbnl2 wild-type and knockout hippocampi (Figure 4C, Table S2). In total, we obtained 121 million 40 nt paired-end reads from three wild-type and three knockout animals, respectively, which were mapped to the mouse reference genome (mm9) or exon junctions (Table S2). We focused on a comprehensive database of ~13,000 cassette exons annotated based on mRNA/EST data, and identified 531 cassette exons with Mbnl2-dependent splicing (FDR ≤0.15, Fisher’s exact test followed by Benjamini correction). Among them, we defined a subset of 209 exons with FDR ≤0.05 and |ΔI| ≥0.1 as a high-confidence set (Table S2). As with splicing microarrays, one of the top candidates was Ndrg4 (Figure 4C).
The exons monitored on microarrays and those analyzed by RNA-seq were compared to evaluate the reliability of each approach. Among the 3,959 exons on the microarrays, 3,222 (81.4%) were also analyzed by our RNA-seq pipeline. In particular, among the 139 high-confidence Mbnl2-dependent exons defined by microarray analysis (|sepscore| ≥0.5 and q-value ≤0.05), 123 (88.5%) were also analyzed by RNA-seq. Conversely, 116 of the 209 (55.5%) high-confidence exons identified from RNA-seq analysis were also analyzed by microarrays. Of the 3,222 exons analyzed by both platforms, 42 exons were identified as high-confidence exons by both platforms (Fig. 4D). The overlap is highly significant (P<1.4×10−32), albeit imperfect, due to inherent platform differences and the relatively limited statistical power of each analysis. Nevertheless, these analyses allowed us to define a combined set of 306 (=139+209−42) Mbnl2-dependent cassette exons derived from 271 genes with high confidence in at least one of the platforms (Table S2). Finally, Gene Ontology (GO) analysis highlighted potential roles for Mbnl2 in neuronal differentiation and development, axon guidance, as well as synaptic functions (Table S2).
Mbnl2 Regulates Alternative Splicing During Postnatal Brain Development
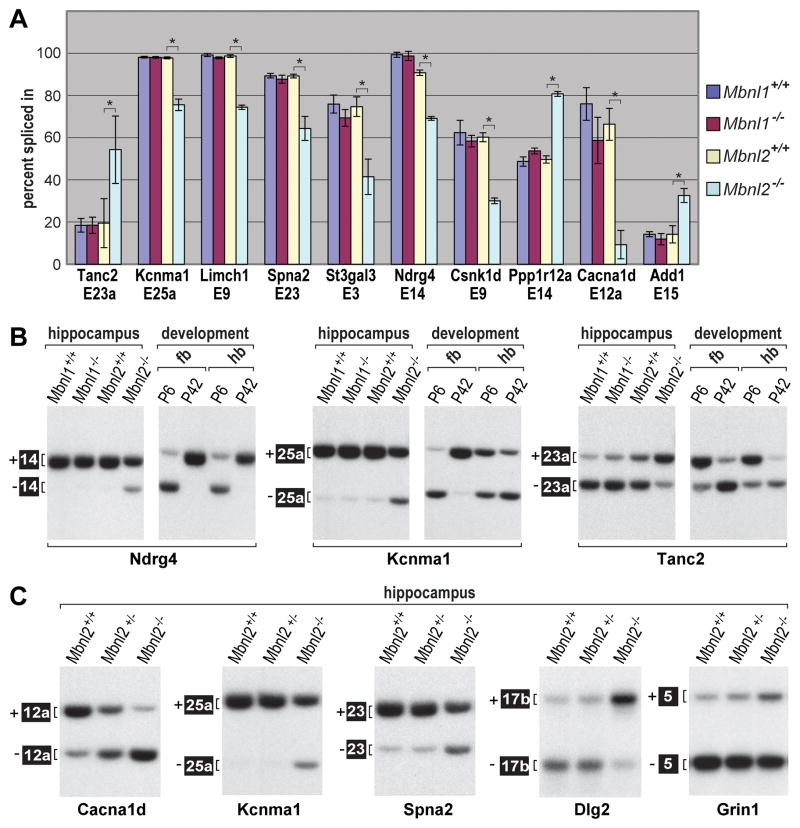
We next determined if these Mbnl2 RNA targets were developmentally regulated. A number of high-scoring splicing targets, as well as previously documented DM1 targets Grin1/Nmdar1 and Mapt, were examined for splicing of relevant exons in the hippocampus of Mbnl1 and Mbnl2 wild-type and knockout sibs by RT-PCR (Figure 5A and Figure S3A). Compared to wild-type sibs, all of these Mbnl2 target exons showed significant changes in alternative splicing in Mbnl2 knockouts. In contrast, the Mbnl2 target exons, except Ryr2, failed to show significant mis-splicing in Mbnl1 knockout brain confirming the reliability of the Mbnl2 targets identified through genome-wide analysis and a non-redundant role of Mbnl2 in CNS splicing regulation. These splicing patterns were then compared to those of the forebrain and hindbrain of P6 neonate and P42 adult WT mice (Figure 5B). Remarkably, the splicing of all the Mbnl2 targets that were examined shifted between P6 and P42 and Mbnl2 knockout adults retained the fetal-like splicing pattern. Furthermore, loss of Mbnl2 mis-regulated these exons by enhancement of fetal exon inclusion (e.g., Tanc2, Ppp1r12a, Add1) or adult exon skipping (e.g., Kcnma1, Csnk1d, Cacna1d). Some of these developmental splicing defects were regional (Figures 5B and S3B). For example, enhanced skipping of Ndrg4 exon 14 was observed throughout the P6 brain while the fetal pattern for Kcnma1 exon 25a was enhanced skipping in the forebrain but an increase in inclusion in the hindbrain (Figure 5B). These results demonstrate that Mbnl2 regulates a distinct set of exons to promote adult splicing patterns during postnatal brain development and this regulation varies in different regions of the brain.
Figure 5. Regulation of Adult Brain Splicing by Mbnl2.
(A) RT-PCR validation of Mbnl2 target exons identified by microarrays and RNA-seq using hippocampal RNAs. Quantification of alternative exon inclusion (percent spliced in) is shown for 10 genes and Mbnl1+/+ (blue), Mbnl1ΔE3/ΔE3 (purple), Mbnl2+/+(yellow) and Mbnl2ΔE2/ΔE2 (turquoise). Data are SEM (n=3) and only the differences between Mbnl2+/+ and Mbnl2ΔE2/ΔE2 were significant (*P<0.05).
(B) Loss of Mbnl2 leads to fetal exon splicing patterns. RT-PCR splicing analysis of select Mbnl2 target RNAs in Mbnl1+/+, Mbnl1ΔE3/ΔE3 (Mbnl1−/−), Mbnl2+/+ and Mbnl2ΔE2/ΔE2 (Mbnl2−/−) were compared to the splicing patterns of wild-type P6 and P42 forebrain (fb) and hindbrain (hb). For some exons (Tanc2 E23a) loss of Mbnl2 completely reproduces the fetal pattern while for others (Ndrg4, Kcnma1) a partial shift is observed.
(C) Splicing screen of epilepsy-associated genes in wild-type (Mbnl2+/+), Mbnl2+/ΔE2 and Mbnl2ΔE2/ΔE2 reveals dysregulation of Cacna1d exon 12a in heterozygous knockout hippocampus.
Since both Mbnl2 heterozygous and homozygous knockouts developed seizures upon PTZ induction, we selected genes/gene families from the splicing microarray or RNA-seq datasets (Tables S1 and S2) that had been previously linked to epilepsy (Klassen et al., 2011) to determine if any of these pre-mRNAs showed splicing dysregulation in heterozygous knockouts. Of eight genes assayed (Mbnl2 targets Tanc2 and Csnk1d were included as controls), two (Cacna1d, Ryr2) showed a significant switch to the fetal pattern in adult Mbnl2+/ΔE2 hippocampus. The Cacna1d (CaV1.3) voltage-gated L-type calcium channel subunit was the most profoundly affected (Figures 5C and S3C). Mis-splicing of these pre-mRNAs in Mbnl2 knockouts was particularly interesting since CUGexp RNAs have the greatest impact on the expression of genes involved in calcium signaling and homeostasis (Osborne et al., 2009).
The Mbnl2 Splicing Regulatory Map
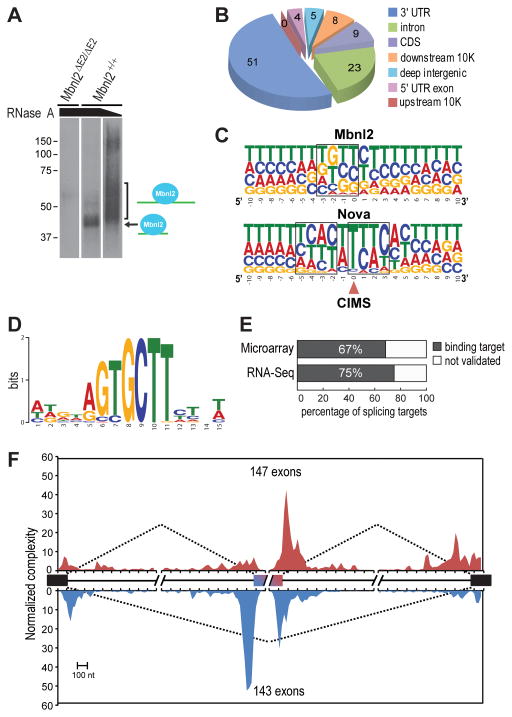
We next used HITS-CLIP to detect target RNAs containing direct binding sites for Mbnl2 in vivo. Following immunopurification of crosslinked RNA-protein complexes from mouse hippocampi, extensive RNase A digestion resulted in the appearance of a major band at ~42 kDa in wild-type that was absent in Mbnl2 knockouts (Figure 6A). At a lower RNase concentration, this distinct band was replaced with a more heterogeneous mixture of RNA-Mbnl2 complexes from which RNA was isolated and subsequently sequenced (Licatalosi et al., 2008).
Figure 6. HITS-CLIP and the Normalized Mbnl2 RNA Splicing Map.
(A) Protein gel autoradiograph of Mbnl2-RNA 32P-labeled complexes following crosslinking, RNase A digestion (high and low concentrations indicated by slanted black bar) and immunopurification with anti-Mbnl2.
(B) Pie chart of unique CLIP RNA tag distribution.
(C) Crosslinking-induced mutation site (CIMS) analysis of Mbnl2 CLIP data. Shown is the base composition of sequences around CIMS for Mbnl2 (top) and Nova (bottom).
(D) De novo motif search using sequences around CIMS (−10 to +10 nt) and MEME (Bailey and Elkan, 1994).
(E) The majority of alternatively spliced genes in Mbnl2ΔE2/ΔE2 knockouts are direct binding targets for Mbnl2.
(F) Normalized complexity Mbnl2 RNA splicing map. Mbnl2 activated (n=147, red) and repressed (n=143, blue) exons are shown.
Three CLIP libraries were prepared from independent biological replicates. Following quality filtering, genomic mapping and removal of potential PCR duplicates, we obtained a stringent set of 703,431 unique CLIP tags that represent independent protein-RNA interactions for further analysis (Table S2). Approximately half (51%) of the unique Mbnl2 CLIP tags were located on annotated 3′ UTRs, making Mbnl2 distinct from other splicing factors such as Nova and PTB, which primarily bind within introns (Figure 6B) (Licatalosi et al., 2008; Llorian et al., 2010; Xue et al., 2009). In addition, there was a substantial number of intron targets (23%), consistent with a role for Mbnl2 in splicing. To identify sites of robust Mbnl2-RNA interaction, we clustered overlapping CLIP tags, and conservatively determined 10,408 peaks, whose peak height is significantly above gene-specific background expected from uniform random distribution (P<0.01, Bonferroni correction; Table S2). These peaks were from 4,792 protein-coding genes, suggesting widespread Mbnl2-RNA interactions.
To determine the precise Mbnl2-RNA interaction sites and refine the Mbnl2 binding motif, we next performed crosslink-induced mutation site (CIMS) analysis to identify protein-RNA crosslink sites (Figure 6C and Table S2) (Zhang and Darnell, 2011). De novo motif analysis using 21-nt sequences around CIMS (−10 to +10 nt) highlighted YGCY (UGCU in particular) as a core element in all top motifs (Figure 6D). The UGCU elements showed a 16-fold enrichment at CIMS compared to flanking sequences (Figure S4A) and UGCU was the most enriched tetramer (Figure S4B). Deletions, specifically at YGCY elements, were found in sequences in or near Mbnl2 target cassette exons (Figure S5). Overall, these data demonstrate that Mbnl2, like Mbnl1, binds to YGCY elements in vivo to regulate splicing.
We next related direct Mbnl2 binding to Mbnl2-dependent splicing and refined the RNA-map of splicing regulation depending on positions of Mbnl2 binding sites. Analysis of the sequenced CLIP tags confirmed that the majority (67–75%) of the targets identified by both microarrays and RNA-seq (FDR <0.05) were direct binding targets of Mbnl2 in vivo (Figure 6E). Finally, we examined the distribution of CLIP tags in 290 (=123+209−42) high-confidence Mbnl2 target cassette exons defined from analysis of microarray or RNA-seq data and also annotated in our alternative splicing database. This set consisted of 147 Mbnl2-activated, and 143 Mbnl2-repressed, cassette exons. An RNA splicing map derived from this set of exons revealed that Mbnl2 binding upstream, within, or near the alternative exon 3′ splice site (3’ss) preferentially inhibited exon inclusion while Mbnl2 binding in the downstream intron, or near the alternative exon 5’ss, generally favored exon inclusion (Figure 6F). Binding of Mbnl2 ~60–70 nt downstream from the 5’ss of alternative exons tended to promote exon inclusion, whereas binding sites overlapping or immediately downstream of the 5’ss repressed exon inclusion.
Mbnl2 Knockouts Model Splicing Shifts in the Human DM1 Brain
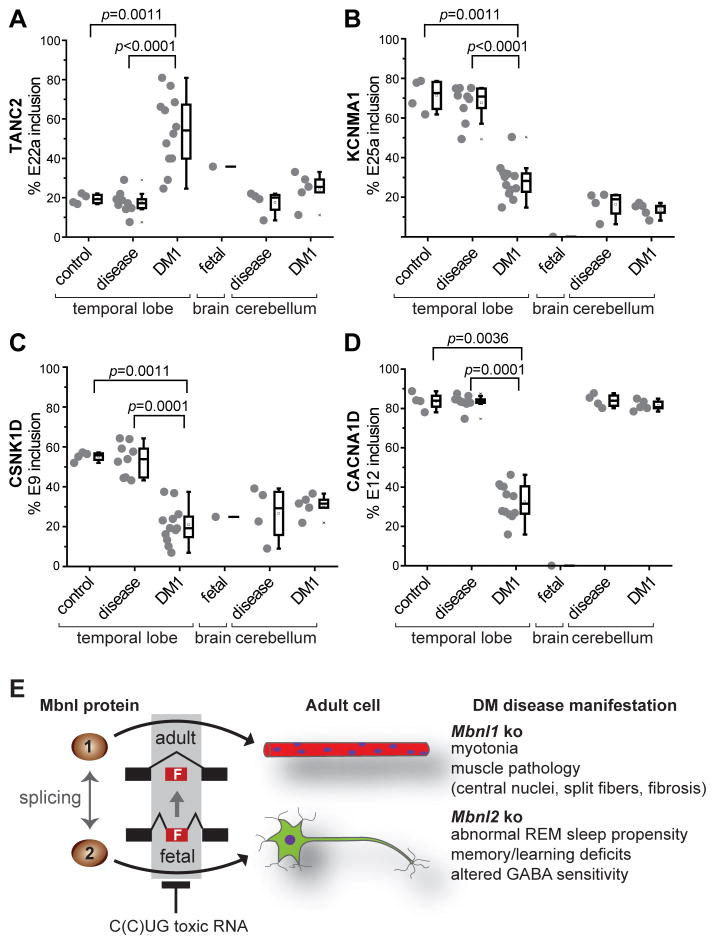
To ascertain if the target exons identified in Mbnl2 knockouts were similarly mis-regulated in the DM1 brain, autopsied human temporal cortex and cerebellar tissues were tested for mis-splicing of exons identified as mouse Mbnl2 targets. Of the 12 target exons examined, 10 were significantly mis-spliced in DM1 adult brain to a fetal pattern compared to normal and other disease controls (Figure 7A-D and Figure S6A). While there was a large variation in the degree of mis-splicing, the transcripts that were the most significantly different between normal and DM1, including CACNA1D, were similarly altered in Mbnl2 knockouts. By contrast, similar splicing trends were not found in the human cerebellum perhaps reflecting the shorter CTG expansion lengths observed in this brain region (Table S5 and Figure S6B) (Lopez Castel et al., 2011). Together, these results argue that Mbnl2 knockout mice provide a useful model for altered splicing in the DM brain.
Figure 7. MBNL2 Brain RNA Splicing Targets Are Dysregulated in DM1.
(A–D) Analysis of DM1 brain for splicing perturbations predicted from the Mbnl2 mouse knockout model. RT-PCR splicing analysis of TANC2, KCNMA1, CSNK1D and CACNA1D using RNAs isolated from normal control temporal cortex (control, n = 4), disease control temporal cortex (disease, n = 9), DM1 temporal cortex (DM1, n=12), fetal control whole brain (fetal, n=1), disease control cerebellum (disease, n=4) and DM1 cerebellum (DM1, n=5) by RT-PCR.
(E) MBNL loss-of-function model for myotonic dystrophy. MBNL1 and MBNL2 function as alternative splicing factors during postnatal development of muscle (red myofibers with blue myonuclei) and brain (green neuron), respectively. Note that MBNL1 and MBNL2 cross-regulate the alternative splicing of key MBNL exons, including a 54 nt exon (MBNL1 exon 7, MBNL2 exon 6) which includes a nuclear localization sequence (Figure S7), so loss of MBNL2 could lead to an increase in nuclear levels MBNL1 and partial restoration of adult splicing patterns since both proteins recognize a YGCY motif.
DISCUSSION
RNA-mediated pathogenesis is emerging as an important disease mechanism in unstable microsatellite disorders (Poulos et al., 2011). The most recent example is a potential role for rGGGGCC repeats in autosomal dominant FTD/ALS (Dejesus-Hernandez et al., 2011; Renton et al., 2011). While the toxic RNA model for DM is supported by considerable experimental evidence, recent studies have suggested that other factors might contribute to disease phenotypes (Sicot et al., 2011; Zu et al., 2011). Thus, it is important to discriminate between the relative effects of toxic RNAs and proteins in unstable microsatellite diseases. The MBNL loss-of-function model for DM allows this distinction because specific disease manifestations, such as myotonia, are replicated in mouse models in the absence of microsatellite expansions (Poulos et al., 2011).
Mbnl2 Controls Splicing of Numerous Alternative Exons during Brain Development
Myotonic dystrophy is classified as a muscular dystrophy but the development and maintenance of normal brain function is also profoundly affected in this disease. Although DM symptoms are proposed to result from dysregulation of alternative splicing, the extent of mis-splicing induced by C(C)UGexp RNAs has been unclear particularly since previous studies have reported only a few mis-splicing events in the DM1 CNS (Jiang et al., 2004; Sergeant et al., 2001). Here, we tested the hypothesis that MBNL2 is an important splicing regulator during brain development and this function is compromised in DM. We generated Mbnl2 knockout mice and discovered that in contrast to Mbnl1, Mbnl2 is not an essential alternative splicing factor during skeletal muscle development. However, Mbnl2 may play a compensatory role when Mbnl1 expression is compromised. The discordance between our results on the effect of Mbnl2 loss on skeletal muscle and a previous report using Mbnl2 gene traps may be attributable to differences in knockout strategy and the fact that prior studies did not evaluate alternative splicing in the CNS (Hao et al., 2008; Lin et al., 2006).
While Mbnl2 knockout mice did not display pronounced muscle pathology, loss of Mbnl2 resulted in widespread splicing abnormalities in the brain. During this study, we uncovered a remarkable similarity between the control of alternative splicing during postnatal development by Mbnl1 in skeletal muscle and Mbnl2 in the brain. Both factors promote adult isoform expression and, similar to Mbnl1, Mbnl2 regulates the developmental splicing of hundreds of alternative cassette exons via the recognition of a YGCY motif in a manner reminiscent of the Nova, Rbfox and PTBP splicing factor families although the binding motifs for these factors are quite different (Du et al., 2010; Li et al., 2007; Licatalosi and Darnell, 2010; Licatalosi et al., 2008; Witten and Ule, 2011; Zhang et al., 2008). Thus, Mbnl2 joins a growing list of alternative splicing factors that control specific pathways in the brain either during development and/or during adult life. In addition to splicing, our HITS-CLIP analysis revealed that about half of Mbnl2 targets are located in annotated 3′ UTRs. Since microarray and RNA-seq analyses did not detect major changes in transcript levels, Mbnl2 may play important roles in RNA localization and/or translation and these pathways could also be affected in the DM brain.
Mbnl2 Knockouts: Model for CNS Disease in Myotonic Dystrophy
Mbnl2 knockout mice show several phenotypes consistent with abnormalities observed in myotonic dystrophy. For example, excessive daytime sleepiness (EDS) is a common and disabling feature of DM type 1 (DM1) (Ciafaloni et al., 2008; Pincherle et al., 2012; Yu et al., 2011). However, the molecular basis of this sleep disturbance is unknown. In some cases with advanced disease, EDS may result from obstructive sleep apnea (Pincherle et al., 2012). DM1 may also have direct effects on sleep regulatory circuits in the CNS and REM sleep changes in patients, including an increase in daytime and nighttime REM sleep propensity and higher frequency of sleep onset REM period(s) (SOREMS) and REM density, have been reported (Bennett et al., 2007; Ciafaloni et al., 2008; Pincherle et al., 2012). Here, we demonstrate related REM sleep changes in Mbnl2 knockout mice, including increased REM sleep amounts and episode numbers. These changes were observed over 24 hours, but were more profound during the active, or dark, period. Mbnl2 knockouts had twice as many REM sleep episodes compared to wild type mice, and a large portion of these episodes had short latencies from the proceeding wake episodes. Profound REM sleep rebound was also seen in Mbnl2 knockout mice after sleep deprivation and, in contrast, there was no apparent changes in wake and NREM sleep parameters in these mutants. Our results indicate that Mbnl2 knockout mice will be useful to study DM1-associated splicing alternations that impact sleep regulatory mechanisms.
Additional phenotypes characteristic of DM include mental retardation in congenital DM1 while childhood through adult onset disease is associated with learning disabilities, autistic behavior, impaired cognitive function, cerebral structural changes and nonverbal episodic memory impairment (Meola and Sansone, 2007; Weber et al., 2010). Interestingly, Mbnl2 knockout mice exhibit impaired learning on a hippocampal-dependent task, a decrease in NMDAR-mediated synaptic transmission, and an impairment of hippocampal synaptic plasticity. Several of the mis-regulated splicing events identified during this study might contribute to these impairments, including Cacna1d (McKinney et al., 2009), Tanc2 (Han et al., 2010), Ndrg4 (Yamamoto et al., 2011) and Grin1 (Shimizu et al., 2000). For example, DM1 patients exhibit increased expression of a splice variant of GRIN1 that includes exon 5 (Jiang et al., 2004), which is thought to contribute to the age-related decline in frontotemporal functions, including memory (Modoni et al., 2008; Romeo et al., 2010; Weber et al., 2010). Removal of exon 5 targets GRIN1 to dendrites (Pal et al., 2003), suggesting that exon 5 inclusion results in decreased dendritic localization. Moreover, Tanc proteins interact with PSD-95, a protein involved in localizing NMDARs at the synapse (Han et al., 2010). Thus, a decrease in dendritic GRIN1 or altered regulation of NMDARs could underlie the decrease in the synaptic NMDAR response, impaired LTP and learning and memory deficits.
While there have been sporadic reports of epilepsy associated with DM, this is not a characteristic overt feature of this disease (Meola and Sansone, 2007). However, DM patients show enhanced sensitivity to barbituates and benzodiazepines which enhance the activity of the GABAA receptor (Harper, 2001). Moreover, the relevance of this hyperexcitability phenotype to DM1 is also supported by our observation that seizures were induced with a GABA antagonist in both Mbnl2 knockouts and the DMSXL transgenic model for DM1. Because the expression of a large CTG expansion in the DMSXL brain is sufficient to increase seizure susceptibility in mice, this study provides the cautionary note that DM may possess some features of an excitability disorder. Finally, we also noted a difference in the latency time to the appearance of the seizure phenotype between males and females which could reflect sex-specific differences in the alternative splicing of seizure-associated genes.
Based on these and additional findings, we propose a modified MBNL combinatorial loss-of function model for DM. MBNL proteins function as alternative splicing factors during postnatal development with MBNL2 the predominant factor in the brain while MBNL1 serves a similar role in skeletal muscle (Figure 7E). Thus, we anticipate that major pathological changes in the DM brain are attributable to toxic RNA expression, MBNL2 sequestration by C(C)UGexp RNAs and dysregulation of specific alternative splicing events required for normal adult CNS function.
EXPERIMENTAL PROCEDURES
Mbnl2 Knockout Mice
The Mbnl2 targeting vector was derived from CHORI clone bMQ-63F6. A 10 kb fragment containing Mbnl2 was subcloned into PL253 (Protocols 1–4, http://web.ncifcrf.gov/research/brb/protocol.aspx). The targeting construct (Figure S1B, Table S3 for PCR primers) was linearized with NotI and electroporated in 129 SvlmJ ES cells followed by selection as described (Kanadia et al., 2003). For constitutive Mbnl2 knockouts, Mbnl2+/con mice were mated to B6.C-Tg(CMV-cre)1Cgn/J mice (JAX). All animal procedures were approved by the University of Florida IACUC. Mouse tissue protein and RNA analyses (Kanadia et al., 2006), immunohistochemistry and X-Gal staining (Emamian et al., 2003), and rotarod analysis (Daughters et al., 2009) were performed as described previously with some modifications (see Supplemental Information).
Sleep/REM Analysis
Implant surgery, EEG/EMG monitoring and EEG data acquisition were performed on 6 month old mice (n=8 for each genotype) as described (Fujiki et al., 2009). Wakefulness was determined by low amplitude and mixed frequency (>4 Hz) EEG with continuous large fluctuation in EMG, and non-REM (NREM) sleep was determined by high amplitude and low frequency (0.5–4 Hz) EEG with no fluctuation in EMG. REM sleep was determined by low amplitude and high frequency EEG (similar to wake stage, but with rhythmic theta waves at 7–9 Hz) with low amplitude EMG. REM theta EEG power was analyzed with fast Fourier transform (FFT) for band frequencies between 4–9 Hz. Sleep deprivation was initiated from ZT 0 for 6 hr with gentle handling.
Morris Water Maze, Electrophysiology and Seizure Susceptibility
Mice were tested for spatial learning/memory using a Morris water maze as described with several modifications (Han et al., 2010). Mice were allowed to swim (1 min) during the training period (4 trials/day for 5 days) and then allowed to rest on the platform. During the examination day, mice were randomly placed in the three non-target quadrants and allowed to swim for 1 min. For electrophysiology, hippocampal slices (~400 μm) were processed and recordings obtained as described (Foster et al., 2008) (see Supplemental Information). Mice (2–5 months) were also tested for seizure susceptibility following injection (40 mg/kg) with pentylenetetrazol. Following injection, the mice were placed in an observational area for 60 min and the time of onset of convulsive behavior and nature/severity of the convulsion were scored according to a modified Racine scale (Luttjohann et al., 2009).
Splicing Microarrays, Paired-end RNA Sequencing and HITS-CLIP
For splicing microarrays and RNA-seq, hippocampal RNAs were obtained from Mbnl2+/+ and Mbnl2ΔE2/ΔE2 mice (2–3 months, n=3 each). Splicing microarray analysis was performed as described (Du et al., 2010) with modifications (see Supplemental Information). For RNA-seq, RNAs were purified and sequencing libraries were constructed using the mRNA-Seq 8-Sample Prep Kit according to the manufacturer’s protocol (Illumina). Libraries were sequenced (40 cycles, both ends) using an Illumina Genome Analyzer IIx. Raw sequence reads were mapped back to the mouse reference genome together with a database of annotated exon junctions compiled from mouse, human, and rat mRNA/EST data. CLIP was performed as reported (Jensen and Darnell, 2008) with modifications (see Supplemental Information).
Human RNA and Splicing Analysis
Temporal cortex and cerebellar autopsy tissues (12 DM1 patients, 9 disease controls) were analyzed (Table S5). This research was approved by the Institutional Ethics Committee and written informed consent for specimen research use was obtained from all patients. RNA was extracted using the ISOGEN procedure (Nippon Gene) and cDNA was synthesized using 1–3 μg of RNA. Random hexamers and cDNA equivalent to 20 ng RNA was PCR-amplified for initial denaturation at 94°C for 10 min and 35 cycles (94°C for 30 s, 55°C for 30 s, 72°C for 30 s) (Table S6). PCR products were analyzed by capillary electrophoresis (Hitachi Electronics). The percentage of each peak was obtained by dividing each signal by the total signal and statistical analysis was performed using the Mann-Whitney U test.
Supplementary Material
Acknowledgments
The authors thank C. Thornton for Mbnl2GT4 mice, L. Ranum for manuscript comments and the Research Resource Network Japan for human brain samples. This work was supported by grants from the NIH (NS058901 to M.S.S; K99GM95713 to C.Z.; NS34389 to R.B.D.; GM084317 to M.A.; AG014979, AG037984, AG036800 to T.C.F.), the McKnight Brain Research Foundation (T.C.F.), the MDA (4280 to M.S.S., 135140 to M.A.), the NCNP Japan (22-7 to K.J.) and the Japanese Ministry of Health, Labour and Welfare (22-118 to T.K.).
Footnotes
ACCESSION NUMBERS
Microarray and RNA-seq data have been deposited in GEO under accession numbers GSE37908 and XXXXX, respectively.
Supplemental information, including supplemental experimental procedures, seven figures and six tables, can be found online at:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- Bland CS, Wang ET, Vu A, David MP, Castle JC, Johnson JM, Burge CB, Cooper TA. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 2010;38:7651–7664. doi: 10.1093/nar/gkq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafaloni E, Mignot E, Sansone V, Hilbert JE, Lin L, Lin X, Liu LC, Pigeon WR, Perlis ML, Thornton CA. The hypocretin neurotransmission system in myotonic dystrophy type 1. Neurology. 2008;70:226–230. doi: 10.1212/01.wnl.0000296827.20167.98. [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove KP, Mazure CM, Staley JK. Evolving knowledge of sex differences in brain structure, function, and chemistry. Biol Psych. 2007;62:847–855. doi: 10.1016/j.biopsych.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES, Kaytor MD, Duvick LA, Zu T, Tousey SK, Zoghbi HY, Clark HB, Orr HT. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- Foster TC, Rani A, Kumar A, Cui L, Semple-Rowland SL. Viral vector-mediated delivery of estrogen receptor-alpha to the hippocampus improves spatial learning in estrogen receptor-alpha knockout mice. Mol Ther. 2008;16:1587–1593. doi: 10.1038/mt.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki N, Cheng T, Yoshino F, Nishino S. Specificity of direct transition from wake to REM sleep in orexin/ataxin-3 transgenic narcoleptic mice. Exp Neurol. 2009;217:46–54. doi: 10.1016/j.expneurol.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goers ES, Purcell J, Voelker RB, Gates DP, Berglund JA. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010;38:2467–2484. doi: 10.1093/nar/gkp1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes-Pereira M, Foiry L, Nicole A, Huguet A, Junien C, Munnich A, Gourdon G. CTG trinucleotide repeat “big jumps”: large expansions, small mice. PLoS Genet. 2007;3:e52. doi: 10.1371/journal.pgen.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Nam J, Li Y, Kim S, Cho SH, Cho YS, Choi SY, Choi J, Han K, Kim Y, et al. Regulation of dendritic spines, spatial memory, and embryonic development by the TANC family of PSD-95-interacting proteins. J Neurosci. 2010;30:15102–15112. doi: 10.1523/JNEUROSCI.3128-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao M, Akrami K, Wei K, De Diego C, Che N, Ku JH, Tidball J, Graves MC, Shieh PB, Chen F. Muscleblind-like 2 (Mbnl2) -deficient mice as a model for myotonic dystrophy. Dev Dyn. 2008;237:403–410. doi: 10.1002/dvdy.21428. [DOI] [PubMed] [Google Scholar]
- Harper PS. Myotonic Dystrophy. 3. London: WB Saunders; 2001. [Google Scholar]
- Holt I, Jacquemin V, Fardaei M, Sewry CA, Butler-Browne GS, Furling D, Brook JD, Morris GE. Muscleblind-like proteins: similarities and differences in normal and myotonic dystrophy muscle. Am J Pathol. 2009;174:216–227. doi: 10.2353/ajpath.2009.080520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet. 2011;12:715–729. doi: 10.1038/nrg3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, Swanson MS. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D, McPherson J, Bourquin T, Lewis L, Villasana D, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145:1036–1048. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshelev M, Sarma S, Price RE, Wehrens XH, Cooper TA. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, Swanson MS, Thornton CA. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Castel A, Nakamori M, Tome S, Chitayat D, Gourdon G, Thornton CA, Pearson CE. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum Mol Genet. 2011;20:1–15. doi: 10.1093/hmg/ddq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttjohann A, Fabene PF, van Luijtelaar G. A revised Racine’s scale for PTZ-induced seizures in rats. Physiol Behav. 2009;98:579–586. doi: 10.1016/j.physbeh.2009.09.005. [DOI] [PubMed] [Google Scholar]
- McKinney BC, Sze W, Lee B, Murphy GG. Impaired long-term potentiation and enhanced neuronal excitability in the amygdala of Ca(V)1.3 knockout mice. Neurobiol Learn Mem. 2009;92:519–528. doi: 10.1016/j.nlm.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola G, Sansone V. Cerebral involvement in myotonic dystrophies. Muscle Nerve. 2007;36:294–306. doi: 10.1002/mus.20800. [DOI] [PubMed] [Google Scholar]
- Modoni A, Silvestri G, Vita MG, Quaranta D, Tonali PA, Marra C. Cognitive impairment in myotonic dystrophy type 1 (DM1): a longitudinal follow-up study. J Neurol. 2008;255:1737–1742. doi: 10.1007/s00415-008-0017-5. [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne RJ, Lin X, Welle S, Sobczak K, O’Rourke JR, Swanson MS, Thornton CA. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal R, Agbas A, Bao X, Hui D, Leary C, Hunt J, Naniwadekar A, Michaelis ML, Kumar KN, Michaelis EK. Selective dendrite-targeting of mRNAs of NR1 splice variants without exon 5: identification of a cis-acting sequence and isolation of sequence-binding proteins. Brain Res. 2003;994:1–18. doi: 10.1016/j.brainres.2003.08.046. [DOI] [PubMed] [Google Scholar]
- Pincherle A, Patruno V, Raimondi P, Moretti S, Dominese A, Martinelli-Boneschi F, Pasanisi MB, Canioni E, Salerno F, Deleo F, et al. Sleep breathing disorders in 40 Italian patients with myotonic dystrophy type 1. Neuromuscul Disord. 2012;22:219–224. doi: 10.1016/j.nmd.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Poulos MG, Batra R, Charizanis K, Swanson MS. Developments in RNA splicing and disease. Cold Spring Harb Perspect Biol. 2011;3:a000778. doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo V, Pegoraro E, Ferrati C, Squarzanti F, Soraru G, Palmieri A, Zucchetta P, Antunovic L, Bonifazi E, Novelli G, et al. Brain involvement in myotonic dystrophies: neuroimaging and neuropsychological comparative study in DM1 and DM2. J Neurol. 2010;257:1246–1255. doi: 10.1007/s00415-010-5498-3. [DOI] [PubMed] [Google Scholar]
- Sergeant N, Sablonniere B, Schraen-Maschke S, Ghestem A, Maurage CA, Wattez A, Vermersch P, Delacourte A. Dysregulation of human brain microtubule-associated tau mRNA maturation in myotonic dystrophy type 1. Hum Mol Genet. 2001;10:2143–2155. doi: 10.1093/hmg/10.19.2143. [DOI] [PubMed] [Google Scholar]
- Shimizu E, Tang YP, Rampon C, Tsien JZ. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science. 2000;290:1170–1174. doi: 10.1126/science.290.5494.1170. [DOI] [PubMed] [Google Scholar]
- Sicot G, Gourdon G, Gomes-Pereira M. Myotonic dystrophy, when simple repeats reveal complex pathogenic entities: new findings and future challenges. Hum Mol Genet. 2011;20:R116–123. doi: 10.1093/hmg/ddr343. [DOI] [PubMed] [Google Scholar]
- Suenaga K, Lee KY, Nakamori M, Tatsumi Y, Takahashi MP, Fujimura H, Jinnai K, Yoshikawa H, Du H, Ares M, Jr, et al. Muscleblind-like 1 knockout mice reveal novel splicing defects in the myotonic dystrophy brain. PLoS One. 2012;7:e33218. doi: 10.1371/journal.pone.0033218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber YG, Roebling R, Kassubek J, Hoffmann S, Rosenbohm A, Wolf M, Steinbach P, Jurkat-Rott K, Walter H, Reske SN, et al. Comparative analysis of brain structure, metabolism, and cognition in myotonic dystrophy 1 and 2. Neurology. 2010;74:1108–1117. doi: 10.1212/WNL.0b013e3181d8c35f. [DOI] [PubMed] [Google Scholar]
- Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Kokame K, Okuda T, Nakajo Y, Yanamoto H, Miyata T. NDRG4 Protein-deficient Mice Exhibit Spatial Learning Deficits and Vulnerabilities to Cerebral Ischemia. J Biol Chem. 2011;286:26158–26165. doi: 10.1074/jbc.M111.256446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Laberge L, Jaussent I, Bayard S, Scholtz S, Raoul M, Pages M, Dauvilliers Y. Daytime sleepiness and REM sleep characteristics in myotonic dystrophy: a case-control study. Sleep. 2011;34:165–170. doi: 10.1093/sleep/34.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.