Abstract
The switch from vegetative to reproductive development in plants necessitates a switch in the developmental program of the descendents of the stem cells in the shoot apical meristem. Genetic and molecular investigations have demonstrated that the plant-specific transcription factor and meristem identity regulator LEAFY (LFY) controls this developmental transition by inducing expression of a second transcription factor, APETALA1, and by regulating the expression of additional, as yet unknown, genes. Here we show that the additional LFY targets include the APETALA1-related factor, CAULI-FLOWER, as well as three transcription factors and two putative signal transduction pathway components. These genes are up-regulated by LFY even when protein synthesis is inhibited and, hence, appear to be direct targets of LFY. Supporting this conclusion, cis-regulatory regions upstream of these genes are bound by LFY in vivo. The newly identified LFY targets likely initiate the transcriptional changes that are required for the switch from vegetative to reproductive development in Arabidopsis.
The plant-specific LEAFY (LFY) protein is necessary and sufficient for the vital switch from vegetative to reproductive development in dicotyledonous plant species (1-9). LFY controls the production of the flowers, which are formed in lieu of secondary inflorescences from the flanks of the shoot apical meristem. Because LFY is required for all of the major features that differentiate flowers from inflorescence branches, it is referred to as a meristem identity gene.
After initiating the meristem identity switch, LFY has a second role in the activation of the floral homeotic genes that specify the identity of organs in the flower (10). The two roles of LFY are separable genetically and molecularly (11, 12). LFY exerts its developmental effects by means of transcriptional regulation; LFY has been shown to be a transcription factor in vivo (13, 14).
Despite the critical importance of this regulator, only three direct targets of LFY have been identified (13-15). Two of the targets, AGAMOUS and APETALA3, are floral homeotic genes that act directly downstream of LFY in flower morphogenesis. Only one known direct LFY target gene product, APETALA1 (AP1), acts in the meristem identity pathway (13).
The ap1-1 mutation partly suppresses the LFY gain-of-function phenotype (9, 16), indicating that AP1 acts downstream of LFY in the floral transition, which culminates in flower formation. Posttranslational activation of a biologically active fusion protein between LFY and the rat glucocorticoid receptor (GR) hormone-binding domain demonstrates that LFY directly activates AP1 in the anlagen of the flower primordia in a protein synthesis-independent fashion (13). In addition, LFY binds to cis-regulatory elements that control AP1 expression (11). Thus, LFY regulates the transition to flower development, at least in part, by inducing AP1 expression in regions of the shoot apical meristem that give rise to flower primordia.
LFY null mutations cause striking defects in the transition to reproductive development; lfy-6 plants, for example, produce a large number of secondary inflorescences until very late in development when defective flowers are formed (1-3). In contrast, the strongest available AP1 mutation has a much weaker effect on inflorescence morphology, implying that other LFY target genes must exist in the meristem identity pathway.
AP1 acts as a floral homeotic gene in addition to its role as a meristem identity regulator (17, 18). The latter function does not depend on LFY, however, because lfy null mutants show strong AP1 expression in flowers from stage three onward (11, 13, 16, 19). The LFY-independent induction of AP1 expression makes it difficult to detect LFY-dependent induction of AP1 at the floral transition by using quantitative methods in entire inflorescences (13). To identify the additional unknown LFY targets that act at the transition to reproductive development together with AP1, it is, therefore, important to first identify a developmental stage at which quantitative induction of AP1 can be observed in response to LFY.
Here we report that quantitative up-regulation of AP1 can be readily observed in 9-day-old seedlings. Using posttranslational activation of LFY-GR, we demonstrate that the closest AP1 homolog, CAULIFLOWER (CAL), is a direct LFY target at this stage in development and that cis-regulatory elements in the putative CAL promoter are bound by LFY. We used microarray analysis to identify direct targets of LFY and chromatin immunoprecipitation (ChIP) to demonstrate in vivo LFY binding to the putative promoter regions of these genes. The predicted function of the proteins encoded by these genes is consistent with their role in the meristem identity switch.
Methods
Plant Growth and Steroid Treatments. Seedlings were germinated on half-strength Murashige and Skoog medium after 7 days of cold treatment at 4°C. Growth was at 21°C in continuous light at a fluence rate of 50 μmol/m2·sec. Steroid treatments were performed essentially as described (see ref. 13 and Supporting Text, which is published as supporting information on the PNAS web site).
RT-PCR. Frozen seedling tissue was ground five times for 10 sec with 1.0-mm glass beads (Biospec Products, Bartlesville, OK) by using a Mini-BeadBeater (Biospec Products), followed by extraction with TRI (total RNA-isolating) reagent (Molecular Research Center) according to the manufacturer's instructions. The resulting RNA was further purified by using RNeasy columns (Qiagen, Valencia, CA). For reverse transcription, 2 μg of the purified RNA was used with the Thermoscript kit (Invitrogen) according to the manufacturer's instructions. We used 0.8 μl of the reverse transcription reaction in a 25-μl PCR with Platinum TAQ (Invitrogen), except for CAL and AT5g60630, for which we used 2 μl of the reverse transcription reaction. PCR conditions are detailed in Supporting Text.
Microarrays. RNA was extracted and purified as described above. Total RNA input was 5 μg. Probe synthesis was performed as described in the GENECHIP Expression Analysis Technical Manual (www.affymetrix.com; Affymetrix, Santa Clara, CA). After first- and second-strand cDNA synthesis, all reactions were tested by PCR for induction and equal template presence by using AP1- and EIF4-specific primers. After fragmentation, 15 μg of cRNA was used for hybridization. Microarray hybridization, data acquisition, and first-pass analysis were performed as described (www.med.upenn.edu/microarr) by the Penn Microarray Facility (University of Pennsylvania, Philadelphia).
Fold up-regulation required for each condition was chosen arbitrarily. Variance per gene was generally less than the randomly chosen cutoffs, allowing for their use as a criterion for prioritizing candidate genes for independent confirmation. Moreover, all putative candidate genes exhibit very low variance within treatment comparisons for all six treatments/genotypes; 96% of identified LFY target genes fall within the arbitrary boundaries chosen for all six comparisons.
ChIP. Seedlings (9-day-old) grown on half-strength Murashige and Skoog plates were treated with dexamethasone or mock solution, as described above, washed in PBS, and crosslinked with 1% formaldehyde for 10 min by using vacuum infiltration. The crosslinking reaction was stopped by addition of 0.1 M glycine. Nuclear extracts were prepared according to ref. 21, lysed as described by the Farnham laboratory (http://mcardle.oncology.wisc.edu/farnham/protocols/chips.html), and diluted 10-fold in lysis dilution buffer (Farnham protocol). Extracts were sonicated in the presence of glass beads (Biospec Products) to achieve a DNA size range between 50 and 1,600 bp with an average of 600 bp for ChIP experiment 1 (ChIP1) and a size distribution from 100 to 3,000 bp with an average of 900 bp in a ChIP experiment 2 (ChIP2). We removed 1/25 of the sample as input. Affinity-purified LFY antiserum (13) was bound to 40 μl of protein A magnetic beads (Dynal Biotech, Lake Success, NY) that had been pretreated with 0.5% BSA in PBS. For ChIP2, twice as much antiserum prebound to beads was used as for ChIP1. After antibody binding (90 min at 4°C), beads were washed twice with PBS/BSA and incubated with the remainder of the nuclear extract overnight at 4°C on a rotating wheel. The immunoprecipitated extracts were washed as described (Farnham protocol), except that 250 mM LiCl was used in the immunoprecipitate wash buffer. Elution, crosslink reversal, and DNA work-up were performed for the input and ChIP samples in parallel according to the location analysis protocol available as supporting information for ref. 22 (http://web.wi.mit.edu/young/origins), except that elution was performed twice and the PCR purification step (Qiagen) was omitted. ChIP PCR was performed as described in Supporting Text.
Results
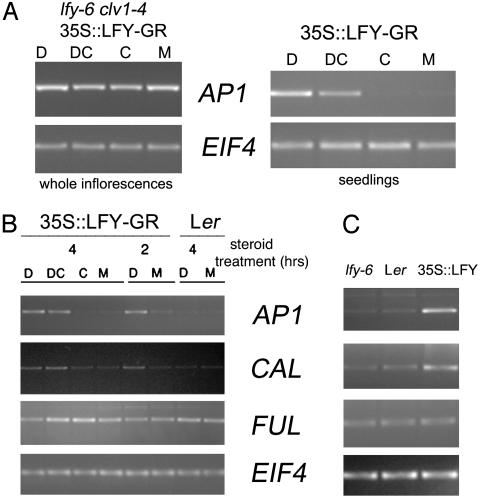
In inflorescences, LFY-independent AP1 expression obscures the LFY-dependent AP1 induction because of the range of developmental stages present in this tissue (ref. 13 and Fig. 1A). To be able to use quantitative methods to identify and test candidate LFY targets, we analyzed earlier developmental stages for up-regulation of AP1 expression after steroid activation of LFY-GR. The highest quantitative up-regulation of AP1 expression after activation of LFY-GR by steroid treatment was observed in 9-day-old seedlings (Fig. 1A and data not shown). Moreover, as shown (13), the observed AP1 up-regulation after dexamethasone treatment of LFY-GR seedlings was direct in that the up-regulation was not affected by cycloheximide and, thus, independent of protein synthesis (Fig. 1A). Because of the absence of AP1 expression from the control samples (cycloheximide- or mock-treated seedlings; Fig. 1A), this stage precedes the floral transition (23, 24). We conclude that 9-day-old LFY-GR seedlings can be used to quantitatively monitor events directly downstream of LFY.
Fig. 1.
(A) RT-PCR analyses of two direct targets of the LFY transcription factor indicate that AP1 is expressed independently of LFY in whole inflorescences (Left). By contrast, API expression depends on LFY and is directly induced by LFY in 9-day-old seedlings (Right). Treatments were as follows: dexamethasone (D), dexamethasone plus the protein synthesis inhibitor cycloheximide (DC), cycloheximide alone (C), and mock (M). The ubiquitously expressed EIF4 gene was amplified to test for equal template presence. Two 4-hr treatments were performed sequentially. (B) Rapid, protein synthesis-independent up-regulation of AP1 and its closest homolog, CAL. Treatments were performed as in A, except durations (indicated above each lane) were shorter. Wild-type Landsberg erecta (Ler) was treated with steroid as a negative control. Genotypes are indicated at the top. Induction of AP1, CAL, and FUL was assayed after LFY-GR activation. Gene names are indicated on the right. The ubiquitously expressed EIF4 gene was amplified to test for equal template presence. (C) Test of up-regulation of the genes used in B in 9-day-old seedlings of three different genotypes: the LFY null mutant lfy-6, the wild type (Ler), and seedlings constitutively overexpressing LFY (35S::LFY). All plants in C were untreated.
LFY controls the switch from inflorescence to flower formation, along with its direct target AP1 and two Arabidopsis transcription factors closely related to AP1, CAL and FUL (25, 26). However, it is unclear whether CAL and FUL act directly downstream of, or in parallel to, LFY. We investigated this question by using LFY-GR seedlings. After LFY activation using two different treatment durations, we observed rapid, quantitative up-regulation of both AP1 and CAL, but not of FUL (Fig. 1B). The observed induction of CAL after LFY-GR activation was independent of protein synthesis and, thus, direct (Fig. 1B). Induction of CAL was dependent on LFY activation because it was not observed in steroid-treated wild-type seedlings (Fig. 1B). Increased CAL expression was observed also in seedlings constitutively overexpressing LFY (Fig. 1C) and is, therefore, not simply due to the presence of the glucocorticoid hormone-binding domain in LFY-GR. FUL expression was not correlated with LFY expression in lfy-6, the wild type, or in seedlings overexpressing LFY (Fig. 1C), suggesting that this AP1-related gene is not regulated by LFY.
lfy-6 mutants exhibit a marked delay in the transition from inflorescence to flower formation (3). This delay is likely due to a defect in the expression of LFY targets that regulate the floral transition, such as AP1 (13) and CAL (this study). However, these cannot be the only targets of LFY because even double mutants between loss-of-function alleles of ap1 and cal delay the transition to flower formation much less than lfy-6 (16, 17, 26). It is, therefore, reasonable to conclude that additional, unidentified LFY targets exist.
To identify the additional LFY targets, we activated LFY-GR in seedlings before the floral transition and compared the transcription profile of these seedlings with that of the seedlings in which LFY-GR was not activated (mock-treated). The two message populations were used to probe whole-genome Arabidopsis microarrays (Affymetrix). The experiment was performed in duplicate by using independently treated seedlings. After steroid treatment of LFY-GR seedlings, 134 genes were up-regulated at least 2-fold (see Fig. 4 A and D, which is published as supporting information on the PNAS web site). Because the LFY-GR protein is synthesized but localized to the cytoplasm before dexamethasone steroid treatment (13), it is possible to activate LFY-GR posttranslationally while inhibiting protein synthesis with cycloheximide. Whole-genome microarrays were probed in duplicate with message populations generated from LFY-GR seedlings treated with dexamethasone plus cycloheximide or with cycloheximide alone. In the dexamethasone- plus cycloheximide-treated plants, 152 genes were up-regulated ≥2-fold, compared with those treated with cycloheximide alone (Fig. 4 B and D). In seedlings treated with steroid in the absence and in the presence of protein synthesis inhibitor, 28 genes were up-regulated >2-fold (Fig. 4D). To rule out the possibility that these genes were up-regulated in response to dexamethasone and not in response to increased levels of nuclear localized LFY-GR protein, we generated message populations from untreated wild-type Ler seedlings and seedlings constitutively overexpressing LFY (35S::LFY). No gross differences in gene expression were observed between the two genotypes (Fig. 4C). Because constitutively increased LFY activity might not produce as large a change in target gene expression as LFY-GR activation produces, because of likely habituation of the seedlings to elevated LFY levels, we arbitrarily lowered the threshold to 1.4-fold up-regulation. This threshold produced a total of 753 up-regulated genes (Fig. 4 C and D), 14 of which were also up-regulated by steroid treatment alone and by steroid treatment plus cycloheximide. These 14 genes are good candidates for direct targets of LFY because they are induced directly (without protein synthesis) after LFY activation and are expressed at elevated levels in plants that constitutively overexpress LFY.
The 14 candidate LFY targets are listed in Table 1 according to their level of induction after LFY activation. We have also included one additional gene (At5g03790; Table 1), which was up-regulated only 1.6-fold by steroid treatment in the presence of protein synthesis inhibitor yet was very highly induced by steroid treatment alone (11-fold). Further analyses (described below) indicate that this gene is indeed a direct target of LFY. The microarray expression data for these candidate LFY targets are indicated in Fig. 4 A-C (genes are numbered as in Table 1). A preliminary functional classification of the candidate LFY target genes reveals that the majority are putative transcription factors or likely involved in signal transduction, consistent with their potential role as direct LFY targets (Table 1). Several other putative targets are implicated in protein modification or sugar metabolism.
Table 1. Functional categories of potential LFY target genes.
| No. | GenBank identifier | GenBank predicted activity | Functional category | D/M | DC/C |
|---|---|---|---|---|---|
| 1 | At5g60630 | Signaling protein? | Signaling | 46 | 35 |
| 2 | At5g49770 | Leucine-rich repeat receptor kinase | Signaling | 27 | 6 |
| 3 | At5g03790 | Homeodomain transcription factor | Transcription | 11 | 1.6 |
| 4 | At3g61250 | myb DNA-binding protein (MYB17) | Transcription | 5 | 3 |
| 5 | At1g16070 | Tubby-related transcription factor | Transcription | 4 | 2.5 |
| 6 | At4g14090 | UDP-glycosyltransferase | Protein modification | 3 | 4.5 |
| 7 | At5g03230 | HMG motif-containing DNA binding | Transcription | 3 | 2 |
| 8 | At4g22780 | EF-1 α-like | Translation | 3 | 3 |
| 9 | At3g43190 | Sucrose synthase | Sugar/amino acid biosynthesis | 3 | 2.5 |
| 10 | At3g52470 | Similar to HIN1 protein | Signaling | 3 | 3.5 |
| 11 | At3g47340 | Glutamine-dependent asparagine synthetase | Sugar/amino acid biosynthesis | 2.5 | 8 |
| 12 | At1g61830 | CHP-rich zinc finger protein | Transcription | 2.5 | 2.5 |
| 13 | At2g44450 | Glycosyl hydrolase family 1 | Sugar/amino acid biosynthesis | 2.5 | 2 |
| 14 | At3g19390 | Cysteine proteinase | Protein modification | 2 | 2.5 |
| 15 | At1g68880 | bZIP family transcription factor | Transcription | 2 | 2 |
Induction ratios (D/M and DC/C) were derived by dividing the mean normalized signal of the treatment condition [dexamethasone (D) or dexamethasone plus cycloheximide (DC)] by the mean normalized control values [mock treatment (M) and cycloheximide treatment (C)], respectively. HMG, high-mobility group; EF, elongation factor; CHP, calcineurin homologous protein; bZIP, basic leucine zipper.
AP1, CAL, and FUL signals were below the threshold of detection in all six message populations (inductions as well as control), presumably because of the low abundance of these messages in entire seedlings. Although the low-intensity signals obtained for these genes were not considered reliable, we noted that AP1 and CAL were induced in steroid-treated seedlings compared with mock-treated seedlings, whereas FUL was not (data not shown), consistent with the results of our RT-PCR analyses.
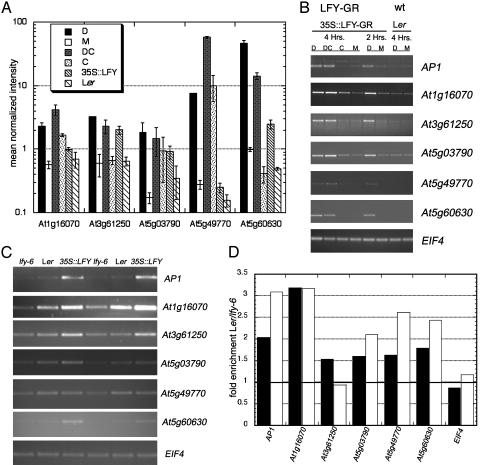
We decided to focus on the five most highly induced candidate LFY targets (Table 1 and Fig. 2A). These genes are all putative transcription factors or likely signal transduction components. Fig. 2A shows the mean normalized intensity of the microarray hybridization signal obtained for each gene by using the different treatments/genotypes. With the exception of At5g03790 (described above), all of these genes were expressed at significantly higher levels in all experimental treatments/genotypes than in the respective controls. At5g03790 had a comparatively high cycloheximide signal, which resulted in a smaller increase in the dexamethasone-plus-cycloheximide treatment compared with cycloheximide treatment alone (Fig. 2A).
Fig. 2.
Test of up-regulation of candidate direct LFY targets. Analysis of the five most highly induced candidate direct LFY targets identified by using microarrays. (A) Mean normalized microarray signal intensity from 9-day-old 35S::LFY-GR seedlings treated with steroid alone (D), steroid plus protein synthesis inhibitor cycloheximide (DC), cycloheximide (C), or mock solution (M). Treatments were performed two times for 4 hr. Shown also is the signal intensity for two untreated genotypes: wild type (Ler) and seedlings constitutively overexpressing LFY (35S::LFY). Error bars indicate SEM, calculated from two independent experiments. (B) Up-regulation of expression of the five candidate direct LFY targets in A was confirmed by using an independent method. RT-PCR was performed on 9-day-old 35S::LFY-GR seedlings, treated as in A except that treatments were performed for 4 or 2 hr. Genotypes used are indicated above. (C) Test of up-regulation of the genes used in B in two biological replicates of three different genotypes: lfy-6 null mutant seedlings, wild-type (Ler) seedlings, and seedlings constitutively overexpressing LFY (35S::LFY). All plants in C were untreated. (D) Quantitation of the message abundance in Ler and lfy-6 from the images in C. The ratio of the Ler/lfy-6 message level is indicated for all genes. The filled and open bars indicate the fold increase in the wild-type Ler/null mutant lfy-6 in the two replicate experiments.
To validate the response of these five genes to LFY activation, we treated seedlings for shorter durations than those used to generate the message populations for the microarray analysis and analyzed gene expression by RT-PCR. All five genes were significantly induced by 4-hr incubation with steroid in the absence and presence of cycloheximide (Fig. 2B), confirming the microarray-based expression data. Moreover, none of the genes were up-regulated in 4-hr steroid-treated wild-type seedlings, indicating that the observed induction was due to LFY activation (Fig. 2B). Induction of all five genes was also detected after a 2-hr steroid treatment, suggesting that up-regulation occurred rapidly after LFY activation. Thus, like AP1 (13) and CAL (Fig. 1B), these five genes are likely direct LFY targets.
To test for up-regulation of these genes in untreated seedlings, we compared their expression levels in lfy-6 mutants, wild-type (Ler), and seedlings constitutively overexpressing LFY (35S::LFY). The expression of all five genes was lowest in lfy-6 and highest in 35S::LFY seedlings (Fig. 2C). Furthermore, for four of the five genes, lfy-6 mutants exhibited reduced expression compared with the wild type (Fig. 2D), indicating that up-regulation of these genes is dependent on endogenous LFY. At3g61250, by contrast, may not depend on endogenous LFY.
During the switch from vegetative to reproductive development, endogenous LFY expression increases, closely followed by an increase in AP1 (23, 24, 27). We examined whether expression of these five genes changed in a similar pattern (see Fig. 5, which is published as supporting information on the PNAS web site). Three of the five LFY targets, as well as CAL, exhibited a marked increase in mRNA expression levels 6-12 days after germination. AP1 expression increased at day 12; CAL, At1g16070, and At5g03790 expression increased at day 8; and At3g61250 expression increased very gradually from day 6 to day 12.
The two genes that did not exhibit a temporal increase in message abundance during the tested developmental timeframe were already expressed strongly at the earliest time point (6 days). Because we used entire seedlings for the expression analysis, it is possible that these two genes are not only expressed in incipient flower primordia in a LFY-dependent fashion but are also expressed in other seedling tissues where they might have additional functions.
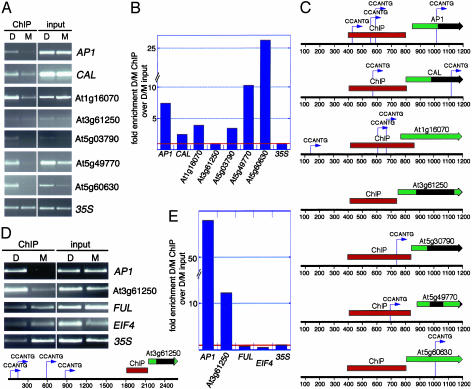
If LFY directly regulates the transcription of the genes identified in this study, it should bind to cis-regulatory sequences in the genomic DNA surrounding these genes. To test for in vivo binding of LFY to cis-regulatory regions, we performed ChIP using highly specific anti-LFY antibodies. Steroid or steroid-free solution was used to treat 35S::LFY-GR seedlings for 4 hr, and then formaldehyde was used to crosslink proteins to DNA. Isolated nuclei were lysed and the chromatin was sonicated to shear the genomic DNA to an average fragment size of 600 bp, followed by incubation with affinity-purified anti-LFY antiserum (13) prebound to protein A magnetic beads and subjected to PCR using gene-specific primers. To test the specificity of this protocol, we examined binding of LFY to a known target sequence (AP1) (11, 14) and a negative control (the 35S promoter). As expected, we observed a marked increase in LFY binding to AP1 sequences in steroid-treated plants, but we observed no difference in the binding of LFY to 35S sequences in steroid compared with mock-treated plants (Fig. 3 A and B). We conclude that ChIP PCR accurately reflects in vivo LFY binding to genomic DNA.
Fig. 3.
ChIP of DNA bound by LFY. (A) Dexamethasone steroid (D) or mock solution (M) treatment of 9-day-old 35S::LFY-GR seedlings was performed for 4 hr, and the seedlings were formaldehyde-crosslinked. Nuclear extracts prior to antibody incubation (input) and after ChIP were subjected to PCR analysis using primers flanking known LFY binding sites (AP1) or upstream of the predicted transcription start (all other genes). Gene names are indicated on the right. (B) Densitometric determination of signal intensity in each ChIP D/M sample was normalized, as described in Supporting Text. (C) The region amplified after ChIP is shown as a red rectangle, and the ORF of each gene is shown as a green rectangle, with the gene name given above. Introns are shown as black rectangles. Consensus LFY binding sites (CCANTG) are indicated by arrows above each sequence. (D) Independent ChIP using a larger genomic DNA fragment and otherwise treated as in A. Gene names are indicated on the right. The consensus LFY binding sites for At3g61250 located upstream of the region shown in C are indicated below the blots. (E) Densitometric analysis of the experiment shown in D.
Next, we tested LFY binding to CAL and to the five LFY targets identified in our microarray experiments (Fig. 3). Because the sequences that regulate expression of these genes are not known, we focused on the genomic region just 5′ to the predicted translation start site. Gene-specific primers that amplify a 400- to 500-bp region upstream of each gene were used to quantify the amount of each DNA sequence associated with LFY in steroid- and mock-treated seedlings (Fig. 3C). CAL and four of the five LFY targets were preferentially bound by LFY in steroid-treated seedlings (Fig. 3 A and B). Furthermore, the amount of DNA recovered in each case correlated very well with the transcriptional induction of this gene. For example, At5g49770 and At5g60630 were the most highly induced (27- and 46-fold, respectively; Table 1; Fig. 2A) and the most strongly bound (10- and 26-fold, respectively; Fig. 3B) of all genes. One gene, At3g61250, was not bound by LFY within the analyzed region (Fig. 3 A and B).
We analyzed a 1,200-bp region centered around the ChIP PCR fragment for presence of the consensus CCANTG LFY-binding motif (11, 14, 28). All regulatory regions that were bound by LFY contained at least one such motif (Fig. 3C). We conclude that LFY not only up-regulates transcription of the newly identified target genes in the absence of protein synthesis but also binds to cis-regulatory elements located in the 5′ genomic region of these genes.
Although one LFY target, At3g61250, was not bound by LFY and contained no LFY-binding motif within the analyzed region (Fig. 3C), several LFY-binding consensus motifs were found in the intergenic region upstream of the analyzed fragment (Fig. 3D). To test whether this region is bound by LFY, we performed ChIP2 by using independently treated seedlings and a sonication regime that yielded larger genomic DNA fragments (average DNA fragment size 900 bp). Binding of LFY to At3g61250 was observed by using these conditions (Fig. 3 D and E), suggesting that the LFY-binding sites of At3g61250 are located further upstream of the predicted translation start site of At3g61250. As expected, 35S, FUL, and EIF4 regulatory elements were not bound by LFY (Fig. 3 D and E); none of these genes is induced by LFY. In addition, very strong LFY-binding to the AP1 regulatory elements was observed in ChIP2, suggesting that additional, distant LFY-binding sites may exist in addition to the known LFY-binding sites in this gene.
Discussion
The developmental regulator LFY controls the switch from the vegetative to the reproductive phase in many plant species (9). Despite the biological importance of this developmental transition, only one direct target of LFY, AP1, has been identified (13). The fact that loss-of-function mutations of AP1 have a much weaker effect on the floral transition than LFY mutations (17, 18) indicates that AP1 cannot be the sole target of LFY. Using a combination of transcription profiling and ChIP, we show here that LFY also directly regulates the transcription of the AP1-related gene CAL and of at least five additional genes. Three of these genes encode putative transcription factors, and two of these genes encode putative signaling molecules.
To identify transcriptional targets of LFY, we took advantage of a form of this protein that can be activated. Studies have shown that LFY-GR is inactive and localized cytoplasmically in the absence of steroid and that steroid activation results in nuclear localization and full biological activity (13). Steroid activation of LFY-GR allows us to compare recent changes in gene expression in plants that do not differ in tissue composition or in developmental stage. LFY-GR was activated in seedlings just before the floral transition to mimic the biological context in which endogenous LFY operates as a meristem identity regulator. The capacity to posttranslationally activate LFY-GR allowed us to identify genes that are transcribed in response to LFY in the absence of protein synthesis.
We chose to study the effect of LFY on gene expression by using whole-genome Arabidopsis arrays (Affymetrix) because Arabidopsis has many large gene families (29), members of which may cross-hybridize to probes on cDNA arrays; the combination of gene-specific 25-nucleotide probes and accompanying mismatch controls improved our ability to discriminate among RNAs from multigene family members. In fact, three of the five genes that we identified are members of large gene families (>50 members) and, where tested, their up-regulation was unique among closely related family members (D.W., unpublished data).
We found that the AP1 homolog CAL and 15 additional genes are up-regulated in 9-day-old seedlings in response to LFY and are likely to be direct targets of this transcription factor. Based on the following evidence, we are confident that CAL and the 5 most highly induced of the 15 genes (described above) are immediate early targets of LFY. First, up-regulation of expression of these genes was observed in the absence of protein synthesis both in microarray experiments and by RT-PCR. Second, up-regulation of their expression was very rapid; elevated expression of all genes was observed after submerging seedlings in steroid solution for only 2 hr. Third, expression of the target genes was correlated with endogenous LFY levels. Fourth, expression of three genes and of CAL increased during the floral transition. The two genes that were expressed uniformly during the floral transition were highly induced by LFY and strongly bound by LFY. Thus, the lack of a temporal increase in their expression may be due to the fact that, as for AP1 (described above), expression of these genes has a LFY-dependent as well as a LFY-independent component. Fifth, sequences near all genes were bound by LFY in vivo, and the amount of bound DNA correlated with the level of nuclear-localized LFY protein. Last, consensus LFY-binding sites were identified in all LFY-bound regions. We conclude, therefore, that these genes are bona fide LFY targets.
CAL is the closest AP1 homolog in Arabidopsis, and mutations in this gene enhance the reproductive transition defect of ap1-1 mutants (17). This observation suggests that CAL is functionally related to AP1 and, like AP1, acts directly downstream of LFY. The data presented here strongly support this conclusion. CAL message is elevated slightly earlier than AP1 message in a developmental time series of the wild type (Fig. 5), suggesting that LFY-dependent activation of CAL may precede that of AP1. It has been proposed that CAL and AP1, in turn, can up-regulate LFY expression (16, 17). Thus, the three genes likely participate in a positive feedback loop, which controls the precise and irreversible transition from inflorescence formation to flower formation in Arabidopsis. A third gene, FUL, closely related to AP1, does not appear to be a direct target of LFY, based on the absence of transcriptional induction and LFY binding. This finding is consistent with previous genetic investigations, which indicated that FUL might act primarily in parallel to LFY (26).
Several LFY targets likely regulate the transcriptional changes that are required for identity switch of the stem cell descendents in the shoot apex at the floral transition. Previous analyses have indicated that transcription factors make up <6% of all genes in Arabidopsis (30). By contrast, they make up 60% of the most highly induced LFY targets, a significant enrichment for this functional category of genes. Among these, At1g16070 encodes a tubby domain containing a putative DNA-binding factor. In mammals, tubby proteins are required for maintenance and function of neuronal cells and link G protein signal transduction to transcriptional regulation (31, 32). Plants have several tubby-like proteins (31), the function of which is unknown. A second transcriptional regulator, At3g61250, encodes a member of the plant-specific R2R3 family of MYB domain transcription factors (33, 34). Several members of this large gene family are involved in developmental regulation (35-37). A third gene, At5g03970, is a putative homeobox-containing transcription factor with an adjacent leucine zipper motif. Proteins in this family are involved in developmental regulation (38-40) and implicated in response to hormone signaling (41).
The two remaining newly identified LFY targets likely play a role in signal transduction. At5g60630 encodes a small serine-glycine-rich protein with predicted transmembrane domains that is thought to be targeted to the secretory pathway. At5g49770 encodes a leucine-rich repeat receptor kinase, which is a member of a very large gene family in plants implicated in developmental signaling, hormone signal transduction, and disease resistance (42-48). The function of the small LRR VIII-1 subfamily to which At5g49770 belongs (49) is not yet understood.
Receptor kinases, signaling molecules, and transcription factor cascades regulate many aspects of the development of higher eukaryotes (50-58). Thus, the predicted function of the newly identified LFY targets fits well with the their proposed function as components of the molecular changes required for the meristem identity switch in Arabidopsis.
In summary, we describe a facile and robust method for identification of direct targets of a developmentally important transcription factor. The combined dual analysis of induction of transcriptional activity of LFY-GR and induction of in vivo binding of LFY-GR after steroid treatment was a powerful method for identification and verification of immediate early target genes. This approach should be useful for investigation of other transcriptional regulators, especially in systems in which genome-wide localization studies can be performed in parallel with transcript profiling (20). The genes that we identified are likely candidates for regulators that act directly downstream of LFY in controlling the onset of reproduction.
Supplementary Material
Acknowledgments
We thank John Wagner, Scott Poethig, and Chang Seob Kwon for critical comments on the manuscript; Yuan Quan for assistance with chromatin immunoprecipitation; and the Penn Microarray Facility for performing microarray hybridization and first-pass data analysis and for advice on data analysis. The entire ATH1 microarray hybridization data set has been deposited in the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/geo). GenBank accession nos. are GSM13778-GSM13785, GSM13789, and GSM13791-GSM13793, and the series accession no. is GSE 911. This article is based on work supported by National Science Foundation Grant 0130804.
Abbreviations: AP1, APETALA1; CAL, CAULIFLOWER; ChIP, chromatin immunoprecipitation; FUL, FRUITFUL; GR, glucocorticoid receptor; Ler, Landsberg erecta; LFY, LEAFY.
Data deposition: The ATH1 microarray hybridization dataset has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (series accession no. GSE 911).
References
- 1.Huala, E. & Sussex, I. M. (1992) Plant Cell 4, 901-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schultz, E. A. & Haughn, G. W. (1991) Plant Cell 3, 771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F. & Meyerowitz, E. M. (1992) Cell 69, 843-859. [DOI] [PubMed] [Google Scholar]
- 4.Ahearn, K. P., Johnson, H. A., Weigel, D. & Wagner, D. R. (2001) Plant Cell Physiol. 42, 1130-1139. [DOI] [PubMed] [Google Scholar]
- 5.Hofer, J., Turner, L., Hellens, R., Ambrose, M., Matthews, P., Michael, A. & Ellis, N. (1997) Curr. Biol. 7, 581-587. [DOI] [PubMed] [Google Scholar]
- 6.Molinero-Rosales, N., Jamilena, M., Zurita, S., Gomez, P., Capel, J. & Lozano, R. (1999) Plant J. 20, 685-693. [DOI] [PubMed] [Google Scholar]
- 7.Pena, L., Martin-Trillo, M., Juarez, J., Pina, J. A., Navarro, L. & Martinez-Zapater, J. M. (2001) Nat. Biotechnol. 19, 263-267. [DOI] [PubMed] [Google Scholar]
- 8.Souer, E., van der Krol, A., Kloos, D., Spelt, C., Bliek, M., Mol, J. & Koes, R. (1998) Development (Cambridge, U.K.) 125, 733-742. [DOI] [PubMed] [Google Scholar]
- 9.Weigel, D. & Nilsson, O. (1995) Nature 377, 495-500. [DOI] [PubMed] [Google Scholar]
- 10.Weigel, D. & Meyerowitz, E. M. (1994) Cell 78, 203-209. [DOI] [PubMed] [Google Scholar]
- 11.Parcy, F., Nilsson, O., Busch, M. A., Lee, I. & Weigel, D. (1998) Nature 395, 561-566. [DOI] [PubMed] [Google Scholar]
- 12.Lee, I., Wolfe, D. S., Nilsson, O. & Weigel, D. (1997) Curr. Biol. 7, 95-104. [DOI] [PubMed] [Google Scholar]
- 13.Wagner, D., Sablowski, R. W. & Meyerowitz, E. M. (1999) Science 285, 582-584. [DOI] [PubMed] [Google Scholar]
- 14.Busch, M. A., Bomblies, K. & Weigel, D. (1999) Science 285, 585-587. [DOI] [PubMed] [Google Scholar]
- 15.Lamb, R. S., Hill, T. A., Tan, Q. K. & Irish, V. F. (2002) Development (Cambridge, U.K.) 129, 2079-2086. [DOI] [PubMed] [Google Scholar]
- 16.Liljegren, S. J., Gustafson-Brown, C., Pinyopich, A., Ditta, G. S. & Yanofsky, M. F. (1999) Plant Cell 11, 1007-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman, J. L., Alvarez, J., Weigel, D., Meyerowitz, E. M. & Smyth, D. R. (1993) Development (Cambridge, U.K.) 119, 721-743. [Google Scholar]
- 18.Schultz, E. A. & Haughn, G. W. (1993) Development (Cambridge, U.K.) 119, 745-765. [Google Scholar]
- 19.Weigel, D. & Meyerowitz, E. M. (1993) Science 261, 1723-1726. [DOI] [PubMed] [Google Scholar]
- 20.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306-2309. [DOI] [PubMed] [Google Scholar]
- 21.Moehs, C. P., McElwain, E. F. & Spiker, S. (1988) Plant Mol. Biol. 11, 507-515. [DOI] [PubMed] [Google Scholar]
- 22.Wyrick, J. J., Aparicio, J. G., Chen, T., Barnett, J. D., Jennings, E. G., Young, R. A., Bell, S. P. & Aparicio, O. M. (2001) Science 294, 2357-2360. [DOI] [PubMed] [Google Scholar]
- 23.Blazquez, M. A., Soowal, L. N., Lee, I. & Weigel, D. (1997) Development (Cambridge, U.K.) 124, 3835-3844. [DOI] [PubMed] [Google Scholar]
- 24.Hempel, F. D., Weigel, D., Mandel, M. A., Ditta, G., Zambryski, P. C., Feldman, L. J. & Yanofsky, M. F. (1997) Development (Cambridge, U.K.) 124, 3845-3853. [DOI] [PubMed] [Google Scholar]
- 25.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1991) Development (Cambridge, U.K.) 112, 1-20. [DOI] [PubMed] [Google Scholar]
- 26.Ferrándiz, C., Gu, Q., Martienssen, R. & Yanofsky, M. F. (2000) Development (Cambridge, U.K.) 127, 725-734. [DOI] [PubMed] [Google Scholar]
- 27.Hempel, F. D., Zambryski, P. C. & Feldman, L. J. (1998) Plant Cell 10, 1663-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong, R. L., Hamaguchi, L., Busch, M. A. & Weigel, D. (2003) Plant Cell 15, 1296-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 30.Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O. J., Samaha, R. R., et al. (2000) Science 290, 2105-2110. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda, A., Nishina, P. M. & Naggert, J. K. (2002) J. Cell Sci. 115, 9-14. [DOI] [PubMed] [Google Scholar]
- 32.Santagata, S., Boggon, T. J., Baird, C. L., Gomez, C. A., Zhao, J., Shan, W. S., Myszka, D. G. & Shapiro, L. (2001) Science 292, 2041-2050. [DOI] [PubMed] [Google Scholar]
- 33.Stracke, R., Werber, M. & Weisshaar, B. (2001) Curr. Opin. Plant Biol. 4, 447-456. [DOI] [PubMed] [Google Scholar]
- 34.Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R. C., Petroni, K., Urzainqui, A., Bevan, M., Martin, C., et al. (1998) Plant J. 16, 263-276. [DOI] [PubMed] [Google Scholar]
- 35.Byrne, M. E., Barley, R., Curtis, M., Arroyo, J. M., Dunham, M., Hudson, A. & Martienssen, R. A. (2000) Nature 408, 967-971. [DOI] [PubMed] [Google Scholar]
- 36.Oppenheimer, D. G., Herman, P. L., Sivakumaran, S., Esch, J. & Marks, M. D. (1991) Cell 67, 483-493. [DOI] [PubMed] [Google Scholar]
- 37.Lee, M. M. & Schiefelbein, J. (1999) Cell 99, 473-483. [DOI] [PubMed] [Google Scholar]
- 38.Ratcliffe, O. J., Riechmann, J. L. & Zhang, J. Z. (2000) Plant Cell 12, 315-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N. & Clark, S. E. (2001) Plant J. 25, 223-236. [DOI] [PubMed] [Google Scholar]
- 40.Talbert, P. B., Adler, H. T., Parks, D. W. & Comai, L. (1995) Development (Cambridge, U.K.) 121, 2723-2735. [DOI] [PubMed] [Google Scholar]
- 41.Sawa, S., Ohgishi, M., Goda, H., Higuchi, K., Shimada, Y., Yoshida, S. & Koshiba, T. (2002) Plant J. 32, 1011-1022. [DOI] [PubMed] [Google Scholar]
- 42.Friedrichsen, D. M., Joazeiro, C. A., Li, J., Hunter, T. & Chory, J. (2000) Plant Physiol. 123, 1247-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeong, S., Trotochaud, A. E. & Clark, S. E. (1999) Plant Cell 11, 1925-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark, S. E., Williams, R. W. & Meyerowitz, E. M. (1997) Cell 89, 575-585. [DOI] [PubMed] [Google Scholar]
- 45.Torii, K. U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R. F. & Komeda, Y. (1996) Plant Cell 8, 735-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., Gardner, J., Wang, B., Zhai, W. X., Zhu, L. H., et al. (1995) Science 270, 1804-1806. [DOI] [PubMed] [Google Scholar]
- 47.Rojo, E., Sharma, V. K., Kovaleva, V., Raikel, N. V. & Fletcher, J. C. (2002) Plant Cell 14, 969-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Becraft, P. W. (2002) Annu. Rev. Cell Dev. Biol. 18, 163-192. [DOI] [PubMed] [Google Scholar]
- 49.Shiu, S. H. & Bleecker, A. B. (2001) Sci. STKE, 22. [DOI] [PubMed]
- 50.Bondos, S. E. & Tan, X. X. (2001) Crit. Rev. Eukaryotic Gene Expression 11, 145-171. [PubMed] [Google Scholar]
- 51.Cripps, R. M. & Olson, E. N. (2002) Dev. Biol. 246, 14-28. [DOI] [PubMed] [Google Scholar]
- 52.Edenfeld, G., Pielage, J. & Klambt, C. (2002) Curr. Opin. Genet. Dev. 12, 473-477. [DOI] [PubMed] [Google Scholar]
- 53.Freeman, M. & Gurdon, J. B. (2002) Annu. Rev. Cell Dev. Biol. 18, 515-539. [DOI] [PubMed] [Google Scholar]
- 54.Hendriks, B. & Reichmann, E. (2002) Biol. Res. 35, 277-286. [DOI] [PubMed] [Google Scholar]
- 55.Mann, R. S. & Carroll, S. B. (2002) Curr. Opin. Genet. Dev. 12, 592-600. [DOI] [PubMed] [Google Scholar]
- 56.Pires-daSilva, A. & Sommer, R. J. (2003) Nat. Rev. Genet. 4, 39-49. [DOI] [PubMed] [Google Scholar]
- 57.Reinke, V. & White, K. P. (2002) Annu. Rev. Genomics Hum. Genet. 3, 153-178. [DOI] [PubMed] [Google Scholar]
- 58.Veenstra, G. J. & Wolffe, A. P. (2001) Trends Biochem. Sci. 26, 665-671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.